Abstract
Early-stage Alzheimer’s disease is characterized by the loss of dendritic spines in the neocortex of the brain. This phenomenon precedes tau pathology, plaque formation, and neurodegeneration and likely contributes to synaptic loss, memory impairment, and behavioral changes in patients. Studies suggest that spine loss is induced by soluble, multimeric Aβ42, whose post-synaptic signaling activates the protein phosphatase calcineurin. We investigated how calcineurin causes spine pathology and found that the cis-trans prolyl isomerase Pin1 is a critical downstream target of Aβ42/calcineurin signaling. In spines, Pin1 interacts with and is dephosphorylated by calcineurin, which rapidly suppresses its isomerase activity. Pin1 knock-out or Aβ42 exposure induced mature spine loss that was prevented by exogenous Pin1. The calcinuerin inhibitor FK506 blocked spine loss in Aβ42-treated wild-type cells but had no effect on Pin1 null neurons. The data implicate Pin1 in spine maintenance and synaptic loss in early Alzheimer’s disease.
Introduction
Early stages of Alzheimer’s Disease (AD) or mild cognitive impairment (MCI) are characterized by memory loss and behavioral changes. These symptoms likely reflect loss of dendritic spines and their synaptic connections induced by soluble, multimeric Aβ42(1, 2). Spine and synaptic loss occurs prior to neurodegeneration or tau pathology suggesting spine preservation strategies could attenuate or prevent AD progression (1). It’s not clear how regulators of spine maintenance are negatively affected during the development of AD by increased, soluble Aβ42.
One protein whose dysfunction is implicated in both spine physiology and AD pathogenesis is the peptidyl-prolyl isomerase, Pin1 (3). Pin1 binds to and catalyzes the cis to trans conversion of amyloid precursor protein (APP), phosphorylated tau (p-tau), and other post-synaptic proteins at phosphoserine (pSer)/pThr-Pro bonds, altering their function and/or catabolism (4). Target binding is mediated through an N-terminal, WW domain whereas isomerization occurs through a C-terminal enzymatic domain. Ser or Thr phosphorylation, typically by proline-directed kinases, such as MAPK, PKC and CDK families, accelerates Pin1-mediated isomerization of peptide bond by approximately 1000-fold (5). Under basal conditions, Pin1 is highly active in neuronal cell bodies, nuclei, axonal growth cones and dendritic spines but can be inhibited by glutamatergic, outside-in signaling that induces phosphorylation of Ser16 within the WW binding domain, preventing target recognition (6). Most relevant to AD, Pin1-mediated isomerization drives APP cleavage toward the α and away from the β/γ secretase pathway, reducing Aβ42 production (7, 8). By binding to and isomerizing p-tau, Pin1 also prevents neurofibrillary tangle formation (4), again suggesting a possible role for its dysfunction in AD pathogenesis.
Consistent with this hypothesis, cortical Pin1 protein content (7) and activity (9) are often reduced or lost in end-stage AD, whereas germ-line Pin1 knockout (KO) accelerates AD pathology in Tg2576 mice, which overproduce Aβ42 (8). The isomerase activity of Pin1 is lost due to oxidation of Cys113, a critical amino acid within its active site (9). Whether alterations in inside-out signaling, especially at synaptic sites, can affect Pin1 activity or steady-state protein amounts during AD evolution is unknown.
Somewhat paradoxically, hippocampal slices from pure-background, germ-line Pin1 KO mice display increased, rather than reduced, long-term potentiation (LTP) (10) and show increased, rather than decreased, hippocampal dendritic spine density (6), phenotypes that are opposite than is predicted from the pathology seen in AD or murine AD models. Pure-background Pin1 KO mice are very difficult to breed with only ~2–5% of the expected viable births. These discrepancies suggested that viable KO mice somehow complemented for the loss of Pin1, masking the true effects of gene ablation. This encouraged us to develop Pin1fl/fl mice so that Pin1 loss could be assessed post-development in vivo and in vitro. Using floxed mice and neuronal cultures derived from them, we found that Pin1 is required to maintain mature dendritic spines, and that post-synaptic Pin1 is inhibited by soluble, multimeric Aβ42-mediated signaling through calcineurin, and that this event causes spine loss. These results suggest that Pin1 can be inactivated early in the evolution of AD by Aβ42 signaling that directly contributes to spine pathology. The results also suggest that prior data from germ-line Pin1 KO mice should be carefully reassessed.
Results
Neocortices from AD patients show progressive losses of Pin1 along with synapses and dendritic spines (11) that can be mimicked in cultured neurons (12) or slice preparations (13) treated with multimeric Aβ42. Unexpectedly, germ-line Pin1 KO cortex tissues exhibit increased dendritic spine density and increased LTP in hippocampal slices (6, 10). To clarify these seemingly contradictory results, we measured total dendritic spine density of primary DIV21 Pin1fl/fl neurons after transfection on DIV7 with Td-tomato and either NLS-GFP (WT) or NLS-GFP-Cre (KO) (fig S1A). Spine counts were reproducibly 35% lower in Pin1 KO versus WT cultures (3.04 ± 0.39 per 10 μm compared to 4.77 ± 0.25 per 10 μm) (Fig. 1A and fig. S1, B and C) with significant reductions in mature mushroom and stubby spines (fig. S1C). To verify the results seen in genetically modified cells, we exposed cultured neurons to recombinant WW domain fused to the trans-activator of transcription (TAT) tag (TAT-WW), which we and others have previously established would enter spines directly, very rapidly, and with minimal toxicity (10, 14, 15). TAT-WW functions as a dominant-negative, preventing endogenous Pin1 from binding to target proteins (16). Spine counts were similarly reduced in WT cells transduced with TAT-WW (fig. S1, D to F) but were restored to normal in KO cells after transduction with recombinant TAT-Pin1 (Fig. 1B and fig. S1, G and H). To control for nonspecific effects of TAT, neurons were also transduced with TAT-GFP. Such cells showed no changes in spine counts or spine types from untreated, control neurons (fig. S1, D to F). Therefore, post-developmental Pin1 loss or inhibition caused loss of mature dendritic spines that could be rescued by exogenous TAT-Pin1 but not TAT-GFP.
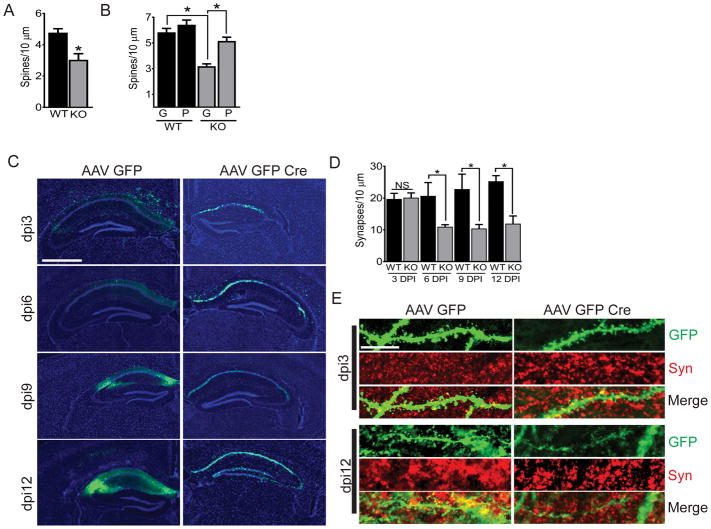
Fig. 1. Pin1 KO causes dendritic spine loss in vitro and vivo.
