Abstract
We cloned and characterized the full-length coding sequence of a small heat shock (sHSP) gene, PfHSP17.2, from Primula forrestii leaves following heat stress treatment. Homology and phylogenetic analysis suggested that PfHSP17.2 is a cytosolic class II sHSP, which was further supported by the cytosolic localization of transient expression of PfHSP17.2 fused with green fluorescent protein reporter. Expression analysis showed that PfHSP17.2 was highly inducible by heat stress in almost all the vegetative and generative tissues and was expressed under salt, cold, and oxidative stress conditions as well. Moreover, the expression of PfHSP17.2 in P. forrestii was detected in certain developmental growth stages. Transgenic Arabidopsis thaliana constitutively expressing PfHSP17.2 displayed increased thermotolerance and higher resistance to salt and cold compared with wild type plants. It is suggested that PfHSP17.2 plays a key role in heat and other abiotic stresses.
Keywords: sHSPs, stress tolerance, Arabidopsis
Introduction
Abiotic stresses such as high temperature, drought, salt, cold, and oxidative damage are the main factors impacting plant growth (Vermeulen et al., 2012; Nelson et al., 2014; Kumar et al., 2015). In response to these stresses, plants have developed a wide range of physiological and biochemical mechanisms to protect themselves from damage (Cabane et al., 1993; Sun et al., 2001; Murakami et al., 2004; Hu et al., 2009; Ahuja et al., 2010, Perez-Clemente et al., 2013; Ruibal et al., 2013). Heat shock proteins (HSPs), which could be induced in almost all organisms by high temperature and other abiotic and biotic stresses, were accumulated in plants as one of the strategies to deal with stressful conditions (Derocher et al., 1991; Lee et al., 1997; Malik et al., 1999; Zhou et al., 2012; Park et al., 2015; Zhai et al., 2016). HSPs are evolutionarily conserved proteins. They are divided into five subfamilies based on their apparent molecular weights, viz. HSP100, HSP90, HSP70, HSP60, and small HSPs (Boston et al., 1996). Among them, small heat shock proteins (sHSPs), which could either protect the plant from damage caused by the stress or help repair the damage, were proved to have vital roles in response to abiotic stresses (Chao et al., 2009; Lin et al., 2010; Wang et al., 2015). According to sequence homology, cellular compartmentation, and immunological properties, sHSPs are classified into 11 different classes: classes I–VI, localized in nucleus or cytoplasm, and the other five positioned in mitochondria, chloroplast, peroxisomes, and endoplasmic reticulum (Waters et al., 2008).
Although sHSPs are highly conserved proteins of around 15 to 42 KDa and characterized by possession of a a-crystalline domain at their C-terminus, their unusual abundance and diversity in plants is considerable and the mechanism of sHSP action is not fully understood. Apart from elevated temperatures, sHSP accumulation could also be triggered by other stress conditions, such as drought, salinity, and cold, and by developmental regulation, such as seed germination, embryogenesis, and fruit development (Coca et al., 1994; zur Nieden et al., 1995; Sun et al., 2001, 2002; Chauhan et al., 2012; Koo et al., 2015). The cytosolic class I and II proteins were found in the absence of stress in maturing seeds of several species such as Nelumbo and wheat (Efeoglu, 2009; Zhou et al., 2012). It has been observed that in maize and other plants, different HSPs express abundantly during pollen development (Magnard et al., 1996; Young et al., 2001; Kumar et al., 2015). Furthermore, overexpression of sHSPs in plants enhance their tolerance to abiotic stresses (Ahn and Zimmerman, 2006; Maimbo et al., 2007; Jiang et al., 2009; Chauhan et al., 2012), whereas plants with reduced expression of sHSP exhibit compromised tolerance to heat resistance (Charng et al., 2006) and disease (Maimbo et al., 2007; Ju et al., 2017).
In our previous study (Hu et al., 2010), Primula forrestii exhibited superior thermal tolerance under heat stress compared with six other Primula species including P. malacoides, P. obconica, P. veris, P. saxatilis, and P. denticulata. Using the suppression subtractive hybridization (SSH) method, we found a sHSP gene that was remarkably up-regulated in the leaves of P. forrestii under high temperature treatment (42 °C for 2 h). In the present study, the full-length cDNA of this gene, termed PfHSP17.2, was isolated and characterized from the heat stressed leaves of P. forrestii. It was significantly induced by other abiotic stresses, such as salt, cold, and oxidative stress, and its expression could also be detected at certain stages of seed development and germination. Transgenic Arabidopsis plants overexpressing PfHSP17.2 displayed an improved salt and temperature tolerance compared with wild type (WT) plants.
Material and Methods
Plant material and growth conditions
Seeds of P. forrestii were harvested from Xiaotangshan experimental fields in Beijing, and the original seeds were collected from Lijiang in the Yuman province in 2007. Seeds were sown in a sterilized soil incubator for germination in a greenhouse maintained at 25 ± 3 °C and at 75-80% relative humidity. Four weeks later, seedlings were transferred into black plastic pots containing 3/4 peat moss and 1/4 perlite, and watered with standard Hoagland solution every 15 d.
Arabidopsis thaliana (ecotype Columbia-0) plants were grown in-soil in the greenhouse (16 h light/8 h dark, 70% relative humidity, 22-25 °C) or in Petri dishes containing Murashige and Skoog (MS) medium with 4.4 gL-1 MS salts, 1% sucrose, and 0.8% agar, pH 5.7. In all experiments, seeds of WT and transgenic lines produced in the same culture cycle were used for analyses.
Full-length PfHSP17.2 cDNA cloning and bioinformatics analysis
Total RNA was isolated from P. forrestii treated with heat stress for 2 h using TRIZOL reagent (Invitrogen) according to the manufacturer’s instructions. cDNA library construction was performed using a SMART cDNA library construction kit (Clontech). The full-length cDNA sequence of the PfHSP17.2 gene was directly cloned by screening the cDNA library using expressed sequence tag (EST) analysis. The functional annotation of ESTs was performed using BLASTX and BLASTN. Protein sequences from species other than P. forrestii were retrieved from GenBank. Then, protein sequences of PfHSP17.2 and other cytosolic class II sHSPs from various plant species were aligned by CLUSTAL W. The phylogenetic tree was constructed by MEGA3 with the neighbor-joining algorithm using default settings.
PfHSP17.2 gene expression profiles in response to abiotic stress
In response to different abiotic stress conditions, the expression profiles of the PfHSP17.2 gene were analyzed using semi-quantitative RT-PCR. Total RNA was extracted from each sample using Plant RNA Extraction kit (TianGen). One μg of RNA was reverse-transcribed in a total volume of 10 μL with 1 μL oligo(dT)18 primer (100 pM), 1.0 μL dNTPs (10 mM), 2 μL 5 M-MLV buffer, 0.25 RNase inhibitor (40 U/μL), and 0.5 μL reverse-transcriptase M-MLV (200 U/μL). The following gene-specific primers were used: Pf2-F2 (5’-GAGAACACGGGGGAC TCTTGACCAT-3’), Pf2-R2 (5’-ATCGGGGAAATTCG AGCTGGTCACC-3’), Actin-F (5’-TCTGGCATCATAC CTTCTACA-3’), and Actin-R (5’-GGATGGCTGGAAG AGGAC-3’). The semi-quantitative RT-PCR was performed in a total volume of 25 μL with the following amplification conditions: 4 min at 94 °C; 30 cycles of 30 s at 94 °C; 40 s at 55 °C; 1 min at 72 °C; and lastly 6 min at 72 °C. PCR products were analyzed by 0.8% agarose gel electrophoresis. All the experiments were performed in triplicate.
Subcellular location analysis
The full-length PfHSP17.2 coding region (without the stop codon) was ligated to the 3’ end of a green fluorescent protein (GFP) to generate a PfHSP17.2–GFP fusion construct, which was driven by the cauliflower mosaic virus (CaMV) 35S promoter. An empty pCAMBIA 1302 was used as a control. The PfHSP17.2–GFP construct and 35S-GFP (control) were introduced into onion epidermal cells by particle bombardment (Bio-Rad, CA, USA). Following bombardment, the cells were incubated in the dark at 25°C for 2–3 days prior to observation. Transiently transformed cells were analyzed using a confocal laser scanning microscope.
