Abstract
Background
A variety of abnormalities in vitamin D metabolism have been reported in patients with active tuberculosis. However, intervention trials have produced inconsistent results. We hypothesized that genetic and epigenetic changes in the key genes of the vitamin D metabolic pathway may partly explain the differences between studies.
Methods
We performed a case-control study followed by a prospective cohort study. We recruited 122 patients with pulmonary tuberculosis and 118 healthy controls. The serum 25-hydroxyvitamin D and 1,25-dihydroxyvitamin D levels were measured. The methylation of the promoter regions of key genes in the vitamin D metabolic pathway (CYP24A1, CYP27A1, CYP27B1, CYP2R1, and VDR) was detected using the Illumina MiSeq platform. The specific methylation profiles were examined as epigenetic biomarkers. The sensitivity, specificity, and receiver operating characteristic (ROC) curves were used to estimate the predictive value of the biomarkers.
Results
The baseline serum 25-hydroxyvitamin D and 1,25-dihydroxyvitamin D concentrations in the cases were significantly lower than those in the controls (51.60 ± 27.25 nmol/L vs. 117.50 ± 75.50 nmol/L, Z = − 8.515, P < 0.001; 82.63 ± 51.43 pmol/L vs. 94.02 ± 49.26 pmol/L, Z = − 2.165, P = 0.03). We sequenced 310 CpG sites in five candidate genes. After Bonferroni correction, there were 55 differentially methylated CpG sites between cases and controls; 41.5% were in the CYP27B1 gene, 31.7% were in the CYP24A1 gene, 14.7% were in the VDR gene, and 12.3% were in the CYP27A1 gene. When we designated the CpG sites that remained significant after the Bonferroni correction as the biomarkers, the area under the curve (AUC) for the cumulative methylation was 0.810 (95% CI 0.754–0.866). There was an interaction between CYP27A1 methylation level and 1,25-dihydroxyvitamin D concentration associated with the risk of TB (ORinteraction = 4.11, 95% CI 1.26–13.36, P = 0.019). The serum 1,25-dihydroxyvitamin D concentration at the end of the intensive treatment stage was related to a patient’s prognosis (P = 0.008). There were 23 CpG sites that were individually related to the treatment outcomes, but the relationships were not significant after the Bonferroni correction.
Conclusion
Both serum vitamin D concentrations and the methylation levels of key genes in the vitamin D metabolic pathway are related to the risk and prognosis of tuberculosis.
Electronic supplementary material
The online version of this article (10.1186/s13148-018-0552-6) contains supplementary material, which is available to authorized users.
Keywords: Tuberculosis, DNA methylation, Vitamin D, Risk, Prognosis
Background
Despite the widespread use of the Bacillus Calmette-Guérin (BCG) vaccine, Mycobacterium tuberculosis (M.tb) infection and active tuberculosis (TB) remain major public health threats [1, 2]. TB is the ninth leading cause of death worldwide and is the leading cause of death from a single infectious agent, ranking above HIV/AIDS. In 2016, 10.4 million people fell ill with TB and there were an estimated 1.3 million TB deaths among HIV-negative people and an additional 374,000 deaths among HIV-positive people [3].
Factors related to the risk of TB include low socioeconomic status, poor nutrition, traditional/cultural traits, tobacco smoking, contacts with sputum smear-positive index patients, and genetic susceptibility [4–8]. Studies have correlated vitamin D deficiency with susceptibility to TB since 1651 when vitamin deficiency was found to be associated with TB for the first time [9]. Moreover, Stead et al. have shown racial differences in the incidence of TB associated with the levels of 25-hydroxyvitamin D (25(OH)D). Recently, studies have reported that hypovitaminosis D results in lower antimycobacterial immunity [10, 11] and is related to increased risk of TB [12, 13]. In vitro studies have revealed that 1,25-dihydroxyvitamin D (1,25(OH)2D) enhances innate immunity by increasing the expression of antimicrobial peptides, including cathelicidin, and inducing the autophagy of infected cells, thus restricting the intracellular growth of M.tb in macrophages [14]. Some studies have shown that vitamin D supplementation during the intensive phase of antimicrobial treatment can increase sputum negative conversion rates [15, 16]. Vitamin D supplementation has been believed to be beneficial in the treatment of patients with TB in observational studies; however, results from clinical trials have been inconclusive [17]. Confounding factors and reverse causation may partly explain the inconsistencies [18], but individual variations in vitamin D metabolism and related variations in immune responses should not be neglected.
Previous studies have mainly focused on genetic polymorphisms of genes in the vitamin D metabolic pathway. With the development of molecular biology, the role of epigenetic traits in gene expression has received greater attention [19]. DNA methylation, which involves the addition of a methyl group to the cytosine in a CpG dinucleotide, is a key epigenetic trait related to a number of biological processes including genomic imprinting, X-chromosome inactivation, aging, and carcinogenesis [20]. Studies have shown that hypermethylation in the promoter region of the cytochrome P450 gene can silence the gene and affect the vitamin D activity [21], but the role of this hypermethylation in the risk of TB has not been systematically studied. Thus, we performed a molecular epidemiological study in a Chinese population aiming to explore the effect of aberrant DNA methylation in the vitamin D metabolic pathway on serum 25(OH)D and 1,25(OH)2D levels and to determine its relation to the risk and prognosis of pulmonary TB.
Methods
Study design and study population
This study used a mixed case-control and prospective cohort design. We recruited 122 patients with pulmonary TB from Zhenjiang and Lianyungang in the province of Jiangsu in China during 2014 and 2016. TB cases were group-matched (by sex and age) with 118 controls from a pool of individuals who participated in community-based health examination programs. Among these controls, individuals with a history of TB, malignancy, diabetes, and HIV were excluded. This study was approved by the Ethics Committee of Nanjing Medical University. After informed consent was obtained from all participants, questionnaires were used to collect demographic data. Venous blood samples were collected for vitamin D measurement and molecular analyses.
Serum 25(OH)D and 1,25(OH)2D measurement
We measured the serum vitamin D using a 25-hydroxyvitamin D kit and a 1,25-dihydroxyvitamin D EIA (Immunodiagnostic Systems Limited, UK). The intra- and interassay coefficient of variation (CV) were < 9% for 25(OH)D and < 20% for 1,25(OH)2D. The minimum detection limits were 12 nmol/L for 25(OH)D and 6 pmol/L for 1,25(OH)2D.
Methylation analysis
We selected five key genes (CYP24A1, CYP27A1, CYP27B1, CYP2R1, and VDR) in the vitamin D metabolic pathway and sequenced the CpG islands in the promoter region of the candidate genes using the Illumina MiSeq platform. The DNA was subjected to sodium bisulfite treatment using an EZ DNA Methylation™-GOLD Kit (Zymo Research, Orange, CA, USA) according to the manufacturer’s protocols. Primers were designed to amplify the regions of interest from the bisulfite-converted DNA (Table 1).
Table 1.