(A) Total spine counts in Pin1fl/fl neurons after transfection with Td-tomato and either NLS-GFP (WT, black) or NLS-GFP-Cre (KO, grey). Neurons were fixed at DIV21. (B) As described in (A), in DIV21 neurons transduced with TAT-GFP (“G”) or TAT-Pin1 (“P”). (C) Representative images of GFP immunofluorescence in hippocampi of AAV-GFP–injected or AAV-GFP-Cre–injected 3-month-old Pin1fl/fl mice at 3, 6, 9 and 12 days post injection (DPI). Sections were counterstained with DAPI. Scale bar, 1000 μm. (D) Total synapse counts at 3, 6, 9, and 12 days DPI from 24 mouse hippocampi injected at 3-months old with either AAV-GFP (WT, black) or AAV-GFP-Cre (KO, grey). (E) Imaging of GFP (green) and antibody to synaptophysin (Syn; red) in dendritic spines from hippocampal neurons described in (C). Shown is a representative composite Z-stack image of three 0.33 μm-thick step sizes. Scale bar, 10 μm. Data are mean ± SEM; N >100 spines from ≥15 images and ≥ 3 coverslips per condition were counted. *P<0.05 by an unpaired t-test (A) or Fisher’s LSD test following two-way analysis of variance (ANOVA) (B and C).
To determine if Pin1 was required for spine stability in vivo, we injected AAV-GFP-Cre or AAV-GFP into the hippocampus of adult Pin1fl/fl mice (Fig. 1C). Synapse numbers in projections from GFP and synaptophysin co-positive CA1 hippocampal neurons were unchanged between the controls and KO mice at 3 days post injection (DPI) but were significantly decreased (~40%) by DPI 6 in Pin1 KO neurons (Fig. 1, D and E). Laser captured, microdissected GFP positive hippocampal cells were analyzed by PCR for recombination. Hippocampi injected with AAV-GFP-Cre showed substantial recombination that was not seen in the thalamus of these animals nor in mice injected with AAV-GFP (fig. S2A). Thus, Pin1 loss both in vitro and in vivo caused spine loss.
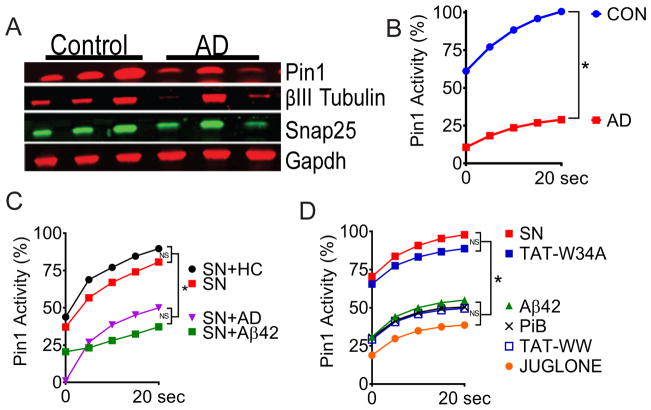
We next assessed Pin1 abundance and isomerase activity in soluble fractions from fresh-frozen, frontal cortex tissue from AD patients and from age-matched individuals that were not diagnosed with cognitive deficit (fig. S2B). The abundance of Pin1 and β-III tubulin protein were consistently reduced in AD samples while the abundance of synaptosomal-associated protein 25 (SNAP25), a commonly used measure of synaptic number (17) was more variable compared to normal, age-matched controls (7) (Fig. 2A). Protease-coupled isomerase assay (18) specific for Pin1 revealed significantly reduced isomerase activity in AD brain lysates compared to controls (Fig. 2B). However, given the amount of immunoreactive protein, Pin1 activity was less than expected in AD samples. These data suggested that Pin1 was somehow inactivated in AD lysates. Pin1 can be oxidized at Cys113, blocking activity (9) or inhibited by outside-in glutamatergic signaling (10). To identify the mechanism, we utilized mouse synaptoneurosomes, a highly enriched preparation of pre- and post-synaptic neuronal connections from brain. Isomerase activity was measured in control synaptoneurosomes or after incubation with soluble AD brain lysates or age-matched control lysates. Control brain lysates had no effect on Pin1 activity but AD lysates significantly suppressed activity (Fig. 2C). We were able to entirely replicate these findings with 100 nM multimerized, soluble Aβ42 (fig. S2C) or with PiB, TAT-WW or juglone, all established Pin1 inhibitors (12) (Fig. 2D). TAT-W34A, that contains an inactivating mutation of the terminal tryptophan in the WW-domain had no effect (Fig. 2D), demonstrating that Pin1 activity in synaptoneurosomes was not affected by TAT-mediated transduction. These data demonstrate that synaptic Pin1 can be inhibited by soluble, multimeric Aβ42 signaling.
Fig. 2. Aβ42 inhibits Pin1 isomerase activity.
(A) Western blotting for Pin1, Snap25, βIII-tubulin and GAPDH in soluble fractions of temporal cortex lysates from 3 Alzheimer’s disease (AD) patients and 3 age-matched control individuals (not diagnosed with dementia; Con). (B) Pin1 activity assay in equal amounts of soluble protein extract from control and AD patient brain tissues described in (A). (C) Pin1 activity assay with murine synaptoneurosomes (SN) that were untreated (red) or treated for 10 min with control brain extract (+HC, black), AD brain extract (+AD, purple), or Aβ42 (green). (D) Pin1 activity assay in SNs that were either untreated (red) or treated with juglone, PiB (, TAT-WW, TAT-W34A, or Aβ42. Data are means of N ≥ 8 replicates per treatment. *P<0.05 by Fisher’s LSD test following two-way analysis of variance (ANOVA) (B, C and D).
Based on these results, we determined the effects of Aβ42 on dendritic spines in Pin1 KO and WT neurons. As expected, Aβ42 significantly reduced total and stubby spine density of WT neurons to the numbers seen in untreated KO cells (Fig. 3A and fig. S3, A and B). Neither Aβ42 nor TAT-GFP had a significant effect on the total spine density of KO neurons (Fig. 3B and fig. S3, C and D). The spines of WT neurons transduced with excess TAT-Pin1 were fully resistant to Aβ42 (Fig. 3A and fig. S3, A and B) while KO cultures treated with TAT-Pin1 were partially sensitive to Aβ42 (Fig. 3B and fig. S3, C and D). Therefore, Pin1 plays a previously unappreciated role in dendritic spine maintenance and its loss pheno-copied spine alterations seen in neurons exposed to multimeric Aβ42. These results strongly suggest that Aβ42 signaling causes spine loss by inhibiting Pin1.