Stress treatments of P. forrestii
Two-month-old plants of P. forrestii were treated with half-strength MS salt solution containing either 200 mm NaCl (salt stress treatment), 20% PEG (drought stress treatment), 0.05% H2O2 (oxidative stress treatment) or 100 μM ABA. The roots, petioles, leaves, and flowers of the treated plants were collected after a 2-h treatment. For heat and cold stress treatment, two-month-old plants of P. forrestii grown in normal conditions were transferred to 42 °C or 4 °C for 12 h. Then, leaves were collected at 0, 10, and 20 min, and 0.5, 1, 1.5, 2, 4, 6, and 12 h after initiation of stress treatment. Plant material was placed directly into liquid nitrogen and stored at -80 °C until use.
Generation of transgenic Arabidopsis plants
The sequencing-confirmed coding region of PfHSP17.2 cDNA was cloned into the pCAMBIA1301 vector under the control of a CaMV 35S RNA promoter. The pCAMBIA1301-35S-PfHSP17.2 recombinant vector was subsequently transferred into Agrobacterium GV3101 and were transformed into Arabidopsis wild type carried out by the floral dip method (Clough and Bent, 1998).
The seeds of T0 generation were harvested and sown in MS. Two-week-old seedlings of T1 plants were screened by hygromycin. Positive plants were transferred into soil and grown in a greenhouse at 25/22 °C (day/night) with a 16-h light/8-h dark photoperiod and 70% relative humidity. The surviving transformants (T1) were confirmed by PCR using the primer pairs Pf2-F1 and Pf2-R1 mentioned above. Actin (At)-F (5’-AGGTAATCAGTAAGGTCACGG-3’) and Actin (At)-R (5’-GGATGGCTGGAAGAGGAC-3’) were used to amplify the Arabidopsis Actin gene fragment as an internal control. T2 seeds were placed on MS agar medium containing hygromycin and the transgenic lines with a 3:1 segregation ratio (resistant:sensitive) were selected to produce T3 seeds. The T3 lines displaying 100% hygromycin resistance were considered homozygous and used for further experiments. The Arabidopsis WT was used as control. All seeds of WT and transgenic plants were collected at the same stage.
Stress treatment of transgenic Arabidopsis plants
For heat stress treatment, wild type and transgenic seedlings were germinated on the same MS medium containing agar in Petri dishes. The Petri dishes were immersed in a water bath at different temperatures for heat tolerance assays. Two 10-day-old transgenic lines and a wild-type control line were exposed to 43 °C for 1 h for the basal thermotolerance assay and to 37 °C for 1 h, 22 °C for 3 h, and 47 °C for 1 h for the acquired thermotolerance assay (modified from Zhu et al., 2009). About 35 plants of each genotype were used. After incubation at 22 °C for 3 d, photographs were taken and the survival rates were calculated.
For the hypocotyl thermotolerance experiment, seedlings were grown on the same MS medium plates. The plates were kept in the dark for 3 d and then transferred to 45 °C for 2 h. Subsequently, the seedlings were incubated under 22 °C. After 3 d recovery, the extent of hypocotyl elongation was measured (Hong and Vierling, 2000).
To evaluate whole plant tolerance to heat stress, 3-week-old wild-type and transgenic seedlings incubated in pots (1 plant per pot) were transferred to 42 ± 1 °C, at relative humidity of 70% for 6 h, and then returned to 22 °C for 5 d. Physiological parameters were assessed immediately after this stress treatment, while photographs were taken after a 5-d recovery (Zhang et al., 2013).
For salt stress treatment, seeds from transgenic and control lines were germinated on MS plates supplemented with 0 mm NaCl, 150 mm NaCl, and 200 mm NaCl respectively. The seeds were stratified by incubation in the dark at 4 °C for 3 d prior to placement under 16-h light/8-h dark conditions with 70% relative humidity and 22-25°C. Germination rates were scored after 10 d. Seedlings were grown in MS medium for 7 d and then transferred to MS medium containing NaCl for 10 d, and the number of lateral roots and the root lengths were measured and calculated (modified from Liu et al., 2011).
In addition, 7-day-old seedlings of wild type and transgenic Arabidopsis from MS agar medium were grown in potted soil for a week. Then, the plants were irrigated with 30 mL of a 300 mm NaCl solution for 7 d and then irrigated again for 3 d with pure water for recovery (Zhang et al., 2008). Physiological parameters were assessed immediately after this stress treatment, while photographs were taken after a 7-d recovery.
For freezing stress treatment, 3-week-old transgenic and wild type Arabidopsis plants were exposed to -10 °C for 2 h, and then returned to normal conditions. Frequency of survival was determined and photographs were taken after 7 d. In addition, 6-week-old transgenic and control line seedlings in each of three independent experiments were incubated at 4 °C for 24 h, removed at desired freezing temperature for 30 h, thawed at 4 °C for 12 h in the dark, and then returned to the original growth condition. Physiological parameters were assessed immediately after this stress treatment (modified from Kong et al., 2011).
Measurement of electrolyte leakage, malondialdehyde (MDA), chlorophyll content, proline content, and peroxidase (POD) activity
The relative electrical conductivity (EC) was measured as described previously (Sharom et al., 1994; Tang, 1999). Samples were separated into two equal groups. The first group was shaken in 5 cm3 double-distilled water at 170 rpm for 2 h at 25 °C, the second group was boiled for 30 min. The relative conductivity (%) was calculated as the ratio of EC of intact leaves and EC of boiled leaves 100.
The extent of lipid peroxidation was estimated by measuring the amount of MDA according to Quan et al. (2004). A total of 0.5 g of fresh material was ground in 5 mL of 5% (w/v) trichloroacetic acid (TCA), and 2 mL of supernatant was mixed with 2 mL of 0.67% (w/v) thiobarbituric acid (TBA) in 5% (w/v) TCA and incubated at 100°C for 30 min. After centrifuging, the optical density was measured at 450, 532, and 600 nm, respectively. The amount of MDA was calculated from the following formula: C = 6.45(A532–A600) -0.56A450.
After freezing treatment, chlorophyll was extracted from individual leaves with 95% ethanol, and chlorophyll contents were determined spectrophotometrically according to Lichtenthaler and Hartmut (1987). Absorbance was recorded at 645, 663, and 470 nm, and chlorophyll and carotenoid content were calculated according to the following formula:
V = volume in mL; X = path length, 1 cm; W = fresh weight in grams.
Proline content was measured following the methods of Bates et al. (1973). Rosette leaves (0.5 g) were excised into 5 mL, 3% aqueous sulfosalicylic acid and incubated at 100 °C for 10 min. The homogenate was centrifuged at 12000 x g for 10 min. The reaction mixture containing 2 mL of supernatant, 2 mL glacial acetic acid, and 2 mL ninhydrin reagent (2.5% ninhydrin in 60% phosphoric acid) was incubated at 100 °C for 30 min and terminated by cooling the tubes on ice. The absorbance was determined at 520 nm.
To measure POD activity, leaf segments (0.5 g) were extracted on ice in 5 mL of 50 mm potassium phosphate buffer, pH 7.0, containing 1 mm ethylenediaminetetraacetic acid (EDTA) and 1% polyvinylpyrrolidone. The homogenate was centrifuged at 15,000 x g for 20 min at a temperature of 4 °C, and the supernatant was immediately used for the following antioxidant enzyme assays. Total POD activity was measured by monitoring the oxidation of 3,3’-dimethoxybenzidine at 470 nm (Zhang et al., 2006).
The experiments were repeated independently at least three times and the results were consistent.
Statistical analysis
Data were tested by analysis of variance using SPSS version 17.0. Significant differences were determined based on p<0.05 or p<0.01.
Results
PfHSP17.2 is a plant cytosolic class II sHSP
To investigate the molecular events of the P. forrestii heat response, a cDNA library was constructed using mRNAs isolated from 2-h heat-treated Primula plants. The full-length PfHSP17.2 cDNA containing 458 bp exhibited high sequence similarities to other cytosolic class II sHSP genes from various species, according to BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi), was predicted to encode a protein of 152 amino acids with a molecular mass of 17.2 kDa, and was designated as PfHSP17.2 (GenBank accession number JX025006).