Primers designed for multiplex PCR
| Gene | Fragment | Forward primer | Reverse primer |
|---|---|---|---|
| CYP24A1 | CYP24A1 _1 | TAGAGGAGGGYGGAGTGGTTT | CACACCCRATAAACTCCRAACTTC |
| CYP24A1 _2 | GGAGATAATTTTTAGGAAGTTATGYGAAGTT | CACTTCAATCCAAACTAAAAATATCTAACTC | |
| CYP27A1 | CYP27A1 _1 | TTGGTTTYGTGGGGGTAGAG | CACCRCRTCCCTCTCCTACAA |
| CYP27A1 _2 | GGAGGGTYGAGTAAAGGTTAGTTAGAT | AAAACCTATCCCRATATAAAACTTCC | |
| CYP27A1 _3 | ATTTTGGGYGGGGGTGTAG | CCCTCCAAAAATCAAATAACTAACC | |
| CYP27A1 _4 | TTTTYGGATTGATTTYGGAGTTAGT | ACTATACRTTTTCCRTACTATATTACTCTTTCC | |
| CYP27A1 _5 | GGTTGAGATTAGATTTYGTAGATGATG | ACCAACTATACCATCCTACTAAATCCT | |
| CYP27B1 | CYP27B1 _1 | GGTTGAGATATGATGTTTAGGAGAAG | TCCCTTCCTACCTACAACTCRTATA |
| CYP27B1 _2 | TTTGGYGTGGGTATAGGTTAAGTTG | CTCACRCAATAAACAATCCRCAAAC | |
| CYP27B1 _3 | GAGTTGTTGYGATAGGAGGGATT | CAACCRACCTCCCACCA | |
| CYP2R1 | CYP2R1 _1 | TTAATGGGAGTATGGTAGGGTTG | AAAAACCCATCRACCRCCTCTA |
| CYP2R1 _2 | GGTAGGGAGGGTYGTTAGGTTG | CAAAACTAAATCRCCTCRAAACCTC | |
| CYP2R1 _3 | TGTAGGGGGAGTTTYGTTTTTGT | CAAACACCRAAAAACCTACTATTAACC | |
| CYP2R1 _4 | GGAAAATTAAGGYGTTTTGAGTTTTA | CACACAAAAAACRCCTTTTAAATATCTAC | |
| VDR | VDR _1 | GTAGTTATTTATAATTTTAGGTTTTAGGAGGTAG | CTCAACCTAATCCCACAAATTAAAA |
| VDR _2 | AGGTGATATYGGGTGGGAGTAAT | CCACCTAAACTAACCAAACCAA | |
| VDR _3 | GGTGTTAGTYGGTAGGYGTTTTTTAG | CATAAAACAAAACACRCTTCTACCCT | |
| VDR _4 | TTTYGATTAATATAGGTTGAAGYGGGTA | CCCACAAATCCAATCCTCTC | |
| VDR _6 | GAATTYGGGAGTAGYGGGAAAG | TACTAAACACTATATTAACRAAACATTTCTCC |
Multiplex PCR was performed using the optimized primer sets. A 20-μl PCR reaction mixture was prepared for each reaction that included 2 μl of template DNA, 3 mM Mg2+, 0.2 mM dNTP, 0.1 μM of each primer, 1× buffer (Takara, Tokyo, Japan), and 1 U of HotStarTaq polymerase (Takara, Tokyo, Japan). The cycling program was 95 °C for 2 min; 11 cycles of 94 °C for 20 s, 63 °C for 40 s with a decreasing temperature step of 0.5 °C per cycle, and 72 °C for 1 min; 24 cycles of 94 °C for 20 s, 65 °C for 30 s, and 72 °C for 1 min; and 72 °C for 2 min.
PCR amplicons from different panels were quantified and pooled, diluted, and subjected to a second round of amplification using the indexed primers. A 20-μl mixture was prepared for each reaction that included 0.3 μM index primer, 0.3 μM forward primer, 0.3 mM dNTP, 1× buffer (New England Biolabs, MA, the USA), 1 U of Q5™ DNA polymerase (New England Biolabs), and 1 μL of diluted template. The cycling program was 98 °C for 30 s; 11 cycles of 98 °C for 10 s, 65 °C for 30 s, and 72 °C for 30 s; and 72 °C for 5 min. The PCR products were separated by agarose gel electrophoresis and purified using a QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany). The libraries from the different samples were quantified and pooled together, then sequenced on the Illumina MiSeq platform according to the manufacturer’s protocols. Sequencing was performed with 2 × 300 bp (overall sequencing read length) paired-end runs.
Statistical analysis
Data were entered with EpiData 3.1 software (Denmark) and analyzed using STATA 12.0 (StataCorp, College Station, TX, USA). Student’s t test and the chi-square test were used to compare the distributions of demographic variables and risk factors between cases and controls. Univariate and multivariate logistic regression models were used to calculate odds ratios (ORs) and 95% confidence intervals (CIs). We analyzed both individual and cumulative methylation levels of the candidate genes. The sensitivity, specificity, and the area under the receiver operating characteristic (ROC) curve were estimated to assess the diagnostic value of the biomarkers. A Spearman correlation was used to estimate the relationship between serum vitamin D levels and methylation status. A Kaplan-Meier curve was used to analyze the effect of methylation level on initial sputum conversion.
Results
General characteristics of study subjects
In total, 122 TB cases (78.7% males and 21.3% females) and 118 controls (72.9% males and 27.1% females) were involved in the analysis. The average age (± standard deviation, SD) was 50.83 ( 20.04) years in cases and 51.97 (± 12.49) years in controls. Due to the frequency matching, there was no significant difference in the distribution of age and sex between the two groups. However, the distribution of marital status was significantly different (P < 0.001). The proportion of those who had ever smoked in the cases was slightly higher than that in the controls (42.6% vs. 39.0%), while the percentage of those who drank alcohol was slightly lower in the cases than in the controls (26.2% vs. 29.7%), although there was not a significant difference (Table 2). Among the cases, 46 (37.7%) were cured, 61 (50.0%) completed treatment, and 15 (12.3%) failed to be treated. We categorized the cases into two groups: successful (cured or completed treatment) and unsuccessful.
Table 2.
General characteristics of cases and controls
| Variables | Controls (n = 118) | Cases (n = 122) | t/χ2 | P |
|---|---|---|---|---|
| Age (years) | ||||
| Mean ± SD | 51.97 ± 12.49 | 50.83 ± 20.04 | 0.530 | 0.597 |
| Sex [n(%)] | 1.104 | 0.293 | ||
| Male | 86 (72.9) | 96 (78.7) | ||
| Female | 32 (27.1) | 26 (21.3) | ||
| Marital status [n(%)] | ||||
| Unmarried | 3 (2.5) | 25 (20.5) | 18.777 | < 0.001 |
| Married | 111 (94.1) | 94 (77.0) | ||
| Widowed/divorced | 4 (3.4) | 3 (2.5) | ||
| Smoking [n(%)] | 0.329 | 0.566 | ||
| Never | 72 (61.0) | 70 (57.4) | ||
| Ever | 46 (39.0) | 52 (42.6) | ||
| Drinking [n(%)] | 0.351 | 0.554 | ||
| Never | 83 (70.3) | 90 (73.8) | ||
| Ever | 35 (29.7) | 32 (26.2) | ||
Serum vitamin D concentrations and methylation levels of CpG sites and the risk of TB
The baseline concentrations of serum 25(OH)D and 1,25(OH)2D were 51.60 ± 27.25 nmol/L and 82.63 ± 51.43 pmol/L among cases, respectively, which were significantly lower than those in the controls (25(OH)D 117.50 ± 75.50 nmol/L, Z = − 8.515, P < 0.001; 1,25(OH)2D 94.02 ± 49.26 pmol/L, Z = − 2.165, P = 0.03). The serum level of 1,25(OH)2D declined during the early phase of antituberculosis treatment, while at the end of the intensive treatment stage, it was 70.81 ± 44.50 pmol/L (Z = − 2.606, P = 0.009).