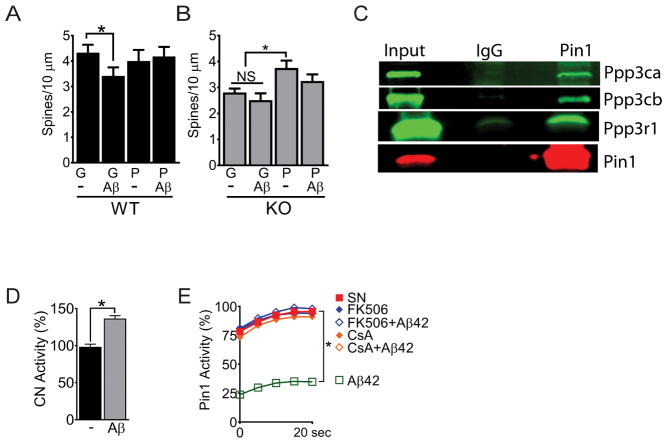
Fig. 3. Calcineurin interacts with Pin1 and mediates Aβ42 signaling.
(A and B) Total spine counts for DIV21 wild-type (WT; A) or DIV21 KO (B) neurons transduced with TAT-GFP (“G”) or TAT-Pin1 (“P”) ± Aβ42 (Aβ). Data are mean ± SEM; N >100 spines from ≥15 images and ≥ 3 coverslips per condition were counted. *P<0.05 by Fisher’s LSD test following two-way analysis of variance (ANOVA). (C) Western blot of atalytic and regulatory subunits of calcineurin inPin1 immunoprecipitations from synaptoneurosomes prepared from P28 mice. (D) Calcineurin (CN) activity in synaptoneurosomes that were either untreated (−) or treated with Aβ42 (Aβ) for 10 min before lysis. Data are mean ± SEM; N = 3 biological replicates. *P < 0.05 by an unpaired t-test. (E) Pin1 activity assay in synaptoneurosomes that were either untreated (SN) or treated with Aβ42, FK506, FK506 + Aβ42, CsA, or CsA + Aβ42 before lysis. Data are means from N ≥ 8 replicates per treatment. *P < 0.05 by Fisher’s LSD test following two-way analysis of variance (ANOVA).
Phosphorylation of Pin1 at Ser16, Ser71 or Ser138 alters activity (16, 19, 20). We immunoprecipitated (IP) Pin1 from synaptoneurosomes after brief treatment (10 min) with Aβ42 and analyzed for interactions and post-translational modifications (PTM) by mass spectrometry (MS). Calcineurin subunits were reproducibly associated with Pin1 by MS (fig. S3E) that was confirmed by IP/IB (Fig. 3C). Aβ42 induced Pin1 dephosphorylation at Ser111, Ser147 and Ser154, sites not previously reported as modified. As seen in primary cortical neurons, dendritic spines and N2A cells (21, 22) we confirmed that Aβ42 quickly increased calcineurin activity in synaptoneurosomes (Fig. 3D). We therefore asked if the calcineurin inhibitors FK506 or cyclosporin A (CsA) could attenuate Aβ42 mediated suppression of Pin1 activity in SN. Indeed, FK506 at 3 nM (~IC50) or CsA at 5 nM (~IC50) completely prevented the suppression of Pin1 isomerase activity by Aβ42 (Fig. 3E). Therefore, calcineurin binds to and likely dephosphorylates Pin1 at Ser111, Ser147 and/or Ser154 in response to Aβ42 signaling, possibly altering Pin1 activity.
Application of FK506 restored spine numbers and complexity in neurons exposed to Aβ42 in vivo (23) and in vitro (24). The dendritic targets of calcineurin that are required for spine maintenance are largely unknown. As expected, FK506 completely rescued spine numbers and maturity in WT cells treated with Aβ42 (Fig. 4A and fig. S4, A and B) but had no effect on spine numbers in Pin1 KO cells irrespective of Aβ42 treatment (Fig. 4A and fig. S4, A and C). Therefore, Aβ42 and calcineurin signaling inhibit Pin1 whose activity is required for spine maintenance. Therapeutics such as FK506 that block calcineurin and preserve Pin1 activity could potentially be effective in attenuating spine losses in early AD.
Fig. 4. Dephosphorylation of Pin1 at Ser111 inhibits Pin1 activity.
(A) Total spine counts for DIV21 WT (black) or KO (grey) neurons ± FK506 (FK) ± Aβ42 (Aβ). (B) Pin1 activity assay in synaptoneurosomes treated with TAT-GFP (GFP), TAT-Pin1-S111D (S111D) or TAT-Pin1-S111A (S111A) ± Aβ42 before lysis. Data are means from N ≥ 8 replicates per treatment. *P<0.05 by Fisher’s LSD test following two-way analysis of variance (ANOVA). (C) Total spine counts in DIV21 wild-type (WT; C) and KO (D) neurons transduced with TAT-GFP (“G”), TAT-Pin1 (“P”), TAT-Pin1-S111A (“A”) or TAT-Pin1-S111D (“D”) ± Aβ42. Data are mean ± SEM; N >100 spines from ≥15 images and ≥ 3 coverslips per condition were counted. *P<0.05 by Fisher’s LSD test following two-way analysis of variance (ANOVA) (A, C, and D).
Ser111 is in very close proximity to the active-site Cys113 (25, 26). Thus, TAT-GFP (TAT-control), TAT-Pin1, TAT-Pin1-S111A (phospho-null) or TAT-Pin1-S111D (phospho-mimetic) recombinant proteins were produced and added to synaptoneurosomes or to Pin1 WT and KO neurons and isomerase activity or spine numbers determined, respectively. TAT-Pin1-S111D transduced synaptoneurosomes were completely resistant to Aβ42, while TAT-Pin1-S111A functioned as a dominant negative similar to C113A (Fig. 4B, fig. S5A). TAT-GFP had no effect on Pin1 activity or responsiveness to Aβ42 (Fig. 4B and fig. S5A). TAT-Pin1-S111A significantly reduced spine numbers in WT cultures to the same degree as Aβ42 or as seen in untreated KO neurons (Fig. 4C and 4D and fig. S5, B to K). Conversely, TAT-Pin1 or TAT-Pin1-S111D had no effect on control WT cells but significantly increased spine counts in WT neurons treated with Aβ42 or untreated KO neurons (Fig. 4, C and D and fig. S5, B to K). However, only TAT-Pin1-S111D was reproducibly able to rescue spine counts in KO cells treated with Aβ42 (Fig. 4D, fig. S4C and S5, H to K). In aggregate, these results suggest that Aβ42 induces calcineurin to dephosphorylate Pin1 at Ser111, rendering it inactive. Pin1-S111D is constitutively active, resistant to Aβ42 signaling and calcineurin-mediated dephosphorylation and supports spine numbers and complexity in both WT and KO neurons.