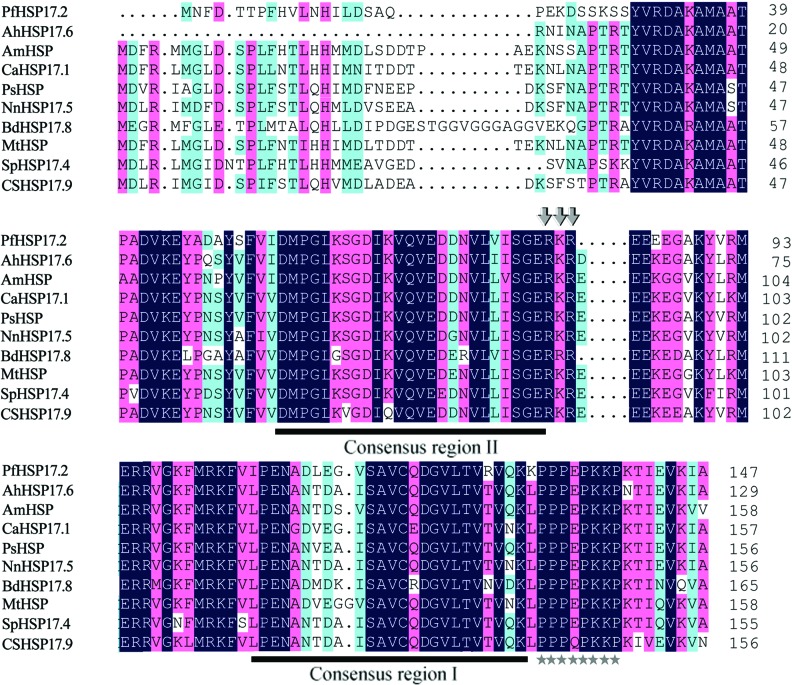
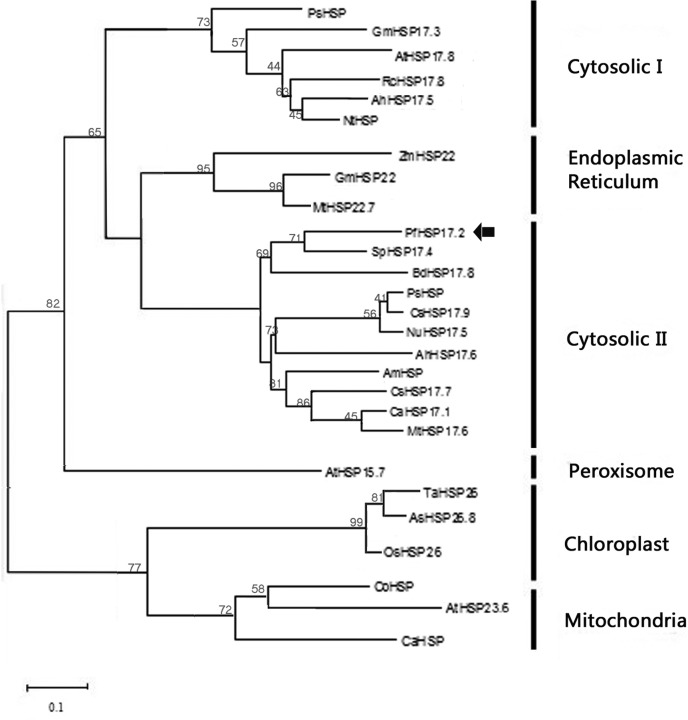
An alignment of the deduced PfHSP17.2 with other plant cytosolic class II sHSPs is shown in Figure 1. PfHSP17.2 shared high sequence identities with AhHSP17.6 (85%), AmHSP (73%), and NnHSP17.5 (71%). PfHSP17.2 contains two conserved heat shock domains, consensus I and II (Narberhaus, 2002). For plant cytosolic class II sHSPs, although a unique domain RDAKAMAATPADV is found at the N-terminal, the sequence of the N-terminal domain is divergent, which may partially account for their functional multiplicity among different plant species. Moreover, a putative nuclear localization signal (RKR) and a polyproline motif (PPPEPKKP) are found at the C-terminal of plant cytosolic class II sHSPs. A phylogenetic tree was constructed based on the similarities of 27 plant sHSPs, which separated the sHSP gene family into six clades, and PfHSP17.2 fell into the cytosolic class II cluster (Figure 2).
Figure 1. Protein sequence multiple alignment of the deduced amino acid sequence of PfHSP17.2 with other plant sHSPs. The two consensus regions in sHSPs are underlined, and identical amino acid residues are highlighted. A putative nuclear localization signal is indicated by arrows and a polyproline motif at the carboxyl end of proteins is marked with asterisks. Their GenBank accession numbers are as follows: Primula forrestii (PfHSP17.2; JX025006); Arachis hypogaea (AhHSP17.6; ACF74271.1); Ammopiptanthus mongolicus (AmHSP; AGS48404); Cicer arietinum (CaHSP17.1; XP_004501443.1); Prunus salicina (PsHSP; ACV93250.1); Nelumbo nucifera (NnHSP17.5; ABK92179.1); Brachypodium distachyon (BdHSP17.8; XP_003564488.1); Medicago truncatula (MtHSP; AES73434.1); Solanum peruvianum (SpHSP17.4; AAT36481.1); Cucumis sativus (CsHSP17.9; XP_004141334.1).
Figure 2. Phylogenetic analysis of the deduced amino acid sequence of PfHSP17.2 and other small sHSPs. The gene abbreviation and GenBank accession numbers are as follows: Primula forrestii (PfHSP17.2; JX025006), Arachis hypogaea (AhHSP17.5; ABC46712.1), Glycine max (GmHSP17.3; NP_001235293.1), Nicotiana tabacum (NtHSP; ADK36668.1), Zea mays (ZmHSP22; NP_001151139.1), Rosa chinensis (RcHSP17.8; ABK32539.1), Arabidopsis thaliana (AtHSP17.8; NP_172220.1), Glycine max (GmHSP22; HSP41_SOYBN), Medicago truncatula (MtHSP22.7; XP_003622351.1), Arabidopsis thaliana (AtHSP15.7; NP_198583.1), Triticum aestivum (TaHSP26; AAC96315.1), Oryza sativa (OsHSP26; BAA78385.1), Agrostis stolonifera (AsHSP26.8; AAN74536.1), Capsicum annuum (CaHSP; ADJ57588.1), Copaifera officinalis (CoHSP; AEX97054.1), Arabidopsis thaliana (AtHSP23.6; NP_194250.1), Arabidopsis thaliana (AtHSP23.5; NP_199957.1); Arachis hypogaea(AhHSP17.6; ACF74271.1); Ammopiptanthus mongolicus (AmHSP; AGS48404); Cicer arietinum (CaHSP17.1; XP_004501443.1); Prunus salicina (PsHSP; ACV93250.1); Nelumbo nucifera (NnHSP17.5; ABK92179.1); Brachypodium distachyon (BdHSP17.8; XP_003564488.1); Medicago truncatula (MtHSP17.6; XP_003603183.1); Solanum peruvianum (SpHSP17.4; AAT36481.1); Cucumis sativus (CsHSP17.9; XP_004141334.1), Prunus salicina (PsHSP; ACV93250.1).
Subcellular localization assay
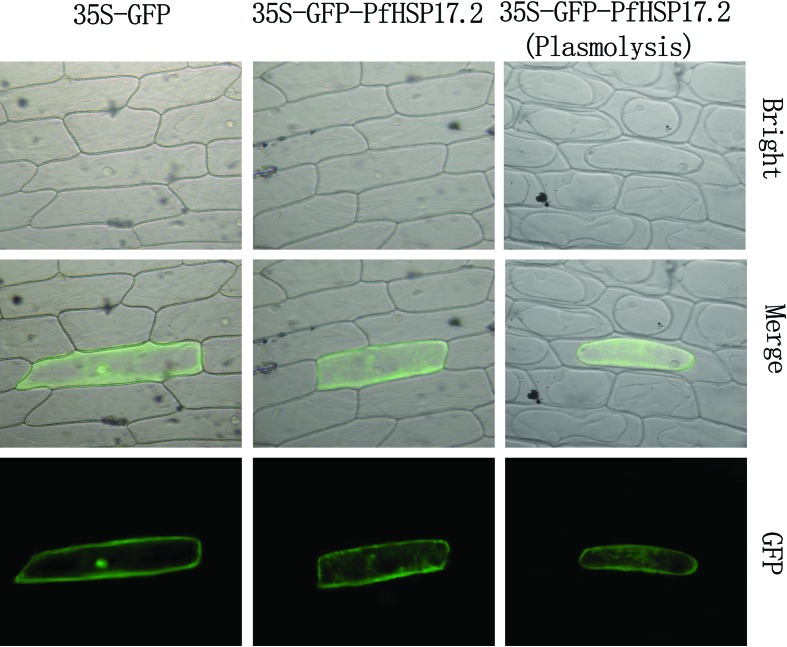
In order to examine the location of PfHSP17.2 in the cell, a GFP-PfHSP17.2 fusion gene construct was transformed into onion epidermal cells. As shown in Figure 3, cultured onion epidermal cells with the GFP-PfHSP17.2 fusion protein had strong fluorescence signals in the cytoplasm. In contrast, cells with the GFP fusion protein alone had signals detected in both the cytoplasm and nucleus. Therefore, PfHSP17.2 was mainly localized in the cytoplasm.
