Abstract
The related NEAT1_1 and NEAT1_2 long noncoding RNAs (lnc RNAs) have been recently implicated in innate immunity against viral infection. We used CRISPR-Cas9 to generate Jurkat CD4+ T cell lines with a knockout (KO) of the NEAT1 gene. Viabilities of NEAT1 KO Jurkat lines were indistinguishable from parental Jurkat cells, as was the induction of CD69 after T cell activation. The KO lines were however more sensitive to the induction of apoptosis than parental Jurkat cells. HIV-1 replication was higher in the KO lines than parental Jurkat cells, demonstrating an anti-HIV function of NEAT1 lncRNAs. We observed a strong down-regulation of NEAT1 lncRNAs following activation of resting peripheral blood mononuclear cells and purified CD4+ T cells. These findings indicate that HIV-1 infection exploits the normal downregulation of anti-viral NEAT1 lncRNAs in activated CD4+ T cells to enhance viral replication.
Keywords: HIV, NEAT1 lncRNA, long non-coding RNA, viral replication
Introduction
It has become appreciated in recent years that long noncoding RNAs (lncRNAs) have important functions in innate immunity (Zhang & Cao, 2016). Two such lncRNAs are the related NEAT1_1 and NEAT1_2 (Nuclear-Enriched Abundant Transcripts) lncRNAs. Both NEAT1 lncRNAs are transcribed from the same promoter but differ at their 3’ ends due to differential transcriptional termination (Wilusz, 2015). NEAT1_1 is 3.7 kb in length and is polyadenylated, while NEAT1_2 is 23 kb in length and is non-polyadenylated. NEAT1 lncRNAs are commonly up-regulated following viral infection, including infections of Japanese encephalitis virus, rabies virus (Saha, Murthy et al., 2006), HSV, influenza virus (Imamura, Imamachi et al., 2014), hantavirus (Ma, Han et al., 2017), and HIV-1 (Zhang, Chen et al., 2013). SiRNA depletions of NEAT1 lncRNAs enhance hantavirus replication (Ma, Han et al., 2017), KSHV replication (Morchikh, Cribier et al., 2017) and HIV-1 gene expression and replication (Budhiraja, Liu et al., 2015;Zhang, Chen et al., 2013), suggesting broad antiviral activity of these lncRNAs.
NEAT1 lncRNAs are found in two distinct nuclear structures – paraspeckles and a recently described ribonucleoprotein (RNP) complex termed the HDP-RNP. Paraspeckles are formed at the sites of transcription of the NEAT1 gene where ~40 proteins use NEAT1 lncRNAs as scaffolds to assemble dynamic RNP structures with diameters of 0.5–1.0 μM (Fox, Nakagawa et al., 2017;Mao, Sunwoo et al., 2011). The HDP-RNP contains HEXIMI1, DNA-PK subunits (DNAPKc, Ku70, and Ku80) and additional proteins also found in paraspeckles. The assembly of NEAT1 lncRNAs in these RNPs is likely to underlie the antiviral activities of the lncRNAs.
A mechanism whereby paraspeckle-associated NEAT1 lncRNAs exert antiviral activity is through sequestration of a paraspeckle protein termed SFPQ. SFPQ is a transcriptional repressor of the IL8 gene and induction of NEAT1 lncRNAs following influenza virus infection increases the number and size of paraspeckles, leading to sequestration of SFPQ in these nuclear structures, away from the IL8 promoter and the resultant induction of IL8 transcription (Imamura, Imamachi et al., 2014). SFPQ has been proposed to be a repressor of two additional antiviral genes, RIG-I and DDX60. Hantavirus infection induces NEAT1 lncRNAs and therefore paraspeckles, and this is associated with induction of RIG-1 and DDX60 mRNAs and proteins, presumably through sequestration of SFPQ from the RIG-I and DDX60 promoters (Ma, Han et al., 2017). The NEAT1 lncRNA HDP-RNP complex is involved in the recognition of infections by DNA viruses through the cGAS-STING-IRF3 pathway (Morchikh, Cribier et al., 2017). The HDP-RNP associates with cGAS and its partner PQBP1. Upon recognition of viral DNA following KSHV infection, the HDP-RNP-cGAS-PQBP1 complex is remodeled, leading to activation of the cGAS-STING antiviral pathway.