We sequenced 310 CpG sites in the promoter regions of the candidate genes (Table 3). The methylation levels of specific sites between cases and controls are shown in Table 4. The correlations for methylation levels in each region with 1,25(OH)2D concentrations were listed in Additional file 1. After Bonferroni correction, there were 55 differentially methylated CpG sites between cases and controls, 41.5% of which were in the CYP27B1 gene, 31.7% of which were in the CYP24A1 gene, 14.7% of which were in the VDR gene, and 12.3% of which were in the CYP27A1 gene. We calculated the cumulative methylation levels by adding the frequencies for all the CpG sites in each gene region and found that the methylation level of CYP27A1_3 was significantly associated with the level of serum 1,25(OH)2D (P = 0.045). To further analyze the cumulative methylation levels by considering multiple CpG sites in each gene, we constructed four models. In model 1, we calculated the cumulative methylation levels by adding the frequencies for all the CpG sites in each gene. In model 2, we calculated the cumulative methylation levels by adding the frequencies for the statistically significant CpG sites in each gene. Model 3 only included hypermethylated CpG sites in the cases and excluded CpG sites with inverse relations between cases and controls. Model 4 only included CpG sites that were statistically significant after the Bonferroni correction.
Table 3.
Sequenced sites of selected genes
| Gene | Fragment | Start/stop | Size(bp) | Number of CpG sites |
|---|---|---|---|---|
| CYP24A1 | CYP24A1_1 | 52790591/52790815 | 224 | 13 |
| CYP24A1_2 | 52790767/52791019 | 252 | 28 | |
| CYP27A1 | CYP27A1_1 | 219646982/219646721 | 261 | 22 |
| CYP27A1_2 | 219646810/219646561 | 249 | 22 | |
| CYP27A1_3 | 219646624/219646403 | 221 | 18 | |
| CYP27A1_4 | 219646465/219646204 | 261 | 8 | |
| CYP27A1_5 | 219646286/219646037 | 249 | 3 | |
| CYP27B1 | CYP27B1_1 | 58160882/58160619 | 263 | 16 |
| CYP27B1_2 | 58160053/58159785 | 268 | 23 | |
| CYP27B1_3 | 58159890/58159688 | 202 | 14 | |
| CYP2R1 | CYP2R1_1 | 14913830/14913634 | 196 | 14 |
| CYP2R1_2 | 14913505/14913273 | 232 | 15 | |
| CYP2R1_3 | 14913339/14913061 | 278 | 24 | |
| CYP2R1_4 | 14913116/14912845 | 271 | 15 | |
| VDR | VDR _1 | 48299590/48299323 | 267 | 18 |
| VDR _2 | 48299412/48299179 | 233 | 19 | |
| VDR _3 | 48299247/48299017 | 230 | 13 | |
| VDR _4 | 48299106/48298885 | 221 | 10 | |
| VDR _6 | 48298733/48298464 | 269 | 15 |
Table 4.
Methylation levels of specific sites between cases and controls
| Gene | Fragment | Methylation level of controls (n = 118) | Methylation level of cases (n = 122) | ||
|---|---|---|---|---|---|
| Mean ± SD | Median | Mean ± SD | Median | ||
| CYP24A1 | CYP24A1_1 | 0.39 ± 0.13 | 0.374 | 0.35 ± 0.12 | 0.335 |
| CYP24A1_2 | 0.48 ± 0.08 | 0.461 | 0.41 ± 0.07 | 0.397 | |
| CYP27A1 | CYP27A1_1 | 0.87 ± 0.23 | 0.818 | 0.75 ± 0.27 | 0.678 |
| CYP27A1_2 | 0.78 ± 0.24 | 0.716 | 0.63 ± 0.24 | 0.557 | |
| CYP27A1_3 | 0.54 ± 0.20 | 0.508 | 0.43 ± 0.18 | 0.398 | |
| CYP27A1_4 | 0.72 ± 0.17 | 0.697 | 0.63 ± 0.12 | 0.606 | |
| CYP27A1_5 | 1.79 ± 0.20 | 1.804 | 1.76 ± 0.01 | 1.784 | |
| CYP27B1 | CYP27B1_1 | 3.22 ± 0.56 | 3.167 | 2.78 ± 0.56 | 2.664 |
| CYP27B1_2 | 1.56 ± 0.33 | 1.493 | 1.40 ± 0.32 | 1.345 | |
| CYP27B1_3 | 0.60 ± 0.14 | 0.578 | 0.53 ± 0.14 | 0.499 | |
| CYP2R1 | CYP2R1_1 | 0.13 ± 0.01 | 0.129 | 0.13 ± 0.01 | 0.126 |
| CYP2R1_2 | 0.15 ± 0.01 | 0.152 | 0.15 ± 0.01 | 0.148 | |
| CYP2R1_3 | 0.22 ± 0.05 | 0.223 | 0.22 ± 0.05 | 0.213 | |
| CYP2R1_4 | 0.13 ± 0.02 | 0.124 | 0.12 ± 0.01 | 0.123 | |
| VDR | VDR _1 | 0.94 ± 0.23 | 0.897 | 0.82 ± 0.23 | 0.781 |
| VDR _2 | 0.37 ± 0.05 | 0.362 | 0.33 ± 0.06 | 0.313 | |
| VDR _3 | 0.21 ± 0.10 | 0.196 | 0.20 ± 0.10 | 0.188 | |
| VDR _4 | 0.13 ± 0.02 | 0.132 | 0.12 ± 0.02 | 0.122 | |
| VDR _6 | 0.26 ± 0.07 | 0.270 | 0.27 ± 0.07 | 0.263 | |
Model 1
We calculated the cumulative methylation levels by adding the frequencies for all the CpG sites in each gene. The cumulative methylation levels of CYP24A1, CYP27A1, CYP27B1, CYP2R1, and VDR were significantly different between cases and controls (P < 0.05, Table 5). The area under the curve (AUC) for each gene is listed in Table 6. The cumulative methylation levels of the CYP27A1 gene showed the highest diagnostic value, with an AUC of 0.739 (95% CI 0.675–0.802), followed by CYP27B1 (AUC 0.735, 95% CI 0.671–0.799). The AUC obtained by combining the cumulative methylation levels of the CpG sites in all genotyped genes was 0.747 (95% CI 0.685–0.809).
Table 5.