Figure 3. Subcellular localization of the PfHSP17.2:GFP fusion protein in onion epidermal cells. Cells were bombarded with DNA-coated gold particles carrying GFP or PfHSP17.2: GFP.
Expression of PfHSP17.2 under different stress treatments
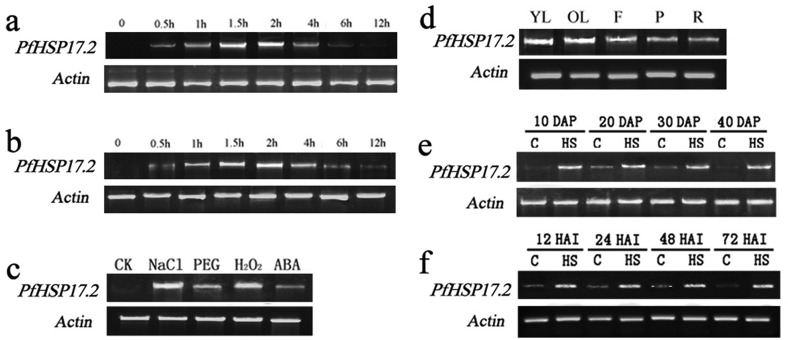
Upon monitoring the expression of wheat PfHSP17.2 by semiquantitative RT-PCR at different time intervals during high temperature stress (42 °C), a transcript abundance was observed after 0.5 h of treatment, and the expression level in the leaves reached its peak at 2 h, began to decline after 4 h, and totally disappeared 12 h later (Figure 4a). Similarly, under a 4 °C cold stress, PfHSP17.2 was expressed after 0.5 h of treatment, reaching the highest expression level 2 h later, and could still be detected after 12 h of treatment (Figure 4b). In addition, the expression of PfHSP17.2 was examined when plants of the same age were subjected to different abiotic stress conditions, including salt stress (NaCl), osmotic stress (PEG), oxidative stress (H2O2) and ABA treatment. Increased PfHSP17.2 transcript abundance was observed in all plant leaves analyzed, but it was slower and less abundant under drought and ABA stress treatment than that under other stressful conditions (Figure 4c). To study the tissue-specific expression pattern of PfHSP17.2 in P. forrestii, its mRNAs were isolated from different tissues of the same heat-treated plant (42 °C for 2 h). The accumulation of PfHSP17.2 was higher in the leaves relative to other tissues (petioles, roots, and flowers) (Figure 4d). Expression of the PfHSP17.2 gene was also monitored in different stages of developing and germinating seeds separately under both thermal stressed and non-stressed conditions (Figure 4e,f). In the developing seeds, the expression of PfHSP17.2 was high at 20 DAP and then decreased to very low levels after 40 DAP. In the germinating seeds, the expression of PfHSP17.2 gradually decreased with the germination process from the initiation of 12 h, and it was hard to detect after 72 h. Taken together, these data indicate that PfHSP17.2 could be specifically expressed in seeds under normal growth conditions and is strongly up-regulated in both developing and germinating seeds under heat stress.
Figure 4. Semi-quantitative RT-PCR of PfHSP17.2 expression in P. forrestii. (a) Heat treatment, time course induction of PfHSP17.2 by heat in P. forrestii; (b) cold treatment, time course induction of PfHSP17.2 by cold in P. forrestii; (c) Transcript levels of PfHSP17.2 in leaves under different treatments for 3 h. CK, no treatment as control (plants only irrigated with half-strength MS salt solution); (d) expression of PfHSP17.2 in different organs after incubation at 42 °C for 2 h; (e) semi-quantitative RT-PCR analysis in developing seeds and germinating seeds (f), respectively. DAP, days after pollination. HAI, hours after imbibition. YL, young leaves (5 d from emergence). OL, old leaves (two weeks from emergence). F, flower. P, petiole. R, root.
Overexpression of PfHSP17.2 in transgenic plants enhances thermotolerance and resistance to salt
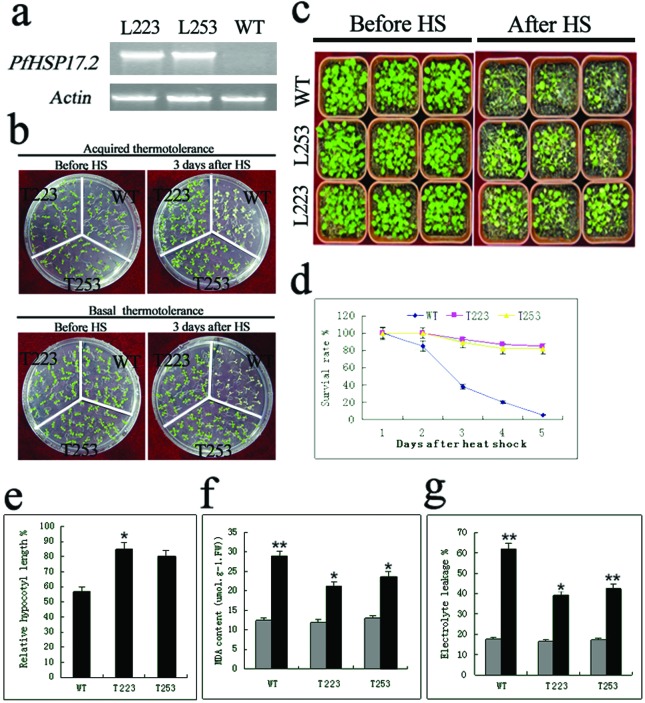
To evaluate the in vivo function of PfHSP17.2, we generated two independent T3 transgenic Arabidopsis lines, T223 and T253, which ectopically expressed PfHSP17.2 under the control of the cauliflower mosaic virus 35S promoter. RT-PCR analysis of the 4-week-old transformants confirmed that PfHSP17.2 was ectopically expressed in the two transgenic lines (Figure 5a).
Figure 5. PCR analysis of PfHSP17.2 transgenic plants and stress tolerance tests. (a) Semi-quantitative RT-PCR analysis of PfHSP17.2 expression in WT and PfHSP17.2 transgenic plants grown at 22°C for 4 weeks; (b) Heat treatments: aquired heat tolerance and basal heat tolerance; (c) 3-week-old plants heat treatment comparison; (d) survival rate after acquired heat treatment; (e) effect of heat stress on hypocotyl elongation of WT and PfHSP17.2-overexpressing plants; (f) MDA; (g) electrolyte leakage. For survival rate (d), hypocotyl elongation (e), MDA (f) and electrolyte leakage (g), error bars indicate SD (n=3). *p<0.05, ** p<0.01. WT, wild type; L223, L253, two transgenic Arabidopsis lines.
To evaluate the effect of PfHSP17.2 on plant HS responses, the acquired and basal thermotolerances were compared between transgenic lines and WT plants. For basal thermotolerance test, all of the WT seedlings were killed after being allowed to recover under standard conditions for 3 d, but nearly 80% of seedlings from both T223 and T253 lines survived (Figure 5b). After a conditioning pretreatment, 100% of transgenic seedlings displayed the acquired thermotolerance, but none of the WT seedlings did (Figure 5b). All of them stopped growing, their cotyledons lost chlorophyll and died after 5 d of recovery.
In order to explore the detailed morphological differences between WT and transgenic plants in thermotolerance, we monitored and compared the development of phenotype before and after heat treatment. Two lines of transgenic seedlings grew better than the WT plants after being subjected to heat stress (42°C), whereas all seedlings grew equally before the treatment (Figure 5c). The survival rate of T223 and T253 transgenic seedlings was obviously higher than that of the WT seedlings (Figure 5d). In addition, a quantitative assay was carried out to identify the thermotolerance of the transgenic plants, according to the measurement of hypocotyl elongation (Hong and Vierling, 2000). After 3 d of recovery at 22 °C, the extent of hypocotyl elongation was measured. As shown in Figure 5e, in the transgenic lines T223 and T253, the relative hypocotyl length was longer than that in the WT plants following heat shock.
To further examine the thermotolerance acquired by the transgenic plants, 4-week-old transgenic and WT seedlings grown in soil were heat-shocked at 42 °C for 12 h. The rosette leaves were taken for assays. The accumulation of MDA is often used as an indicator of lipid peroxidation (Smirnoff, 1995). At normal temperature, no significant difference in MDA content and relative electrical conductivity was detected among the WT and the transgenic lines. However, after heat stress, higher MDA content was detected in WT than in transgenic plants (Figure 5f). The relative electrical conductivity of transgenic plants was significantly lower than that of WT after high temperature stress (Figure 5g), which indicates less membrane damage in transgenic lines.