The generation of a NEAT1 knockout (KO) mouse line indicates that NEAT1 lncRNAs are not essential for cell viability (Nakagawa, Naganuma et al., 2011). However, fertility of the KO mice is impaired due to a defect in formation of the corpus luteum (Nakagawa, Shimada et al., 2014) and mouse embryo fibroblasts (MEFs) from NEAT1 KO mice are more sensitive to induction of apoptosis than MEFs from wild type mice (Hirose, Virnicchi et al., 2014). The role of NEAT1 lncRNAs in immune system function, either adaptive or innate, has not been examined in the NEAT1 KO mice.
To investigate NEAT1 lncRNAs and HIV-1 infection, we generated NEAT1 KO Jurkat CD4+ T cell lines. Although the KO Jurkat cell lines were more sensitive than parental Jurkat cells to the induction of apoptosis by cycloheximide treatment, the KO lines possess growth rates and respond to T cell activation similar to parental cells. We found that HIV-1 replicates to higher levels in the KO lines than parental Jurkat cells, demonstrating an anti-HIV function in Jurkat CD4+ T cells. We also observed that NEAT1 lncRNA levels are strongly down-regulated following activation of peripheral blood lymphocytes (PBMCs) and resting CD4+ T cells isolated from blood donors. Productive HIV-1 infection in CD4+ T cells requires cellular activation (Zack, Arrigo et al., 1990). Our findings indicate that the down-regulation of the antiviral NEAT1 lncRNAs in activated CD4+ T cells contributes to high levels of HIV-1 replication in these cells.
Materials and Methods.
Cell lines.
Jurkat and its derivatives (J3C9, J3E5) were cultured in RPMI supplemented with 10% fetal bovine serum (FBS); 293T cells were cultured in DMEM supplemented with 10% FBS.
Primary cells.
Resting CD4 T cells were isolated from healthy donors (Gulf Coast Regional Blood Center, Houston, TX) using the RosetteSep human CD4+T cell enrichment cocktail (STEMCELL technologies, catalogue number (cat. no. 15062); activated cells were removed using CD30 Microbeads (Miltenyi Biotec, cat. no. 130–051-401). Cells were activated with phytohemagglutinin (PHA) (1 μg/mL) and cultured in RPMI supplemented with 10% fetal bovine serum. PBMCs from HIV-positive donors were obtained from a specimen bank established by Dr. Roberto Arduino (UT Houston Health Science Center). The cryopreserved PBMCs were thawed and half were left untreated, while the other half was treated with 1μg/ml PHA for 2 days. Purified CD4+ T cells and PBMCs were cultures in RPMI supplemented with 10% FBS and IL-2 (30 U/ml).
RNA isolation.
Cytoplasmic RNA was isolated from cultured cells by the PARIS™ Kit Protein and RNA isolation system (Thermo Fisher Scientific, cat. no. AM1921) Total RNA was isolated from cultured cells using miRNeasy Mini Kit (Qiagen, cat. no. 217004), and Real-time RT-qPCR was performed using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, cat. no. 4368814) and Powerup SYBR Green Master Mix (Applied Biosystem, cat. no. A25743) using primers for unspliced Gag and spliced viral RNAs, and pre-GAPDH and processed GAPDH RNAs as controls. Primers for unspliced viral RNA: Gag1-qPCR 5’ GGGACCCAGCCATAAAGC3’; Gag2-qPCR 5’ GCTGAATTTGTTACTTGGCTCA 3’; Primers for spliced viral RNA: Spliced-F 5’ GTCTCTCTGGTTAGACCAG 3’; Spliced-R 5’ TTGGGAGGTGGGTTGCTTTGATAGAG 3’; Primers for unspliced GAPDH: Pre-GAPDH-F 5’ CCACCAACTGCTTAGCACC 3’ Pre-GAPDH-R 5’CTCCCCACCTTGAAAGGAAAT 3’
HIV-1 NL4–3 virus production and infection.
pNL4–3 plasmid (AIDS Reagent Program, cat. no. 114) was transfected into 293T cell using lipofectamine 2000 (Thermo Fisher Scientific, cat. no.52758). Viruses were collected 48 hours post-transfection and 0.45um filter filtered viruses were transduced into Jurkat cells in the presence of polybrene.
Antibodies.
Goat-anti-HIV gp160B (cat. no.188), mouse-anti-HIV-1 gag (cat. no. 3539), and rabbit-anti-Rev (cat. no. 2949) were obtained from the NIH AIDS Reagent Program. Rabbit anti RBM14 (cat. no. ab70636) was from Abcam, rabbit-anti-PARP-1 (cat. no. 9542p) was from Cell Signaling, p54nrb (cat. no. 611278) was from BD Transduction Laboratories and β-actin (cat. no. 611278 A2103) from Sigma-Aldrich.
