Abstract
Cancer is one of the most serious diseases that cause an enormous number of deaths all over the world. Tumor metabolism has great discrimination from that of normal tissues. Exploring the tumor metabolism may be one of the best ways to find biomarkers for cancer detection, diagnosis and to provide novel insights into internal physiological state where subtle changes may happen in metabolite concentrations. Nuclear Magnetic Resonance (NMR) technique nowadays is a popular tool to analyze cell extracts, tissues and biological fluids, etc, since it is a relatively fast and an accurate technique to supply abundant biochemical information at molecular levels for tumor research. In this review, approaches in tumor metabolism are discussed, including sample collection, data profiling and multivariate data analysis methods etc. Some typical applications of NMR are also summarized in tumor metabolism.
Keywords: High resolution Magic Angle Spinning, Metabolomics, Multivariate data analysis, NMR technique, Tumor metabolism
Introduction
Cancer nowadays is a major public health problem in the world. The occurrence of cancer is increasing because of the growth and aging of the population, as well as increasing formed risk factors such as dirty air, polluted water, overweight, smoking, too much pressure in life.1 Based on GLOBOCAN estimates, about 14.1 million new cancer cases and 8.2 million deaths occurred in 2012.2 In China, it has been estimated that there are up to 3 million new cases and over 2 million deaths. More seriously, the cancer death rate will continue to increase without effective therapeutic options. Fortunately, some diagnostic methods and techniques are already widely used in clinic, such as chest radiograph,3, 4 fibreoptic bronchoscopy,5 Computed Tomography (CT) scan,6, 7 Magnetic Resonance Imaging (MRI)8, 9 and positron-emission tomography (PET),10 etc. Although they can provide helpful information about the size and location of the tumors, combining two or even more of these methods, they provide little information about metabolic profiling of the tumors.11, 12 It is vitally crucial for analyzing biomarkers at the molecular level for tumor diagnosis, monitoring and treatment.13, 14, 15 Therefore, some effective complementary methods are especially valuable and challenging to investigate tumor metabolic profiling.
Metabolomics is a fast growing field of research downstream of transcriptomics, genomics, which mainly involves the multicomponent analysis of cell extracts, tissues and biological fluids.16, 17, 18, 19, 20 It provides a snapshot of the metabolic dynamics that reflect the response of living systems to both pathophysiological stimuli and/or genetic alteration.21, 22, 23 It has been reported that tumor metabolism differs from that of normal tissue.24, 25 Exploring the tumor metabolome may be the best way to reveal the phenotypic changes relative to biological function, especially where subtle changes in metabolite concentrations can be tractable.26 In general, the primary analytical techniques used in metabolomics are Mass Spectrometry (MS)27, 28, 29 and Nuclear Magnetic Resonance (NMR).30, 31, 32 NMR is a non-destructive and non-invasive technique that can provide complete structural analysis of an extensive range of organic molecules in complex mixtures,33, 34 thus, can be used to differentiate metabolism between tumor and healthy tissues, aiming at finding possible biomarkers of presence and/or degree of different cancers such as brain,35 prostate,36 breast,37 liver,38 renal39 and esophageal40 cancer.
NMR technique analytical platform
NMR has been employed as an excellent tool in exploring tumor metabolism due to its advantages, such as non-invasive, non-destructive, highly reproducible, providing real-time detection for biological samples near the physiological environment,41, 42 offering both qualitative and quantitative information of compounds in complex mixtures precisely and exquisite spectral editing technique make the method flexible and efficient.43, 44 NMR technique can uses not only 1H spectra but also other nuclei like 13C, 15N and 31P etc. to obtain helpful information about the tumor metabolic pathways.45, 46, 47 These spectroscopies have pretty wide spectral width, meaning low possibility of signal overlapping, but their low sensitivity also limit their application, although some metabolites contain phosphorus and nitrogen.48 Because of high sensitivity and almost all compounds contain proton in biological tissues, 1H-NMR is the most widely used method to differentiate metabolism between tumor and normal tissues.32, 49
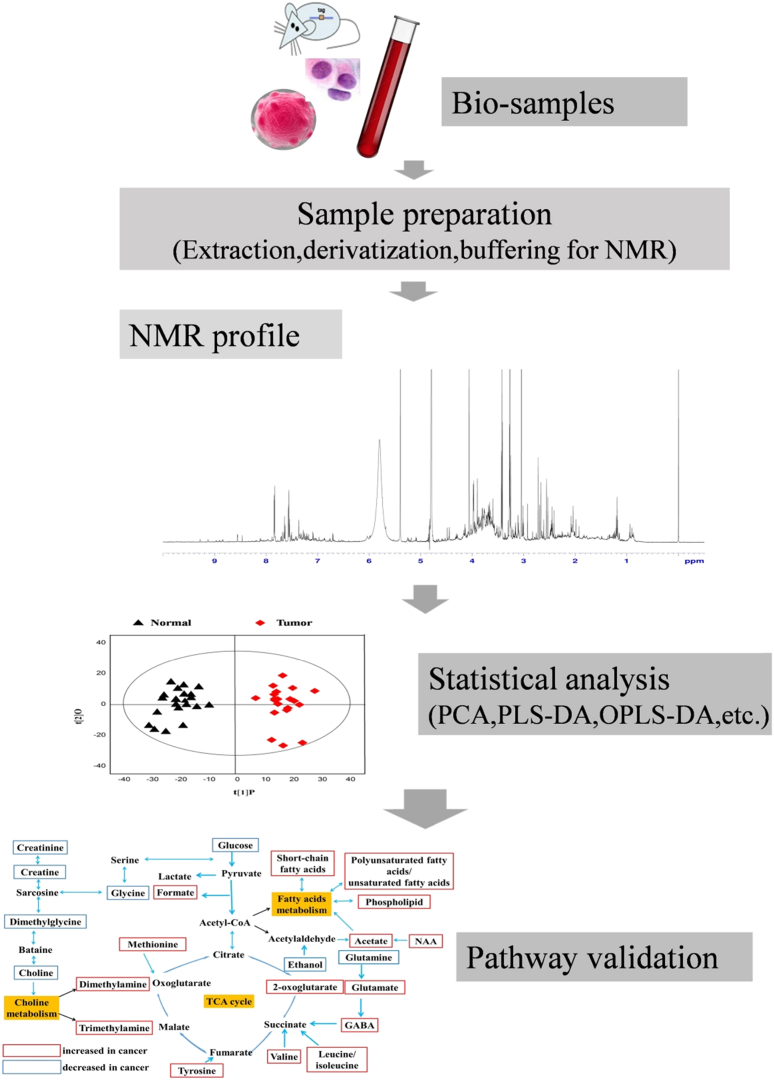
Fig. 1 shows a standard workflow of tumor metabolism studies by NMR, including sample collection, data profiling, pattern recognition and final validation.50 NMR spectroscopy combined with pattern recognition technique is a vitally helpful tool in tumor metabolic profiling as it has the potential to obtain comprehensive characteristics and pathways between tumor and healthy tissues.36, 51, 52, 53 Usually, final results may supply abundant biochemical information at molecular levels for further tumor research, aiming at effectively promoting the process of early diagnosis and treatment of tumor control.