Discussion
One of the earliest pathologic events in evolving Alzheimer’s disease is the loss of synapses and dendritic spines (27). These changes presumably underlie initial memory and behavioral deficits and occur before neuronal losses or neurofibrillary tangle formation (27). The work presented here demonstrates that a signaling cascade initiated by soluble Aβ42 leads to calcineurin activation and Pin1 inhibition, negatively affecting dendritic spine health. Furthermore, the data suggests that calcineurin inhibitors such as FK506 could attenuate spine loss due to Aβ42 signaling, potentially reducing early AD symptoms. As spine loss can accelerate neuronal death (25), spine sparing strategies could also slow AD development.
Pin1 can regulate Aβ42 production and p-tau accumulation (3, 7, 8, 28) . Discrepancies between spine counts and LTP in germ-line Pin1 KO and AD model mice that lose Pin1 over time led us to create and study Pin1fl/fl mice. Post-natal Pin1 loss caused a significant decrease in spine density that could be rapidly rescued by the transduction of exogenous Pin1. Corroborating data were seen in Pinfl/fl adult mice after intrahippocampal delivery of AAV-CRE-GFP, confirming that Pin1 is required for dendritic spine maintenance in mature neurons. The differences in spine density phenotype in germ-line and conditional knockout cells suggest that Pin1 has distinct functions in dendritic spine development and spine maintenance. At this point, we do not know whether other observed phenotypes in germline KO mice including impaired numbers and differentiation of neural progenitor cells, growth cone abnormalities, and misorganization of global brain architecture (6, 29–31) accurately reflect Pin1’s role in neurodevelopment.
Pin1 activity is sensitive to outside-in glutamatergic signaling (10) but regulation by Aβ42 signaling has not been previously shown. These effects were observed with multimeric Aβ42, a phenotype similar to the suppressive effects on LTP in slices, dendritic spine density, and neurotoxicity in culture (1, 2, 30). Multimeric, soluble Aβ42 causes a variety of post-synaptic signaling abnormalities including alteration of Ca2+ homeostasis involving both intracellular and extracellular stores (32). Calcineurin is a calcium/calmodulin dependent protein phosphatase highly expressed in normal brain and aberrantly activated in AD patients (33–35), mouse models and cultured neurons (21, 35, 36). Increased calcineurin activity disrupts synaptic networks, reduces LTP (37), increases long-term depression (LTD) (38) and decreases memory (34, 39–41). Based on our data, Aβ42 mediated effects on calcineurin can be modeled and studied in synaptoneurosomes, permitting detailed biochemical analysis.
One of the downstream effects of Aβ42-mediated calcineurin activation is Pin1 inactivation. These effects are likely direct as Pin1 interacts with multiple calcineurin subunits and the loss of Pin1 activity can be prevented by mechanistically distinct calcineurin inhibitors FK506 and cyclosporine. FK506 binds FKBP12 to form a complex that inhibits calcineurin activity while CsA forms a complex with cyclophilin A to block calcineurin activity (42–44). FK506 has been shown to rescue Aβ42-induced dendritic spines loss, calcium dysregulation, hippocampal LTP and behavioral deficiencies in mice (23, 32, 36, 45). A retrospective study showed that transplant patients receiving FK506 developed markedly less AD than expected (36). Patient’s immunosuppressed with the mTOR inhibitor rapamycin showed the expected incidence of AD, suggesting that calcineurin inhibition, rather than peripheral immuno-suppression, was required to reduce AD incidence in transplant patients. Here, FK506 was able to rescue Aβ42-mediated dendritic spine loss only in Pin1 replete cultures. Whether Pin1 is also involved in upstream Aβ42 signaling that activates calcineurin remains to be determined. If so, our data would predict that Ser111 phosphorylated Pin1 would oppose calcineurin activation by Aβ42. Altogether, these data demonstrate that FK506 rescued Aβ42-mediated dendritic spine loss in a Pin1 dependent manner.
The rapid kinetics of Aβ42-mediated effects are most consistent with changes in post-translational modifications (PTMs) of Pin1. Phosphorylation at Ser16 by PKMζ or PKA (10, 16) or Ser71 by DAPK1 (19) inhibited Pin1 activity while MLK3 mediated, Ser138 phosphorylation had the opposite effect (20). Polo-like kinase 1 (PLK1) phosphorylation at Ser65 stabilized Pin1 protein without alteration of isomerization activity (46). The identification of Ser111 phosphorylation as required for activity has not been described. This amino acid is within the catalytic pocket of Pin1 and very close to active site Cys113 (26). Phospho-null Pin1-S111A mutants showed dominant negative activity as previously shown for C113A mutants while phospho-mimetic Pin1-S111D was constitutively active, fully resistant to Aβ42-mediated inhibition and supported spine counts irrespective of Aβ42 treatment. Based on the work here and the data of others, we propose that the gradual accumulation of multimeric Aβ42 in evolving AD increases calcineurin activity, causing a progressive reduction of Pin1 activity in post-synaptic dendritic spines. Presumably Pin1 interacts with and is required for the function of protein(s) essential for spine maintenance and complexity. As Pin1 also regulates Aβ42 production (8), we also propose Pin1 inhibition accelerates Aβ42 production, further suppressing Pin1. The potential for calcineurin inhibition to actively interfere with this feed-forward loop makes it an intriguing therapeutic concept.
Materials and Methods
Animals
Pin1fl/fl mice were created using traditional methods (fig. S2A). Pin1fl/fl mice were maintained on a C57/Bl6 background and were bred by crossing homozygous Pin1fl/fl mice with homozygous Pin1fl/fl mice. Mice were fed a standard chow (Teklad Global 16% Rodent Diet, #2016) ad libitum and were housed in a standard 12-hour light/dark cycle. All animal use was done in accordance with protocols approved by The University of Texas Southwestern Medical Center Institutional Use and Animal Care Committee.
Synaptoneurosome Preparation
Pin1fl/fl mice in a C57Bl/6 background were used for all synaptoneurosome preparations per the method described in (12). Briefly, mouse cortices were harvested from mice that were heavily sedated with 2,2,2-tribromoethanol (Avertin). Using a 5 ml homogenizer, 2 brains were processed at a time with 10 strokes of pestle A then pestle B. Combined homogenate was then prepared per the method described in (10). SN preps were aliquoted and treated with juglone, PiB, CsA, concatamerized Aβ42, FK506 or TAT proteins purified per (10). The reactions were stopped with a final concentration of 1% Triton-X. Samples were snap-frozen and briefly stored at −80°C until assayed.