Cumulative methylation levels of multiple CpG sites in each gene between cases and controls using different models
| Gene | Model 1 | Model 2 | Model 3 | Model 4 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cumulative methylation level | t | P | Cumulative methylation level | t | P | Cumulative methylation level | t | P | Cumulative methylation level | t | P | |||||
| Controls (n = 118) | Cases (n = 122) | Controls (n = 118) | Cases (n = 122) | Controls (n = 118) | Cases (n = 122) | Controls (n = 118) | Cases (n = 122) | |||||||||
| CYP24A1 | 0.87 ± 0.16 | 0.76 ± 0.17 | 5.120 | < 0.001 | 0.46 ± 0.08 | 0.38 ± 0.08 | 7.929 | < 0.001 | 0.46 ± 0.08 | 0.38 ± 0.08 | 7.929 | < 0.001 | 0.28 ± 0.06 | 0.23 ± 0.05 | 7.699 | < 0.001 |
| CYP27A1 | 4.70 ± 0.74 | 4.19 ± 0.68 | 5.537 | < 0.001 | 2.40 ± 0.57 | 1.96 ± 0.51 | 6.182 | < 0.001 | 2.40 ± 0.57 | 1.96 ± 0.51 | 6.182 | < 0.001 | 0.59 ± 0.17 | 0.47 ± 0.16 | 5.803 | < 0.001 |
| CYP27B1 | 5.37 ± 0.97 | 4.72 ± 0.95 | 5.271 | < 0.001 | 4.95 ± 0.92 | 4.31 ± 0.90 | 5.417 | < 0.001 | 4.95 ± 0.92 | 4.31 ± 0.90 | 5.417 | < 0.001 | 3.05 ± 0.61 | 2.57 ± 0.61 | 5.998 | < 0.001 |
| CYP2R1 | 0.63 ± 0.06 | 0.61 ± 0.06 | 2.118 | 0.035 | 0.08 ± 0.02 | 0.07 ± 0.02 | 2.203 | 0.029 | 0.07 ± 0.01 | 0.06 ± 0.01 | 6.207 | < 0.001 | – | – | – | – |
| VDR | 1.92 ± 0.32 | 1.74 ± 0.32 | 4.203 | < 0.001 | 1.12 ± 0.22 | 0.97 ± 0.23 | 4.909 | < 0.001 | 1.04 ± 0.21 | 0.88 ± 0.22 | 5.812 | < 0.001 | 0.40 ± 0.07 | 0.33 ± 0.08 | 6.237 | < 0.001 |
Table 6.
Diagnostic values for TB of selected genes using different models
| Gene | Model 1 | Model 2 | Model 3 | Model 4 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AUC | 95% CI | P | AUC | 95% CI | P | AUC | 95% CI | P | AUC | 95% CI | P | |
| CYP24A1 | 0.707 | 0.641–0.772 | < 0.001 | 0.794 | 0.737–0.851 | < 0.001 | 0.794 | 0.737–0.851 | < 0.001 | 0.793 | 0.736–0.850 | < 0.001 |
| CYP27A1 | 0.739 | 0.675–0.802 | < 0.001 | 0.760 | 0.699–0.821 | < 0.001 | 0.760 | 0.699–0.821 | < 0.001 | 0.747 | 0.685–0.809 | < 0.001 |
| CYP27B1 | 0.735 | 0.671–0.799 | < 0.001 | 0.739 | 0.675–0.803 | < 0.001 | 0.739 | 0.675–0.803 | < 0.001 | 0.747 | 0.684–0.810 | < 0.001 |
| CYP2R1 | 0.578 | 0.506–0.650 | 0.037 | 0.611 | 0.540–0.683 | 0.003 | 0.743 | 0.679–0.806 | < 0.001 | – | – | – |
| VDR | 0.664 | 0.596–0.732 | < 0.001 | 0.709 | 0.644–0.775 | < 0.001 | 0.740 | 0.677–0.803 | < 0.001 | 0.758 | 0.696–0.819 | < 0.001 |
AUC area under the curve, CI confidence interval
Model 2
We calculated the cumulative methylation levels by adding the frequencies for the statistically significant CpG sites in each gene. The cumulative methylation levels of CYP24A1, CYP27A1, CYP27B1, CYP2R1, and VDR remained significantly different between cases and controls (P < 0.05). As shown in Table 6, the AUC of each gene was higher than that in model 1. CYP24A1 showed the highest diagnostic value, with an AUC of 0.794 (95% CI 0.737–0.851). The AUC obtained by combining the cumulative methylation levels of the CpG sites in model 2 was 0.805 (95% CI 0.749–0.860).
Model 3
We further excluded CpG sites with inverse relations between cases and controls and only included 164 hypermethylated CpG sites in the cases for analysis. The cumulative methylation levels of CYP24A1, CYP27A1, CYP27B1, CYP2R1, and VDR remained significantly different between cases and controls (P < 0.001). The CpG sites of the CYP24A1, CYP27A1, and CYP27B1 genes included in model 3 were the same as those in model 2. For the CYP2R1 and VDR genes, the AUC increased to 0.743 (95% CI 0.679–0.806) and 0.740 (95% CI: 0.677–0.803), respectively (Table 6). The AUC obtained by combining the cumulative methylation levels of all of the aforementioned CpG sites was 0.838 (95% CI 0.789–0.888).
Model 4
We included 55 methylated CpG sites that remained statistically significant after the Bonferroni correction. As shown in Table 5, CYP24A1, CYP27A1, CYP27B1, and VDR had significantly different methylation levels between cases and controls (P < 0.001). The AUC obtained by combining the cumulative methylation levels of all the aforementioned CpG sites was 0.810 (95% CI 0.754–0.866).
Interaction between serum vitamin D and methylation levels
As shown in Table 7, the methylation levels of the CYP24A1, CYP27A1, CYP27B1, and VDR genes were significantly associated with the levels of serum 25(OH)D (P < 0.05). To explore the interaction between vitamin D and methylation in the risk of TB, we categorized the genes into hyper- and hypomethylated genes based on the ROC curves and divided the serum vitamin D levels into low and high levels based on the median. There was a significant interaction between CYP27A1 methylation levels and 1,25(OH)2D concentrations in model 1 and model 4 and between the methylation levels of VDR and CYP2R1 and 1,25(OH)2D concentrations in model 3 (P < 0.05, Table 8). Based on model 4, the ORinteraction was 4.11 (95% CI 1.26–13.36, P = 0.019) (Table 9).
Table 7.
Correlation analysis between cumulative methylation levels and 25-hydroxyvitamin D levels
| Gene | Model 1 | Model 2 | Model 3 | Model 4 | ||||
|---|---|---|---|---|---|---|---|---|
| r | P | r | P | r | P | r | P | |
| CYP24A1 | 0.213 | < 0.001 | 0.287 | < 0.001 | 0.287 | < 0.001 | 0.303 | < 0.001 |
| CYP27A1 | 0.294 | < 0.001 | 0.279 | < 0.001 | 0.279 | < 0.001 | 0.227 | < 0.001 |
| CYP27B1 | 0.195 | 0.002 | 0.201 | 0.002 | 0.201 | 0.002 | 0.210 | 0.001 |
| CYP2R1 | 0.021 | 0.742 | 0.078 | 0.232 | 0.231 | < 0.001 | – | – |
| VDR | 0.177 | 0.006 | 0.202 | 0.002 | 0.238 | < 0.001 | 0.308 | < 0.001 |
r correlation coefficient
Table 8.
Interaction analysis of CpG island methylation levels and 1,25-dihydroxyvitamin D levels in TB risk
| Gene | Model 1 | Model 2 | Model 3 | Model 4 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P | |
| VDR | 1.99 | 0.68–5.84 | 0.209 | 2.72 | 0.89–8.30 | 0.078 | 3.51 | 1.11–11.12 | 0.033 | 2.45 | 0.78–7.68 | 0.125 |
| CYP27B1 | 2.39 | 0.69–8.34 | 0.171 | 2.60 | 0.68–10.01 | 0.164 | 2.60 | 0.68–10.01 | 0.164 | 2.19 | 0.67–7.18 | 0.197 |
| CYP24A1 | 1.79 | 0.51–6.29 | 0.367 | 2.52 | 0.75–8.47 | 0.136 | 2.52 | 0.75–8.47 | 0.136 | 2.39 | 0.69–8.29 | 0.169 |
| CYP2R1 | 1.19 | 0.40–3.47 | 0.756 | 2.06 | 0.71–5.98 | 0.184 | 4.34 | 1.30–14.49 | 0.017 | – | – | – |
| CYP27A1 | 3.49 | 1.10–11.08 | 0.034 | 2.47 | 0.77–7.98 | 0.129 | 2.47 | 0.77–7.98 | 0.129 | 4.11 | 1.26–13.36 | 0.019 |
Table 9.