Fig. 1.
Tumor metabolism studies by NMR.
Sample preparation
NMR-based metabolomics has been performed to study a range of different biofluids types including plasma,54 serum,55 urine,56 cerebrospinal fluid,57 saliva,58 and feces.59 Intact tissues and cells samples can also be analyzed by NMR technique.60, 61, 62 To validate those data, sample collection, storage and processing procedures are extremely critical.63, 64 For example, some deuterated buffer should be added to adjust the pH value to avoid the systematic bias by different pH values.65, 66 Typically, urine, serum and plasma samples should be stored in appropriate conditions after collection to reduce sample degradation from multiple freeze/thaw cycles.67, 68 Biofluids and tissue samples should be stored at or below −70 °C.69 Blood plasma or serum samples have high protein content which will obscure the resonances of small molecule metabolites. Therefore, it is necessary to physically remove proteins prior to analysis, which is done by precipitation or extractant or to weaken the intensity of protein resonances in the 1H-NMR spectrum due to their faster rates of T2 relaxation.70, 71 In addition, samples collected from a group of volunteers should be careful to minimize and account for effects from factors such as gender, age, diet, fasting state, exercise, and physical activity.72, 73, 74
Data analysis
NMR and MS both have been employed as common analytical tools in tumor metabolism research. NMR data can be quantitatively analyzed more directly, while MS technique has higher sensitivity. One-dimensional NMR spectroscopy of biological samples usually has hundreds of peaks, so it is a big challenge to obtain all its useful information.75, 76, 77 Therefore, it is vitally helpful to combine the pattern recognition. Several multivariate statistical analysis methods can be employed to discriminate metabolites between complex systems such as principle component analysis (PCA), hierarchical cluster analysis (HCA), partial least squares (PLS), discriminant function analysis (DFA) and artificial neural networks (ANN).78, 79, 80
As a kind of unsupervised methods, PCA is the most widely used statistical analysis methods in metabonomics which can reveal outliers, groups and trends in the original dataset effectively.81, 82 It reduces the multidimensional data space into a much lower dimensional model space while converting the dataset in two matrices, so called scores and loadings. The result shows how samples related to each other and performs the variable contribution to the classification results. PCA can identify the largest variation in the dataset, but the latent variables responsible for the class separation may not be in the direction of the largest variation.83, 84 Instead, orthogonal partial least squares discriminant analysis (OPLS-DA), a very powerful supervised method in metabolomics is a regression model that reflects the correlation between multivariate data and dependent variables with class information. In OPLS-DA, a single component is used as a predictor to describe the separation between classes, where the other components are orthogonal variations to reflect the separation within classes. OPLS-DA emphasizes separations between classes, reduces separation within class at the same time.85, 86, 87, 88 Therefore, an OPLS-DA scores plot will have tighter clusters with a larger separation between groups compared to PCA.89 Usually, supervised and unsupervised multivariate statistical techniques are commonly used together to provide a deep explanation of the different metabolite concentrations between experimental groups.
NMR databases
The great challenge of NMR-based metabolomics is to recognize obtained peaks which can be assigned to different compounds.15 Nowadays, there are a number of public databases available to promote the identification of metabolites including the Human Metabolome Database (HMDB),90 Biological Magnetic Resonance Data Bank,91 NMR database of Linkoeping, Magnetic Resonance Metabololomics Database and Madison-qingdao Metabolomics Consortium Database (MMCD).
These databases can be time-saving in metabolite identification and facilitate the validation of relevant biological pathways. HMDB, the most comprehensive curative human metabolite database in the world, provides a comprehensive compound description, names, structural information, reference NMR spectra, biofluids concentrations, pathway information and other public databases.92
Applications of NMR technique in tumor metabolism
In the following sections, we summarized some typical applications of NMR in various tumor metabolomics. To facilitate systematization and comparison of those studies, Table 1 shows an overview of the main cancer-related metabolic findings in different cancer types.
Table 1.
Main cancer-related metabolic findings in different cancer types unveiled by NMR technique.
| Cancer type | Sample | Metabolic changes in cancer VS controls | Reference |
|---|---|---|---|
| Colorectal | Serum | (+) acetate, acetoacetate, 3-hydroxybutyrate, lactate,pyruvate (−) glucose, myo-inositol,taurine, dimethylglycine |
Ludwig et al93 |
| Fecal extracts | (+)acetate,valerate, propionate, butyrate (−)β-glucose, Gln, glutamate |
Amiot et al94 | |
| Biopsies | (+)taurine, glutamate, aspartate, lactate (−)myo-inositol, β-glucose |
Piotto et al95 | |
| Liver | Serum | (+) acetate, N-acetylglycoproteins,Gln, glycerol, α-ketoglutarate,1-methylhistidine, Phe, pyruvate, Tyr (−) acetoacetate, Cho, LDL, VLDL, Val |
Gao et al96 |
| Urine | (+)carnitine, creatine (−) acetone, creatinine,glycine, hippurate, TMAO |
Shariff et al97 | |
| Lung | Plasma | (+) lactate, VLDL, LDL, pyruvate (−) acetate, alanine, citrate, formate, Gln, HDL, His, methanol, Tyr, Val |
Rocha et al54 |
| Urine | (+) N-acetylglutamine, citrate, creatinine, 3-hydroxyisobutyrate, 3-hydroxyisovalerate (−) hippurate, trigonellinamide, trigonelline |
Carrola et al98 | |
| Breast | Tissues | Changes in Cho,creatine, β-glucose, GPC, glycine, myo-inositol, PCho,taurine |
Bathe et al99 |
| Serum | Metastatic vs early disease: (+)acetoacetate, glycerol, pyruvate, mannose, glutamate, 3-hydroxybutyrate, N-acetylglycoproteins, (−)His, alanine,betaine |
Elodie et al100 | |
| Pancreatic | Serum | (+)soleucine, triglyceride,leucine,creatinine (−)3-hydroxybutyrate, 3-hydroxyisovalerate, lactate, TMAO |
Ouyang et al55 |
| Plasma | (+)N-acetyl glycoprotein, DMA, VLDL,acetone (−)lactate, 3-hydroxybutyrate, HDL, LDL, citrate, glutamate, alanine,Gln, His, isoleucine, lysine, Val |
Lin et al101 |
Abbreviations: Cho, choline; Tyr, tyrosine; Val, valine; His, Histidine; Phe,phenylalanine; PCho, phosphocholine; GPC, glycerolphosphocholine; Gln,glutamine; DMA,dimethylamine; HDL, high density lipoprotein; LDL, low density lipoprotein; TMAO,trimethylamine N-oxide; VLDL, very low density lipoprotein; (+) increased in cancer, (−) decreased in cancer relatively to control.