Neuronal Culture
Cortexes from Pin1fl/fl P1 pups were dissected, roughly chopped with a razor blade, and then placed in a 1:1 solution of 0.25% trypsin:DPBS with 10 U of DNase per ml of dissection solution. After 30 mins, the digestion was stopped by the addition of MEM with 20% fetal calf serum. Cells were centrifuged for 5 mins at 1000 RPM, supernatant was aspirated and the cells were washed again with the stop solution. Media was aspirated and the cell pellet resuspended in 2 ml of Neurobasal-A supplemented with B27, L-glutamine, penicillin and streptomycin, and 5% fetal calf serum. Cells were then filtered through a 70 μm filter, a 40 μm filter, and then plated on coverslips coated with poly-D lysine. At DIV7, cultures were transfected with Td-Tomato and NLS-GFP (wild-type, WT) plasmids or Td-Tomato and NLS-GFP-Cre (knock-out, KO) plasmids using a calcium phosphate method (47). The Td-Tomato and NLS-GFP-Cre plasmids were used as previously published (48).
DIV21 cultures were treated with 100 nM of TAT-Pin1 (WT), TAT-Pin1S111A, TAT-Pin1S111D, TAT-WW, TAT-W34A or TAT-GFP for 3 hrs. 100 nM Aβ42 or vehicle was added to the culture for an additional hr. 5 nM FK506 was added 15 mins before Aβ42. Cells were fixed in 4% paraformaldehyde (PFA,) 4% sucrose and analyzed for spine morphology. A Z stack was captured using a 100X objective on a Zeiss Axiovert LSM510 confocal microscope or a 63X objective with 2.73 zoom on a Leica DM6000B confocal microscope. An entire experimental data set was analyzed on one microscope. At least 15 Z stacks per group were analyzed using NeuronStudio (49) with the modifications described (48). Briefly, dendrites were traced and the build spine function was used to identify spines. Computer generated spine classifications were then manually reviewed by the user and data was exported to Excel and Graphpad Prism where statistical analysis was performed. Graphing of data was performed in Graphpad Prism® v7.01. For image presentation, representative confocal Z stacks were opened in Image J, converted to a Z project with max intensity and saved as a .tif file. Images were then rotated and cropped in Photoshop for insertion in the figure.
Pin1 Isomerase Activity Assays
1 mg of Pin1 substrate (Suc-Ala-Glu-Pro-Phe-pNA, Peptides International) was suspended in 15 μl DMSO, then diluted 1:100 in 1M lithium chloride in trifluoroethanol and allowed to incubate a minimum of 30 mins at RT. Chymotrypsin was suspended at 100 mg per 1.67 ml 0.001 N HCL. Final concentration of assay buffer was 50 mM Hepes, pH 7, 100 mM NaCl, pH 7, 2 mM DTT, 0.04 mg/ml BSA, 0.03 mg/mL chymotrypsin. Frozen SN lysates were thawed on ice. Analysis was performed on a V630 Spectrophotometer by Jasco with a multi-well accessory at 390 nm. To perform the PPIase Assay on SN preps, 100 μl SN lysate was premixed with 1 ml assay buffer and incubated at least 30 mins in a 25°C heat block. 110 μl was pipetted into the 8 wells in the multi-well accessory unit and individually assessed. Each well was blanked while containing the original sample, then 5 μl of Pin1 substrate was added and the sample was immediately analyzed every 0.1 seconds for 60 seconds. The human brain samples were run with 2 ug per assay. Data was exported to Graphpad Prism® v7.01 for graphing and analysis. All isomerase assays were run with a BSA only control to assess spontaneous isomerization of the substrate. The BSA values were subtracted from the test samples using baseline subtraction. For graphing purposes, the samples were then normalized so that the BSA was set to zero and the SN samples were set to 100%.
Human brain samples and western blots
Soluble fraction from human brain samples were prepared according to the method of Shankar et al. (2, 50). 35 μg of human brain lysate was run on an Any kD™ gel (Bio-Rad, 456-9034) and transferred to nitrocellulose, blotted, and scanned using a Licor Odyssey and analyzed using Image Studio lite. Antibodies used were Pin1 (SC-46660, Santa Cruz), Snap25 (ab24737, Abcam), βIII Tubuliln (801202, Biolegends) and Gapdh (60004-1-Ig, Protein Tech.)
Aβ42 preparation and concatamerization
Human Aβ42 was purchased from Peninsula Laboratories (San Carlos, CA) or Bachem America (Torrance, CA) and diluted to 100 μM in 200 mM HEPES, pH 8.0. The solution was gently agitated for 48 hrs at room temperature, aliquoted, and stored at −80°C. The resultant Aβ42 was analyzed by western blot for concatamerization (50) (fig S2C). Human amyloid B (82E1, Immuno-Biological Laboratories) and B Amyloid 17–24 (4G8, Covance) were used to detect oligomers.
XL-Bead Preparation
Pin1 antibody (MAB2294, R&D Systems) or normal mouse IgG (SC-2025, Santa Cruz) were immobilized on Protein G Sepharose® Fast Flow Beads (P3296, Sigma). Beads were equilibrated in PBS for 1 hour with multiple buffer changes. Antibody was added at 4 μg per 20 μl bead slurry in PBS and allowed to bind overnight at 4°C. Beads were washed 3X with fresh 0.2 M sodium borate. Freshly made dimethyl pimelimidate dihydrochloride was then used at 5.5 mg/ml in 0.2 M sodium borate for 40 mins at RT to cross-link the antibody to the beads. Beads were rinsed and incubated for 2 hours at RT with 0.2 M ethanolamine. Beads were washed 3X with PBS and stored up to 4 weeks.
Lysate Preparation
Synaptoneurosome lysates were made as described above except no Triton-X was added post-treatment. Samples were instead pelleted at 5,000 RPM for 7.5 mins at 4°C, at which point pellets were washed with chilled DPBS with calcium and magnesium (Corning, 21-0303-CV.) Pellets were then snap frozen and subsequently stored at −80°C. Pellet pairs from several preparations were thawed on ice and combined with IP Buffer: 1X stimulation buffer (10 mM Tris, pH 7, 0.5 mM Na2HPO4, 0.4 mM KH2PO4, 4 mM NaHCO3, 80 mM NaCl and 0.25% Triton-X) plus Halt™ Protease & Phosphatase Inhibitor (Thermo, 1861280.) Samples underwent 3 freeze/thaw cycles followed by a 30 min incubation on ice with occasional mixing. Samples were then centrifuged (15,000 RPM, 15 mins, 4°C). The supernatant was kept while pellets were suspended in additional IP buffer and the spin was repeated. Supernatants from both spins were combined and concentration measured with a Pierce™ BCA Protein Assay Kit (23227, Thermo).
