Abstract
Mobile elements have significantly impacted genome structure of most organisms. The continued activity of the mobile element, LINE-1 (L1), through time has contributed to the accumulation of over half a million L1 copies in the human genome. Most copies in the human genome belong to evolutionary older extinct L1s. Here we apply our previous published approach to “revive” the extinct L1 PA13A; an L1 family that was active about 60 million year ago (mya). The reconstructed L1PA13A is retrocompentent in culture, but shows a significantly lower level of activity in HeLa cells when compared to the modern L1 element (L1PA1) and a 40 million year old L1PA8. L1 elements code for two proteins (ORF1p and ORF2p) that are necessary for retrotransposition. Using PA13A-PA1 and PA13A-PA8 L1 chimeric elements, we determined that both the ORF1p and ORF2p contribute to the observed decrease in retrotransposition efficiency of L1PA13A. The lower retrotransposition rate of L1PA13A is consistent in both human and rodent cell lines. However, in rodent cells, the chimeric element L1PA:1–13 containing the modern L1PA1 ORF1p shows a recovery in the retrotransposition rate, suggestive that the L1PA13A ORF2p efficiently drives retrotransposition in these cells. The functionality of the L1PA13A ORF2p was further confirmed by demonstrating its ability to drive Alu retrotransposition in rodent cells. The variation in L1PA13A retrotransposition rates observed between rodent and human cells are suggestive that cellular environment significantly affects retrotransposition efficiency, which may be mediated through an interaction with ORF1p. Based on these observations, we speculate that the observed differences between cell lines may reflect an evolutionary adaptation of the L1 element to its host cell.
Keywords: LINE-1, L1, L1 families, ORF1 protein, ORF2 protein, retrotransposition
1. Introduction
The LINE-1 (L1) element belongs to Class I elements also known as retroelements or retrotransposons. L1s mobilize using an RNA intermediate to generate new genomic copies in a process termed retrotransposition (Boeke et al, 1985). During this process, the L1 RNA uses a target primed reverse transcription (TPRT) step to generate the new copy at the site of insertion (Luan et al, 1993). L1 is the only currently active autonomous element present in the human genome (Belancio et al, 2009). The human L1 is about 6 kb long and contains two open reading frames encoding the proteins ORF1p and ORF2p. Both proteins are required for L1 retrotransposition (Moran et al, 1996). ORF1p is thought to function as a nucleic acid chaperone (Martin and Bushman 2001); while ORF2p provides both the endonuclease and reverse transcriptase activities needed to drive insertions (Feng et al, 1996; Singer et al, 1993). Both ORF1p and ORF2p interact with the L1 RNA to form an RNP complex (Kolosha and Martin, 1997; Doucet et al., 2010). The L1 proteins also drive other non-autonomous retroelements such as Alu (Dewannieux et al, 2003). Although, Alu, parasitizes the L1 machinery for its own retrotransposition, Alu only strictly requires ORF2p for its own mobilization (Dewannieux et al, 2003; Wallace et al, 2008).
The general structure of L1 elements is relatively conserved throughout mammalian L1 evolutionary history. However, the ORF1p (particularly the N-terminus) shows poor sequence conservation between L1 elements from different species [reviewed in (Martin 2006)]. Furthermore, evolutionary studies have consistently shown that ORF1p has undergone significant adaptive evolution (Khan et al, 2006). In conjunction, all these observations point to a potential role of ORF1p that requires an interaction with cellular components for proper function.