Immunoblots.
Cells were lysed in RIPA buffer (50mM Tris.Cl pH8, 150mM NaCl, 1%NP-40, 0.5%DOC, 0.1%SDS) plus 5mM DTT and protease inhibitor cocktail (Sigma-Aldrich cat. no. P8340) on ice for 30 minutes. The soluble portion of extracts was loaded on an SDS-PAGE followed by transfer to nitrocellulose membranes. After blocking with 5% BSA, membranes were probed with antibodies using standard protocols.
CRISPR-CAS9 knockouts.
A Multiplex CRISPR/Cas9 Assembly System kit (Addgene kit 10000000055) was used to make NEAT1 KO cell lines. Six guide RNAs (gRNAs) were designed with web-based software (two guide RNAs at the promoter region, two at the 5’ end and two at the 3’ end of NEAT1_2 sequences). Location of the gRNAs is shown in Figure 1. Six gRNA expression cassettes were inserted into pX330A-1×6. The resulting plasmid together with a GFP expression plasmid was transfected into Jurkat cells by electroporation at 1600V for 10 ms for three times. After three days, GFP-positive cells were sorted by flow cytometry and deposited into 96 wells at one cell per well. Two weeks later, cells in wells were collected, lysed with Solution for Blood (Sigma-Aldrich L3298) and neutralized by Neutralization Solution for Blood (Sigma-Aldrich, N9784). The resulting lysates were used directly for PCR reactions.
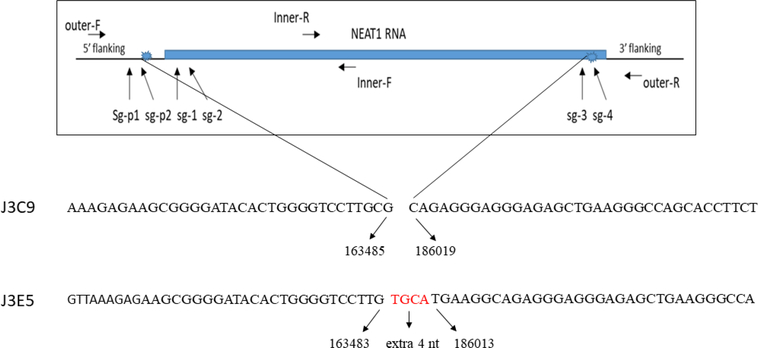
Figure 1. Strategy to delete NEAT1 gene.
Positions of guide RNAs used to delete the NEAT1 gene are indicated. Positions of PCR primers to verify deletion of NEAT1 gene in genomic DNA are indicated (primer sequences are indicated in Materials and Methods). The genomic sequence of deletions for the J3C9 and J3E5 lines are shown at the bottom of the figure; deletions in both NEAT1 alleles in both cell lines were identical. The sequence reference numbers are from Homo sapiens genomic DNA, chromosome 11:cloneRP11–86708.
Primers
Sequence of oligonucleotides used to construct guide RNAs for plasmid construction are listed; nucleotides in smaller font were used as annealing sequences in plasmid constructions and are not NEAT1 sequences.
sgRNA-P1: caccgCAAGCGACCCCGGCGGCTCC; sgRNA-P1c: aaacGGAGCCGCCGGGGTCGCTTGc;
sgRNA-p2: caccgATACACTGGGGTCCTTGCGT; sgRNA-p2c: aaacACGCAAGGACCCCAGTGTATc;
sgRNA-1: caccgCTGAGCGAGCCCGGGTGCGC; sgRNA-1c: aaacGCGCACCCGGGCTCGCTCAGc;
sgRNA-2: caccgTATGTAATTTTCGCTCGGCC; sgRNA-2c: aaacGGCCGAGCGAAAATTACATAc;
sgRNA-3: caccgACGGACCCGCCGTGGGCCCA; sgRNA-3c: aaacTGGGCCCACGGCGGGTCCGTc;
sgRNA-4 : caccGCTTGTGCCTCTGCTCGTGA; sgRNA-4c aaacTCACGAGCAGAGGCACAAGC
PCR Primers to verify the NEAT1 KO in genome:
NEAT1-inner-F 5’ TCAGGACACAACATGGCTCTC 3’; NEAT1-inner-R 5’ CTGGGCAAGGGCTGAATCTT 3’;
NEAT1 outer F 5’ TGTCCCTCGGCTATGTCAGA3’; NEAT-outer-R 5’ CCATGGGCTGCACTCAGTAA 3’
Primers to quantify NEAT1 RNA levels: NEAT-1F 5’ GGGCCATCAGCTTTGAATAA 3’; NEAT-1R 5’ CTTGAAGCAAGGTTCCAAGC 3’; NEAT1–2F 5’ CAGTTAGTTTATCAGTTCTCCCATCCA 3’; NEAT1–2R 5’ GTTGTTGTCGTCACCTTTCAACTCT 3’
Fluorescent in situ hybridization (FISH).