Immunoprecipitation and immunoblotting
Freshly made lysates were pre-cleared with normal mouse IgG-XL-beads for 2 hours at 4°C. After a quick spin, the pre-cleared lysates were transferred to new tubes and were incubated overnight with Pin1-XL-beads. Beads were washed 3X with IP buffer and stored in 2X SDS-PAGE loading buffer. Mass spectrometry samples were run on an Any kD™ gel (456-9034, Bio-Rad) and underwent a Pierce® Silver Stain for Mass Spectrometry kit (24600, Thermo). A visible band around the size of Pin1 (18 kD) was excised and sent for post-translational modification analysis or the entire sample was run 10 mm into a protein gel and excised for analysis of Pin1-interacting proteins by the UT Southwestern Proteomics Core. Protein gel pieces were reduced and alkylated with DTT (20 mM) and iodoacetamide (27.5 mM). A 0.1 μg/μL solution of trypsin in 50 mM triethylammonium bicarbonate (TEAB) was added to completely cover the gel, allowed to sit on ice, and then 50 μL of 50 mM TEAB was added and the gel pieces were digested overnight (Pierce). Following solid-phase extraction cleanup with an Oasis HLB μelution plate (Waters), the resulting peptides were reconstituted in 10 uL of 2% (v/v) acetonitrile (ACN) and 0.1% trifluoroacetic acid in water. 5 uL were injected onto an Orbitrap Fusion Lumos mass spectrometer (Thermo Electron) coupled to an Ultimate 3000 RSLC-Nano liquid chromatography systems (Dionex). Samples were injected onto a 75 μm i.d., 50-cm long EasySpray column (Thermo), and eluted with a gradient from 1–28% buffer B over 60 min. Buffer A contained 2% (v/v) ACN and 0.1% formic acid in water, and buffer B contained 80% (v/v) ACN, 10% (v/v) trifluoroethanol, and 0.1% formic acid in water. The mass spectrometer operated in positive ion mode with a source voltage of 2.4 kV and an ion transfer tube temperature of 275 °C. MS scans were acquired at 120,000 resolution in the Orbitrap and up to 10 MS/MS spectra were obtained in the ion trap for each full spectrum acquired using higher-energy collisional dissociation (HCD) for ions with charges 2–7. Dynamic exclusion was set for 25 s after an ion was selected for fragmentation.
Raw MS data files were converted to a peak list format and analyzed using the central proteomics facilities pipeline (CPFP), version 2.0.3 (51, 52). Peptide identification was performed using the X!Tandem (53) and open MS search algorithm (OMSSA) (54) search engines against the mouse protein database from Uniprot, with common contaminants and reversed decoy sequences appended (55). Fragment and precursor tolerances of 20 ppm and 0.6 Da were specified, and three missed cleavages were allowed. Carbamidomethylation of Cys was set as a fixed modification and oxidation of Met was set as a variable modification. An additional requirement of two unique peptide sequences per protein was used for protein identification. For phosphorylation analysis, phosphorylation of Ser, Thr, and Tyr were also set as variable modifications, and phosphorylation sites were localized using ModLS PTM Localization6 and confirmed by manual interpretation.
WB-IP interaction samples were run on an Any kD™ gel and transferred to nitrocellulose membrane using TransTurbo blot technology. Blots were blocked in 5% BSA in TBS. Membranes were scanned by a Licor Odyssey. Antibodies to Pin1 (MAB2294) and Ppp3r1 (AF1348) were purchased from R&D Systems, antibody to Ppp3ca (A300-908A-M) was from Bethyl Laboratories, and antibody to Ppp3cb (PA5-15581) was from Thermo Fisher Scientific.
Calcineurin Activity Assay
Lysates were made the same day as the Calcineurin Cellular Activity Assay Kit, Colorimetric (Millipore, 207007) was performed per the company protocol. Plates were read at A620 by a Tecan plate reader Spark 10.
Hippocampal AAV1 Stereotactic Injection
AAV1.CMV.PI.eGFP.WPRE.bGH and AAV1.CMV.HI.eGFP-Cre.WPRE.SV40 were purchased from the University of Pennsylvania vector core and injected bilaterally into the CA1 of 8 week old Pin1fl/fl mice in the UT Southwestern Neuro-models facility. In total, 24 mice were used with three mice per condition per time point. Mice were sacrificed at 3, 6, 9, or 12 days post injection (D.P.I.). Mice were deeply anesthetized and perfused with 4% PFA. Brains were postfixed in PFA and then transferred through a series of sucrose gradients. 20 μm sections were then cut with a vibrotome, stained with DAPI and mounted. They were then examined for GFP in the hippocampus. For synapse counts in AAV-injected mice, sections were immuno-stained for synaptophysin (17785-1-AP, Protein Tech) and stained with DAPI. The DAPI staining was used to localize to the CA1 region of the hippocampus. Images were then taken in the apical distal region of the CA1 for synapse analysis. Confocal 0.33 μm Z stacks of a least 6 sections were taken and spines counted and colocalization of GFP and synaptophysin was analyzed with ImageJ using the NeuronJ and SynaptJ plugins. Laser capture microdissection was used to isolate tissue from the brains of AAV injected mice. Regions of infected (green cells) from the hippocampus and uninfected (green negative) cells from the thalamus were used for DNA genotyping analysis
Statistical Analysis
Experiments were replicated at least three times. Data are mean ± SEM. Statistical analysis was performed with Prism (GraphPad) software. Statistical differences between means were analyzed by either an unpaired T-test or Fisher’s LSD test following two-way analysis of variance. Statistical significance was defined as P < 0.05.
Supplementary Material
Fig. S1. Pin1 loss causes spine loss.
Fig. S2. Pin1 successfully recombined in mouse hippocampus after AAV-GFP-CRE.
Fig. S3. Pin1 reconstitution restores mature spine counts in presence of Aβ42.
Fig. S4. FK506 restoration of spines requires Pin1.
Fig. S5. Pin1-S111D restores spine counts in wild-type or KO cells treated with Aβ42.
Acknowledgments
We thank the UTSW Neuropathology Core of the Alzheimer Disease Center (P30 AG12300) for the brain tissue; the UTSW Proteomics Core (CPRIT RP120613) for the mass spectrometry analysis; the UTSW Live Cell Imaging Facility, a Shared Resource of the Harold C. Simmons Cancer Center, (supported in part by an NCI Cancer Center Support Grant, 1P30 CA142543-01), the UTSW Whole Brain Microscopy Facility (supported by the Texas Institute for Brain Injury and Repair) and the Neuro-Models Facility Core (supported by the Texas Institute for Brain Injury and Repair and the Haggerty Center for Research on Brain Injury and Repair in Stroke) for technical assistance; and members of the UTSW AD research group for their comments and suggestions. We would like to specially thank John Shelton of UTSW Histo-Pathology Core for his assistance with laser microdissection of cells in the brain.
Funding: This work was supported by the Senator Betty and Dr. Andy Andujar Endowment and the NIH (P01HL88594 to J.S.M. and R01AG055577 to I.B. and E.T.K.).