Crossover analysis of CYP27A1 methylation levels and 1,25-dihydroxyvitamin D levels in the risk of TB
| 1,25(OH)2Da | Cumulative methylation levelb | Cases | Controls | OR | 95% CI |
|---|---|---|---|---|---|
| Low | High | 23 | 50 | 0.30 | 0.14–0.66 |
| High | High | 21 | 42 | 0.33 | 0.15–0.73 |
| Low | Low | 52 | 9 | 3.78 | 1.48–9.62 |
| High | Low | 26 | 17 | 1 | |
| ORinteraction = 4.11 (95% CI 1.26–13.36, P = 0.019) | |||||
aCutoff point of 1,25(OH)2D 86 pmol/L
bCutoff point of cumulative methylation level of CYP27A1 0.48
Treatment outcomes of patients with TB
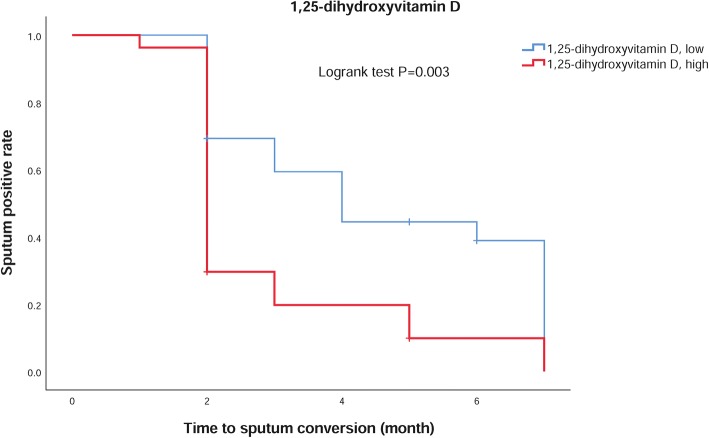
The baseline serum 1,25(OH)2D concentration was related to the treatment outcome (Z = − 2.655, P = 0.008). Patients with high serum 1,25(OH)2D concentrations had a decreased risk of treatment failure (adjusted relative risk (RR) 0.07, 95% CI 0.01–0.39). As shown in Fig. 1, the sputum conversion rate was significantly higher in patients with higher serum 1,25(OH)2D levels (χ2 = 8.85, P = 0.003).
Fig. 1.
Time to sputum conversion in TB patients and 1,25-dihydroxyvitamin D concentration
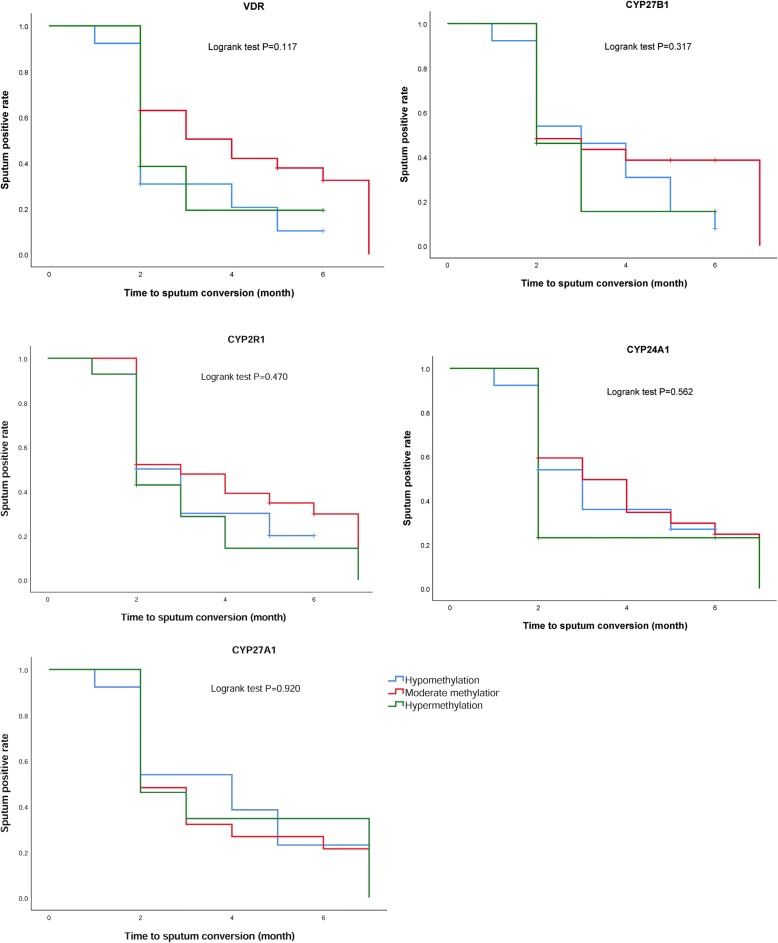
There were 23 CpG sites that were significantly related to the treatment outcomes (P < 0.05). The percentage of differentially methylated sites was 14.6% in the CYP24A1 gene, 2.7% in the CYP27A1 gene, 5.7% in the CYP27B1 gene, 5.9% in the CYP2R1 gene, and 10.7% in the VDR gene. However, no CpG sites remained significant after the Bonferroni correction. The cumulative methylation levels were categorized into three groups based on quartile (hypomethylation P0–P25; moderate methylation P25–P75; hypermethylation P75–P100.). No significant relation was found between cumulative methylation level and sputum bacterium conversion (Fig. 2). We conducted crossover analysis and found no evidence of an interaction between CYP27A1 methylation level and 1,25(OH)2D concentration in sputum bacterium conversion (Table 10).
Fig. 2.
Time to sputum conversion in TB patients and cumulative methylation levels of genes in the vitamin D metabolic pathway. The cumulative methylation levels are categorized into three groups based on quartile. Hypomethylation, P0–P25. Moderate methylation, P25–P75. Hypermethylation, P75–P100
Table 10.
Crossover analysis of CYP27A1 methylation levels and 1,25-dihydroxyvitamin D levels in the sputum conversion of patients with TB
| 1,25(OH)2Da | Cumulative methylation levelb | Sputum conversion | OR | 95% CI | |
|---|---|---|---|---|---|
| Yes | No | ||||
| Low | High | 9 | 3 | 0.27 | 0.02–3.09 |
| High | High | 10 | 4 | 0.23 | 0.02–2.39 |
| Low | Low | 11 | 4 | 0.25 | 0.02–2.61 |
| High | Low | 11 | 1 | 1 | |
aCutoff point of 1,25(OH)2D 79 pmol/L
bCutoff point of cumulative methylation level of CYP27A1 0.435
Discussion
Vitamins are being revisited for their role in pathogenicity and for their antimycobacterial properties. Vitamin C and vitamin D have been shown to possess antimycobacterial properties [9]. Previous studies have reported an association between vitamin D and immunity against TB [22, 23]. High levels of vitamin D can decrease the reactivation of latent TB and reduce the severity of active TB [24–27]. Vitamin D deficiency is believed to be a risk factor for the acquisition of TB infection [28, 29]. However, vitamin D supplementation to improve outcomes in TB patients has resulted in contradictory results [30, 31] that may be partly attributable to individual variation in vitamin D metabolic capacity and immunity. Previous studies have suggested that activation or silencing of certain signaling pathways plays a role in TB development [32]. Genetic information is carried not only in DNA sequences but also in epigenetic variations [33, 34]. In this study, we used next-generation sequencing to quantify the methylation levels of five vitamin D metabolic pathway genes and observed a significant association with the risk of TB.
