Abstract
The majority of cancer-related deaths are caused by tumor recurrence, metastasis and therapeutic resistance. During the late stages of tumor progression, multiple factors are involved, including the downregulation and/or loss of function of metastasis suppressors. Epithelial protein lost in neoplasm (EPLIN), an actin-binding protein, was initially identified as a putative tumor suppressor that is frequently downregulated in epithelial tumors. Recent evidence indicates that EPLIN may negatively regulate epithelia-to-mesenchymal transition (EMT), a crucial process by which cancer cells acquire invasive capabilities and therapeutic resistance. Importantly, downregulation of EPLIN is associated with clinical metastasis in a variety of solid tumors, suggesting that EPLIN could be a suppressor of metastasis. In this review, I will discuss the regulation and function of EPLIN in human cancer cells and explore the clinical significance of EPLIN in metastatic disease.
Keywords: Actin cytoskeleton, Chemoresistance, Epithelial-to-mesenchymal transition, EPLIN, Metastasis suppressor, Tumor suppressor
Tumor progression is a complicated, multi-step process driven by a series of genetic and epigenetic alterations. At early stages of tumorigenesis, the inactivation and/or loss of growth-retarding tumor suppressor genes (such as PTEN and p53) promote uncontrolled cell division and transformation into cancer cells. During metastasis, malignant tumors invade other organs and spread to distant sites, usually accompanied by the inactivation and/or loss of metastasis suppressor genes (such as E-cadherin), which specifically suppress metastasis without affecting primary tumor growth.1, 2, 3, 4, 5, 6 Identification of the distinct roles of these suppressor genes can provide a better understanding of tumor progression and help the development of precise therapy to prevent and treat metastasis, the major cause of cancer-related deaths. Recent studies have demonstrated epithelial protein lost in neoplasm (EPLIN) as an important safeguard against tumor progression in multiple cancer types. Here, I will review the current literature on the regulation and function of EPLIN in human cancer cells.
Sequence and structure of EPLIN
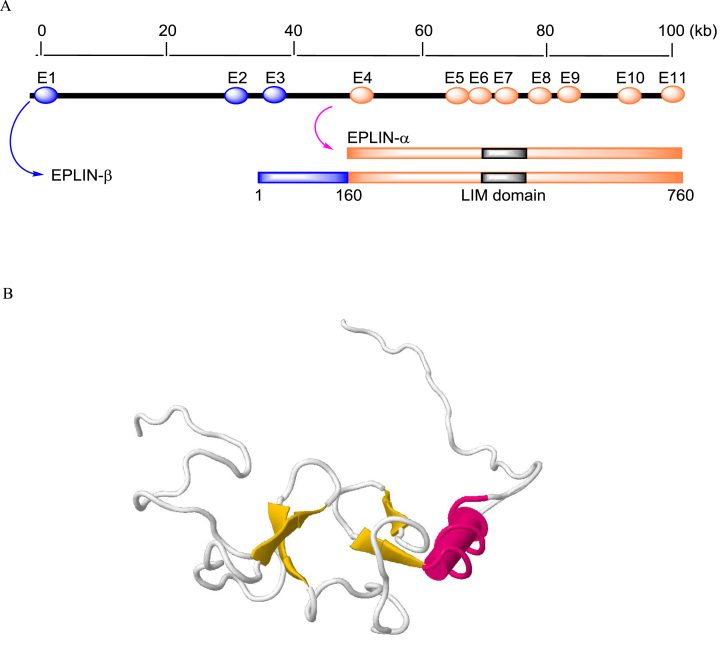
EPLIN, or LIM domain and actin binding 1 (LIMA1), was first described by Maul et al in 1999.7 The human EPLIN gene is located on chromosome 12q13.12 and encompasses 107.79 kb of DNA. The longest transcript isoform contains 11 exons. Two isoforms of EPLIN, i.e., EPLIN-α and EPLIN-β, can be generated from an alternative RNA processing event; the mRNA of the β isoform requires all 11 exons, whereas the EPLIN-α mRNA only requires exons 4–11. The resulting EPLIN-α protein has 600 amino acid (aa) residues and the reading frame of EPLIN-β is extended by an additional 160 aa7, 8 (Fig. 1A). Both isoforms contain a single centrally located LIM domain that can form two closely spaced zinc-binding subdomains9 and allow EPLIN to dimerize with itself or associate with other proteins (Fig. 1B). EPLIN also contains at least two actin-binding domains at both the amino- and carboxyl-termini, although the carboxyl-terminal domain appears to be more efficient in filament binding.10, 11
Fig. 1.
(A) The genomic and protein structures of human EPLIN isoforms. (B) Three-dimensional structure of LIM domain of human EPLIN (PDB ID = 2D8Y), generated with FirstGlance in Jmol.