Stellaris RNA Fish probes were purchased from Biosearch Technologies. Cells grown in 24-well culture dishes with coverslips were fixed and processed for FISH exactly as described in the Stellaris FISH Protocols (Biosearch Technologies). Cells were analyzed using the DeltaVision (Deconvolution) Image Restoration Microscope in the Baylor College of Medicine Integrated Microscopy Core Laboratory.
Flow Cytometry.
Parental Jurkats or J3C9 and J3E5 KO cells were cultured for 24 hours with DMSO, PMA/IO, or CD3/CD28 mabs. Cells were then washed and stained for viability (Zombie Violet dye) and CD69 (APC) expression (Biolegend). Data was acquired with a Gallios Flow Cytometer and analyzed with Kaluza software (Beckman-Coulter).
Results
Validation of NEAT1 lncRNA knockout Jurkat cell lines.
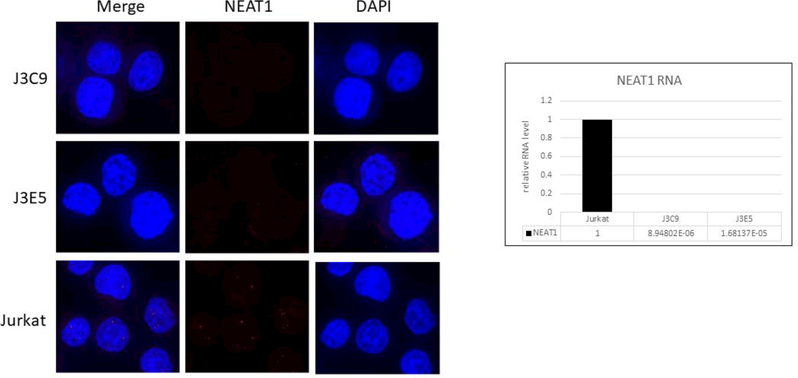
We used CRISPR-Cas9 methodology to generate clonal Jurkat cell lines with deletions of the NEAT1 gene (see Fig. 1). Two NEAT1 KO lines, J3C9 and J3E5, were utilized in the present study. Fluorescence in situ hybridization (FISH) and Real-Time qRT PCR were used to verify the absence of NEAT1 lncRNAs in the KO lines. In FISH analysis, multiple discrete nuclear puncta were visible with the NEAT1 lncRNA probe in parental Jurkat cells, whereas no such puncta were visible in the KO lines (Fig 2A). In agreement with the FISH data, NEAT1 RNA sequences were essentially below the limit of detection in qRT-PCR assays in either KO cell line (Fig. 2A). These data demonstrate that the J3C9 and J3E5 NEAT1 KO lines do not express NEAT1 lncRNAs.
Figure 2. Jurkat NEAT1 KO cell lines.
A. Fluorescent in situ hybridization was used to examine NEAT1 lncRNAs in NEAT1 KO lines J3C9 and J3E5 and parental Jurkat cells. B. Real-Time RT-PCR assays were used to quantify relative expression of NEAT1 lncRNAs in NEAT1 KO lines J3C9 and J3E5. PCR primers for NEAT1 lncRNAs were in shared sequences between NEAT1_1 and NEAT1_2 and detect both RNAs. The fold-change values for real-time RT-PCR assays are shown on the bottom of the bar graphs. The Ct values were approximately 16 for β−2-microglobin and 36 or great for NEAT1 KO RNA samples.
NEAT1 KO cell lines are not impaired for T cell activation but are sensitive to induction of apoptosis.