Vitamin D has two main active metabolites: 25(OH)D and 1,25(OH)2D [35]. It binds to the vitamin D receptor on the membrane or cell nucleus to begin its activity in the transcription process [36]. In the present study, we observed that TB patients had significantly lower serum 1,25(OH)2D concentrations than controls and that the concentrations decreased after the initiation of antituberculosis therapy. 1,25(OH)2D is the biologically active form of vitamin D [37] and is associated with treatment outcomes of TB patients. The higher the 1,25(OH)2D concentration, the greater the likelihood of successful treatment and the higher the sputum negative conversion rate. Our findings support the hypothesis that we can assist TB treatment by increasing sunlight radiation and vitamin D intake. In a randomized double-blind placebo-controlled trial among 120 Mongolian children, vitamin D supplementation seemed to prevent the conversion of the tuberculin skin test [38]. However, another report suggested that the time to sputum conversion was not reduced with vitamin D supplementation [17].
VDR is essential for adequate immune function [22]. One study examined the methylation statuses of 17 CpGs in VDR and determined their relation to TB [39]. Our findings showed that hypomethylation of the VDR promoter may be a potential biomarker of TB, but its impact on VDR expression remains to be studied. CYP2R1 is a hepatic microsomal enzyme responsible for the 25-hydroxylation of vitamin D that is highly conserved among species ranging from fish to humans [40, 41]. CYP24A1 encodes 24-hydroxylase, which initiates the metabolism of both 25(OH)D and 1,25(OH)2D [40]. Zhou et al. conducted an intervention trial with vitamin D supplementation in postmenopausal women and reported that the baseline DNA methylation levels of CYP2R1 and CYP24A1 were negatively correlated with the concentrations of active 25(OH)D metabolite after vitamin D supplementation and that subjects with high DNA methylation levels needed higher vitamin D supplementation to reach optimal serum levels [42]. The CYP27A1 gene codes for 27-hydroxylase [43]. The methylation of CYP27A1 has been reported to be associated with the balance of bile acid in nonalcoholic fatty liver disease and in drug metabolism [44]. CYP27B1 catalyzes the de novo production of 1,25(OH)2D from accumulated 25(OH)D; 1,25(OH)2D is delivered to the cells via the vitamin D-binding protein (DBP), which is encoded by GC. The liganded VDR-transcription factor complex binds to vitamin D response elements (VDREs) in cathelicidin antimicrobial peptide (CAMP), activating CAMP expression [45]. The methylation level of CYP27B1 is elevated in primary lymphoma and leukemia cells [46].
There are several limitations in this study. First, we only measured the vitamin D levels at the baseline and at the end of the intensive treatment stage of TB patients; long-term continuous monitoring for the whole course of the disease may provide more information. Second, the regulation of gene transcription is complex, including genetic variation, modulation of the interactions of control factors with the transcriptional machinery, and epigenetic modification. These regulatory pathways do not function independently. Previous studies have reported methylation variation in association with genetic variation across individuals [47]. Genetic and epigenetic mechanisms may interact and together affect biological processes and disease development [48]. In this study, we only measured the methylation of the promoter regions of key genes in the vitamin D metabolic pathway. We noticed that the difference in methylation was subtle between cases and controls, although the P value was significant. Whether this subtle methylation difference could alter vitamin D metabolism and change macrophage M.tb killing capacity is not clear and needs more exploration. Other factors, such as genetic polymorphisms, sunlight exposure, food intake, and drug supplementation, can also affect vitamin D levels and the risk of TB [45]. Third, immune responses downstream of the vitamin D metabolic pathway should also be considered. Vitamin D has no direct antimycobacterial action, but its active metabolite 1,25(OH)2D modulates host responses to M.tb infection [49]. 1,25(OH)2D has been shown to induce antimycobacterial activity in macrophages in vitro, upregulate protective innate host responses, and trigger antimicrobial peptides such as cathelicidin [50]. M.tb entry into the body is mediated by macrophage toll-like receptors, which induce antibacterial autophagy by upregulating and activating VDR and increasing 1a-hydroxylase. Calcitriol activation of VDR induces CAMP gene expression and the subsequent production of cathelicidin, which disrupts the bacterial cell membrane and induces autophagy in monocytes [51]. Exogenous 1,25(OH)2D induces a superoxide burst and enhances phagolysosome fusion in M.tb-infected macrophages [49]. Simultaneously, the T cells secrete IFN-g, which promotes antimicrobial peptide expression, autophagy, phagosomal maturation, and antimicrobial action against M.tb within macrophages [51]. In this study, we did not evaluate the immune responses downstream of the vitamin D metabolic pathway. These immune responses should be considered in future studies.
In conclusion, our results suggest that the methylation levels of the CYP24A1, CYP27A1, CYP27B1, CYP2R1, and VDR genes in the metabolic pathway of vitamin D are related to the risk and prognosis of TB. Investigating the role of abnormal metabolism of vitamin D is important for the prevention and control of TB. Individualized vitamin D intervention based on epigenetic traits of key genes in its metabolic pathway will be valuable for the prevention and control of tuberculosis.
Additional file
Spearman’s correlation for methylation levels in each region with 1,25-dihydroxyvitamin D levels. (DOC 37 kb)
Acknowledgements
We would like to thank all the participants for their contribution.
Funding
This work was supported by the National Natural Science Foundation of China (81473027, 81673249), the National Key R&D Program of China (2017YFC0907000), the Social Development Project in Jiangsu Province (BE2015694), Scientific Research Innovation Project for Graduate Students in Jiangsu Province (KYCX17_1293), the Six Talent Peaks Project in Jiangsu Province (2014-YY-023), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Abbreviations
- 1,25(OH)2D
1,25-dihydroxyvitamin D
- 25(OH)D
25-hydroxyvitamin D
- AUC
Area under the curve
- BCG
Bacillus Calmette-Guérin
- CAMP
Cathelicidin antimicrobial peptide
- CI
Confidence interval
- CV
Coefficient of variation
- DBP
Vitamin D-binding protein
- M.tb
Mycobacterium tuberculosis
- OR
Odds ratio
- ROC
Receiver operating characteristic
- RR
Relative risk
- SD
Standard deviation
- TB
Tuberculosis
- VDREs
Vitamin D response elements
Authors’ contributions
MW and JW conceived, initiated, and led the study. MW, WK, BH, ZL, HS, PS, and JW analyzed the data with input from all the authors. MW, WK, and JW prepared the manuscript. All authors reviewed and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Nanjing Medical University. After informed consent was obtained from all participants, questionnaires were used to collect demographic data.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kaushal D, Foreman TW, Gautam US, Alvarez X, Adekambi T, Rangel-Moreno J, Golden NA, Johnson AM, Phillips BL, Ahsan MH, et al. Mucosal vaccination with attenuated Mycobacterium tuberculosis induces strong central memory responses and protects against tuberculosis. Nat Commun. 2015;6:8533. doi: 10.1038/ncomms9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu G, Mao X, Wang J, Pan H. Clustering and recent transmission of Mycobacterium tuberculosis in a Chinese population. Infect Drug Resist. 2018;11:323–330. doi: 10.2147/IDR.S156534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO. Global Tuberculosis report 2017. 2017; http://www.who.int/tb/publications/global_report/en/. Accessed 1 Aug 2018.