Footnotes
Author Contributions: NRS, MAO, ETK, IB and JSM designed the project. NRS and MAO performed the experiments and analyzed the data along with JSM. JH generated and maintained the Pinfl/fl mice. NRS, MAO and JSM wrote the manuscript with contributions from ETK and IB. All authors commented on and approved the final version.
Competing Interests: IB is a consultant for Teva Pharmaceutical Industries. The other authors declare that they have no competing interests.
Data and materials availability: The mass spectrometry data has been deposited to MassIVE data respository (https://massive.ucsd.edu), accession ID MSV000082066. All other data necessary for interpretation have been in shown in the manuscript or supplemental materials.
References and Notes
- 1.Li S, et al. Soluble oligomers of amyloid Beta protein facilitate hippocampal long-term depression by disrupting neuronal glutamate uptake. Neuron. 2009;62:788–801. doi: 10.1016/j.neuron.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shankar GM, et al. Amyloid-beta protein dimers isolated directly from Alzheimer’s brains impair synaptic plasticity and memory. Nature medicine. 2008;14:837–842. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu PJ, Wulf G, Zhou XZ, Davies P, Lu KP. The prolyl isomerase Pin1 restores the function of Alzheimer-associated phosphorylated tau protein. Nature. 1999;399:784–788. doi: 10.1038/21650. [DOI] [PubMed] [Google Scholar]
- 4.Kimura T, et al. Isomerase Pin1 stimulates dephosphorylation of tau protein at cyclin-dependent kinase (Cdk5)-dependent Alzheimer phosphorylation sites. The Journal of biological chemistry. 2013;288:7968–7977. doi: 10.1074/jbc.M112.433326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu KP, Zhou XZ. The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nature reviews. Molecular cell biology. 2007;8:904–916. doi: 10.1038/nrm2261. [DOI] [PubMed] [Google Scholar]
- 6.Antonelli R, et al. Pin1 Modulates the Synaptic Content of NMDA Receptors via Prolyl-Isomerization of PSD-95. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2016;36:5437–5447. doi: 10.1523/JNEUROSCI.3124-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liou YC, et al. Role of the prolyl isomerase Pin1 in protecting against age-dependent neurodegeneration. Nature. 2003;424:556–561. doi: 10.1038/nature01832. [DOI] [PubMed] [Google Scholar]
- 8.Pastorino L, et al. The prolyl isomerase Pin1 regulates amyloid precursor protein processing and amyloid-beta production. Nature. 2006;440:528–534. doi: 10.1038/nature04543. [DOI] [PubMed] [Google Scholar]
- 9.Chen CH, et al. Pin1 cysteine-113 oxidation inhibits its catalytic activity and cellular function in Alzheimer’s disease. Neurobiology of disease. 2015;76:13–23. doi: 10.1016/j.nbd.2014.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Westmark PR, et al. Pin1 and PKMzeta sequentially control dendritic protein synthesis. Science signaling. 2010;3:ra18. doi: 10.1126/scisignal.2000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scheff SW, Price DA. Alzheimer’s disease-related alterations in synaptic density: neocortex and hippocampus. Journal of Alzheimer’s disease: JAD. 2006;9:101–115. doi: 10.3233/jad-2006-9s312. [DOI] [PubMed] [Google Scholar]
- 12.Westmark PR, Westmark CJ, Jeevananthan A, Malter JS. Preparation of synaptoneurosomes from mouse cortex using a discontinuous percoll-sucrose density gradient. Journal of visualized experiments: JoVE. 2011 doi: 10.3791/3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivanov AD, Tukhbatova GR, Salozhin SV, Markevich VA. NGF but not BDNF overexpression protects hippocampal LTP from beta-amyloid-induced impairment. Neuroscience. 2015;289:114–122. doi: 10.1016/j.neuroscience.2014.12.063. [DOI] [PubMed] [Google Scholar]
- 14.Farias GG, et al. Wnt-5a/JNK signaling promotes the clustering of PSD-95 in hippocampal neurons. The Journal of biological chemistry. 2009;284:15857–15866. doi: 10.1074/jbc.M808986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sui WH, et al. Myosin Va mediates BDNF-induced postendocytic recycling of full-length TrkB and its translocation into dendritic spines. Journal of cell science. 2015;128:1108–1122. doi: 10.1242/jcs.160259. [DOI] [PubMed] [Google Scholar]
- 16.Lu PJ, Zhou XZ, Liou YC, Noel JP, Lu KP. Critical role of WW domain phosphorylation in regulating phosphoserine binding activity and Pin1 function. The Journal of biological chemistry. 2002;277:2381–2384. doi: 10.1074/jbc.C100228200. [DOI] [PubMed] [Google Scholar]
- 17.Tafoya LC, et al. Expression and function of SNAP-25 as a universal SNARE component in GABAergic neurons. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2006;26:7826–7838. doi: 10.1523/JNEUROSCI.1866-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nature structural biology. 2000;7:639–643. doi: 10.1038/77929. [DOI] [PubMed] [Google Scholar]
- 19.Lee TH, et al. Death-associated protein kinase 1 phosphorylates Pin1 and inhibits its prolyl isomerase activity and cellular function. Molecular cell. 2011;42:147–159. doi: 10.1016/j.molcel.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rangasamy V, et al. Mixed-lineage kinase 3 phosphorylates prolyl-isomerase Pin1 to regulate its nuclear translocation and cellular function. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:8149–8154. doi: 10.1073/pnas.1200804109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang M, Zhang P, Zhao Y, Jin A, Liu X. Abeta mediates Sigma receptor degradation via CaN/NFAT pathway. American journal of translational research. 2016;8:3471–3481. [PMC free article] [PubMed] [Google Scholar]
- 22.Wu HY, et al. Amyloid beta induces the morphological neurodegenerative triad of spine loss, dendritic simplification, and neuritic dystrophies through calcineurin activation. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2010;30:2636–2649. doi: 10.1523/JNEUROSCI.4456-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rozkalne A, Hyman BT, Spires-Jones TL. Calcineurin inhibition with FK506 ameliorates dendritic spine density deficits in plaque-bearing Alzheimer model mice. Neurobiology of disease. 2011;41:650–654. doi: 10.1016/j.nbd.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller EC, et al. Differential modulation of drug-induced structural and functional plasticity of dendritic spines. Molecular pharmacology. 2012;82:333–343. doi: 10.1124/mol.112.078162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dorostkar MM, Zou C, Blazquez-Llorca L, Herms J. Analyzing dendritic spine pathology in Alzheimer’s disease: problems and opportunities. Acta neuropathologica. 2015;130:1–19. doi: 10.1007/s00401-015-1449-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ranganathan R, Lu KP, Hunter T, Noel JP. Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell. 1997;89:875–886. doi: 10.1016/s0092-8674(00)80273-1. [DOI] [PubMed] [Google Scholar]