Colorectal cancer
Colorectal cancer (CRC) is one of the most prevalent tumor types. Understanding the metabolic profile of colorectal tumor is important for therapeutic approaches and molecular diagnosis. Amiot et al94 investigated the fecal metabolic phenotype of patients with advanced colorectal tumor and controls using 1H-NMR and multivariate modeling. PCA results showed that advanced colorectal tumor demonstrated increased fecal concentrations of four short-chain fatty acids (valerate, acetate, propionate and butyrate) and decreased signals relating to β-glucose, Gln, and glutamate. The prediction accuracy of the method is higher than that of the guaiac-fecal occult blood test and the Wif-1 methylation test. Beatriz et al102 performed high resolution Magic Angle Spinning (HR-MAS) NMR to analyze metabolites in intact tumor samples and samples of adjacent mucosa obtained from colorecal cancer patients. The results indicated marked biochemical differences between the two types of tissues by PCA and OPLS-DA. Moreover, metabolic profiles were able to differentiate tumors of different T-and N-stages (T stage has greater weight than the N stage and the former affects survival more significantly) on the basis of tumor node metastasis (TNM) classification. Bertini et al used NMR to profile the serum metabolome in patients with metastatic colorectal cancer (mCRC) and determine whether a disease marker may exist that is strong enough to predict overall survival (OS).103 In the validation set, supervised predictive models allowed a separation of 96.7% of patients from the healthy controls. Wang et al104 applied PCA, PLS-DA and OPLS-DA to analyze the 1H-NMR profiling data to identify the distinguishing metabolites of rectal cancer. Results showed excellent separation among the different stages of rectal cancer tissues and normal controls. These modified metabolites revealed disturbance of energy, amino acids, ketone body and choline metabolism, which may be correlated with the progression of human rectal cancer. Piotto et al105 applied HR-MAS NMR to characterize the metabolic fingerprint of both tumoral and normal tissue samples obtained from a cohort of patients affected by primary colorectal cancer. By analyzing the data using PLS-DA revealed that tumor tissue samples are richer in taurine, glutamate, aspartate and lactate whereas normal tissues contain a higher amount of myo-inositol and β-glucose. The statistical model was subsequently used to perform a blind test on tumor and healthy tissue.
Breast cancer
Breast cancer (BC) is the globally highest incidence and mortality form of all malignant diseases in women and its recurrence rate is also very high. More than three quarters of BC patients can achieve a high survival rate if the cancer is diagnosed at its early stage. It is imperative to identify markers for BC diagnosis and management including prediction, early diagnosis and individualized treatment. Li et al examined 31 breast tissue samples (13 cancer, 9 benign and 9 normal) obtained by core needle biopsy using multivariate modeling.106 Although it was impossible to distinguish the benign tumors and normal tissues, cancer and non-cancer samples can be discriminated well with OPLS-DA on the NMR spectra. A subsequent blind test showed 69% sensitivity and 94% specificity in the prediction of the tumor status. Beathe et al compared the metabolic profiles of tissues collected from 85 breast cancer patients, where the concentration levels of GPC, PC and choline were monitored carefully.99 The concentrations of choline and glycine were much higher in tumor samples larger than 2 cm compared with smaller tumors samples, indicating that HR-MAS NMR could be an efficient tool for BC diagnosis with the ability to identify cancer stages. Then they performed Electronic Reference to access In vivo Concentrations (ERETIC) applying with HR-MAS NMR to quantify metabolites in intact BC samples.107 The nine significant discriminatory metabolites were b-glucose, lactate, glycine, myo-inositol, taurine, GPC, PC, choline and creatine. Elodie et al attempted to identify metabolic serum changes associated with advanced metastatic breast cancer (MBC) in comparison to the localized early disease (EBC).100 The results clearly distinguished EBC and MBC samples with 89.8% sensitivity and 79.3% specificity, where higher levels of serum concentrations of acetoacetate, 3-hydroxybutyrate, glycerol, pyruvate, mannose, N-acetylglycoproteins, glutamate and lower concentration of histidine, alanine and betaine metabolites were observed in MBC tissues. Giskeodegard et al conducted biopsies which were excised during surgery and analyzed by HR-MAS NMR from BC patients.108 The data were analyzed by PLS-DA, probabilistic neural networks (PNNs) and Bayesian belief networks (BBNs). It was also found that estrogen and progesterone receptor status can be successfully predicted to improve predictions of the hormonal therapy of breast carcinomas. Vincent et al employed a combination of NMR and MS methods to build and validate a model for early BC recurrence detection based on a set of 257 retrospective serial samples.109 Utilizing the model, over 55% patients recurrence could be detected as early as 13 months before the recurrence was diagnosed.
Lung cancer
Lung cancer is one of the most serious health problems, also the most common cause of cancer death. There has been a large amount of research in the field. Claudia et al applied PCA and HCA followed by 1H-NMR resulted in good discrimination between tumor and non-involved tissues, showing that inherently different metabolic profiling characterize the two tissue types.54 In a similar study, NMR was utilized to measure metabolites in urine from lung cancer patients and control healthy group.110 The multivariate modeling of urinary profiles discriminate the two groups, where the Monte Carlo Cross Validation of the classification model highlighted 93% sensitivity, 94% specificity and an overall classification rate of 93.5%. Liu et al conducted a new algorithm named PSO-SVWL-PLSDA to rectify the weakened efficacy by the high similarity of metabolic profiles combined with 1H-NMR metabonomics.111 Data clustering in discriminative metabolites for the lung cancer diagnosis were lactate, glucose, threonine, valine, taurine, trimethylamine, Gln, glycoprotein, proline and lipid. Chen et al performed the metabonomic characteristics of 51 lung tissues from 17 patients with lung cancer using the 1H-NMR and the multivariate data analysis methods.112 The findings clearly disclosed metabonomic characteristics of lung cancer tissues at various sites. Durate et al searched for the metabolic markers of lung cancer in urine by combining analysis of the NMR data including PCA, PLS-DA, and OPLS-DA.113 The obtained results showed a high level of sensitivity and 100% specificity. Moreover, PLS-DA of a subset of tumour samples allowed adenocarcinomas to be discriminated from carcinoid tumors and epidermoid carcinomas, showing differences in metabolite levels between these histological types. Jordan et al investigated discrimination between tissue and serum metabolites for squamous cell carcinoma (SCC) and adenocarcinoma (AC) in the lung tumors by HR-MAS NMR.114 The results disclosed the potential to differentiate between the tested lung cancer types and controls. Rocha et al investigated the variations in the metabolic profile of blood plasma from lung cancer patients and control group by NMR-based metabonomics.115 PLS-DA modeling of CPMG spectra from plasma, subjected to Monte Carlo Cross Validation provided the potential of this approach to screen and diagnose for lung cancer. The distinguishing metabolites between the patients and the control group were lower levels of HDL, higher levels of VLDL and LDL. The patients' plasma had significantly high level of lactate and pyruvate but significantly low levels of citrate, formate, acetate, glucose, Gln, alanine, tyrosine and valine.