Regulation of EPLIN expression
EPLIN is highly expressed in placenta, kidney, pancreas, prostate, ovary, spleen and heart (UniProtKB/Swiss-Prot). In human cancer cells, the two EPLIN isoforms appear to be differentially expressed in a highly context-dependent manner.7 For example, EPLIN-α is prevalently presented in human breast cancer cells and head and neck cancer cells, whereas EPLIN-β is the major isoform in some human prostate cancer cells. EPLIN-α and -β seem to be equally expressed by several melanoma cells.12, 13, 14, 15
The regulatory mechanism for the differential expression of EPLIN-α and -β remains largely elusive. The promoter region of human EPLIN-α includes a serum response factor (SRF) binding site, whereas the EPLIN-β promoter contains putative binding sites for Oct-1, Sp1, and AP1. Serum stimulation or transient expression of several Rho-family small GTPases both activate SRF and induced the transcription of EPLIN-α but not EPLIN-β in NIH3T3 fibroblasts, suggesting that EPLIN-α is the primary response gene in these cells.8 Similarly, a consensus SRF binding site was identified only in the murine EPLIN-α promoter, which is responsible for the induction of EPLIN-α in response to the binding of SRF and its coactivator MAL/myocardin-related transcription factor (MRTF).16
EPLIN can be regulated at post-translational levels. In prostate cancer cells, we have demonstrated that epidermal growth factor (EGF) could activate the phosphorylation, ubiquitination, and degradation of human EPLIN proteins through an extracellular signal-regulated kinase 1/2 (ERK1/2)-dependent signaling cascade. Two serine residues (serine 362 and serine 604) were identified as putative ERK1/2 phosphorylation sites in human EPLIN proteins, and point mutations at these residues (serine to alanine) rendered resistance to EGF-induced protein turnover. Interestingly, a putative PEST sequence (RASSLSESSPPK) with a PEST score of +5.89 (PEST scores greater than +5 are considered significant) was identified in human EPLIN protein, which also could contribute to the post-translational regulation of EPLIN.17
Expression of EPLIN in human cancers
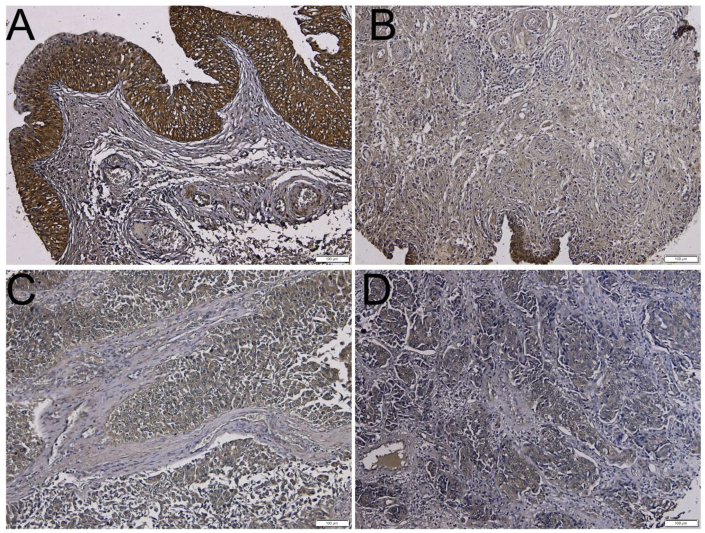
In several established human cancer cell lines, EPLIN was found to be reduced in a majority of oral (8/8), prostate (7/7) and breast (5/6) cancer cells. Downregulation of EPLIN-α, compared to EPLIN-β, appears to be more significant.7 However, the expression profile of EPLIN in human cancer tissues was not examined until a decade later.14 Using an antibody specific to EPLIN-α, Jiang et al performed immunohistochemical (IHC) staining on cancerous and normal mammary tissues. Compared to normal tissues, breast cancer tissues express a lower level of EPLIN-α, and grade 2/3 tumors have significantly decreased EPLIN-α compared with grade 1. Importantly, reduced levels of EPLIN-α are associated with poor prognosis and increased incidence of recurrence and mortality. In human esophageal cancers, EPLIN-α transcripts were expressed at lower levels in tumor tissues compared to normal tissue. EPLIN-α is reduced in grade 3–5 tumors when compared to grade 2 tumors. Lower level of EPLIN-α is also associated with local advanced esophageal cancer, including TNM stages 2–4, as well as lymphatic metastasis. Significantly, patients who died of esophageal cancer have lower levels of EPLIN-α compared to those who remained disease-free.15 In epithelial ovarian cancer, a similar pattern of EPLIN-α reduction was also observed, suggesting that EPLIN-α is a potential prognostic marker.18 We examined the IHC expression of EPLIN in a human bladder tissue microarray (TMA) and found that EPLIN expression is reduced in cancerous tissues when compared with that in normal tissues (Fig. 2).