- 27.Shankar GM, Walsh DM. Alzheimer’s disease: synaptic dysfunction and Abeta. Molecular neurodegeneration. 2009;4:48. doi: 10.1186/1750-1326-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galas MC, et al. The peptidylprolyl cis/trans-isomerase Pin1 modulates stress-induced dephosphorylation of Tau in neurons. Implication in a pathological mechanism related to Alzheimer disease. The Journal of biological chemistry. 2006;281:19296–19304. doi: 10.1074/jbc.M601849200. [DOI] [PubMed] [Google Scholar]
- 29.Nakamura K, et al. Prolyl isomerase Pin1 regulates neuronal differentiation via beta-catenin. Molecular and cellular biology. 2012;32:2966–2978. doi: 10.1128/MCB.05688-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shankar GM, et al. Natural oligomers of the Alzheimer amyloid-beta protein induce reversible synapse loss by modulating an NMDA-type glutamate receptor-dependent signaling pathway. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2007;27:2866–2875. doi: 10.1523/JNEUROSCI.4970-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sosa LJ, et al. Protein interacting with NIMA (never in mitosis A)-1 regulates axonal growth cone adhesion and spreading through myristoylated alanine-rich C kinase substrate isomerization. Journal of neurochemistry. 2016;137:744–755. doi: 10.1111/jnc.13612. [DOI] [PubMed] [Google Scholar]
- 32.Demuro A, et al. Calcium dysregulation and membrane disruption as a ubiquitous neurotoxic mechanism of soluble amyloid oligomers. The Journal of biological chemistry. 2005;280:17294–17300. doi: 10.1074/jbc.M500997200. [DOI] [PubMed] [Google Scholar]
- 33.Liu F, et al. Truncation and activation of calcineurin A by calpain I in Alzheimer disease brain. The Journal of biological chemistry. 2005;280:37755–37762. doi: 10.1074/jbc.M507475200. [DOI] [PubMed] [Google Scholar]
- 34.Taglialatela G, Rastellini C, Cicalese L. Reduced Incidence of Dementia in Solid Organ Transplant Patients Treated with Calcineurin Inhibitors. Journal of Alzheimer’s disease: JAD. 2015;47:329–333. doi: 10.3233/JAD-150065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu HY, et al. Distinct dendritic spine and nuclear phases of calcineurin activation after exposure to amyloid-beta revealed by a novel fluorescence resonance energy transfer assay. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2012;32:5298–5309. doi: 10.1523/JNEUROSCI.0227-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dineley KT, et al. Amyloid-beta oligomers impair fear conditioned memory in a calcineurin-dependent fashion in mice. Journal of neuroscience research. 2010;88:2923–2932. doi: 10.1002/jnr.22445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ikegami S, et al. A facilitatory effect on the induction of long-term potentiation in vivo by chronic administration of antisense oligodeoxynucleotides against catalytic subunits of calcineurin. Brain research. Molecular brain research. 1996;41:183–191. doi: 10.1016/0169-328x(96)00094-0. [DOI] [PubMed] [Google Scholar]
- 38.Mulkey RM, Endo S, Shenolikar S, Malenka RC. Involvement of a calcineurin/inhibitor-1 phosphatase cascade in hippocampal long-term depression. Nature. 1994;369:486–488. doi: 10.1038/369486a0. [DOI] [PubMed] [Google Scholar]
- 39.Malleret G, et al. Inducible and reversible enhancement of learning, memory, and long-term potentiation by genetic inhibition of calcineurin. Cell. 2001;104:675–686. doi: 10.1016/s0092-8674(01)00264-1. [DOI] [PubMed] [Google Scholar]
- 40.Weitlauf C, Winder D. Calcineurin, synaptic plasticity, and memory. Scientific World Journal. 2001;1:530–533. doi: 10.1100/tsw.2001.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhuo M, et al. A selective role of calcineurin aalpha in synaptic depotentiation in hippocampus. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:4650–4655. doi: 10.1073/pnas.96.8.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu J, et al. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 43.Steiner JP, et al. Neurotrophic actions of nonimmunosuppressive analogues of immunosuppressive drugs FK506, rapamycin and cyclosporin A. Nature medicine. 1997;3:421–428. doi: 10.1038/nm0497-421. [DOI] [PubMed] [Google Scholar]
- 44.Tedesco D, Haragsim L. Cyclosporine: a review. J Transplant. 2012;2012:230386. doi: 10.1155/2012/230386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Overk CR, Rockenstein E, Florio J, Cheng Q, Masliah E. Differential calcium alterations in animal models of neurodegenerative disease: Reversal by FK506. Neuroscience. 2015;310:549–560. doi: 10.1016/j.neuroscience.2015.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eckerdt F, et al. Polo-like kinase 1-mediated phosphorylation stabilizes Pin1 by inhibiting its ubiquitination in human cells. The Journal of biological chemistry. 2005;280:36575–36583. doi: 10.1074/jbc.M504548200. [DOI] [PubMed] [Google Scholar]
- 47.Jiang M, Chen G. High Ca2+-phosphate transfection efficiency in low-density neuronal cultures. Nature protocols. 2006;1:695–700. doi: 10.1038/nprot.2006.86. [DOI] [PubMed] [Google Scholar]
- 48.Sun S, et al. Reduced synaptic STIM2 expression and impaired store-operated calcium entry cause destabilization of mature spines in mutant presenilin mice. Neuron. 2014;82:79–93. doi: 10.1016/j.neuron.2014.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodriguez A, Ehlenberger DB, Dickstein DL, Hof PR, Wearne SL. Automated three-dimensional detection and shape classification of dendritic spines from fluorescence microscopy images. PloS one. 2008;3:e1997. doi: 10.1371/journal.pone.0001997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shankar GM, Welzel AT, McDonald JM, Selkoe DJ, Walsh DM. Isolation of low-n amyloid beta-protein oligomers from cultured cells, CSF, and brain. Methods in molecular biology. 2011;670:33–44. doi: 10.1007/978-1-60761-744-0_3. [DOI] [PubMed] [Google Scholar]
- 51.Trudgian DC, Mirzaei H. Cloud CPFP: a shotgun proteomics data analysis pipeline using cloud and high performance computing. J Proteome Res. 2012;11:6282–6290. doi: 10.1021/pr300694b. [DOI] [PubMed] [Google Scholar]
- 52.Trudgian DC, et al. CPFP: a central proteomics facilities pipeline. Bioinformatics. 2010;26:1131–1132. doi: 10.1093/bioinformatics/btq081. [DOI] [PubMed] [Google Scholar]
- 53.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 54.Geer LY, et al. Open mass spectrometry search algorithm. Journal of proteome research. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 55.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nature methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Pin1 loss causes spine loss.
Fig. S2. Pin1 successfully recombined in mouse hippocampus after AAV-GFP-CRE.
Fig. S3. Pin1 reconstitution restores mature spine counts in presence of Aβ42.
Fig. S4. FK506 restoration of spines requires Pin1.
Fig. S5. Pin1-S111D restores spine counts in wild-type or KO cells treated with Aβ42.