Liver cancer
Hepatocellular carcinoma (HCC) is a leading cancer worldwide in terms of incidence and mortality, for the lack of surveillance of patients. Yang et al compared metabolite levels in sera of HCC tumor and non-involved adjacent liver tissues by HR-MAS NMR.116 The discriminating candidate biomarkers were lactate, amino acids including glutamate, glutamine, Gly, leucine and alanine, Cho, PC, GPC, triglycerides, glucose, phosphorylethanolamine and glycogen. Gao et al monitored the changes in endogenous metabolites of liver cirrhosis (LC) and HCC using single blood samples.96 Results indicated a higher level of acetate, pyruvate, Gln, a-ketoglutarate, glycerol, tyrosine, 1-methylhistidine and phenylalanine, together with lower level of LDL, isoleucine, Val, acetoacetate, creatine, Cho and unsaturated lipids in HCC patients. Moreover, pathway analysis suggested altered energy metabolism with changes in the tricarboxylic acid (TCA) cycle in LC and HCC patients. In a similar study, Nahon et al compared the metabolic profiles from 154 cirrhotic patients with and without HCC.117 The result indicated that serum NMR spectra combined with the OPLS analysis model provided an evident discrimination between cirrhotic and large HCC tumors. Perturbations observed in the synthesis of glutaminolysis, citrate cycle, phospholipid and glycolysis metabolism have potential to assess pathological hepatic lesions. Debora et al investigated the metabolic profiles from primary HCC, cirrhotic tissues (CIR), hepatic metastases from colorectal carcinomas and non-cirrhotic normal tissues by 1H-NMR combined with pattern-recognition and visualization techniques.117 Results indicated clear metabolic differences in the studied grades and the tissue signals of lactate and the glucose were primarily discriminate the different histological samples, which can be of great benefit to liver tumor diagnosis. Antonio et al obtained NMR spectroscopy of 51 needle biopsies (14 primary nodules, 14 recurrent, and 23 paired cirrhotic specimens) to elucidate the metabolite changes associated with HCC.118 The results clearly differentiated primary HCC from recurrences using PLS-DA models and disclosed alterations in choline metabolism.
Pancreatic cancer
Pancreatic cancer is a malignant tumor. Once diagnosed, surgical cure is no longer an efficient option for most patients, thus early detection of pancreatic cancer is vital for its treatment. OuYang et al studied the serum collected from pancreatic cancer patients with NMR.55 Results showed good distinction between cancer and benign group. The analysis were revealed by higher level of soleucine, leucine and creatinine and lower values of 3-hydroxybutyrate, 3-hydroxyisovalerate, lactate, triglyceride and TMAO from pancreatic cancer patients compared to control group. These metabolite changes could be used as metabolic markers for the early detection of pancreatic cancer. Oliver et al analyzed the sera from pancreatic cancer patients and healthy volunteers by 1H-NMR followed by statistical analyses.119 The diagnostic model distinguished benign from malignant pancreatic lesions accurately via excluding patients with overt diabetes mellitus. Lin et al used 1H-NMR spectra to profile the plasma metabolites obtained from 19 pancreatic cancer patients.101 The levels of N-acetyl glycoprotein, DMA, VLDL and acetone in the PC group increased, whereas lactate, 3-hydroxybutyrate, HDL, LDL, alanine, glutamate, Gln, His, isoleucine, lysine and valine were found to be reduced.
Conclusions and future perspectives
NMR technique has become one of the most powerful tools in biology and medicine research. On the basis of the combination of 1H-NMR spectroscopy with pattern recognition methods of multivariate data analysis, metabolomics can provide comprehensive characteristics of the major metabolic pathways to evaluate the status of the tumor more precisely. Therefore, NMR application in tumor metabolomics research could have a great potential to provide valuable information about early diagnosis, treatment options, processes and prognosis estimate of cancer and other serious diseases. Nowadays, NMR sensitivity has been greatly improved by higher magnetic field, cryoprobe, fast 2D NMR experiments, polarization transfer etc., so NMR technique can be more widely applied in tumor metabolomics studies.
Conflicts of interest
All authors have none to declare.
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics. CA A Cancer J Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics. CA A Cancer J Clin. 2012;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Oken M.M., Hocking W.G., Kvale P.A. Screening by chest radiograph and lung cancer mortality: the prostate, lung, colorectal, and ovarian (plco) randomized trial. JAMA. 2011;306(17):1865–1873. doi: 10.1001/jama.2011.1591. [DOI] [PubMed] [Google Scholar]
- 4.Oken M.M., Marcus P.M., Hu P. Baseline chest radiograph for lung cancer detection in the randomized prostate, lung, colorectal and ovarian cancer screening trial. J Natl Cancer Inst. December 21, 2005;97(24):1832–1839. doi: 10.1093/jnci/dji430. [DOI] [PubMed] [Google Scholar]
- 5.Karahalli E., Yilmaz A., Türker H., Özvaran K. Usefulness of various diagnostic techniques during fiberoptic bronchoscopy for endoscopically visible lung cancer: should cytologic examinations be performed routinely? Respiration. 2001;68(6):611–614. doi: 10.1159/000050581. [DOI] [PubMed] [Google Scholar]
- 6.Berrington de González A., Mahesh M., Kim K. Projected cancer risks from computed tomographic scans performed in the United States in 2007. Archives Intern Med. 2009;169(22):2071–2077. doi: 10.1001/archinternmed.2009.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berrington de González A., Kim K.P., Berg C.D. Low-dose lung computed tomography screening before age 55: estimates of the mortality reduction required to outweigh the radiation-induced cancer risk. J Med Screen. September 1, 2008;15(3):153–158. doi: 10.1258/jms.2008.008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Padhani A.R., Liu G., Mu-Koh D. Diffusion-weighted magnetic resonance imaging as a cancer biomarker: consensus and recommendations. Neoplasia. 2009;11(2):102–125. doi: 10.1593/neo.81328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chenevert T.L., McKeever P.E., Ross B.D. Monitoring early response of experimental brain tumors to therapy using diffusion magnetic resonance imaging. Clin Cancer Res. September 1, 1997;3(9):1457–1466. [PubMed] [Google Scholar]