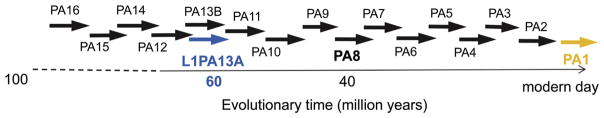
The presence of L1 retroelements in mammalian genomes can be traced back over 100 million years (Burton et al, 1986; Furano 2000; Lander et al, 2001). In the human genome, a few dominant L1 elements amplified over discreet time periods (Figure 1) leading to distinct human L1 lineages or L1 families (Smit 1996). The ongoing activity of these elements led to the accumulation of over half a million L1 copies in the human genome contributing to ~17% of its mass (Lander et al, 2001). However, most of the L1 copies are either truncated elements or fossils of previously active (i.e. extinct) L1 elements. Determination of the amount of sequence divergence of these ‘fossil’ sequences allows for the estimation of the activity of the different dominant L1 families during particular evolutionary periods of the human genome (Khan et al, 2006). Current analyses indicate that only ~80–100 of the L1s (all belonging to the modern L1 family L1Hs a.k.a. L1PA1) are full-length and currently retrotranspositionally competent (Brouha et al, 2003). Whole genome sequence evaluation of the older L1 families (>30 million years old) demonstrate that the majority of the copies are highly deteriorated making it unlikely to finding a potentially active full length copy in the human genome.
Figure 1. Representative schematic of the evolution of the L1PA lineage.
During most of the primate evolution of the PA lineage, the L1 families (represented as arrows) have evolved as a single lineage where one dominant family is active until replaced by the younger subfamily. The youngest and currently active family is L1PA1 (yellow) and the higher the numbers the older the L1 family. The L1PA13A family (blue) is estimated to have been active ~60 million years ago.
Our previously published work demonstrates that, with some careful scrutiny, the use of L1 family consensus sequences are a practical sequence source for the reconstruction of extinct elements (Wagstaff et al, 2012). In this manuscript, we present the data of the reconstruction and the functional evaluation of a 60 million year old L1 element, L1PA13A.
2. Materials and Methods
2.1 Constructs
pBS-L1PA1CHmneo (Addgene # 51288) (Wagstaff et al, 2011), pBS-L1PA8CHmneo (Addgene # 69608)(Wagstaff et al, 2012) vectors contain the codon optimized ORF1 and ORF2 of the consensus sequence of each family tagged with the mneoI cassette have been previously described (Figure 2A). All bicistronic L1 constructs were built using pBS-L1PA1CHmneo or pBS-L1PA8CHmneo as base by substituting the ORF1 and ORF2 coding sequences with the corresponding synthesized L1 PA13A sequences using available restriction sites, see Figure 3A (Wagstaff et al, 2011). All new constructs were sequence verified. The pAluYa5-neoTET, contains a the tagged AluYa5 (Kroutter et al, 2009)(Addgene # 51283). pBudORF2CH expressed the ORF2p from the L1RP, an L1 PA1 element (Wagstaff et al, 2011) pBudORF2PA13A –myc was generated by cloning the L1PA13A ORF2 sequence into the HindIII-BamHI sites of the pBudCE4.1 vector in a manner that removes the stop codon of the ORF2 so the expressed protein will contain the myc-his tag at the carboxy terminus.
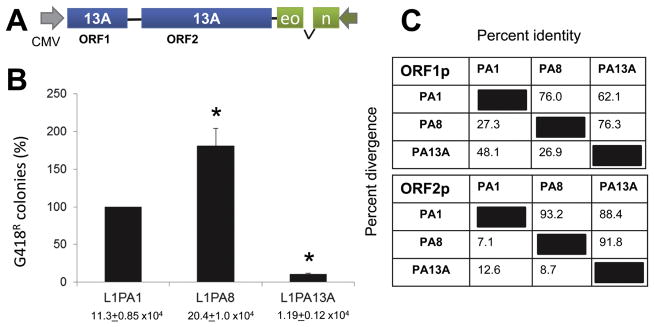
Figure 2.
A. The L1PA13A construct contains the codon optimized ORF1 and ORF2 sequences (blue) separated the L1RP inter-ORF sequence and driven by a CMV promoter. The 3′ end contains a neomycin indicator cassette (green) designed to monitor retrotransposition in culture. The indicator cassette contain an inverted neomycin resistance gene disrupted by an intron will splice only from a transcript generated by the CMV; where only retrotransposed L1 sequences derived from the spliced RNA will confer G418 resistance. B. The relative retrotransposition efficiencies of the tagged L1PA1, L1PA8 and L1PA13A L1 elements in HeLa cells are shown. Columns represent the G418R colony means normalized relative to the L1PA1 (100%) with the standard deviation shown as error bars. Asterisks indicate significantly different results Student’s paired t-test relative to L1PA1 (P=0.001; n ≥ 6). Retrotransposition frequencies are indicated below each column. C. The amino acid sequences of the ORF1p and ORF2p from L1PA1, L1PA8 and L1PA13A families were analyzed using DNASTAR MegAlign Version 10 by ClustalW (Weighted) method to determine percent identity and divergence.