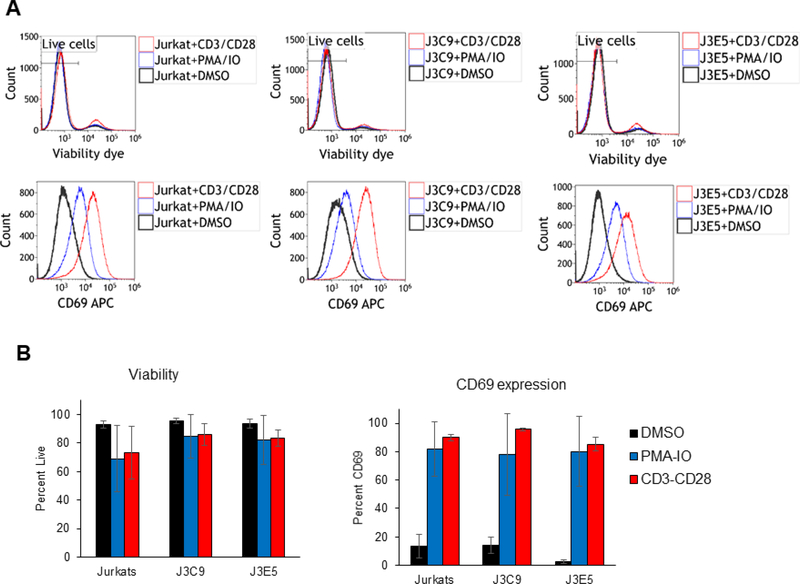
We used flow cytometry to examine whether the NEAT1 KO cell lines may have altered viability or defects in a T cell activation response. Exponentially growing cultures were stained with a viability dye and flow cytometry analysis demonstrated that the J3C9 and J3E5 cell lines have similar viabilities as parental Jurkat cells (Fig. 3A). Cell cultures were treated with anti-CD3/CD28 beads, PMA + ionomycin, or control DMSO and 24 hours later induction of the CD69 T cell activation marker was examined. The induction of CD69 was indistinguishable between the KO cell lines and parental Jurkat cells (Fig. 3A). The bar graphs in Figure 3B summarize cell viability and CD69 induction data from three independent experiments (Fig. 3B).These data indicate that loss of NEAT1 lncRNAs has no effect on Jurkat cell viability and NEAT1 lncRNAs are not required for a T cell activation response.
Figure 3. Flow cytometry analysis of Jurkat NEAT1 KO cell lines.
A. NEAT1 KO lines J3C9, J3E5, and parental Jurkat cells were culture for 24 hours in the presence of DMSO, PMA/ionomycin, or anti-CD3/CD28 beads. Flow cytometry was used to evaluate CD69 expression and viability of cells (stained with Zombie Violet dye); a representative experiment is shown. B. The bar graphs summarize the data from three independent experiments.
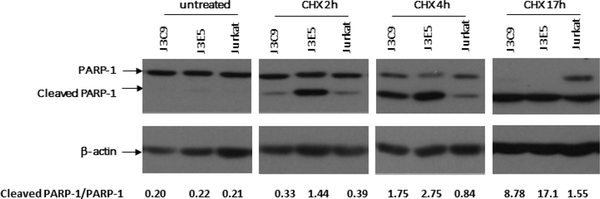
Mouse embryo fibroblasts from NEAT1 KO mice are more sensitive to inducers of apoptosis than wild type mice (Hirose, Virnicchi et al., 2014). To determine if the Jurkat NEAT1 KO lines are sensitive to induction of apoptosis, cultures were treated with cycloheximide and cell extracts were prepared at two, four, and 17 hours post-treatment. Apoptosis was monitored by cleavage of PARP-1 in an immunoblot (Fig. 4). Both the J3C9 and J3E5 lines demonstrated enhanced PARP-1 cleavage relative to the parental Jurkat cells. These data agree with previous studies of mouse KO cells and indicate that loss of NEAT1 lncRNAs can sensitize cells to apoptosis.
Figure 4. NEAT1 KO cell lines are sensitive to induction of apoptosis.
NEAT1 KO lines J3C9, J3E5, and parental Jurkat cells were treated with cycloheximide (50μg/ml) for the indicated time before cell extracts were prepared. PARP-1 and β-actin were analyzed in an immunoblot. Quantitation of PARP-1 cleavage (ratio of cleaved/uncleaved) is shown at the bottom of the figure.
HIV-1 replication is enhanced in NEAT1 KO cell lines.