- 10.Juweid M.E., Cheson B.D. Positron-emission tomography and assessment of cancer therapy. N Engl J Med. 2006;354(5):496–507. doi: 10.1056/NEJMra050276. [DOI] [PubMed] [Google Scholar]
- 11.Yi C.A., Lee K.S., Lee H.Y. Coregistered whole body magnetic resonance imaging-positron emission tomography (MRI-PET) versus PET-computed tomography plus brain MRI in staging resectable lung cancer. Cancer. 2013;119(10):1784–1791. doi: 10.1002/cncr.28000. [DOI] [PubMed] [Google Scholar]
- 12.Erasmus J.J., Sabloff B.S. CT, positron emission tomography, and MRI in staging lung cancer. Clin Chest Med. 2008;29(1):39–57. doi: 10.1016/j.ccm.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 13.Sugimoto M., Wong D.T., Hirayama A., Soga T., Tomita M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics. 2010;6(1):78–95. doi: 10.1007/s11306-009-0178-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Claudino W.M., Goncalves P.H., di Leo A., Philip P.A., Sarkar F.H. Metabolomics in cancer: a bench-to-bedside intersection. Crit Rev Oncol Hematol. 2012;84(1):1–7. doi: 10.1016/j.critrevonc.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 15.Patel S., Ahmed S. Emerging field of metabolomics: big promise for cancer biomarker identification and drug discovery. J Pharm Biomed Anal. 2015;107:63–74. doi: 10.1016/j.jpba.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 16.Yizhak K., Benyamini T., Liebermeister W., Ruppin E., Shlomi T. Integrating quantitative proteomics and metabolomics with a genome-scale metabolic network model. Bioinformatics. June 15, 2010;26(12):i255–i260. doi: 10.1093/bioinformatics/btq183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicholson J.K., Wilson I.D. High resolution proton magnetic resonance spectroscopy of biological fluids. Prog Nucl Magnetic Reson Spectrosc. 1989/01/01;21(4):449–501. [Google Scholar]
- 18.Nambiar P.R., Gupta R.R., Misra V. An “Omics” based survey of human colon cancer. Mutat Res Fundam Mol Mech Mutagen. 11/10/2010;693(1–2):3–18. doi: 10.1016/j.mrfmmm.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 19.Raamsdonk L.M., Teusink B., Broadhurst D. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat Biotechnol. 2001;19(1):45–50. doi: 10.1038/83496. [DOI] [PubMed] [Google Scholar]
- 20.Bino R.J., Hall R.D., Fiehn O. Potential of metabolomics as a functional genomics tool. Trends Plant Sci. 2004;9(9):418–425. doi: 10.1016/j.tplants.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 21.Kell D.B. Systems biology, metabolic modelling and metabolomics in drug discovery and development. Drug Discov Today. 2006;11(23):1085–1092. doi: 10.1016/j.drudis.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Nicholson J.K., Lindon J.C., Holmes E. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999/01/01;29(11):1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 23.Dunn W.B., Broadhurst D.I., Atherton H.J., Goodacre R., Griffin J.L. Systems level studies of mammalian metabolomes: the roles of mass spectrometry and nuclear magnetic resonance spectroscopy. Chem Soc Rev. 2011/01;40(1):387–426. doi: 10.1039/b906712b. [DOI] [PubMed] [Google Scholar]
- 24.Bathen T.F., Sitter B., Sjøbakk T.E., Tessem M.-B., Gribbestad I.S. Magnetic resonance metabolomics of intact tissue: a biotechnological tool in cancer diagnostics and treatment evaluation. Cancer Res. 2010/09;70(17):6692–6696. doi: 10.1158/0008-5472.CAN-10-0437. [DOI] [PubMed] [Google Scholar]
- 25.Yang J., Xu G., Zheng Y. Diagnosis of liver cancer using HPLC-based metabonomics avoiding false-positive result from hepatitis and hepatocirrhosis diseases. J Chromatogr B. 2004;813(1):59–65. doi: 10.1016/j.jchromb.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 26.Griffin J.L., Shockcor J.P. Metabolic profiles of cancer cells. Nat Rev Cancer. 2004;4(7):551–561. doi: 10.1038/nrc1390. [DOI] [PubMed] [Google Scholar]
- 27.Woo H.M., Kim K.M., Choi M.H. Mass spectrometry based metabolomic approaches in urinary biomarker study of women's cancers. Clin Chim Acta. 2009;400(1):63–69. doi: 10.1016/j.cca.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 28.Zürbig P., Mischak H. 2008. Capillary Electrophoresis Coupled to Mass Spectrometry for Biomarker Discovery and Diagnosis of Kidney Diseases. [DOI] [PubMed] [Google Scholar]
- 29.Kolch W., Neusüß C., Pelzing M., Mischak H. Capillary electrophoresis–mass spectrometry as a powerful tool in clinical diagnosis and biomarker discovery. Mass Spectrom Rev. 2005;24(6):959–977. doi: 10.1002/mas.20051. [DOI] [PubMed] [Google Scholar]
- 30.Zhang J., Wei S., Liu L. NMR-based metabolomics study of canine bladder cancer. Biochimica Biophysica Acta (BBA) – Mol Basis Dis. 2012;1822(11):1807–1814. doi: 10.1016/j.bbadis.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 31.Gebregiworgis T., Powers R. Application of NMR metabolomics to search for human disease biomarkers. Comb Chem High Throughput Screen. 2012;15(8):595–610. doi: 10.2174/138620712802650522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zira A.N., Theocharis S.E., Mitropoulos D., Migdalis V., Mikros E. 1H NMR metabonomic analysis in renal cell carcinoma: a possible diagnostic tool. J Proteome Res. 2010;9(8):4038–4044. doi: 10.1021/pr100226m. [DOI] [PubMed] [Google Scholar]
- 33.Yang C., Richardson A.D., Osterman A., Smith J.W. Profiling of central metabolism in human cancer cells by two-dimensional NMR, GC-MS analysis, and isotopomer modeling. Metabolomics. 2008;4(1):13–29. [Google Scholar]
- 34.de Graaf R.A., Behar K.L. Quantitative 1H NMR spectroscopy of blood plasma metabolites. Anal Chem. 2003;75(9):2100–2104. doi: 10.1021/ac020782+. [DOI] [PubMed] [Google Scholar]
- 35.Wilson M., Davies N.P., Brundler M.-A., McConville C., Grundy R.G., Peet A.C. High resolution magic angle spinning 1H NMR of childhood brain and nervous system tumours. Mol Cancer. 2009;8(1):1–11. doi: 10.1186/1476-4598-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeFeo E.M., Wu C.-L., McDougal W.S., Cheng L.L. A decade in prostate cancer: from NMR to metabolomics. Nat Rev Urol. 2011;8(6):301–311. doi: 10.1038/nrurol.2011.53. [DOI] [PubMed] [Google Scholar]
- 37.Denkert C., Bucher E., Hilvo M. Metabolomics of human breast cancer: new approaches for tumor typing and biomarker discovery. Genome Med. 2012;4(4):37. doi: 10.1186/gm336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li C.W., Kuo Y.C., Chen C.Y. Quantification of choline compounds in human hepatic tumors by proton MR spectroscopy at 3T. Magnetic Reson Med. 2005;53(4):770–776. doi: 10.1002/mrm.20412. [DOI] [PubMed] [Google Scholar]