Figure 3. Both ORF1p and ORF2p contribute to the observed low retrotransposition rate of L1PA13A.
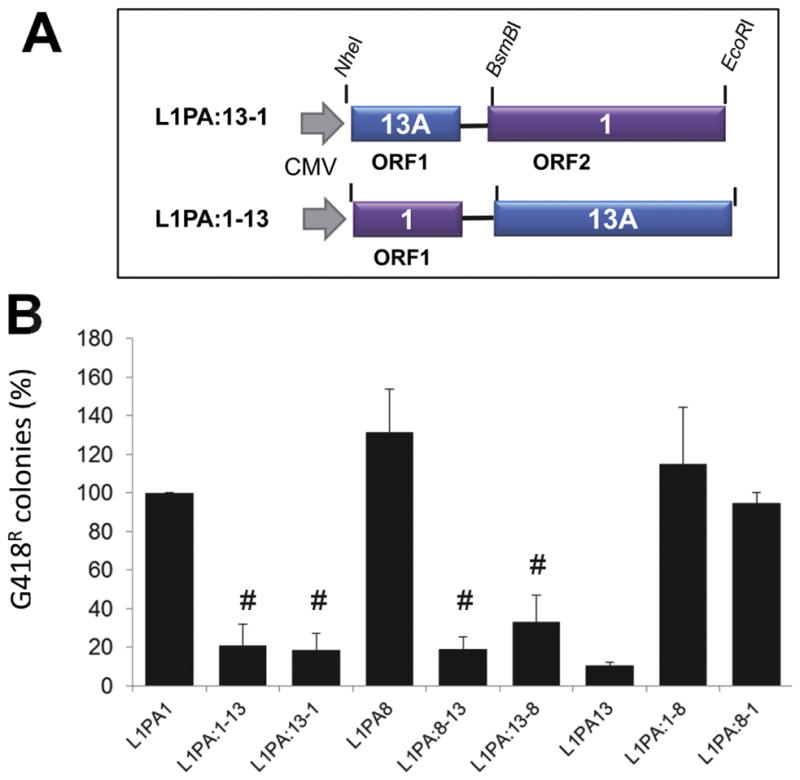
A. Representative schematic of the chimeric L1s with the unique restriction sites used in the building of the constructs is shown. B. The relative retrotransposition efficiencies of the tagged chimeric L1 elements in HeLa cells are shown. Columns represent the G418R colony means normalized relative to the L1PA1 (100%) with the standard deviation shown as error bars. # indicates results that are NOT significantly different from L1PA13 Student’s paired t-test (P>0.1; n ≥ 4).
2.2 Reconstruction of the L1PA13A
The codon optimized NheI- BsmBI flanked L1 PA13A ORF1 plus the inter ORF region and the BsmBI-EcoRI flanked ORF2 consensus sequences were synthesized by GenScript. Codon optimization of the sequences was performed using Primo Optimum 3.4 (http://www.changbioscience.com/primo/primoo.html). The sequences were cloned into the L1PA1CHmneo base to create pBS-L1PA13ACHmneo (Addgene # 69613).