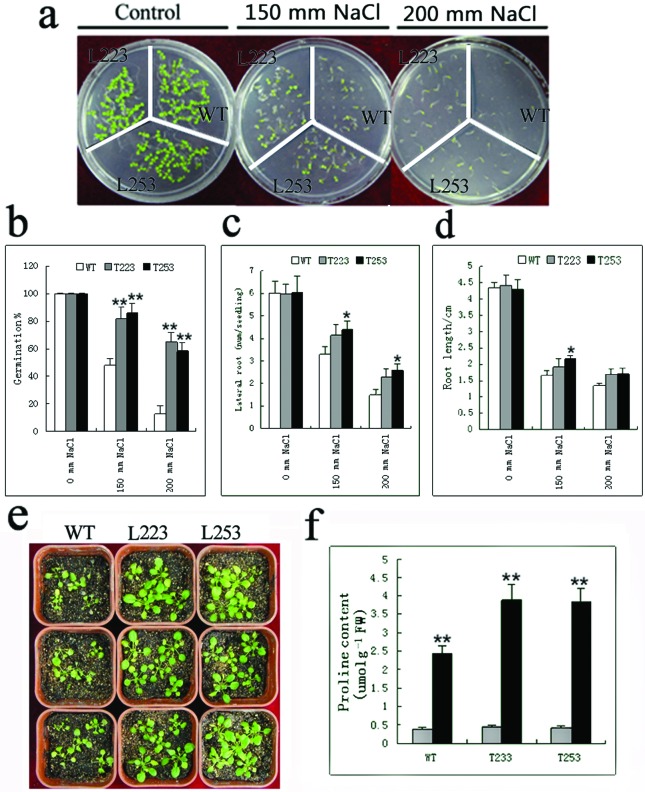
For salt stress treatment, seeds from two homozygous PfHSP17.2-overexpressing lines and the WT Arabidopsis were germinated on MS plates supplemented with different concentrations of NaCl. On MS plates without NaCl, the transgenic lines did not show any significant difference from WT plants during germination. Compared with the wild-type Arabidopsis, the transgenic lines showed a significantly higher germination rate on the medium containing 150 or 200 mm NaCl (Figure 6a). More than 85% germination was found at 150 mm NaCl, compared to 50% for WT. At a high\er concentration of NaCl (200 mm) in the medium, both T223 and T253 lines still showed a germination rate of 60%, by contrast the WT seeds only germinated less than 20% (Figure 6b).
Figure 6. Performance of PfHSP17.2-overexpressing transgenic plants under salt stress. (a) Germination on different salt MS (0 mm NaCl, 150 mm NaCl, 200 mm NaCl); (b) germination rate; (c) root number; (d) root length; (e) phenotypic comparison of the plants under salt stress; (f) proline content in WT and transgenic plants in response to salt stress. Error bars indicate SD (n=3). *p<0.05, ** p<0.01. WT, wild type; L223, L253, two transgenic Arabidopsis lines.
The sensitivity of seedling growth to salt stress was also examined. Seedlings grown on MS medium for 7 d were transferred to MS medium supplemented with 0, 150, and 200 mm NaCl for 10 d. Without salt treatment, all the transgenic plants showed no differences in growth compared to control plants. On medium containing 200 mm NaCl, the control seedlings showed less lateral root number than the transgenic plants (Figure 6c). However, no significant difference in root length was detected among the WT and two transgenic lines on MS medium with or without NaCl except for T253 in 150 mm NaCl (Figure 6d). In addition, 2-week-old WT and transgenic seedlings were irrigated with 300 mm NaCl solution for 7 d. The two lines of transgenic plants recovered more robustly than the WT plants after being shifted to normal conditions (Figure 6e). Proline accumulation is an important factor in determining stress tolerance and can play a major role in osmotic adjustment and may also have a number of other protective roles (Voetberg and Sharp, 1991; Liu and Zhu, 1997). At normal condition, no significant difference in proline content was detected among the WT and transgenic lines, but a significantly greater increase in the proline content was detected in the transgenic lines compared with that in the WT plants (Figure 6f), suggesting that transgenic plants were more tolerant to the salt treatment.
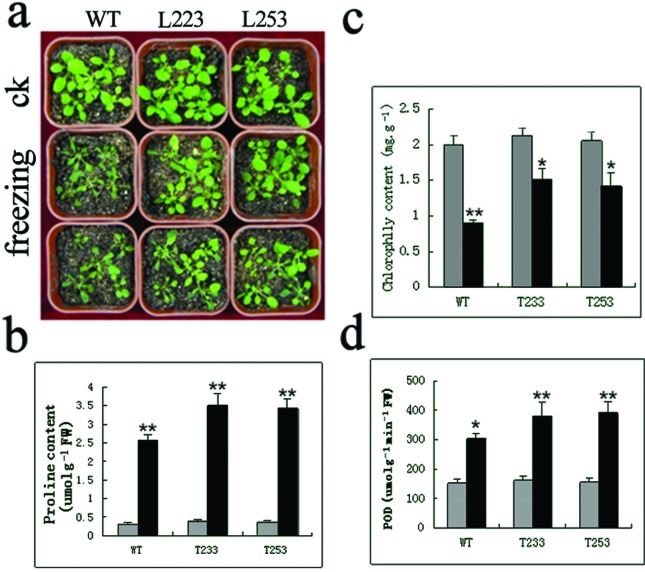
To test freezing tolerance of transgenic lines, after a 2 h -10°C treatment the leaves of WT plants were wilted and showed similar symptoms to that after flooding, whereas the leaves of transgenic plants turned yellow, and returned to normal growth in a short term. When returned to normal condition, transgenic plants recovered quickly, whereas the WT showed obvious tissue damage (Figure 7a). To further examine the cold tolerance acquired by the transgenic plants, 4-week-old transgenic and WT seedlings grown in soil were exposed to 4 °C for 12 h. An obviously higher proline content was detected in the transgenic lines compared with that in the WT plants (Figure 7b). Chlorophyll content assessment is one of the most useful and widely applied techniques for evaluating the effects of environmental stresses (Xue et al., 2010; Kong et al., 2011; Ning et al., 2010). The total chlorophyll content in WT and two selected transgenic lines (T223 and T253) was remarkably decreased after stress. However, the transgenic plants’ decrease was lower than that in WT (Figure 7c), suggesting that transgenic plants are more tolerant to the chilling treatment. It has been determined previously that the activities of antioxidant enzymes in plants are correlated with increased tolerance to cold stresses (Kong et al., 2011; Li et al., 2012). As shown in Figure 7d, significantly higher POD activity was detected in the transgenic plants than in WT plants in response to cold conditions.
Figure 7. Performance of PfHSP17.2-overexpressing transgenic plants under cold stress. (a) Cold tolerance of the PfHSP17.2 transformants; (b) proline content; (c) chlorophyll content; (d) POD changes. Error bars indicate SD (n=3). *p<0.05, ** p<0.01. WT, wild type; L223, L253, two transgenic Arabidopsis lines.
Discussion
There have been many studies focusing on the function of sHSPs, which work as molecular chaperons and bind to unfolded proteins, prevent aggregation, induce correct refolding, and facilitate correct cell function under stress conditions (Al-Whaibi, 2011). Nevertheless, their biological functions have still not been fully understood. In the present study, we isolated and cloned a full-length cDNA of PfHSP17.2 from heat-treated P. forrestii leaves. Based on alignment and phylogenetic analysis with sHSP sequences of other plants, PfHSP17.2 was determined to be a member of the plant cytosolic class II sHSPs contained in two conserved domains, which indicated it might have similar functions as the members in cytosolic class II sHSP (Kaur et al., 2015; Kumar et al., 2015). Subcellular localization assay revealed that GFP-PfHSP17.2 is localized only in the cytoplasm (Figure 3), similarly to other cytosolic sHSPs (Scharf et al., 2001; Zhou et al., 2012).
Previous studies have described a correlation between sHSP expression and adaptation to various stress conditions in plants. A peroxisomes sHSP, AtHSP15.7, was strongly induced by oxidative stress as well as heat (Ma et al., 2006). The rose sHSP, RcHSP17.8, was triggered by heat stress, drought, cold, and salt (Jiang et al., 2009). ThHSP18.3 from Tamarix hispida was found to be highly expressed under salt, drought, heavy metals, and both low and high temperatures (Gao et al., 2012). The obvious induction of NnHSP17.5 in Nelumbo nucifera resulted from heat stress and oxidative stress (Zhou et al., 2012). However, an Arabidopsis cytosolic class II sHSP, AtHSP17.6A, was induced by osmotic stress but not oxidative stress (Sun et al., 2001). ZmHSP16.9C was obviously accumulated in response to heat and oxidative stress, but could not be induced by NaCl, PEG, and cold conditions (Sun et al., 2012). Similarly, the expressions of OsHSP16.9A, OsHSP16.9B, and OsHSP16.9C were not detected under salt, drought, and cold stresses (Guan et al., 2004; Sarkar et al., 2009).