- 39.Tosi M.R., Tugnoli V., Bottura G., Lucchi P., Battaglia A., Giorgianni P. Biochemical characterization of human renal tumors by in vitro nuclear magnetic resonance. J Mol Struct. 5/30/ 2001;565–566:323–327. [Google Scholar]
- 40.Zhang X., Xu L., Shen J. Metabolic signatures of esophageal cancer: NMR-based metabolomics and UHPLC-based focused metabolomics of blood serum. Biochimica Biophysica Acta (BBA) – Mol Basis Dis. 2013;1832(8):1207–1216. doi: 10.1016/j.bbadis.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 41.Dumas M.-E., Maibaum E.C., Teague C. Assessment of analytical reproducibility of 1H NMR spectroscopy based metabonomics for large-scale epidemiological research: the INTERMAP Study. Anal Chem. 2006;78(7):2199–2208. doi: 10.1021/ac0517085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gil A.M., de Pinho P.G., Monteiro M.S., Duarte I.F. NMR metabolomics of renal cancer: an overview. Bioanalysis. 2015/09/01;7(18):2361–2374. doi: 10.4155/bio.15.167. [DOI] [PubMed] [Google Scholar]
- 43.Liu M., Nicholson J.K., Lindon J.C. High-resolution diffusion and relaxation edited one-and two-dimensional 1H NMR spectroscopy of biological fluids. Anal Chem. 1996;68(19):3370–3376. doi: 10.1021/ac960426p. [DOI] [PubMed] [Google Scholar]
- 44.Martin-Pastor M. Experiments for the editing of singlet peaks and simplification of 1H NMR spectra of complex mixtures. J Agric food Chem. 2014;62(5):1190–1197. doi: 10.1021/jf4044869. [DOI] [PubMed] [Google Scholar]
- 45.Katz-Brull R., Margalit R., Bendel P., Degani H. Choline metabolism in breast cancer; 2H-, 13C-and 31P-NMR studies of cells and tumors. Magnetic Reson Mater Phys Biol Med. 1998;6(1):44–52. doi: 10.1007/BF02662511. [DOI] [PubMed] [Google Scholar]
- 46.Daly P.F., Lyon R.C., Faustino P.J., Cohen J.S. Phospholipid metabolism in cancer cells monitored by 31P NMR spectroscopy. J Biol Chem. 1987;262(31):14875–14878. [PubMed] [Google Scholar]
- 47.Shanaiah N., Desilva M.A., Gowda G.N., Raftery M.A., Hainline B.E., Raftery D. Class selection of amino acid metabolites in body fluids using chemical derivatization and their enhanced 13C NMR. Proc Natl Acad Sci. 2007;104(28):11540–11544. doi: 10.1073/pnas.0704449104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emwas A.-H.M., Salek R.M., Griffin J.L., Merzaban J. NMR-based metabolomics in human disease diagnosis: applications, limitations, and recommendations. Metabolomics. 2013;9(5):1048–1072. [Google Scholar]
- 49.Duarte I.F., Gil A.M. Metabolic signatures of cancer unveiled by NMR spectroscopy of human biofluids. Prog Nucl Magnetic Reson Spectrosc. 2012;62:51–74. doi: 10.1016/j.pnmrs.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 50.Ah Zhang, Sun H., Qiu S., Xj Wang. NMR-based metabolomics coupled with pattern recognition methods in biomarker discovery and disease diagnosis. Magnetic Reson Chem. 2013;51(9):549–556. doi: 10.1002/mrc.3985. [DOI] [PubMed] [Google Scholar]
- 51.Tiziani S., Lopes V., Günther U.L. Early stage diagnosis of oral cancer using 1H NMR–based metabolomics. Neoplasia. 2009;11(3) doi: 10.1593/neo.81396. 269–IN210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jordan K.W., Cheng L.L. NMR-based metabolomics approach to target biomarkers for human prostate cancer. Expert Rev Proteomics. 2007;4(3):389–400. doi: 10.1586/14789450.4.3.389. [DOI] [PubMed] [Google Scholar]
- 53.DeBerardinis R.J., Lum J.J., Hatzivassiliou G., Thompson C.B. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7(1):11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 54.Rocha C.M., Carrola J., Barros A.S. Metabolic signatures of lung cancer in biofluids: NMR-based metabonomics of blood plasma. J Proteome Res. 2011;10(9):4314–4324. doi: 10.1021/pr200550p. [DOI] [PubMed] [Google Scholar]
- 55.OuYang D., Xu J., Huang H., Chen Z. Metabolomic Profiling of serum from human pancreatic cancer patients using 1H NMR spectroscopy and principal component analysis. Appl Biochem Biotechnol. 2011;165(1):148–154. doi: 10.1007/s12010-011-9240-0. [DOI] [PubMed] [Google Scholar]
- 56.Slupsky C.M., Steed H., Wells T.H. Urine metabolite analysis offers potential early diagnosis of ovarian and breast cancers. Clin Cancer Res. 2010;16(23):5835–5841. doi: 10.1158/1078-0432.CCR-10-1434. [DOI] [PubMed] [Google Scholar]
- 57.Dunne V.G., Bhattachayya S., Besser M., Rae C., Griffin J.L. Metabolites from cerebrospinal fluid in aneurysmal subarachnoid haemorrhage correlate with vasospasm and clinical outcome: a pattern-recognition 1H NMR study. NMR Biomed. 2005;18(1):24–33. doi: 10.1002/nbm.918. [DOI] [PubMed] [Google Scholar]
- 58.Wei J., Xie G., Zhou Z. Salivary metabolite signatures of oral cancer and leukoplakia. Int J Cancer. 2011;129(9):2207–2217. doi: 10.1002/ijc.25881. [DOI] [PubMed] [Google Scholar]
- 59.Monleon D., Morales J.M., Barrasa A., Lopez J.A., Vazquez C., Celda B. Metabolite profiling of fecal water extracts from human colorectal cancer. NMR Biomed. 2009;22(3):342–348. doi: 10.1002/nbm.1345. [DOI] [PubMed] [Google Scholar]
- 60.Tomlins A.M., Foxall P.J., Lindon J.C. High resolution magic angle spinning 1H nuclear magnetic resonance analysis of intact prostatic hyperplastic and tumour tissues. Anal Commun. 1998;35(3):113–115. [Google Scholar]
- 61.Beckonert O., Coen M., Keun H.C. High-resolution magic-angle-spinning NMR spectroscopy for metabolic profiling of intact tissues. Nat Protoc. 2010;5(6):1019–1032. doi: 10.1038/nprot.2010.45. [DOI] [PubMed] [Google Scholar]
- 62.Somashekar B.S., Kamarajan P., Danciu T. Magic angle spinning NMR-based metabolic profiling of head and neck squamous cell carcinoma tissues. J proteome Res. 2011;10(11):5232–5241. doi: 10.1021/pr200800w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sykes B.D. Urine stability for metabolomic studies: effects of preparation and storage. Metabolomics. 2007;3(1):19–27. [Google Scholar]
- 64.Bernini P., Bertini I., Luchinat C., Nincheri P., Staderini S., Turano P. Standard operating procedures for pre-analytical handling of blood and urine for metabolomic studies and biobanks. J Biomol NMR. 2011;49(3–4):231–243. doi: 10.1007/s10858-011-9489-1. [DOI] [PubMed] [Google Scholar]
- 65.Brown S.A., Simpson A.J., Simpson M.J. Evaluation of sample preparation methods for nuclear magnetic resonance metabolic profiling studies with Eisenia fetida. Environ Toxicol Chem. 2008;27(4):828–836. doi: 10.1897/07-412.1. [DOI] [PubMed] [Google Scholar]