- 4.Maro I, Lahey T, MacKenzie T, Mtei L, Bakari M, Matee M, Pallangyo K, von Reyn CF. Low BMI and falling BMI predict HIV-associated tuberculosis: a prospective study in Tanzania. Int J Tuberc Lung Dis. 2010;14(11):1447–1453. [PMC free article] [PubMed] [Google Scholar]
- 5.Rhines AS. The role of sex differences in the prevalence and transmission of tuberculosis. Tuberculosis (Edinb) 2013;93(1):104–107. doi: 10.1016/j.tube.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 6.Melsew YA, Doan TN, Gambhir M, Cheng AC, McBryde E, Trauer JM. Risk factors for infectiousness of patients with tuberculosis: a systematic review and meta-analysis. Epidemiol Infect. 2018;146(3):345–353. doi: 10.1017/S0950268817003041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng Y, Wang F, Pan H, Qiu S, Lu J, Wu L, Wang J, Lu C. Obesity-associated gene FTO rs9939609 polymorphism in relation to the risk of tuberculosis. BMC Infect Dis. 2014;14:592. doi: 10.1186/s12879-014-0592-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu J, Pan H, Chen Y, Tang S, Feng Y, Qiu S, Zhang S, Wu L, Xu R, Peng X, et al. Genetic polymorphisms of IFNG and IFNGR1 in association with the risk of pulmonary tuberculosis. Gene. 2014;543(1):140–144. doi: 10.1016/j.gene.2014.03.042. [DOI] [PubMed] [Google Scholar]
- 9.Tyagi G, Singh P, Varma-Basil M, Bose M. Role of vitamins B, C, and D in the fight against tuberculosis. Int J Mycobacteriol. 2017;6(4):328–332. doi: 10.4103/ijmy.ijmy_80_17. [DOI] [PubMed] [Google Scholar]
- 10.Venturini E, Facchini L, Martinez-Alier N, Novelli V, Galli L, de Martino M, Chiappini E. Vitamin D and tuberculosis: a multicenter study in children. BMC Infect Dis. 2014;14:652. doi: 10.1186/s12879-014-0652-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keflie TS, Nolle N, Lambert C, Nohr D, Biesalski HK. Vitamin D deficiencies among tuberculosis patients in Africa: a systematic review. Nutrition. 2015;31(10):1204–1212. doi: 10.1016/j.nut.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Nnoaham KE, Clarke A. Low serum vitamin D levels and tuberculosis: a systematic review and meta-analysis. Int J Epidemiol. 2008;37(1):113–119. doi: 10.1093/ije/dym247. [DOI] [PubMed] [Google Scholar]
- 13.Zeng J, Wu G, Yang W, Gu X, Liang W, Yao Y, Song Y. A serum vitamin D level <25nmol/l pose high tuberculosis risk: a meta-analysis. PLoS One. 2015;10(5):e0126014. doi: 10.1371/journal.pone.0126014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Selvaraj P, Harishankar M, Afsal K, Vitamin D. Immuno-modulation and tuberculosis treatment. Can J Physiol Pharmacol. 2015;93(5):377–384. doi: 10.1139/cjpp-2014-0386. [DOI] [PubMed] [Google Scholar]
- 15.Nursyam EW, Amin Z, Rumende CM. The effect of vitamin D as supplementary treatment in patients with moderately advanced pulmonary tuberculous lesion. Acta Med Indones. 2006;38(1):3–5. [PubMed] [Google Scholar]
- 16.Martineau AR, Timms PM, Bothamley GH, Hanifa Y, Islam K, Claxton AP, Packe GE, Moore-Gillon JC, Darmalingam M, Davidson RN, et al. High-dose vitamin D(3) during intensive-phase antimicrobial treatment of pulmonary tuberculosis: a double-blind randomised controlled trial. Lancet. 2011;377(9761):242–250. doi: 10.1016/S0140-6736(10)61889-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia J, Shi L, Zhao L, Xu F. Impact of vitamin D supplementation on the outcome of tuberculosis treatment: a systematic review and meta-analysis of randomized controlled trials. Chin Med J. 2014;127(17):3127–3134. [PubMed] [Google Scholar]
- 18.Scragg R. Limitations of vitamin D supplementation trials: why observational studies will continue to help determine the role of vitamin D in health. J Steroid Biochem Mol Biol. 2018;177:6–9. doi: 10.1016/j.jsbmb.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 19.Manjrekar J. Epigenetic inheritance, prions and evolution. J Genet. 2017;96(3):445–456. doi: 10.1007/s12041-017-0798-3. [DOI] [PubMed] [Google Scholar]
- 20.Guastafierro T, Bacalini MG, Marcoccia A, Gentilini D, Pisoni S, Di Blasio AM, Corsi A, Franceschi C, Raimondo D, Spano A, et al. Genome-wide DNA methylation analysis in blood cells from patients with Werner syndrome. Clin Epigenetics. 2017;9:92. doi: 10.1186/s13148-017-0389-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu H, Wang X, Shi H, Su S, Harshfield GA, Gutin B, Snieder H, Dong Y. A genome-wide methylation study of severe vitamin D deficiency in African American adolescents. J Pediatr. 2013;162(5):1004–1009. doi: 10.1016/j.jpeds.2012.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wahyunitisari MR, Mertaniasih NM, Amin M, Artama WT, Koendhori EB. Vitamin D, cell death pathways, and tuberculosis. Int J Mycobacteriol. 2017;6(4):349–355. doi: 10.4103/ijmy.ijmy_120_17. [DOI] [PubMed] [Google Scholar]
- 23.Tessema B, Moges F, Habte D, Hiruy N, Yismaw S, Melkieneh K, Kassie Y, Girma B, Melese M, Suarez PG. Vitamin D deficiency among smear positive pulmonary tuberculosis patients and their tuberculosis negative household contacts in Northwest Ethiopia: a case-control study. Ann Clin Microbiol Antimicrob. 2017;16(1):36. doi: 10.1186/s12941-017-0211-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eklund D, Persson HL, Larsson M, Welin A, Idh J, Paues J, Fransson SG, Stendahl O, Schon T, Lerm M. Vitamin D enhances IL-1beta secretion and restricts growth of Mycobacterium tuberculosis in macrophages from TB patients. Int J Mycobacteriol. 2013;2(1):18–25. doi: 10.1016/j.ijmyco.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Harishankar M, Selvaraj P. Influence of Cdx2 and TaqI gene variants on vitamin D3 modulated intracellular chemokine positive T-cell subsets in pulmonary tuberculosis. Clin Ther. 2017;39(5):946–957. doi: 10.1016/j.clinthera.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 26.Huang SJ, Wang XH, Liu ZD, Cao WL, Han Y, Ma AG, Xu SF. Vitamin D deficiency and the risk of tuberculosis: a meta-analysis. Drug Des Devel Ther. 2017;11:91–102. doi: 10.2147/DDDT.S79870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Azam F, Shaheen A, Arshad R. Frequency of hypovitaminosis D and its associated risk factors in newly diagnosed pulmonary tuberculosis patients. Pak J Med Sci. 2016;32(2):480–484. doi: 10.12669/pjms.322.8996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iftikhar R, Kamran SM, Qadir A, Haider E, Bin UH. Vitamin D deficiency in patients with tuberculosis. J Coll Physicians Surg Pak. 2013;23(10):780–783. [PubMed] [Google Scholar]