Fig. 2.
IHC expression of EPLIN in human bladder cancer vs. normal tissues in a bladder TMA (obtained from Creative-Bioarray, New York). A & B: normal bladder tissues; C: grade 1, stage II, T2N0M0; D: grade 3, stage II, T2aN0M0.
Expression of EPLIN in metastatic tumors
We searched Gene Expression Omnibus and Oncomine databases for the expression profile of EPLIN in a number of epithelial cancers.12 Analyses on four independent sets of microarray data on clinical prostate cancer19, 20, 21, 22 revealed that EPLIN transcripts were expressed at a similar level in primary tumors and normal prostatic tissues, but were remarkably reduced in metastatic tumors. We further collected matched pairs of prostate cancer tissue specimens from primary tumors and lymph node metastases and examined the IHC staining of EPLIN. Intriguingly, the expression of EPLIN proteins was significantly reduced in lymph node metastases when compared with that in primary tumors. Similar patterns were also observed in several other epithelial cancers, in which the EPLIN immunointensity was markedly reduced in the lymph node metastases compared with matched primary tumors from the breast, colon, rectum, and head and neck.22 Steder et al reported that low EPLIN expression in melanoma was correlated with increased Breslow depth (>4 mm) of invasion versus high EPLIN levels at <1 mm. There was a statistically significant loss of EPLIN expression in melanoma metastases compared to the primary tumors. Consistent with our findings, an inverse association between EPLIN transcripts and the metastatic potential or tumor grade of prostate, colon, and head and neck cancer was observed.13
Cellular functions of EPLIN
EPLIN is an actin-binding protein involved in the regulation of actin dynamics
Biochemical studies showed that EPLIN regulates actin cytoskeleton and cellular architecture via several parallel mechanisms.10, 11 First, the two actin-binding domains allow EPLIN-α to efficiently cross-link and bundle actin filaments. Second, by binding to more than one actin subunit, EPLIN may lower the monomer dissociation rate constant at the pointed end of actin filaments and stabilize them from depolymerization. Third, EPLIN-α inhibits actin nucleation by Arp2/3 complex by increasing the lag at the outset of polymerization. Thus, by influencing both assembly (nucleation) and disassembly (stability) of filamentous actin (F-actin), EPLIN inhibits actin turnover and limits the dynamic remodeling of the actin cytoskeleton.
EPLIN is indispensable for the maintenance of epithelial phenotypes
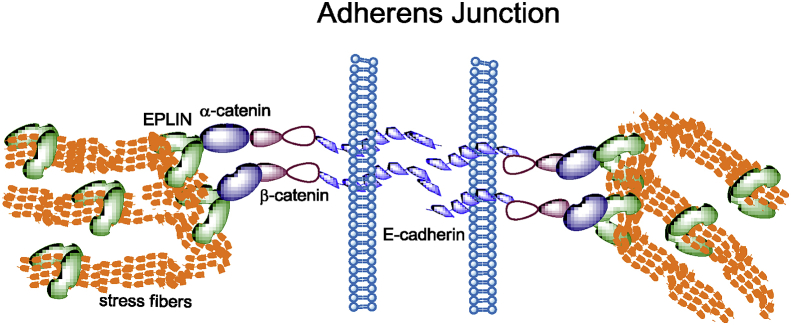
Peptide tandem mass spectrometry and co-immunoprecipitation analyses identified EPLIN as a binding partner of α-catenin. The physical interaction with α-catenin is required for the localization of EPLIN at cell–cell adjunctions and in the formation of a complex with E-cadherin cytoplasmic domain. Further, EPLIN depletion disrupts the apical organization of F-actin and the association between the cortical actin bundles and the cadherin–catenin complex, suggesting that EPLIN is indispensable for the stabilization of the circumferential actin belt at adherens junctions (AJs).23 Indeed, the formation of a cadherin-β-catenin-α-catenin-EPLIN-F-actin complex is essential to the maintenance of apical–basal polarity in epithelial cells (Fig. 3). At the AJs, EPLIN can respond to mechanical forces and is involved in the remodeling of the junctional architecture of epithelial cells.24
Fig. 3.
EPLIN links the cadherin-β-catenin-α-catenin complex to actin cytoskeletons, which is essential to the maintenance of apical–basal polarity in epithelial cells.