- 66.Reily M.D., Robosky L.C., Manning M.L., Butler A., Baker J.D., Winters R.T. DFTMP, an NMR reagent for assessing the near-neutral pH of biological samples. J Am Chem Soc. 2006;128(38):12360–12361. doi: 10.1021/ja063773h. [DOI] [PubMed] [Google Scholar]
- 67.Grimes J.H., O'Connell T.M. The application of micro-coil NMR probe technology to metabolomics of urine and serum. J Biomol NMR. 2011;49(3–4):297–305. doi: 10.1007/s10858-011-9488-2. [DOI] [PubMed] [Google Scholar]
- 68.Canelas A.B., Ras C., Ten Pierick A., van Dam J.C., Heijnen J.J., Van Gulik W.M. Leakage-free rapid quenching technique for yeast metabolomics. Metabolomics. 2008;4(3):226–239. [Google Scholar]
- 69.Briscoe C.J., Hage D.S. Factors affecting the stability of drugs and drug metabolites in biological matrices. Bioanalysis. 2009 doi: 10.4155/bio.09.20. [DOI] [PubMed] [Google Scholar]
- 70.Tiziani S., Emwas A.-H., Lodi A. Optimized metabolite extraction from blood serum for 1H nuclear magnetic resonance spectroscopy. Anal Biochem. 2008;377(1):16–23. doi: 10.1016/j.ab.2008.01.037. [DOI] [PubMed] [Google Scholar]
- 71.Psychogios N., Hau D.D., Peng J. The human serum metabolome. PLoS One. 2011;6(2):e16957. doi: 10.1371/journal.pone.0016957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rasmussen L.G., Savorani F., Larsen T.M., Dragsted L.O., Astrup A., Engelsen S.B. Standardization of factors that influence human urine metabolomics. Metabolomics. 2011;7(1):71–83. [Google Scholar]
- 73.Beger R.D. A review of applications of metabolomics in cancer. Metabolites. 2013;3(3):552–574. doi: 10.3390/metabo3030552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Emwas A.-H., Luchinat C., Turano P. Standardizing the experimental conditions for using urine in NMR-based metabolomic studies with a particular focus on diagnostic studies: a review. Metabolomics. 2015;11(4):872–894. doi: 10.1007/s11306-014-0746-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang J., Zhang S., Li Z. 1H-NMR-based metabolomics of tumor tissue for the metabolic characterization of rat hepatocellular carcinoma formation and metastasis. Tumor Biol. 2011;32(1):223–231. doi: 10.1007/s13277-010-0116-7. [DOI] [PubMed] [Google Scholar]
- 76.Jonsson P., Johansson A.I., Gullberg J. High-throughput data analysis for detecting and identifying differences between samples in GC/MS-based metabolomic analyses. Anal Chem. 2005/09/01;77(17):5635–5642. doi: 10.1021/ac050601e. [DOI] [PubMed] [Google Scholar]
- 77.Madsen R., Lundstedt T., Trygg J. Chemometrics in metabolomics—a review in human disease diagnosis. Anal Chim Acta. 2010;659(1):23–33. doi: 10.1016/j.aca.2009.11.042. [DOI] [PubMed] [Google Scholar]
- 78.Kohl S.M., Klein M.S., Hochrein J., Oefner P.J., Spang R., Gronwald W. State-of-the art data normalization methods improve NMR-based metabolomic analysis. Metabolomics. 2012;8(1):146–160. doi: 10.1007/s11306-011-0350-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu S.-Y., Zhang R.-L., Kang H., Fan Z.-J., Du Z. Human liver tissue metabolic profiling research on hepatitis B virus-related hepatocellular carcinoma. World J Gastroenterology WJG. 2013;19(22):3423–3432. doi: 10.3748/wjg.v19.i22.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Snow P.B., Smith D.S., Catalona W.J. Artificial neural networks in the diagnosis and prognosis of prostate cancer: a pilot study. J Urol. 1994;152(5 Pt 2):1923–1926. doi: 10.1016/s0022-5347(17)32416-3. [DOI] [PubMed] [Google Scholar]
- 81.Lehnhardt F.G., Bock C., Röhn G., Ernestus R.I., Hoehn M. Metabolic differences between primary and recurrent human brain tumors: a 1H NMR spectroscopic investigation. NMR Biomed. 2005;18(6):371–382. doi: 10.1002/nbm.968. [DOI] [PubMed] [Google Scholar]
- 82.Gribbestad I.S., Petersen S.B., Fjøsne H.E., Kvinnsland S., Krane J. 1H NMR spectroscopic characterization of perchloric acid extracts from breast carcinomas and non-involved breast tissue. NMR Biomed. 1994;7(4):181–194. doi: 10.1002/nbm.1940070405. [DOI] [PubMed] [Google Scholar]
- 83.Bylesjö M., Rantalainen M., Cloarec O., Nicholson J.K., Holmes E., Trygg J. OPLS discriminant analysis: combining the strengths of PLS-DA and SIMCA classification. J Chemom. 2006;20(8-10):341–351. [Google Scholar]
- 84.Goodpaster A.M., Romick-Rosendale L.E., Kennedy M.A. Statistical significance analysis of nuclear magnetic resonance-based metabonomics data. Anal Biochem. 2010;401(1):134–143. doi: 10.1016/j.ab.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 85.Westerhuis J.A., van Velzen E.J., Hoefsloot H.C., Smilde A.K. Multivariate paired data analysis: multilevel PLSDA versus OPLSDA. Metabolomics. 2010;6(1):119–128. doi: 10.1007/s11306-009-0185-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Molteni C.G., Cazzaniga G., Condorelli D.F., Fortuna C.G., Biondi A., Musumarra G. Successful application of OPLS-DA for the discrimination of wild-type and mutated cells in acute lymphoblastic leukemia. QSAR Comb Sci. 2009;28(8):822–828. [Google Scholar]
- 87.Boccard J., Rutledge D.N. A consensus orthogonal partial least squares discriminant analysis (OPLS-DA) strategy for multiblock Omics data fusion. Anal Chim Acta. 2013;769:30–39. doi: 10.1016/j.aca.2013.01.022. [DOI] [PubMed] [Google Scholar]
- 88.Ramadan Z., Jacobs D., Grigorov M., Kochhar S. Metabolic profiling using principal component analysis, discriminant partial least squares, and genetic algorithms. Talanta. 2006;68(5):1683–1691. doi: 10.1016/j.talanta.2005.08.042. [DOI] [PubMed] [Google Scholar]
- 89.Worley B., Halouska S., Powers R. Utilities for quantifying separation in PCA/PLS-DA scores plots. Anal Biochem. 2013;433(2):102–104. doi: 10.1016/j.ab.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wishart D.S., Tzur D., Knox C. HMDB: the human metabolome database. Nucleic acids Res. 2007;35(suppl 1):D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ulrich E., Akutsu H., Doreleijers J. BioMagResBank. Nucleic Acids Res. 2008;36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Forsythe I.J., Wishart D.S. Exploring human metabolites using the human metabolome database. Curr Protoc Bioinforma. 2009:14.18.11–14.18.45. doi: 10.1002/0471250953.bi1408s25. [DOI] [PubMed] [Google Scholar]
- 93.Ludwig C., Ward D., Martin A. Fast targeted multidimensional NMR metabolomics of colorectal cancer. Magnetic Reson Chem MRC. 2009;47:S68–S73. doi: 10.1002/mrc.2519. [DOI] [PubMed] [Google Scholar]