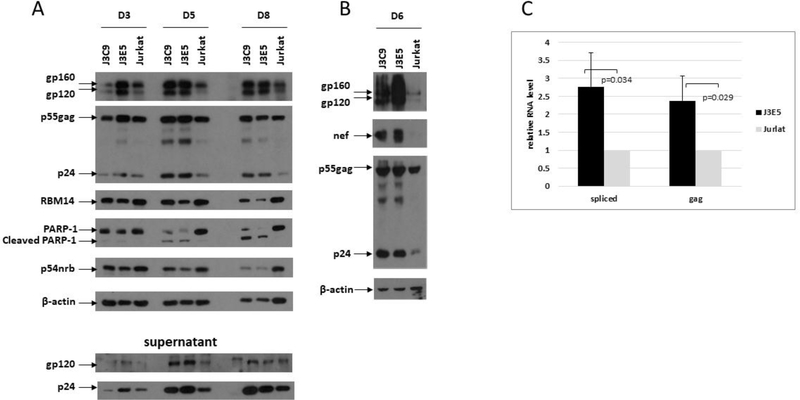
To examine HIV-1 replication in NEAT1 KO lines, cultures were infected with HIV NL4–3 and an immunoblot was used to evaluate infections at day three, five, and eight post-infection (Fig. 5A). PARP-1 cleavage was notably higher in infections in the KO lines, in agreement with induction of apoptosis by cycloheximide shown in Figure 3. Despite higher levels of apoptosis in the KO cell lines, HIV-1 replication was enhanced in both the J3C9 and J3E5 cell lines relative to parental Jurkats. Envelope expression (gp160/120) and Gag expression (p55 and p24) was higher in cell extracts in the KO lines, as was as the levels of p24 in culture supernatants. The data shown in Figure 5A is representative of more than three biological repeat experiments. In an additional experiment, Nef, Envelope (gp160/120), and Gag expression (p55 and p24) were examined at day six of infection with HIV NL4–3 (Fig. 5B). Similar to the data in Figure 5A, Nef and the other viral proteins were expressed at higher levels in the KO cell lines relative to parental Jurkat cells.
Figure 5. HIV-1 replicates to higher levels in NEAT1 KO cell lines.
A.NEAT1 KO cell lines J3C9, J3E5, and parental Jurkat cells were infected with HIV-1 NL4–3. Cell extracts were prepared and culture supernatants were collected at the indicated days post-infection. The indicated proteins were examined in an immunoblot. B. NEAT1 KO cell lines J3C9, J3E5, and parental Jurkat cells were infected with HIV-1 NL4–3. Cell extracts were prepared at day six post-infection. The indicated proteins were examined in an immunoblot. C. Cytoplasmic RNA was isolated from NL4–3-infected J3C9, J3E5, and parental Jurkat cells at day 5 post-infection; spliced and unspliced Gag viral transcripts were quantified by Real-time RT-PCR. Data from four independent experiments are shown and indicated p-values were determined with a Student’s t-test.
Cytoplasmic RNA was extracted from NL4–3 infected cells at day five post-infection and spliced and unspliced viral RNA was quantified by Real-Time qPCR (Fig. 5C). There was a >2-fold increase in level of both spliced and unspliced viral RNA in the KO lines, consistent with the immunoblot data. The data in Figure 5 indicate that NEAT1 lncRNAs possess an anti-viral function for HIV-1.
NEAT1 lncRNAs are down-regulated in activated PBMCs and CD4+ T cells.
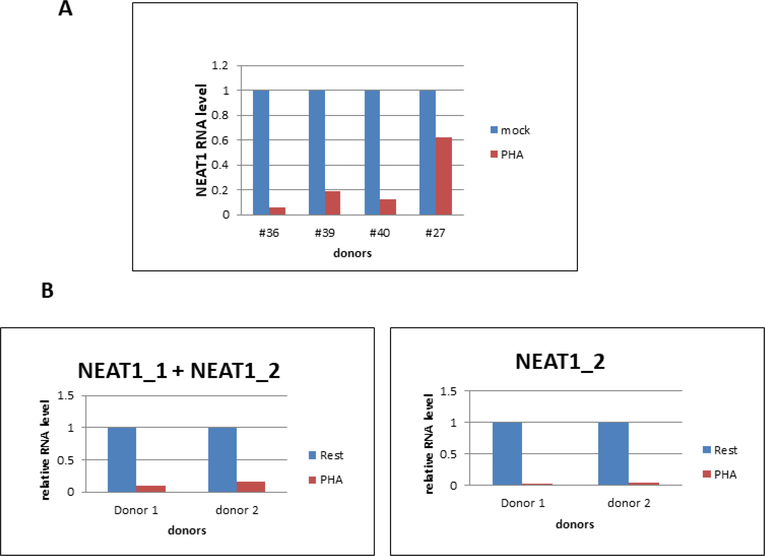
We also evaluated NEAT1 lncRNA expression in primary cells relevant to HIV-1 replication. We obtained PBMCs from a tissue bank of HIV-infected donors and incubated cells with and without PHA stimulation. RNA was extracted at 48 hours post-treatment, and levels of NEAT1 lncRNAs were quantified by Real-Time RT-PCR (Fig. 6A). The PCR primers used in this assay targeted sequences in common with NEAT1_1 and NEAT1_2 lncRNAs. PHA stimulation reduced NEAT1 lncRNA expression in PBMCs from all four donors, ranging from a ~40% reduction in donor 27 to > 90% reduction in donor 36.