Endothelial AJs are required for the integrity of vascular endothelium and is also most important for transmitting mechanical signals directly to the actin cytoskeleton. VE-cadherin, an endothelial-specific member of the cadherin protein family, plays an indispensable role in the endothelial AJs.25 Similar to its function in epithelial cells, EPLIN directly interacts with α-catenin anchored to the VE-cadherin-β-catenin complex within the endothelial AJs. By connecting the VE-cadherin-β-catenin-α-catenin complex to the actin cortical ring and promoting vinculin junctional recruitment, EPLIN reinforces the cohesion of cell–cell junctions and stabilizes the capillary structures in endothelium.26, 27, 28 Therefore, the EPLIN-α-catenin interaction may provide a mechanosensory machinery that is essential to vascular morphogenesis and mechanotransduction.
EPLIN has an important role in the regulation of cytokinesis
Small-interfering RNA (siRNA) depletion of EPLIN from Hela and MCF-7 cells induces cytokinesis failure, suggesting that the loss of EPLIN may induce aneuploidy and contribute to genomic instability.29 The function of EPLIN in the regulation of membrane ingression and cleavage furrow formation seems to be dependent on its association with the actin-myosin II contractile ring and septin filaments, two cytoskeletal systems required for a complete cytokinesis. During the final stages of ingression, EPLIN is required for the recruitment of RhoA and Cdc42 at the cleavage furrow. Both EPLIN-α and -β localize to the cleavage furrow, suggesting the two isoforms may have similar effects on cytokinesis. In another study, Sundvold et al reported that EPLIN can recruit a putative lipid transporter ACAT-related protein required for viability 1 (Arv1) to the cleavage furrow at the onset of telophase, which subsequently recruits myosin heavy chain 9 (MYH9) and myosin light chain 9 (MYL9) by interacting with IQmotif-containing GTPase-activating protein (IQGAP1). Interestingly, EPLIN-β seems to have a stronger interaction with Arv1 when compared with the EPLIN-α isoform, suggesting the additional 160 aa at the N terminus may contribute to the physical association between the two proteins.30
EPLIN is a tumor suppressor in certain epithelial cancer cells
Ectopic expression of EPLIN-β or EPLIN-α suppresses the in vitro growth of U2-OS osteosarcoma cells, and overexpression of EPLIN-α demonstrated a similar inhibitory effect on growth and proliferation in MDA MB231 (breast cancer), PC-3 (prostate cancer) and KYSE150 (esophageal cancer) cells. The subcutaneous growth of MDA MB231 and PC-3 tumors in mice was also retarded by the forced expression of EPLIN-α.7, 14, 15, 31 Conversely, EPLIN-α depletion in ovarian cancer cells increased growth, invasion, adhesion and migration in vitro.18 These results, combined with the fact that EPLIN is frequently downregulated in certain human cancer cell lines and primary tumors, have led to the hypothesis that EPLIN may act as a tumor suppressor in certain epithelial cancers. Nonetheless, these studies are mostly observational, and the mechanism by which EPLIN controls the growth and proliferation of cancer cells remains largely unknown.
EPLIN is a suppressor of epithelial-to-mesenchymal transition (EMT)
Acquisition of migratory and invasive capabilities by cancer cells at the primary site resembles epithelial-to-mesenchymal transition (EMT), a process in which epithelial cells lose polarity and gain motility through downregulation of epithelial markers, disruption of the cadherin/catenin adhesion complex and re-expression of mesenchymal molecules.32 By conducting quantitative proteomics in lineage-derived human prostate cancer cell lines exhibiting classical EMT, we observed a significant reduction of EPLIN-β in highly invasive and mesenchymal-like cells. Western blotting analyses confirmed that both EPLIN-α and -β isoforms were abundantly expressed in epithelial-like and low-invasive ARCaPE cells, but markedly reduced in mesenchymal and highly invasive ARCaPM cells. A similar association between EPLIN expression and invasive phenotypes was observed in other experimental models of prostate cancer and squamous cell carcinoma of the head and neck. These data indicated that EPLIN remains to be highly expressed in certain epithelial cancer cells, and its downregulation is associated with increased invasiveness. EPLIN depletion using siRNAs and short hairpin RNA (shRNA) approaches promoted EMT and significantly increased the in vitro migratory and invasive capabilities in various cancer cell lines, including ARCaPE, LNCaP, PC-3 and MCF-7.12 Consistently, Steder et al observed that the invasive capabilities of melanoma cells were increased by shRNA knockdown of EPLIN-β or EPLIN-α/-β, or were inhibited by the overexpression of EPLIN-α or EPLIN-β.13
Previous studies have shown that overexpression of EPLIN in cancer cells usually reduced growth and proliferation, whereas EPLIN depletion resulted in increased proliferation.7, 14, 15, 31 On the contrary, however, we observed that EPLIN knockdown in ARCaPE cells induced cell cycle arrest and inhibited the in vitro proliferation and in vivo growth in athymic nude mice (12 and unpublished results). EPLIN depletion also significantly enhanced cell resistance to treatment with docetaxel and doxorubicin (by ∼8-fold and ∼4.4-fold, respectively).12 These results indicated that, at least in some prostate cancer cells, EPLIN has a more complicated function than simply being a tumor suppressor.