In this study, the expression of PfHSP17.2 was found rapidly induced in leaves of P. forrstii in response to various abiotic stresses, including heat shock (42 °C), cold (4°C), salt (NaCl), and oxidative stress (H2O2) (Figure 4a-c), indicating that PfHSP17.2 functions as a stress response of heat as well as cold and salt. However, the expression of PfHSP17.2 was not obviously affected by ABA treatment compared with its accumulation under other stresses mentioned above (Figure 4c). It is possible that, like PfHSP17.1 (Zhang et al., 2013), PfHSP17.2 functions in an ABA-independent manner. Furthermore, the up-regulation of PfHSP17.2 in the leaf of P. forrestii reached an expression peak within 2 h in response to both 42 °C (Figure 4a) and 4 °C temperature stresses (Figure 4b), reflecting the instantaneous and transient feature of heat shock response. However, the expression of PfHSP17.2 could still be detected after 12 h of cold stress, while it was hard to find under 6 h of heat stress, which indicates that PfHSP17.2 may have different mechanisms in response to different stressful conditions. In addition, different expression patterns of PfHSP17.2 were observed in leaves, petioles, and roots (Figure 4d), which suggests that PfHSP17.2 has tissue-specific expression patterns, and may have different functions in different tissues in response to the same stress. As shown in Figure 4d, in response to heat treatment, PfHSP17.2 may mainly function in leaves while PtHSP17.8 was significantly accumulated in stem and roots (Li et al., 2016).
Numerous studies have previously described that sHSPs were not generally present in vegetable tissues under normal conditions, but were found at specific developmental stages, such as seed development and germination, pollen development, and fruit maturation (Waters et al., 1996; He and Yang, 2013). A Nelumbo cytosolic class II sHSP gene (NnHSP17.5) was strongly expressed after 15 days of pollination. Arabidopsis cytosolic class I sHSPs were suggested to play a role in early stages of seed development (Dafny-Yelin et al., 2008). A wheat chloroplastic small heat shock protein (sHSP26) was involved in seed maturation and germination. Similarly, the expression of PfHSP17.2 was detected specifically in seeds under the normal physiological condition at certain stages of seed development and germination (Figure 4e,f), which is in accordance with the observations in other studies (Wehmeyer et al., 1996; Sun et al., 2001; Kaur et al., 2015). However, it is unlike OsHSP18.2, which is markedly increased at the late maturation stage and is highly abundant in dry seeds, and sharply decreases after germination, indicating it may participate in seed vigor and longevity (Kaur et al., 2015). Our data suggest that PfHSP17.2 may play an important role during seed maturation and subsequent seed germination, and can thereby contribute to seed viability and vigor.
Evidence for plant sHSP stress-related functions in vivo has been demonstrated by the over-expression of specific sHSPs in various plants (Sanmiya et al., 2004; Perez et al., 2009; Xue et al., 2010; Zhou et al., 2012). Transgenic Arabidopsis that constitutively expressed RcHSP17.8 exhibited increased tolerance to heat, salt, osmotic, and drought stress (Jiang et al., 2009). Overproduction of sHSP17.7 in rice enhanced drought tolerance in transgenic seedlings (Murakami et al., 2004). In Arabidopsis, overexpression of wheat chloroplastic sHSP26 resulted in improved heat tolerance (Chauhan et al., 2012). A Gossypium arboreum sHSP, GHSP26, enhanced drought tolerance in transformed cotton plants (Maqbool et al., 2010). Overexpression of PtHSP17.8 enhanced tolerance to heat and salt stresses in Arabidopsis (Li et al., 2016).
In this study, the PfHSP17.2 coding sequence was introduced into A. thaliana by Agrobacterium-mediated transformation and overexpressed under control of the CaMV 35S promoter. Under normal growth conditions, no apparent phenotypic difference was noted in the transgenic lines T223 and T253, compared with the WT line. However, significant differences in heat stress tolerance were indicated by phenotypic changes (Figure 5b,c).
As important physiology indexes, relative electrical conductivity, proline, and chlorophyll contents, MDA and antioxidative enzymes such as POD activities could be used as indicators to evaluate stress tolerance in plants (Kar and Mishra, 1976). Transgenic plants displayed an improved thermal tolerance compared with the WT Arabidopsis. This response was also characterized by higher survival rate calculation, longer hypocotyl elongation measurement, less MDA, and electrolyte leakage in transgenic lines (Figure 5). These results demonstrated that transgenic Arabidopsis have higher heat stress tolerance than WT plants.
For salt stress, according to the measurements of the germination rate, the numbers of lateral roots, the length of roots, and proline content, two transgenic lines showed improved salt tolerance (Figure 6). In the cold stress experiment, the transgenic plants had more biomass after stress (Figure 7a), and maintained higher levels of chlorophyll content, proline content, and POD, suggesting that transgenic Arabidopsis plants have a higher level of tolerance to low temperature. As a molecular chaperon, PfHSP17.2 might not be involved in the biosynthesis of proline, chlorophyll content, or antioxidative enzymes directly, so they were not significantly changed in transgenic lines (T223 and T253) under normal conditions, despite the fact that PfHSP17.2 was driven by a constitutive 35S promoter. However, the over-accumulated PfHSP17.2 could protect enzymes and proteins such as proline, to prevent their destruction or degradation under stress conditions in transgenic Arabidopsis, thus enhancing the stress tolerance of transgenic plants.
In summary, our results demonstrate that a plant cytosolic class II sHSP gene, PfHSP17.2, was remarkably induced in P. forrstii leaves under high temperature treatment and can also be significantly up-regulated under cold, salt, and oxidative stress conditions, as well as in the seed development and germination processes. Moreover, the overexpression of PfHSP17.2 in Arabidopsis may contribute to the enhanced tolerance to thermal, salt, and cold stresses. Further investigation are needed to gain more information on the function and regulatory mechanisms of PfHSP17.2 in plant-stress responses.
Acknowledgments
This work was supported by the Ministry of Science and Technology (No.2012BAD01B07).