Figure 6. NEAT1 lncRNAs are down-regulated following T cell activation.
A. PBMCs from four HIV-infected individuals (obtained from a PBMC bank established by Dr. Roberto Arduino, UT Houston Health Science Center) were cultured with and without PHA for 48 hours, RNA was extracted and NEAT1 lncRNA levels quantified with Real-Time RT-qPCR. PCR primers were in shared sequences between NEAT1_1 and NEAT1_2. B. Resting CD4+ T cells isolated from two healthy blood donors were cultured with and without PHA for 48 hours, RNA was extracted and NEAT1_1 and NEAT1_2 lncRNA levels quantified with Real-Time qPCR. In the left panel, PCR primers were in shared sequences between NEAT1_1 and NEAT1_2; in the right panel, PCR primers were in sequences unique to NEAT1_2.
We purified resting CD4+ T cells from two healthy donors and incubated with and without PHA for 48 hours. Real-Time RT-PCR assays for sequences in common to both NEAT1_1 and NEAT1_2 lncRNAs demonstrated an 80–90% reduction in the lncRNAs in both donors following T cell activation (Fig. 6B). We also used PCR primers unique to the longer NEAT1_2 transcript and observed a similar >90% reduction specifically for NEAT1_2 in both donors. These data in PBMCs and CD4+ T cells indicate that T cell activation results in strong down-regulation of NEAT1 lncRNA levels.
Discussion
The analysis of NEAT1 KO Jurkat cell lines in this study has shown that NEAT 1 lncRNAs possess an anti-HIV activity. This confirms previous siRNA experiments which reported that siRNA depletions of NEAT1 lncRNAs enhance HIV-1 gene expression and replication (Zhang, Chen et al., 2013;Budhiraja, Liu et al., 2015). Our observation that NEAT1 lncRNAs are strongly down-regulated when resting CD4+ T cells are activated is notable, as it has long known that HIV-1 replication in CD4+ T cells requires cellular activation (Zack, Arrigo et al., 1990). It appears that in activated CD4+ T cells, HIV-1 exploits the natural reduction of these anti-viral lncRNAs to increase replication. Given the increased sensitivity of NEAT1 KO Jurkat cell lines to apoptosis (Fig. 4), it is notable that induction of apoptosis in the promonocytic cell line U1 or CD4+ T cell line ACH-2 can result in reactivation of latent HIV-1 (Khan, Hand et al., 2015). It is therefore possible that the reduction of NEAT1 lncRNAs following activation of resting CD4+ T cells can increase the sensitivity of HIV-infected cells to apoptosis and this may enhance viral replication under some conditions.
The functional consequences in activated CD4+ T cells of the down-regulation of NEAT1 lncRNAs are unclear. Paraspeckle formation is dependent upon NEAT1 lncRNAs and these nuclear structures function in the nuclear retention of cellular mRNAs that contain inverted Alu repeats in their 3’ untranslated sequences (Prasanth, Prasanth et al., 2005;Chen & Carmichael, 2009). As there are approximately 300 cellular transcripts with inverted Alu repeats in their 3’ UTR (Yulug, Yulug et al., 1995), the immense increase in transcription following activation of resting CD4+ T cells is likely to include many mRNAs with inverted Alu repeats. It is conceivable that down-regulation of NEAT1 lncRNAs in activated CD4+ T cells and the subsequent reduction in paraspeckles may allow this class of mRNA to be rapidly exported from the nucleus and contribute to the phenotype of activated cells.
Our previous study of RBM14, a paraspeckle-associated protein, led us to propose that paraspeckles may retain unspliced or incompletely spliced HIV-1 transcripts in the nucleus where they are either degraded or exported to the cytoplasm by the viral Rev protein (Budhiraja, Liu et al., 2015). In the present study, we observed a large increase in the levels of unspliced viral RNA in the NEAT1 KO lines relative to parental Jurkat cells (Fig. 4B). In the absence of NEAT1 lncRNAs and paraspeckles, unspliced HIV-1 transcripts may both escape degradation in the nucleus and more efficiently exit the nucleus, and this may increase viral replication levels.