2.3 Cell Culture
The following cell lines were used: HeLa (ATCC CCL2), Baby Hamster Kidney: BHK (ATCC CCL10), and Chinese Hamster Ovary UV20: CHOUV20 (ATCC CRL1862). HeLa were grown in MEM supplemented with 10% FBS, non-essential amino acids and sodium pyruvate and the rodent cells in DMEM supplemented with 10% fetal bovine serum (FBS) (ThermoFisher Scientific). A total of 1 × 105 cells were seeded in T25 flasks. The following day transient transfection of 0.3 μg of the neo tagged L1 plasmids was performed using the Lipofectamine Plus (ThermoFisher Scientific) transfection protocol following the manufacturer’s recommended protocol. Transient Alu retrotransposition assays were performed as previously described (Ade and Roy-Engel 2016; Kroutter et al, 2009). Transfected cells were grown under selection media for 14 days and stained for at least 30 minutes with a crystal violet staining solution (0.2% crystal violet in 5% acetic acid and 2.5% isopropanol).
2.4 Identity, divergence and phylogenetic tree analyses
Multiple alignments were performed using performed using Lasergene version 10 Core suite, MegAlign (DNASTAR, Madison, WI, USA). Sequence distances and phylogenetic trees were generated after alignment of the protein sequences using Clustal W in MegAlign.
3. Results
3.1 Reconstruction of the L1PA13A
Reconstruction of the L1PA13A followed our previously published approached used for the reconstruction of L1PA4 and L1 PA8 (Wagstaff et al, 2012). The available ORF1 and ORF2 sequences of the L1PA13A consensus (Khan et al, 2006) were used to perform a BLAT query of the human genome (UCSC Genome Browser, hg19 Assembly: http://genome.ucsc.edu/cgi-bin/hgBlat). The top 35 ORF1 and top 16 ORF2 sequences labeled as “L1PA13” by BLAT were retrieved (Supplemental Data) and further scrutinized to eliminate highly deteriorated sequences. A total of 27 ORF1 and 14 ORF2 sequences were used to generate the sequence alignment and consensus following our previously used methodology (Wagstaff et al, 2012). Based on the alignment and concordance with other human L1 PA family consensus sequences and primate sequences, we modified two potential ambiguous amino acids in the ORF1 sequence and 13 in the ORF2 sequence. A list of the modified amino acids is provided in Table 1. Based on the careful evaluation of the alignments and evolutionary comparisons, we performed our experimental analyses under the presumption that the modified consensus sequences represent the best estimation of the L1PA13A proteins active 60 million years ago. The codon optimized sequences of the modified consensus of the ORF1 and ORF2 L1PA13A were synthesized and cloned into our base L1 retrotransposition vector (Figure 2A) used to evaluate activity.
Table 1.
Changes to the L1PA13A consensus sequence.
| Position | Consensus AA | Modified AA | Support for choice of modification |
|---|---|---|---|
| ORF1 56 | M | L | Most common residue + conserved in younger L1PAs |
| ORF1 173 | S | T | Most common residue + conserved in younger L1PAs |
|
| |||
| ORF2 152 | V | I | Most common residue + polymorphic site |
| ORF2 210 | H | S | Most common residue + highly conserved in primates |
| ORF2 213 | I | L | Most common residue + highly conserved in primates |
| ORF2 225 | T | N | Most common residue + highly conserved in primates |
| ORF2 314 | F | I | Most common residue + highly conserved in primates |
| ORF2 353 | Q | R | Most common residue + polymorphic site |
| ORF2 361 | K | E | Most common residue + highly conserved in primates |
| ORF2 904 | S | N | Most common residue + conserved in older L1PAs (11–16) |
| ORF2 920 | A | P | Most common residue + highly conserved in primates |
| ORF2 1090 | K | N | Most common residue |
| ORF2 1168 | L | I | Most common residue + highly conserved in primates |
| ORF2 1189 | H | Y | Most common residue + highly conserved in primates |
| ORF2 1217 | V | I | Most common residue + most common in primates |
3.2 L1PA13A shows low retrotransposition efficiency
The reconstructed full-length L1PA13A element proved to be retrocompetent in HeLa cells (Fig. 2B). As previously observed, L1PA8 is highly efficient showing higher retrotransposition rates than the modern L1PA1 (Wagstaff et al, 2012). In contrast, the older L1PA13A shows a significantly lower retrotransposition rate of only 10% of L1PA1 activity.