- 94.Amiot A., Dona A.C., Wijeyesekera A. 1H NMR spectroscopy of fecal extracts enables detection of advanced colorectal neoplasia. J Proteome Res. 2015/09/04;14(9):3871–3881. doi: 10.1021/acs.jproteome.5b00277. [DOI] [PubMed] [Google Scholar]
- 95.Piotto M., Moussallieh F.-M., Dillmann B. Metabolic characterization of primary human colorectal cancers using high resolution magic angle spinning 1H magnetic resonance spectroscopy. Metabolomics. 2009;5 [Google Scholar]
- 96.Gao H., Lu Q., Liu X. Application of 1H NMR-based metabonomics in the study of metabolic profiling of human hepatocellular carcinoma and liver cirrhosis. Cancer Sci. 2009;100(4):782–785. doi: 10.1111/j.1349-7006.2009.01086.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shariff M.I., Gomaa A.I., Cox I.J. Urinary metabolic biomarkers of hepatocellular carcinoma in an Egyptian population: a validation study. J Proteome Res. 2011;10(4):1828–1836. doi: 10.1021/pr101096f. [DOI] [PubMed] [Google Scholar]
- 98.Carrola J., CuM Rocha, AnS Barros. Metabolic signatures of lung cancer in biofluids: NMR-based metabonomics of urine. J Proteome Res. 2010;10(1):221–230. doi: 10.1021/pr100899x. [DOI] [PubMed] [Google Scholar]
- 99.Sitter B., Lundgren S., Bathen T.F., Halgunset J., Fjosne H.E., Gribbestad I.S. Comparison of HR MAS MR spectroscopic profiles of breast cancer tissue with clinical parameters. NMR Biomed. 2006;19(1):30–40. doi: 10.1002/nbm.992. [DOI] [PubMed] [Google Scholar]
- 100.Jobard E., Pontoizeau C., Blaise B.J., Bachelot T., Elena-Herrmann B., Trédan O. A serum nuclear magnetic resonance-based metabolomic signature of advanced metastatic human breast cancer. Cancer Lett. 2/1/2014;343(1):33–41. doi: 10.1016/j.canlet.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 101.Zhang L., Jin H., Guo X. Distinguishing pancreatic cancer from chronic pancreatitis and healthy individuals by 1H nuclear magnetic resonance-based metabonomic profiles. Clin Biochem. 2012;45(13–14):1064–1069. doi: 10.1016/j.clinbiochem.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 102.Jiménez B., Mirnezami R., Kinross J. 1H HR-MAS NMR spectroscopy of tumor-induced local metabolic “field-effects” enables colorectal cancer staging and prognostication. J Proteome Res. 2013/02/01;12(2):959–968. doi: 10.1021/pr3010106. [DOI] [PubMed] [Google Scholar]
- 103.Bertini I., Cacciatore S., Jensen B.V. Metabolomic NMR fingerprinting to identify and predict survival of patients with metastatic colorectal cancer. Cancer Res. January 1, 2012;72(1):356–364. doi: 10.1158/0008-5472.CAN-11-1543. [DOI] [PubMed] [Google Scholar]
- 104.Wang H., Wang L., Zhang H. 1H NMR-based metabolic profiling of human rectal cancer tissue. Mol Cancer. 2013;12(1):1–12. doi: 10.1186/1476-4598-12-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Piotto M., Moussallieh F.-M., Dillmann B. Metabolic characterization of primary human colorectal cancers using high resolution magic angle spinning 1H magnetic resonance spectroscopy. Metabolomics. 2008;5(3):292–301. [Google Scholar]
- 106.MuLan L., Yonghyun S., Nariya C. An HR-MAS MR metabolomics study on breast tissues obtained with core needle biopsy. PLoS One. 2011;6(10):1–8. doi: 10.1371/journal.pone.0025563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sitter B., Bathen T.F., Singstad T.E. Quantification of metabolites in breast cancer patients with different clinical prognosis using HR MAS MR spectroscopy. NMR Biomed. 2010;23(4):424–431. doi: 10.1002/nbm.1478. [DOI] [PubMed] [Google Scholar]
- 108.Giskeødegård G.F., Grinde M.T., Sitter B. Multivariate modeling and prediction of breast cancer prognostic factors using mr metabolomics. J Proteome Res. 2010/02/05;9(2):972–979. doi: 10.1021/pr9008783. [DOI] [PubMed] [Google Scholar]
- 109.Asiago V.M., Alvarado L.Z., Shanaiah N. Early detection of recurrent breast cancer using metabolite profiling. Cancer Res. November 1, 2010;70(21):8309–8318. doi: 10.1158/0008-5472.CAN-10-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Carrola J., Rocha C.M., Barros A.S. Metabolic signatures of lung cancer in biofluids: NMR-based metabonomics of urine. J Proteome Res. 2011/01/07;10(1):221–230. doi: 10.1021/pr100899x. [DOI] [PubMed] [Google Scholar]
- 111.Liu Y.-F., Xu S., Gong H., Cui Y.-F., Song D.-D., Zhou Y.-P. Partial least-squares discriminant analysis optimized by particle swarm optimization: application to 1H nuclear magnetic resonance analysis of lung cancer metabonomics. J Chemom. 2015;29(10):537–546. [Google Scholar]
- 112.Chen W., Zu Y., Huang Q. Study on metabonomic characteristics of human lung cancer using high resolution magic-angle spinning 1H NMR spectroscopy and multivariate data analysis. Magnetic Reson Med. 2011;66(6):1531–1540. doi: 10.1002/mrm.22957. [DOI] [PubMed] [Google Scholar]
- 113.Duarte I.F., Rocha C.M., Barros A.S. Can nuclear magnetic resonance (NMR) spectroscopy reveal different metabolic signatures for lung tumours? Virchows Arch. 2010;457(6):715–725. doi: 10.1007/s00428-010-0993-6. [DOI] [PubMed] [Google Scholar]
- 114.Jordan K.W., Adkins C.B., Su L. Comparison of squamous cell carcinoma and adenocarcinoma of the lung by metabolomic analysis of tissue–serum pairs. Lung Cancer. 2010;68(1):44–50. doi: 10.1016/j.lungcan.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rocha C.M., Barros A.S., Gil A.M. Metabolic profiling of human lung cancer tissue by 1H high resolution magic angle spinning (HRMAS) NMR spectroscopy. J Proteome Res. 2010/01/04;9(1):319–332. doi: 10.1021/pr9006574. [DOI] [PubMed] [Google Scholar]
- 116.Yang Y., Li C., Nie X. Metabonomic studies of human hepatocellular carcinoma using high-resolution magic-angle spinning 1H NMR Spectroscopy in conjunction with multivariate data analysis. J Proteome Res. 2007/07/01;6(7):2605–2614. doi: 10.1021/pr070063h. [DOI] [PubMed] [Google Scholar]
- 117.Nahon P., Amathieu R., Triba M.N. Identification of serum proton NMR metabolomic fingerprints associated with hepatocellular carcinoma in patients with alcoholic cirrhosis. Clin Cancer Res. December 15, 2012;18(24):6714–6722. doi: 10.1158/1078-0432.CCR-12-1099. [DOI] [PubMed] [Google Scholar]
- 118.Solinas A., Chessa M., Culeddu N. High resolution-magic angle spinning (HR-MAS) NMR-based metabolomic fingerprinting of early and recurrent hepatocellular carcinoma. Metabolomics. 2013;10(4):616–626. [Google Scholar]
- 119.Bathe O.F., Shaykhutdinov R., Kopciuk K. Feasibility of Identifying pancreatic cancer based on serum metabolomics. Cancer Epidemiol Biomark Prev. January 1, 2011;20(1):140–147. doi: 10.1158/1055-9965.EPI-10-0712. [DOI] [PubMed] [Google Scholar]