The acquired invasiveness upon EPLIN depletion was accompanied by the changes of EMT markers at molecular levels, including reduced E-cadherin and increased vimentin. In prostate cancer cells, EPLIN can affect multiple genes, including those involved in the regulation of EMT, Wnt/β-catenin signaling, actin cytoskeleton, invasion and metastasis, apoptosis, adhesion and extracellular matrix remodeling, and growth factor signaling.12 Among them, zinc finger E-box-binding homeobox 1 (ZEB1), versican, matrix metalloproteinase-7 (MMP-7), Bcl-2A, fibroblast growth factor 5 (FGF5), cAMP-responsive element-binding protein (CREB) and myeloid cell leukemia-1 (Mcl-1) were increased upon EPLIN depletion, and the inhibitor of differentiation 2 (ID2), myosin light chain kinase (MYLK) and insulin-like growth factor-binding protein-3 (IGFBP-3) were reduced. EPLIN depletion also resulted in a remarkable reduction of several microRNAs (miRNAs), including miR-205, miR-200b and miR-429, whose downregulation is thought to be the essential feature of EMT and acquisition of cancer stem cell properties. In melanoma cells, it appears that EPLIN can significantly affect a signaling axis consisting of insulin-like growth factor receptor (IGF-1R)-Akt-Stat3. Overexpression of EPLIN induced a substantial decrease in p-Akt and p-Stat3, which is associated with downregulation of Slug and upregulation of E-cadherin.13
Conclusion
EPLIN was discovered more than a decade ago, but our understanding of this important protein remains rudimentary. Early studies have been focused on the actin-binding properties of EPLIN and its role in the control of the actin cytoskeleton. EPLIN is indispensable for stabilization of the circumferential actin belt, therefore playing an essential role in the maintenance of apical–basal polarity in both epithelial and endothelial cells. The function of EPLIN during cytokinesis can be attributed to its association with the actin-myosin II contractile ring and septin filaments, thereby regulating membrane ingression and formation of the cleavage furrow. Given the inhibitory effect of EPLIN on the remodeling of the actin cytoskeleton, the reduction of EPLIN expression in epithelial cancer cells may result in increased migratory and invasive capabilities. Recent studies, including those from our laboratory, indicated a broader role of EPLIN in the control of cancer cell behaviors. In addition to its function as a structural protein, it appears that EPLIN is actively involved in cancer cell signaling, presumably by interacting with multiple proteins implicated in cancer progression (Table 1; Fig. 4). In prostate cancer and melanoma cells, it has become clear that EPLIN depletion promotes EMT and activates oncogenic pathways, which is associated with dramatic changes in morphology and cellular behaviors (such as chemoresistance). Significantly, EPLIN downregulation correlates with clinical metastasis in several solid tumors. These results suggest that EPLIN may have distinct functions in various stages of tumor progression: it may act as a tumor suppressor gene during tumorigenesis, but can further serve as a metastasis suppressor gene to prevent and retard the invasion and dissemination of primary cancer cells. Further exploration of the function of EPLIN as a regulator of metastasis and therapeutic resistance can lay foundations to design novel preventative and therapeutic strategies for metastatic disease.
Table 1.
EPLIN-interacting proteins and potential functions.