Sequence comparisons of the amino acid sequences of the ORF1p and the ORF2p show that L1PA8 shares 76.0% and 93.2% identity to the L1PA1 proteins and 76.3% and 91.8% identity to the L1 PA13A proteins, respectively (Figure 2C). This is suggestive that the number of evolutionary changes in these proteins that occurred during the time period from L1PA13A to L1PA8 and the changes from L1PA8 to L1PA1 are similar. However, the changes that occurred during the evolutionary time from L1PA13A to L1PA8 seem to have significantly impacted the retrotransposition efficiency of L1PA13A in HeLa cells in culture.
3.3 Both the ORF1p and ORF2p contribute to the decrease in retrotransposition efficiency of L1PA13A in HeLa cells
L1 requires both ORF1p and ORF2p for retrotransposition (Moran et al, 1996). To determine if the reconstructed ORF1, ORF2 or both sequences are the main determinants of the reduced retrotransposition rate of L1PA13A, we created chimeric L1 elements where the two ORFs correspond to different families (Figure 3A). If the ORF1p or the ORF2p of the L1PA13A is the main limiting factor, it will decrease retrotransposition efficiency of the chimeric L1. The retrotransposition results demonstrate that all the chimeric constructs that contain an LPA13A ORF sequence show significantly lower retrotransposition rates in HeLa cells (Figure 3B).
Our previous data showed that mouse human L1 chimeras were retrocompetent. However, when either the human ORF1 or the ORF2 sequences were swapped for the mouse orthologous sequence we observed about a 40% reduction in retrotransposition efficiency (Wagstaff et al, 2011). This observation is suggestive that chimeric elements may show reduced retrotransposition rates due to a potential reduction of compatibility between ORF1p and ORF2p. However, the retrotransposition data show the L1PA8-L1PA1 chimeric elements (L1PA:1–8 and L1PA8:1) retain retrotransposition rates comparable to their parental constructs (Figure 3B). This is suggestive that chimeric elements from the same lineage (i.e. human) may be less likely to be incompatible. Thus, the lower efficiency in the mouse-human chimeras may reflect cross species incompatibilities, while the reduced activity of the L1A13A chimeras could be an indication that the HeLa cells may not be as supportive of older elements. Overall, our data indicate that both the L1PA13A ORF1p and ORF2p contribute to the reduced activity observed in HeLa cells.
3.4 Cellular environment influences in retrotransposition efficiency of the L1PA13A proteins
Host evolution has been proposed to have directly influenced the L1 evolution through negative selective forces (Boissinot and Furano 2001). Because “modern” human cells may be less supportive of the ancient L1PA13A, we proceeded to test the different constructs in cell lines from other species. Evaluation of the retrotransposition efficiency of the L1 constructs in rodent cells showed that the chimeric L1 with the L1PA1 ORF1 and the L1PA13A ORF2 (L1PA:1–13) construct showed retrotransposition rates comparable to the L1PA1 and higher than the parental L1PA13A (Figure 4). These data support the hypothesis that the proteins of the different L1 families have evolved to adapt to their cellular environment.
Figure 4. L1PA13A retrotransposition is consistently low in rodent cell lines.