Footnotes
Associate Editor: Marcio de Castro Silva Filho
References
- Ahn YJ, Zimmerman JL. Introduction of the carrot HSP17.7 into potato (Solanum tuberosum L.) enhances cellular membrane stability and tuberization in vitro. Plant Cell Environ. 2006;29:95–104. doi: 10.1111/j.1365-3040.2005.01403.x. [DOI] [PubMed] [Google Scholar]
- Ahuja I, de Vos RC, Bones AM, Hall RD. Plant molecular stress responses face climate change. Trends Plant Sci. 2010;15:664–674. doi: 10.1016/j.tplants.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Al-Whaibi MH. Plant heat-shock proteins: a mini review. J King Saud Univ Sci. 2011;23:139–150. [Google Scholar]
- Bates LS, Waldren RP, Teare ID. Rapid determination of free proline for water-stress studies. Plant Soil. 1973;39:205–207. [Google Scholar]
- Boston RS, Viitanen PV, Vierling E. Molecular chaperones and protein folding in plants. Plant Mol Biol. 1996;32:191–222. doi: 10.1007/BF00039383. [DOI] [PubMed] [Google Scholar]
- Cabane M, Calvet P, Vincens P, Boudet AM. Characterization of chilling-acclimation-related proteins in soybean and identification of one as a member of the heat shock protein (HSP 70) family. Planta. 1993;190:346–353. doi: 10.1007/BF00196963. [DOI] [PubMed] [Google Scholar]
- Chao YY, Hsu YT, Kao CH. Involvement of glutathione in heat shock–and hydrogen peroxide–induced cadmium tolerance of rice (Oryza sativa L.) seedlings. Plant Soil. 2009;318:37–45. [Google Scholar]
- Charng Y, Liu H, Liu N, Hsu F, Ko S. Arabidopsis Hsa32, a novel heat shock protein, is essential for acquired thermotolerance during long recovery after acclimation. Plant Physiol. 2006;140:1297–1305. doi: 10.1104/pp.105.074898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan H, Khurana N, Nijhavan A, Khurana JP, Khurana P. The wheat chloroplastic small heat shock protein (sHSP26) is involved in seed maturation and germination and imparts tolerance to heat stress. Plant Cell Environ. 2012;35:1912–1931. doi: 10.1111/j.1365-3040.2012.02525.x. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Coca MA, Almoguera C, Jordano J. Expression of sunflower low-molecular-weight heat-shock proteins during embryogenesis and persistence after germination: Localization and possible functional implications. Plant Mol Biol. 1994;25:479–492. doi: 10.1007/BF00043876. [DOI] [PubMed] [Google Scholar]
- Dafny-Yelin M, Tzfira T, Vainstein A, Adam Z. Non-redundant functions of sHSP-Cis in acquired thermotolerance and their role in early seed development in Arabidopsis . Plant Mol Biol. 2008;67:363–373. doi: 10.1007/s11103-008-9326-4. [DOI] [PubMed] [Google Scholar]
- Derocher AE, Helm KW, Lauzon LM, Vierling E. Expression of a conserved family of cytoplasmic low molecular weight heat shock proteins during heat stress and recovery. Plant Physiol. 1991;96:1038–1047. doi: 10.1104/pp.96.4.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeoglu B. Heat shock proteins and heat shock response in plants G.U. J Sci. 2009;22:67–75. [Google Scholar]
- Gao C, Jiang B, Wang Y, Liu G, Yang C. Overexpression of a heat shock protein (ThHSP18.3) from Tamarix hispida confers stress tolerance to yeast. Mol Biol Rep. 2012;39:4889–4897. doi: 10.1007/s11033-011-1284-2. [DOI] [PubMed] [Google Scholar]
- Guan JC, Jinn TL, Yeh CH, Feng SP, Chen YM, Lin CY. Characterization of the genomic structures and selective expression profiles of nine class I small heat shock protein genes clustered on two chromosomes in rice (Oryza sativa L.) Plant Mol Biol. 2004;56:795–809. doi: 10.1007/s11103-004-5182-z. [DOI] [PubMed] [Google Scholar]
- He D, Yang P. Proteomics of rice seed germination. Front Plant Sci. 2013;4:246. doi: 10.3389/fpls.2013.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SW, Vierling E. Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc Natl Acad Sci U S A. 2000;97:4392–4397. doi: 10.1073/pnas.97.8.4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Hu G, Han B. Genome-wide survey and expression profiling of heat shock proteins and heat shock factors revealed overlapped and stress specific response under abiotic stresses in rice. Plant Sci. 2009;176:583–590. doi: 10.1016/j.plantsci.2009.01.016. [DOI] [PubMed] [Google Scholar]
- Hu W, Zhang Q, Pan H, Dong L. Preliminary study on physiological indexes of heat-resistance of Primula . Chin Agric Sci Bull. 2010;26:158–163. [Google Scholar]
- Jiang C, Xu J, Zhang H, Zhang X, Shi J, Li M, Ming F. A cytosolic class I small heat shock protein, RcHSP17.8, of Rosa chinensis confers resistance to a variety of stresses to Escherichia coli, yeast and Arabidopsis thaliana . Plant Cell Environ. 2009;32:1046–1059. doi: 10.1111/j.1365-3040.2009.01987.x. [DOI] [PubMed] [Google Scholar]
- Ju Y, Tian H, Zhang R, Zuo L, Jin G, Qian X, Ding X, Li X, Chu Z. Overexpression of Oshsp18.0-CI enhances resistance to bacterial leaf streak in rice. Rice. 2017;10:12. doi: 10.1186/s12284-017-0153-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar M, Mishra D. Catalase, peroxidase, and polyphenol oxidase activities during rice leaf senescence. Plant Physiol. 1976;57:315–319. doi: 10.1104/pp.57.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H, Petla BP, Kamble NU, Singh A, Rao V, Salvi P, Ghosh S, Majee M. Differentially expressed seed aging responsive heat shock protein Oshsp18.2 implicates in seed vigor, longevity and improves germination and seedling establishment under abiotic stress. Front Plant Sci. 2015;6:713. doi: 10.3389/fpls.2015.00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Pan J, Zhang M, Xing X, Zhou Y, Liu Y, Li D. ZmMKK4, a novel group C mitogen-activated protein kinase kinase in maize (Zea mays), confers salt and cold tolerance in transgenic Arabidopsis . Plant Cell Environ. 2011;34:1291–1303. doi: 10.1111/j.1365-3040.2011.02329.x. [DOI] [PubMed] [Google Scholar]
- Koo HJ, Park SM, Kim KP, Suh MC, Lee MO, Lee SK, Xia X, Hong CB. Small heat shock proteins can release light dependence of tobacco seed during germination. Plant Physiol. 2015;167:1030–1038. doi: 10.1104/pp.114.252841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Lavania D, Singh AK, Negi M, Siddiqui MH, Al-Whaibi MH, Grover A. Identification and characterization of a small heat shock protein 17.9-CII gene from faba bean (Vicia faba L.) Acta Physiol Plant. 2015;37:1–13. [Google Scholar]
- Lee GJ, Roseman AM, Saibil HR, Vierling E. A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J. 1997;16:659–671. doi: 10.1093/emboj/16.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 1987;148:350–382. [Google Scholar]
- Li J, Zhang J, Jia H, Li Y, Xu X, Wang L, Lu M. The Populus trichocarpa PtHSP17.8 involved in heat and salt stress tolerances. Plant Cell Rep. 2016;35:1587–1599. doi: 10.1007/s00299-016-1973-3. [DOI] [PubMed] [Google Scholar]
- Li M, Ji L, Yang X, Meng Q, Guo S. The protective mechanisms of CaHSP26, in transgenic tobacco to alleviate photoinhibition of PSII during chilling stress. Plant Cell Rep. 2012;31:1969–1979. doi: 10.1007/s00299-012-1309-x. [DOI] [PubMed] [Google Scholar]
- Lin YL, Chao YY, Kao CH. Exposure of rice seedlings to heat shock protects against subsequent Cd-induced decrease in glutamine synthetase activity and increase in specific protease activity in leaves. J Plant Physiol. 2010;167:1061–1065. doi: 10.1016/j.jplph.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Liu J, Zhu JK. Proline accumulation and salt-stressinduced gene expression in a salt-hypersensitive mutant of Arabidopsis . Plant Physiol. 1997;114:591–596. doi: 10.1104/pp.114.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Wang Y, Wang N, Dong YY, Fan XD, Liu XM, Yang J, Li HY. Cloning of a vacuolar H+-pyrophosphatase gene from the halophyte Suaeda corniculata whose heterologous overexpression improves salt, saline-alkali and drought tolerance in Arabidopsis . J Integr Plant Biol. 2011;53:731–742. doi: 10.1111/j.1744-7909.2011.01066.x. [DOI] [PubMed] [Google Scholar]
- Ma CL, Haslbeck M, Babujee L, Jahn O, Reumann S. Identification and characterization of a stress-inducible and a constitutive small heat-shock protein targeted to the matrix of plant peroxisomes. Plant Physiol. 2006;141:47–60. doi: 10.1104/pp.105.073841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnard JL, Vergne P, Dumas C. Complexity and genetic variability of heat-shock protein expression in isolated maize microspores. Plant Physiol. 1996;111:1085–1096. doi: 10.1104/pp.111.4.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maimbo M, Ohnishi K, Hikichi Y, Yoshioka H, Kiba A. Induction of a small heat shock protein and its functional roles in Nicotiana plants in the defense response against Ralstonia solanacearum . Plant Physiol. 2007;145:1588–1599. doi: 10.1104/pp.107.105353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik MK, Slovin JP, Hwang CH, Zimmerman JL. Modified expression of a carrot small heat shock protein gene, Hsp17.7, results in increased or decreased thermotolerance. Plant J. 1999;20:89–99. doi: 10.1046/j.1365-313x.1999.00581.x. [DOI] [PubMed] [Google Scholar]