| Protein | Interaction | Function | Reference(s) |
|---|---|---|---|
| Actin | The two actin-binding domains of EPLIN cross-link and bundle actin filaments | EPLIN inhibits actin turnover and limits the dynamic remodeling of the actin cytoskeleton | 7, 10, 11 |
| α-catenin | The binding of EPLIN and α-catenin requires the N- and C-terminal regions of EPLIN and the vinculin homology (VH)3-C-terminal region of α-catenin | EPLIN links the cadherin–catenin complex to F-actin, stabilizing the adhesion belt | 23 |
| Septin 2 (Nedd5) | EPLIN is associated with Septin 2 during interphase and mitosis | EPLIN contributes to Septin 2 cleavage furrow localization during late stages of cytokinesis | 29 |
| Myosin IIb | EPLIN is associated with myosin IIb of the contractile ring | EPLIN enhances the accumulation of actin and active myosin II at the cleavage furrow during the final stages of ingression | 27, 29, 30, 33, 34, 35 |
| Supervillin | The C-terminal region of supervillin binds EPLIN | EPLIN and supervillin are concentrated at the cleavage furrow during the early stages of cytokinesis and may be required for cell division | 35 |
| Paxillin | EPLIN is localized in peripheral actin bundles at focal adhesions and formed a protein complex with paxillin | EPLIN may stabilize focal adhesions by interacting with paxillin | 31, 36 |
| ERK1/2 | ERK binds and phosphorylates mouse EPLIN at Ser360, Ser602, and Ser692 | EPLIN phosphorylation is required for PDGF-induced stress fiber disassembly, membrane ruffling and cell migration | 37 |
| PTEN | PTEN is associated with actin remodeling complex including EPLIN | It is unclear if EPLIN is required for the PTEN regulation of cell size checkpoint | 38 |
| PINCH-1 (Lims1) | PINCH-1 recruits EPLIN to the integrin adhesion sites in keratinocytes | Regulates the spreading and migration on collagen and fibronectin | 34 |
| Arv1 | EPLIN recruits Arv1 to the cleavage furrow in early telophase | Optimizes furrow ingression during cell division | 30 |
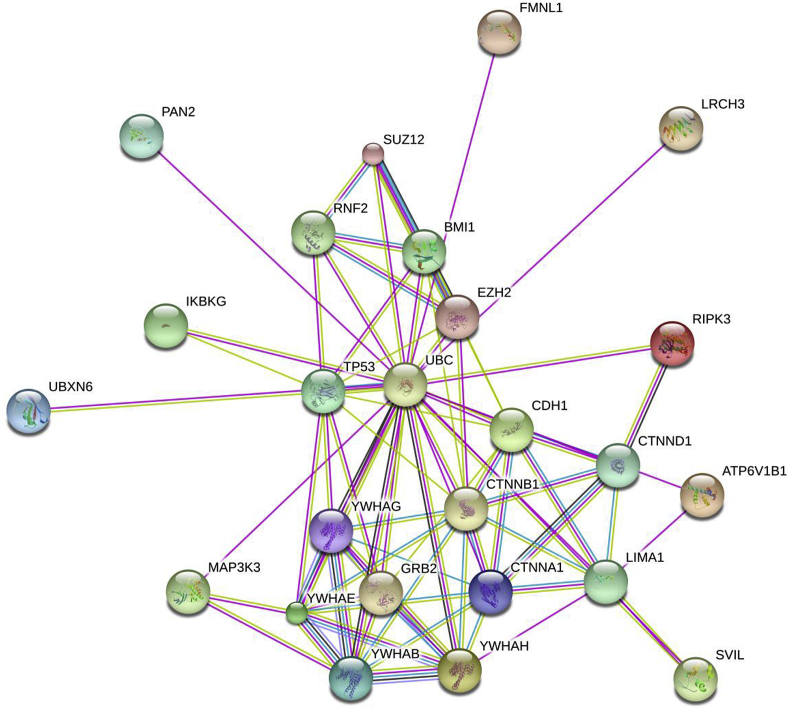
Fig. 4.
Potential EPLIN-interacting proteins, predicted using STRING program. Abbreviations: ATP6V1B1: ATPase, H+ transporting, lysosomal 56/58 kDa, V1 subunit B1; BMI1: BMI1 polycomb ring finger oncogene; CDH1: cadherin 1, type 1, E-cadherin (epithelial); CTNNA1: catenin (cadherin-associated protein), alpha 1; CTNNB1: catenin (cadherin-associated protein), beta 1; CTNND1: catenin (cadherin-associated protein), delta 1; EZH2: enhancer of zeste homolog 2; FMNL1: formin-like 1; GRB2: growth factor receptor-bound protein 2; IKBKG: inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma; LIMA1: LIM domain and actin binding 1, or EPLIN; LRCH3: leucine-rich repeats and calponin homology (CH) domain containing 3; MAP3K3: mitogen-activated protein kinase kinase kinase 3; PAN2: PAN2 poly(A) specific ribonuclease subunit homolog; RIPK3: receptor-interacting serine–threonine kinase 3; RNF2: ring finger protein 2; SUZ12: suppressor of zeste 12 homolog, or Polycomb group (PcG) protein; SVIL: supervillin; TP53: tumor protein p53; UBC: ubiquitin C; UBXN6: UBX domain protein 6; YWHAB: tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide; YWHAE: tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide; YWHAG: tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide; YWHAH: tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide.
Conflicts of interest
The author declares no conflict of interest.
Acknowledgements
This work was supported by National Cancer Institute grants 1R21CA164612-01A1, 1R41CA186498-01A1 and 1R41CA206725-01A1, Georgia Research Alliance Ventures grant, University of Georgia/Augusta University Cancer Research Initiative Award, and Georgia Cancer Center Startup Fund (D. Wu). We thank Dr. Rhea-Beth Markowitz for editorial assistance.