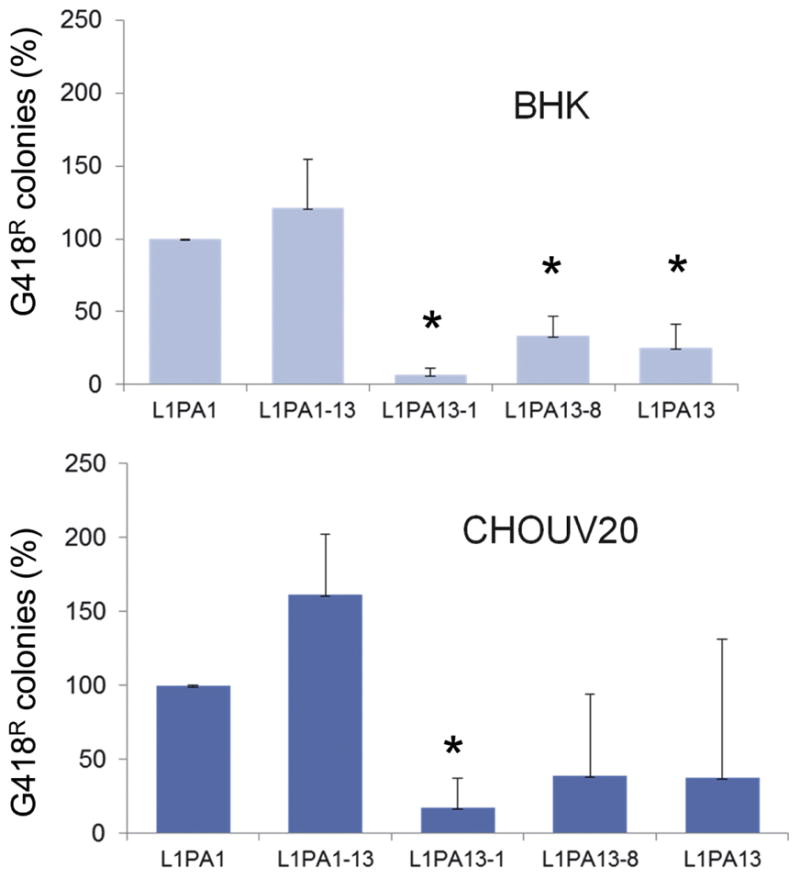
The relative retrotransposition efficiencies of the tagged chimeric L1 elements in rodent cells (Baby Hamster Kidney (BHK) and Chinese Hamster Ovary UV20 (CHOUV20) are shown. Columns represent the G418R colony means normalized relative to the L1PA1 (100%) with the standard deviation shown as error bars. Asterisks indicate significantly different results Student’s paired t-test relative to L1PA1 (P=0.01; n ≥ 4).
3.5 The ORF1 protein is the dominant protein contributing to the observed lower retrotransposition rate of L1PA13A
Previous analyses propose that the rapid evolution of the ORF1 sequence could reflect an adaptation of the L1 to its cellular host (Boissinot et al, 2004; Khan et al, 2006). Our data in rodent cells indicate that the L1PA13A ORF2p is likely fully functional with comparable retrotransposition efficiency to L1PA1. To confirm this observation, we evaluated the ability of the L1PA13A ORF2p to drive Alu retrotransposition. Because Alu elements only strictly require the ORF2p to drive their retrotransposition (Wallace et al, 2008), this provides the ideal assay to evaluate the L1PA13A ORF2p. Evaluation of the retrotransposition efficiency of Alu driven by the L1PA13A ORF2p expression construct in human and rodent cells is shown in Figure 5. Again, we observed that in HeLa cells, the Alu retrotransposition efficiency driven by the L1PA13A ORF2p is about 10% of what is observed for the Alu driven by L1PA1 ORF2p. In contrast, the Alu retrotransposition rates in rodent cells are comparable. These data indicate that the cellular environment affects the L1PA13A ORF2p efficient activity. Furthermore, the lower retrotransposition rates observed for the L1PA13A in rodents may be mediate through the ORF1p.
Figure 5. The ORF2p L1PA13A supports Alu retrotransposition.