- Maqbool A, Abbas W, Rao AQ, Irfan M, Zahur M, Bakhsh A, Riazuddin S, Husnain T. Gossypium arboreum GHSP26 enhances drought tolerance in Gossypium hirsutum. Biotechnol Prog. 2010;26:21–25. doi: 10.1002/btpr.306. [DOI] [PubMed] [Google Scholar]
- Murakami T, Matsuba S, Funatsuki H, Kawaguchi K, Saruyama H, Tanida M, Sato Y. Over-expression of a small heat shock protein, sHSP17.7, confers both heat tolerance and UV-B resistance to rice plants. Mol Breed. 2004;13:165–175. [Google Scholar]
- Narberhaus F. a-Crystallin-type heat shock proteins: Socializing minichaperones in the context of a multichaperone network. Microbiol Mol Biol Rev. 2002;66:64–93. doi: 10.1128/MMBR.66.1.64-93.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson GC, Mensbrugghe D, Ahammad H, Blanc E, Calvin K, Hasegawa T, Havlik P, Heyhoe E, Kyle P, Lotze-Campen H. Agriculture and climate change in global scenarios: why don’t the models agree. Agr. Econ. 2014;45:85–101. [Google Scholar]
- Ning J, Li X, Hicks LM, Xiong L. A Raf-like MAPKKK gene DSM1 mediates drought resistance through reactive oxygen species scavenging in rice. Plant Physiol. 2010;152:876–890. doi: 10.1104/pp.109.149856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SM, Kim KP, Joe MK, Lee MO, Koo HJ, Hong CB. Tobacco class I cytosolic small heat shock proteins are under transcriptional and translational regulations in expression and heterocomplex prevails under the high-temperature stress condition in vitro. Plant Cell Environ. 2015;38:767–776. doi: 10.1111/pce.12436. [DOI] [PubMed] [Google Scholar]
- Perez-Clemente RM, Vives V, Zandalinas SI, Lopez-Climent MF, Munoz V, Gomez-Cadenas A. Biotechnological approaches to study plant responses to stress. BioMed Res Int. 2013;2013:654120. doi: 10.1155/2013/654120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez DE, Hoyer JS, Johnson AI, Moody ZR, Lopez J, Kaplinsky NJ. BOBBER1 is a noncanonical Arabidopsis small heat shock protein required for both development and thermotolerance. Plant Physiol. 2009;151:241–252. doi: 10.1104/pp.109.142125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan RD, Shang M, Zhang H, Zhao YX, Zhang JR. Improved chilling tolerance by transformation with betA gene for the enhancement of glycinebetaine synthesis in maize. Plant Sci. 2004;166:141–149. [Google Scholar]
- Ruibal C, Castro A, Carballo V, Szabados L, Vidal S. Recovery from heat, salt and osmotic stress in Physcomitrella patens requires a functional small heat shock protein PpHsp16.4. BMC Plant Biol. 2013;13:174–191. doi: 10.1186/1471-2229-13-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanmiya K, Suzuki K, Egawa Y, Shono M. Mitochondrial small heat-shock protein enhances thermotolerance in tobacco plants. FEBS Lett. 2004;557:265–268. doi: 10.1016/s0014-5793(03)01494-7. [DOI] [PubMed] [Google Scholar]
- Sarkar NK, Kim YK, Grover A. Rice sHsp genes: Genomic organization and expression profiling under stress and development. BMC Genomics. 2009;10:393. doi: 10.1186/1471-2164-10-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharom M, Willemot C, Thompson JE. Chilling injury induces lipid phase changes in membranes of tomato fruit. Plant Physiol. 1994;105:305–308. doi: 10.1104/pp.105.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf KD, Siddique M, Vierling E. The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing α-crystallin domains (Acd proteins) Cell Stress Chaperones. 2001;6:225–237. doi: 10.1379/1466-1268(2001)006<0225:tefoat>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnoff N. In: Smirnoff N (ed) Environment and Plant Metabolism: Flexibility and Acclimation. BiosScientific Publishers; Oxford: 1995. Antioxidant systems and plant response to the environment; pp. 217–243. [Google Scholar]
- Sun L, Liu Y, Kong X, Zhang D, Pan J, Zhou Y, Yang X. ZmHSP16. 9, a cytosolic class I small heat shock protein in maize (Zea mays), confers heat tolerance in transgenic tobacco. Plant Cell Rep. 2012;31:1473–1484. doi: 10.1007/s00299-012-1262-8. [DOI] [PubMed] [Google Scholar]
- Sun W, Bernard C, Van de Cotte B, Van Montagu M, Verbruggen N. At-HSP17.6A, encoding a small heat-shock protein in Arabidopsis, can enhance osmotolerance upon overexpression. Plant J. 2001;27:407–415. doi: 10.1046/j.1365-313x.2001.01107.x. [DOI] [PubMed] [Google Scholar]
- Sun W, Van Montagu M, Verbruggen N. Small heat shock proteins and stress tolerance in plants. Biochim Biophys Acta. 2002;1577:1–9. doi: 10.1016/s0167-4781(02)00417-7. [DOI] [PubMed] [Google Scholar]
- Tang ZC. In: Tang ZC, Modern Experimental Protocols in Plant Physiology. Beijing Science Press; Beijing: 1999. The measurement of electrical conductivity; pp. 302–308. [Google Scholar]
- Vermeulen S, Zougmore R, Wollenberg E, Thornton P, Nelson G, Kristjanson P, Kinyangi J, Jarvis A, Hansen J, Challinor A. Climate change, agriculture and food security: A global partnership to link research and action for low-income agricultural producers and consumers. Curr Opin Environ Sustain. 2012;4:128–133. [Google Scholar]
- Voetberg GS, Sharp RE. Growth of the maize primary root at low water potentials III. Roles of increased proline deposition in osmotic adjustment. Plant Physiol. 1991;96:1125–1130. doi: 10.1104/pp.96.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Yu X, Mao Y, Liu Y, Liu G, Liu Y, Niu X. Overexpression of a small heat-shock-protein gene enhances tolerance to abiotic stresses in rice. Plant Breed. 2015;134:384–393. [Google Scholar]
- Waters ER, Aevermann BD, Sanders-Reed Z. Comparative analysis of the small heat shock proteins in three angiosperm genomes identifies new subfamilies and reveals diverse evolutionary patterns. Cell Stress Chaperones. 2008;13:127–142. doi: 10.1007/s12192-008-0023-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters ER, Lee GJ, Vierling E. Evolution, structure and function of the small heat shock proteins in plants. J Exp Bot. 1996;47:325–338. [Google Scholar]
- Wehmeyer N, Hernandez LD, Finkelstein RR, Vierling E. Synthesis of small heat-shock proteins is part of the developmental program of late seed maturation. Plant Physiol. 1996;112:747–757. doi: 10.1104/pp.112.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Peng R, Xiong A, Li X, Zha D, Yao Q. Over-expression of heat shock protein gene hsp26 in Arabidopsis thaliana enhances heat tolerance. Biol Plant. 2010;54:105–111. [Google Scholar]
- Young TE, Ling J, Geisler-Lee J, Tanguay RL, Caldwell C, Gallie DR. Developmental and thermal regulation of the maize heat shock protein, HSP101. Plant Physiol. 2001;127:777–791. [PMC free article] [PubMed] [Google Scholar]
- Zhai M, Sun Y, Jia C, Peng S, Liu Z, Yang G. Over-expression of jrshsp17.3, gene from Juglans regia, confer the tolerance to abnormal temperature and nacl stresses. J Plant Biol. 2016;59:549–558. [Google Scholar]
- Zhang A, Jiang M, Zhang J, Tan M, Hu X. Mitogen-activated protein kinase is involved in abscisic acid-induced antioxidant defense and acts downstream of reactive oxygen species production in leaves of maize plants. Plant Physiol. 2006;141:475–487. doi: 10.1104/pp.105.075416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Gao Y, Pan H, Hu W, Zhang QX. Cloning and characterisation of a Primula, heat shock protein gene, PfHSP17.1, which confers heat, salt and drought tolerance in transgenic Arabidopsis thaliana . Acta Physiol Plant. 2013;35:3191–3200. [Google Scholar]
- Zhang XX, Liu SK, Takano T. Two cysteine proteinase inhibitors from Arabidopsis thaliana, AtCYSa and AtCYSb, increasing the salt, drought, oxidation and cold tolerance. Plant Mol Biol. 2008;68:131–143. doi: 10.1007/s11103-008-9357-x. [DOI] [PubMed] [Google Scholar]
- Zhou YL, Chen HH, Chu P, Li Y, Tan B, Ding Y, Tsang EWT, Jiang LW, Wu KQ, Huang SZ. NnHSP17.5, a cytosolic class II small heat shock protein gene from Nelumbo nucifera, contributes to seed germination vigor and seedling thermotolerance in transgenic Arabidopsis. Plant Cell Rep. 2012;31:379–389. doi: 10.1007/s00299-011-1173-0. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Wang Z, Jing Y, Wang L, Liu X, Liu Y, Deng X. Ectopic over-expression of BhHsf1, a heat shock factor from the resurrection plant Boea hygrometrica, leads to increased thermotolerance and retarded growth in transgenic Arabidopsis and tobacco. Plant Mol Biol. 2009;71:451–467. doi: 10.1007/s11103-009-9538-2. [DOI] [PubMed] [Google Scholar]
- zur Nieden U, Neumann D, Bucka A, Nover L. Tissue-specific localization of heat-stress proteins during embryo development. Planta. 1995;196:530–538. [Google Scholar]