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Buchanan C., Lago Huvelle M.A., Peters M.G. Metastasis suppressors: basic and translational advances. Curr Pharm Biotechnol. 2011;12(11):1948–1960. doi: 10.2174/138920111798376914. [DOI] [PubMed] [Google Scholar]
- 2.Fatemian T., Chowdhury E.H. Targeting oncogenes and tumor suppressors genes to mitigate chemoresistance. Curr Cancer Drug Targets. 2014;14(7):599–609. doi: 10.2174/156800961407140926104458. [DOI] [PubMed] [Google Scholar]
- 3.Oren M. Tumor suppressors. Teaming up to restrain cancer. Nature. 1998;391(6664):233–234. doi: 10.1038/34551. [DOI] [PubMed] [Google Scholar]
- 4.Steeg P.S. Metastasis suppressors alter the signal transduction of cancer cells. Nat Rev Cancer. 2003;3(1):55–63. doi: 10.1038/nrc967. [DOI] [PubMed] [Google Scholar]
- 5.Thiolloy S., Rinker-Schaeffer C.W. Thinking outside the box: using metastasis suppressors as molecular tools. Semin Cancer Biol. 2011;21(2):89–98. doi: 10.1016/j.semcancer.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 6.Weinberg R.A. 2nd ed. Garland Science; New York, NY: 2014. The Biology of Cancer. [Google Scholar]
- 7.Maul R.S., Chang D.D. EPLIN, epithelial protein lost in neoplasm. Oncogene. 1999;18(54):7838–7841. doi: 10.1038/sj.onc.1203206. [DOI] [PubMed] [Google Scholar]
- 8.Chen S., Maul R.S., Kim H.R., Chang D.D. Characterization of the human EPLIN (Epithelial Protein Lost in Neoplasm) gene reveals distinct promoters for the two EPLIN isoforms. Gene. 2000;248(1–2):69–76. doi: 10.1016/s0378-1119(00)00144-x. [DOI] [PubMed] [Google Scholar]
- 9.Perez-Alvarado G.C., Miles C., Michelsen J.W. Structure of the carboxy-terminal LIM domain from the cysteine rich protein CRP. Nat Struct Biol. 1994;1(6):388–398. doi: 10.1038/nsb0694-388. [DOI] [PubMed] [Google Scholar]
- 10.Song Y., Maul R.S., Gerbin C.S., Chang D.D. Inhibition of anchorage-independent growth of transformed NIH3T3 cells by epithelial protein lost in neoplasm (EPLIN) requires localization of EPLIN to actin cytoskeleton. Mol Biol Cell. 2002;13(4):1408–1416. doi: 10.1091/mbc.01-08-0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maul R.S., Song Y., Amann K.J., Gerbin S.C., Pollard T.D., Chang D.D. EPLIN regulates actin dynamics by cross-linking and stabilizing filaments. J Cell Biol. 2003;160(3):399–407. doi: 10.1083/jcb.200212057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang S., Wang X., Osunkoya A.O. EPLIN downregulation promotes epithelial-mesenchymal transition in prostate cancer cells and correlates with clinical lymph node metastasis. Oncogene. 2011;30(50):4941–4952. doi: 10.1038/onc.2011.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steder M., Alla V., Meier C. DNp73 exerts function in metastasis initiation by disconnecting the inhibitory role of EPLIN on IGF1R-AKT/STAT3 signaling. Cancer Cell. 2013;24(4):512–527. doi: 10.1016/j.ccr.2013.08.023. [DOI] [PubMed] [Google Scholar]
- 14.Jiang W.G., Martin T.A., Lewis-Russell J.M., Douglas-Jones A., Ye L., Mansel R.E. Eplin-alpha expression in human breast cancer, the impact on cellular migration and clinical outcome. Mol Cancer. 2008;7(1):71. doi: 10.1186/1476-4598-7-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y., Sanders A.J., Zhang L., Jiang W.G. EPLIN-alpha expression in human oesophageal cancer and its impact on cellular aggressiveness and clinical outcome. Anticancer Res. 2012;32(4):1283–1289. [PubMed] [Google Scholar]
- 16.Leitner L., Shaposhnikov D., Descot A., Hoffmann R., Posern G. Epithelial Protein Lost in Neoplasm alpha (Eplin-alpha) is transcriptionally regulated by G-actin and MAL/MRTF coactivators. Mol Cancer. 2010;9:60. doi: 10.1186/1476-4598-9-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang S., Wang X., Iqbal S. Epidermal growth factor promotes protein degradation of epithelial protein lost in neoplasm (EPLIN), a putative metastasis suppressor, during epithelial-mesenchymal transition. J Biol Chem. 2013;288(3):1469–1479. doi: 10.1074/jbc.M112.438341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu R., Martin T.A., Jordan N.J., Ruge F., Ye L., Jiang W.G. Epithelial protein lost in neoplasm-alpha (EPLIN-alpha) is a potential prognostic marker for the progression of epithelial ovarian cancer. Int J Oncol. 2016;48(6):2488–2496. doi: 10.3892/ijo.2016.3462. [DOI] [PubMed] [Google Scholar]