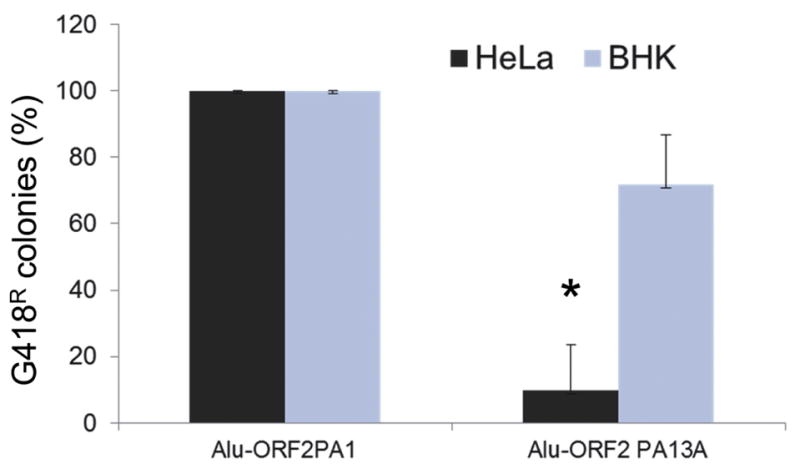
The relative retrotransposition efficiencies of the tagged Alu in human (HeLa) and rodent cells (BHK) are shown. Columns represent the G418R colony means normalized relative to the L1PA1 (100%) with the standard deviation shown as error bars. Asterisks indicate significantly different results Student’s paired t-test relative to L1PA1 (P=0.001; n = 4).
4. Discussion and Conclusions
Our data further confirms that the use of verified L1 consensus sequences of ancient elements is a valid approach to revive extinct elements. However, this approach does not ensure that a reconstructed ancient element will show high levels of retrotransposition activity when evaluated using the available cell assay systems. Although at this time we are unable to provide the exact reason for the observed low retrotransposition of the L1PA13A, we propose several possibilities.
First, unrecognized errors in the consensus sequence used for the reconstruction led to the creation of an imperfect representation of the LPA13A element. However, the observation that the chimeric L1PA1-L1PA13A (L1PA:1–13) and the L1PA13A ORF2p efficiently supported retrotransposition under a different cellular environment (i.e. rodent cells), suggests that at least the ORF2p is likely an acceptable representation of what was active 60 million years ago. Although the presence of the L1PA13A ORF1 sequence in all L1 constructs tested reduced retrotransposition efficiency, the larger number of sequences used in the alignment strongly support the consensus created. Thus, it is unlikely that the ORF1 consensus contains unrecognized errors.
Secondly, the actual L1PA13A may have had a lower retrotransposition rate. In the period that L1PA13A was active (between 55 and 65 mya), three other different L1PA families (L1PA12, L1PA13B, and L1PA14) amplified at about the same time (Khan et al, 2006). Furthermore, Khan et al note that the copy number from each of these L1 families is relative low when compared to the younger L1PA families, suggestive of limited L1 amplification and possibly reflective of lower retrotransposition rates.
Lastly, the use of a human cell line, such as HeLa, from a “modern” human does not appropriately reflect the cellular environment that existed 60 million years ago. The observation that the chimeric L1 with evolutionary different components showed differential effects on L1 retrotransposition efficiency in cell lines from different species suggests that the reduced retrotransposition efficiency of the L1PA13A may be due to a lack of adaptation to the cellular environment provided by the HeLa cells. In rodent cells, the ORF1p appears to be the main determining component determining retrotransposition efficiency. This is not surprising, as the ORF1 sequence has been shown to have undergone selective pressure that drove rapid evolution (Khan et al, 2006), which likely reflects the adaptation of the ORF1p to the host (cellular environment). Thus, it is possible that the L1PA13A ORF1p is less efficient as it lacks the “adaptive” changes that are present in the modern ORF1p (L1PA1).
Overall, our data shows that the use of reconstructed ancient elements provides new avenues to study the evolutionary adaptation of mobile elements to its cellular environment. Future studies on L1PA13A could provide a better understanding of what cellular components are missing that are need for efficient retrotransposition.
Supplementary Material
Supplemental List 1. BLAT matches to consensus ORF1 protein of L1PA13A that are “called” as L1PA13 by BLAT.
Supplemental List 2. BLAT matches to consensus ORF2 protein of L1PA13A that are “called” as L1PA13 by BLAT.
Highlights.
Successful reviving of an ancient extinct human retrotransposon.
Cellular environment differentially affects the retrotransposition efficiency possibly through an ORF1p interaction
Acknowledgments
The data generated in this publication was supported by Grants Number P20RR020152/P20 GM103518 (GM) and R01GM079709A to AMR-E and SCS supplement R01GM079709A to AMR-E from the National Institutes of Health (NIH). The contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH.