Finally, a block to nuclear export of HIV-1 RNA contributes to latent infection (Lassen, Ramyar et al., 2006). It is therefore possible that NEAT1 lncRNAs may be a feasible target for HIV-1 cure strategies that involve reactivation of latent virus. The NEAT1 KO mouse shows that NEAT1 lncRNAs are not essential, and this study indicates that loss of NEAT1 lncRNAs does not affect CD4+ T cell viability. Methods such as siRNA that reduce the level of NEAT1 lncRNAs in cells harboring latent infection may enhance nuclear export of viral RNAs and increase expression of viral antigens, allowing the immune system to recognize and clear these cells.
Acknowledgements
This work was supported by National Institutes of Health grant AI24866 to APR. We thank Dr. Roberto Arduino for the patient PBMC samples.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Budhiraja S, Liu H, Couturier J, Malovannaya A, Qin J, Lewis DE, Rice AP, 2015. Mining the human complexome database identifies RBM14 as an XPO1-associated protein involved in HIV-1 Rev function. J Virol 89, 3557–3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, Carmichael GG, 2009. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol.Cell 35, 467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Nakagawa S, Hirose T, Bond CS, 2017. Paraspeckles: Where Long Noncoding RNA Meets Phase Separation. Trends in Biochemical Sciences 43, 124–135. [DOI] [PubMed] [Google Scholar]
- Hirose T, Virnicchi G, Tanigawa A, Naganuma T, Li R, Kimura H, Yokoi T, Nakagawa S, Benard M, Fox AH, Pierron G, 2014. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol.Biol Cell 25, 169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K, Kato A, Kawaguchi Y, Sato H, Yoneda M, Kai C, Yada T, Suzuki Y, Yamada T, Ozawa T, Kaneki K, Inoue T, Kobayashi M, Kodama T, Wada Y, Sekimizu K, Akimitsu N, 2014. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol.Cell 53, 393–406. [DOI] [PubMed] [Google Scholar]
- Khan SZ, Hand N, Zeichner SL, 2015. Apoptosis-induced activation of HIV-1 in latently infected cell lines. Retrovirology. 12, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassen KG, Ramyar KX, Bailey JR, Zhou Y, Siliciano RF, 2006. Nuclear retention of multiply spliced HIV-1 RNA in resting CD4+ T cells. PLoS Pathog. 2, e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Han P, Ye W, Chen H, Zheng X, Cheng L, Zhang L, Yu L, Wu X, Xu Z, Lei Y, Zhang F, 2017. The Long Noncoding RNA NEAT1 Exerts Antihantaviral Effects by Acting as Positive Feedback for RIG-I Signaling. Journal of Virology 91, e2250–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YS, Sunwoo H, Zhang B, Spector DL, 2011. Direct visualization of the cotranscriptional assembly of a nuclear body by noncoding RNAs. Nat.Cell Biol 13, 95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morchikh M, Cribier A, Raffel R, Amraoui S, Cau J, Severac D, Dubois E, Schwartz O, Bennasser Y, Benkirane M, 2017. HEXIM1 and NEAT1 Long Non-coding RNA Form a Multi-subunit Complex that Regulates DNA-Mediated Innate Immune Response. Mol.Cell 67, 387–399. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Naganuma T, Shioi G, Hirose T, 2011. Paraspeckles are subpopulationspecific nuclear bodies that are not essential in mice. J Cell Biol 193, 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Shimada M, Yanaka K, Mito M, Arai T, Takahashi E, Fujita Y, Fujimori T, Standaert L, Marine JC, Hirose T, 2014. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development. 141, 4618–4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL, 2005. Regulating gene expression through RNA nuclear retention. Cell 123, 249–263. [DOI] [PubMed] [Google Scholar]
- Saha S, Murthy S, Rangarajan PN, 2006. Identification and characterization of a virusinducible non-coding RNA in mouse brain. J Gen.Virol. 87, 1991–1995. [DOI] [PubMed] [Google Scholar]
- Wilusz JE, 2015. Long noncoding RNAs: Re-writing dogmas of RNA processing and stability. Biochim.Biophys.Acta. 1859, 128–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yulug IG, Yulug A, Fisher EM, 1995. The frequency and position of Alu repeats in cDNAs, as determined by database searching. Genomics 27, 544–548. [DOI] [PubMed] [Google Scholar]
- Zack JA, Arrigo SJ, Weitsman SR, Go AS, Haislip A, Chen IS, 1990. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell 61, 213–222. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Chen CY, Yedavalli VS, Jeang KT, 2013. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. MBio 4, e00596–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cao X, 2016. Long noncoding RNAs in innate immunity. Cell Mol.Immunol. 13, 138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]