- 19.Varambally S., Yu J., Laxman B. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8(5):393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 20.Chandran U.R., Ma C., Dhir R. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer. 2007;7:64. doi: 10.1186/1471-2407-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu Y.P., Landsittel D., Jing L. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22(14):2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 22.Lapointe J., Li C., Higgins J.P. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci USA. 2004;101(3):811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abe K., Takeichi M. EPLIN mediates linkage of the cadherin catenin complex to F-actin and stabilizes the circumferential actin belt. Proc Natl Acad Sci USA. 2008;105(1):13–19. doi: 10.1073/pnas.0710504105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taguchi K., Ishiuchi T., Takeichi M. Mechanosensitive EPLIN-dependent remodeling of adherens junctions regulates epithelial reshaping. J Cell Biol. 2011;194(4):643–656. doi: 10.1083/jcb.201104124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dorland Y.L., Huveneers S. Cell-cell junctional mechanotransduction in endothelial remodeling. Cell Mol Life Sci. 2017;74(2):279–292. doi: 10.1007/s00018-016-2325-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chervin-Petinot A., Courcon M., Almagro S. Eplin interacts with alpha-catenin and actin filaments in endothelial cells and in vitro stabilizes the vascular capillary network. J Biol Chem. 2012;287(10):7556–7572. doi: 10.1074/jbc.M111.328682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abu Taha A., Schnittler H.J. Dynamics between actin and the VE-cadherin/catenin complex: novel aspects of the ARP2/3 complex in regulation of endothelial junctions. Cell Adh Migr. 2014;8(2):125–135. doi: 10.4161/cam.28243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Citi S., Guerrera D., Spadaro D., Shah J. Epithelial junctions and Rho family GTPases: the zonular signalosome. Small GTPases. 2014;5(4):1–15. doi: 10.4161/21541248.2014.973760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chircop M., Oakes V., Graham M.E. The actin-binding and bundling protein, EPLIN, is required for cytokinesis. Cell Cycle. 2009;8(5):757–764. doi: 10.4161/cc.8.5.7878. [DOI] [PubMed] [Google Scholar]
- 30.Sundvold H., Sundvold-Gjerstad V., Malerod-Fjeld H., Haglund K., Stenmark H., Malerod L. Arv1 promotes cell division by recruiting IQGAP1 and myosin to the cleavage furrow. Cell Cycle. 2016;15(5):628–643. doi: 10.1080/15384101.2016.1146834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanders A.J., Martin T.A., Ye L., Mason M.D., Jiang W.G. EPLIN is a negative regulator of prostate cancer growth and invasion. J Urol. 2011;186(1):295–301. doi: 10.1016/j.juro.2011.03.038. [DOI] [PubMed] [Google Scholar]
- 32.Nieto M.A., Huang R.Y., Jackson R.A., Thiery J.P. Emt: 2016. Cell. 2016;166(2):21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 33.Gulino-Debrac D. Mechanotransduction at the basis of endothelial barrier function. Tissue Barriers. 2013;1(2):e24180. doi: 10.4161/tisb.24180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohoka A., Kajita M., Ikenouchi J. EPLIN is a crucial regulator for extrusion of RasV12-transformed cells. J Cell Sci. 2015;128(4):781–789. doi: 10.1242/jcs.163113. [DOI] [PubMed] [Google Scholar]
- 35.Smith T.C., Fang Z., Luna E.J. Novel interactors and a role for supervillin in early cytokinesis. Cytoskeleton (Hoboken) 2010;67(6):346–364. doi: 10.1002/cm.20449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanders A.J., Ye L., Mason M.D., Jiang W.G. The impact of EPLINalpha (Epithelial protein lost in neoplasm) on endothelial cells, angiogenesis and tumorigenesis. Angiogenesis. 2010;13(4):317–326. doi: 10.1007/s10456-010-9188-7. [DOI] [PubMed] [Google Scholar]
- 37.Han M.Y., Kosako H., Watanabe T., Hattori S. Extracellular signal-regulated kinase/mitogen-activated protein kinase regulates actin organization and cell motility by phosphorylating the actin cross-linking protein EPLIN. Mol Cell Biol. 2007;27(23):8190–8204. doi: 10.1128/MCB.00661-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J.S., Xu X., Li H. Mechanistic analysis of a DNA damage-induced, PTEN-dependent size checkpoint in human cells. Mol Cell Biol. 2011;31(13):2756–2771. doi: 10.1128/MCB.01323-10. [DOI] [PMC free article] [PubMed] [Google Scholar]