Abbreviations
- DSBs
Double Strand Breaks
- FBS
Fetal Bovine Serum
- LINE-1
Long Interspersed Element
- mya
million years ago
- neo
neomycin
- ORF
Open Reading Frame
- RT
reverse transcriptase
- UTR
untranslated region
Footnotes
Declaration of Interest
None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ade C, Roy-Engel AM. SINE Retrotransposition: Evaluation of Alu Activity and Recovery of De Novo Inserts. Methods Mol Biol. 2016;1400:183–201. doi: 10.1007/978-1-4939-3372-3_13. [DOI] [PubMed] [Google Scholar]
- Belancio VP, et al. LINE dancing in the human genome: transposable elements and disease. Genome Med. 2009;1:97. doi: 10.1186/gm97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J, et al. Ty elements transpose through an RNA intermediate. Cell. 1985;40:491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- Boissinot S, Furano AV. Adaptive evolution in LINE-1 retrotransposons. Mol Biol Evol. 2001;18:2186–2194. doi: 10.1093/oxfordjournals.molbev.a003765. [DOI] [PubMed] [Google Scholar]
- Boissinot S, et al. Different rates of LINE-1 (L1) retrotransposon amplification and evolution in New World monkeys. J Mol Evol. 2004;58:122–130. doi: 10.1007/s00239-003-2539-x. [DOI] [PubMed] [Google Scholar]
- Brouha B, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci U S A. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton FH, et al. Conservation throughout mammalia and extensive protein-encoding capacity of the highly repeated DNA long interspersed sequence one. J Mol Biol. 1986;187:291–304. doi: 10.1016/0022-2836(86)90235-4. [DOI] [PubMed] [Google Scholar]
- Dewannieux M, et al. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- Feng Q, et al. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Furano AV. The biological properties and evolutionary dynamics of mammalian LINE-1 retrotransposons. Prog Nucleic Acid Res Mol Biol. 2000;64:255–294. doi: 10.1016/s0079-6603(00)64007-2. [DOI] [PubMed] [Google Scholar]
- Khan H, et al. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 2006;16:78–87. doi: 10.1101/gr.4001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroutter EN, et al. The RNA Polymerase Dictates ORF1 Requirement and Timing of LINE and SINE Retrotransposition. PLoS Genet. 2009;5:e1000458. doi: 10.1371/journal.pgen.1000458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Luan DD, et al. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- Martin SL. The ORF1 Protein Encoded by LINE-1: Structure and Function During L1 Retrotransposition. J Biomed Biotechnol. 2006;2006:45621. doi: 10.1155/JBB/2006/45621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SL, Bushman FD. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol Cell Biol. 2001;21:467–475. doi: 10.1128/MCB.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran JV, et al. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- Singer MF, et al. LINE-1: a human transposable element. Gene. 1993;135:183–188. doi: 10.1016/0378-1119(93)90064-a. [DOI] [PubMed] [Google Scholar]
- Smit AF. The origin of interspersed repeats in the human genome. Curr Opin Genet Dev. 1996;6:743–748. doi: 10.1016/s0959-437x(96)80030-x. [DOI] [PubMed] [Google Scholar]
- Wagstaff BJ, et al. Evolutionary conservation of the functional modularity of primate and murine LINE-1 elements. PLoS One. 2011;6:e19672. doi: 10.1371/journal.pone.0019672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff BJ, et al. Molecular Reconstruction of extinct LINE-1 elements and their interaction with non-autonomous elements. Mol Biol Evol. 2012 doi: 10.1093/molbev/mss202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace N, et al. LINE-1 ORF1 protein enhances Alu SINE retrotransposition. Gene. 2008;419:1–6. doi: 10.1016/j.gene.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental List 1. BLAT matches to consensus ORF1 protein of L1PA13A that are “called” as L1PA13 by BLAT.
Supplemental List 2. BLAT matches to consensus ORF2 protein of L1PA13A that are “called” as L1PA13 by BLAT.