- 29.Talat N, Perry S, Parsonnet J, Dawood G, Hussain R. Vitamin d deficiency and tuberculosis progression. Emerg Infect Dis. 2010;16(5):853–855. doi: 10.3201/eid1605.091693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharma V, Mandavdhare HS, Kumar A, Sharma R, Sachdeva N, Prasad KK, Rana SS. Prevalence and clinical impact of vitamin D deficiency on abdominal tuberculosis. Ther Adv Infect Dis. 2017;4(3):83–86. doi: 10.1177/2049936117709722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi R, Jeong BH, Koh WJ, Lee SY. Recommendations for optimizing tuberculosis treatment: therapeutic drug monitoring, pharmacogenetics, and nutritional status considerations. Ann Lab Med. 2017;37(2):97–107. doi: 10.3343/alm.2017.37.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Esterhuyse MM, Weiner J, 3rd, Caron E, Loxton AG, Iannaccone M, Wagman C, Saikali P, Stanley K, Wolski WE, Mollenkopf HJ, et al. Epigenetics and proteomics join transcriptomics in the quest for tuberculosis biomarkers. MBio. 2015;6(5):e01187–e01115. doi: 10.1128/mBio.01187-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He L, Gao L, Shi Z, Li Y, Zhu L, Li S, Zhang P, Zheng G, Ren Q, Li Y, et al. Involvement of cytochrome P450 1A1 and glutathione S-transferase P1 polymorphisms and promoter hypermethylation in the progression of anti-tuberculosis drug-induced liver injury: a case-control study. PLoS One. 2015;10(3):e0119481. doi: 10.1371/journal.pone.0119481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shnorhavorian M, Schwartz SM, Stansfeld B, Sadler-Riggleman I, Beck D, Skinner MK. Differential DNA methylation regions in adult human sperm following adolescent chemotherapy: potential for epigenetic inheritance. PLoS One. 2017;12(2):e0170085. doi: 10.1371/journal.pone.0170085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zerwekh JE. Blood biomarkers of vitamin D status. Am J Clin Nutr. 2008;87(4):1087S–1091S. doi: 10.1093/ajcn/87.4.1087S. [DOI] [PubMed] [Google Scholar]
- 36.Norval M, Coussens AK, Wilkinson RJ, Bornman L, Lucas RM, Wright CY. Vitamin D status and its consequences for health in South Africa. Int J Environ Res Public Health. 2016;13(10):1019. doi: 10.3390/ijerph13101019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hussein H, Ibrahim F, Boudou P. Evaluation of a new automated assay for the measurement of circulating 1,25-dihydroxyvitamin D levels in daily practice. Clin Biochem. 2015;48(16–17):1160–1162. doi: 10.1016/j.clinbiochem.2015.05.018. [DOI] [PubMed] [Google Scholar]
- 38.Verrall AJ, Netea MG, Alisjahbana B, Hill PC, van Crevel R. Early clearance of Mycobacterium tuberculosis: a new frontier in prevention. Immunology. 2014;141(4):506–513. doi: 10.1111/imm.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andraos C, Koorsen G, Knight JC, Bornman L. Vitamin D receptor gene methylation is associated with ethnicity, tuberculosis, and TaqI polymorphism. Hum Immunol. 2011;72(3):262–268. doi: 10.1016/j.humimm.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang TJ, Zhang F, Richards JB, Kestenbaum B, van Meurs JB, Berry D, Kiel DP, Streeten EA, Ohlsson C, Koller DL, et al. Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet. 2010;376(9736):180–188. doi: 10.1016/S0140-6736(10)60588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng JB, Levine MA, Bell NH, Mangelsdorf DJ, Russell DW. Genetic evidence that the human CYP2R1 enzyme is a key vitamin D 25-hydroxylase. Proc Natl Acad Sci U S A. 2004;101(20):7711–7715. doi: 10.1073/pnas.0402490101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Y, Zhao LJ, Xu X, Ye A, Travers-Gustafson D, Zhou B, Wang HW, Zhang W, Lee Hamm L, Deng HW, et al. DNA methylation levels of CYP2R1 and CYP24A1 predict vitamin D response variation. J Steroid Biochem Mol Biol. 2014;144:207–214. doi: 10.1016/j.jsbmb.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nie S, Chen G, Cao X, Zhang Y. Cerebrotendinous xanthomatosis: a comprehensive review of pathogenesis, clinical manifestations, diagnosis, and management. Orphanet J Rare Dis. 2014;9:179. doi: 10.1186/s13023-014-0179-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schioth HB, Bostrom A, Murphy SK, Erhart W, Hampe J, Moylan C, Mwinyi J. A targeted analysis reveals relevant shifts in the methylation and transcription of genes responsible for bile acid homeostasis and drug metabolism in non-alcoholic fatty liver disease. BMC Genomics. 2016;17:462. doi: 10.1186/s12864-016-2814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meyer V, Saccone DS, Tugizimana F, Asani FF, Jeffery TJ, Bornman L. Methylation of the vitamin D receptor (VDR) gene, together with genetic variation, race, and environment influence the signaling efficacy of the toll-like receptor 2/1-VDR pathway. Front Immunol. 2017;8:1048. doi: 10.3389/fimmu.2017.01048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wjst M, Heimbeck I, Kutschke D, Pukelsheim K. Epigenetic regulation of vitamin D converting enzymes. J Steroid Biochem Mol Biol. 2010;121(1–2):80–83. doi: 10.1016/j.jsbmb.2010.03.056. [DOI] [PubMed] [Google Scholar]
- 47.Schultz MD, He Y, Whitaker JW, Hariharan M, Mukamel EA, Leung D, Rajagopal N, Nery JR, Urich MA, Chen H, et al. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature. 2015;523(7559):212–216. doi: 10.1038/nature14465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olsson AH, Volkov P, Bacos K, Dayeh T, Hall E, Nilsson EA, Ladenvall C, Ronn T, Ling C. Genome-wide associations between genetic and epigenetic variation influence mRNA expression and insulin secretion in human pancreatic islets. PLoS Genet. 2014;10(11):e1004735. doi: 10.1371/journal.pgen.1004735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martineau AR, Honecker FU, Wilkinson RJ, Griffiths CJ. Vitamin D in the treatment of pulmonary tuberculosis. J Steroid Biochem Mol Biol. 2007;103(3–5):793–798. doi: 10.1016/j.jsbmb.2006.12.052. [DOI] [PubMed] [Google Scholar]
- 50.Wejse C, Gomes VF, Rabna P, Gustafson P, Aaby P, Lisse IM, Andersen PL, Glerup H, Sodemann M. Vitamin D as supplementary treatment for tuberculosis: a double-blind, randomized, placebo-controlled trial. Am J Respir Crit Care Med. 2009;179(9):843–850. doi: 10.1164/rccm.200804-567OC. [DOI] [PubMed] [Google Scholar]
- 51.Turnbull ER, Drobniewski F. Vitamin D supplementation: a comprehensive review on supplementation for tuberculosis prophylaxis. Expert Rev Respir Med. 2015;9(3):269–275. doi: 10.1586/17476348.2015.1042458. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Spearman’s correlation for methylation levels in each region with 1,25-dihydroxyvitamin D levels. (DOC 37 kb)
Data Availability Statement
All data generated or analyzed during this study are included in this published article.