Abstract
The purpose of this study is to measure the expression of microRNA-4463 and microRNA-6087 between normal persons and patients with hepatocellular carcinoma (HCC), and to clarify the meaning of them in the prognosis evaluation in HCC. Forty-five samples from healthy people and patients, who had been diagnosed with hepatocellular carcinoma before any treatment, were collected to study respectively. Real-time PCR was used to detect the expression of miRNA-4463 and miRNA-6087 in the serum of control group and hepatocellular carcinoma patients. The expression of miR-4463 in the serum of HCC patients was significantly higher than that in control group (P < 0.05), and the expression level was independent of gender, tumor size, cell types, stages, alanine aminotransferase (ALT), aspartate aminotransferase (AST), total bilirubin (TBIL) and HBsAg status (P > 0.05). But there was a significant difference of different level of AFP in HCC (P < 0.05), and the difference between the group of AFP lower than 400 ug/l and the control group is statistically significant (P < 0.05). Besides, the survival time had showed a significant difference at the high and low expression levels (P < 0.05). But the expression level of miRNA-6087 was no difference in HCC and control group. The disorder of miRNA-4463 occurred in HCC, even the AFP level doesn't rises. What's more, patients who get the high level of miRNA-4463 seem to have a shorter survival time. And it contributes great to the prognostic evaluation. This is the first study to illustrate the potential significance of miRNA-4463 in the prognosis in HCC.
Keywords: Hepatocellular carcinoma, malignant tumors, microRNA-4463, microRNA-6087, miR-4463, Prognostic
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors in the worldwide, and lays on the third leading cancer-related deaths,1, 2, 3 coming with the epidemiological characteristics of high degree of malignancy, high metastasis rate, high recurrence rate and low survival rate.4 HCC is a huge threat for human's lives and healthy, and becomes a huge challenge for the maintenance of human public health.5, 6, 7 Hepatocellular carcinoma is the imbalance in the regulation of gene transcription level, which leads to the formation of clonal abnormal proliferation of new organisms. The etiopathology of HCC is complicated such as the activation of many oncogenes, and the inactivation of tumor suppressor genes.8 Because of the lack of effective means of early diagnosis, most patients were diagnosed in the advanced stage with distant metastases or intrahepatic metastasis. In spite of given aggressive treatment, such as liver transplantation, molecular biological treatment, the five-year survival rate is within 30%.9 Because of the poor prognosis, and the short survival time, early detection and diagnosis are of great at significance. And early diagnosis could help to receive timely effective treatment, finally to improve the prognosis and prolong the survival period.
MicroRNA (miRNA) is a single stranded, endogenous non-coding small molecule RNA.10 A number of studies have confirmed that miRNAs may play an important role in the diagnosis and targeted therapy of cancer.11, 12, 13 The abnormal expression of some specific miRNAs had been demonstrated in specimens of liver.14, 15 Studies have shown that many miRNAs were participated in the multiplication and metastasis of HCC, which having important significance for the treatment and the prognosis. This study means to make clear that the relationship of the expression level of miR-4463 with pathology and physiology of HCC. And we had tried to analysis the correlation between the expression of miR-4463 and the clinical characteristics. But there was no related research of the expression of miR-4463 in HCC.
Materials and methods
Case group and control group
Forty-five samples with HCC were collected from First Affiliated Hospital and Nanhua Hospital of University of South China, without any treatment. All patients were diagnosed by two methods of ultrasound, CT and histopathology, which combined with related-history and the level of serum AFP.16 Forty-five normal samples were obtained from the physical examination center in Nanhua Hospital of University of South China. All participators had written informed consent before the study, which was permitted by the Ethics Committee of University of South China. 2 ml peripheral blood was obtained from all the patients and volunteers. Then the upper serum were obtained after centrifugation and stored in −80 °C.
The HCC group includes 12 females and 33 males (age 31–80 years, mean 57.64 ± 11.52 years). And the control group was made up of 18 females and 27 males (aged 28–72 years, mean 45.22 ± 11.28 years).
RNA extraction
200 ul serum and 1000 ul QIAzol Lysis Reagent were mixed in tube. Then 7 ul miRNeasy Serum/Plasma Spike-In Control (Qiagen, Germany) was added and mixed thoroughly. 200 ul chloroform was added to the tube and shook vigorously for 15 s and incubated for 2 min, which followed by centrifugation for 15 min at 12000× g at 4 °C. Then 600 ul of the upper aqueous phase was transferred to a new collection tube, and 900 ul ethanol was put into the tube and mixed thoroughly. Each 700 ul of the mixture was added into an RNeasy MinElute spin column which with a 2 ml collection tube, and centrifuged. Then 700 ul Buffer RWT was put into the RNeasy MinElute spin column, and centrifuged. Then 500 ul Buffer RPE was added into the RNeasy MinElute spin column, and centrifuged. 500 ul of 80% ethanol was put into the RNeasy MinElute spin column, and centrifuged. A new 2 ml collection tube was placed into the RNeasy MinElute spin column, and centrifuged, as to dry the spin column membrane. A new 1.5 ml collection tube was placed into the RNeasy MinElute spin column. Then 14 ul RNase-free water was added directly to the center of the spin column membrane, and centrifuged. The leftover was the total RNA.
cDNA synthesis
The reagents of RNA reverse transcription to cDNA had used miScript Reverse Transcriptase Kit (Qiagen, Germany). We had mixed the 3 ul purified RNA, 4 ul of 5× miScript HISpec Buffer, 2 ul of 10× miScript Nucleics Mix, 9 ul of RNase-free water and 2 ul of miScript Reverse Transcriptase Mix. In brief, the final volume in the tube was 20 ul. Incubated for 60 min at 37 °C, 5 min at 95 °C and 30 min at 4 °C. Finally 200 ul RNase-free water had been added into the tube, and stored the whole at −20 °C.
Real-time PCR
MiScript SYBR Green PCR Kit (Qiagen, Germany) was used for miRNA quantification. Each reaction includes 5 ul of 2× QuantiTect SYBR Green PCR master Mix, 1 ul of 10× miScript Universal Primer, 1 ul of 10× Has_miR-39 miScript Primer Assay or 1 ul of miR-4463 miScript Primer Assay, 2 ul of RNase-free water and 1 ul of Diluted reverse transcription reaction. So the total volume was 10 ul. The 48-well plate had been used. And each sample was provided with multiple holes and repeated three times to make sure the accuracy of the finally result. There were two steps for the reaction, first one was PCR initial activation at 95 °C for 15 min, followed by 40 cycles of 94 °C for 15 s, 55 °C for 30 s, and 70 °C for 30 s.
The cycle threshold (Ct) value means the cycle number at which fluorescence is crossed a threshold. The expression of miRNA is normalized by calculating ΔCT value, which is compared with the reference CT value (ΔCT = CTIncRNA−CTreference). The relative amount of each miRNA-4463 in HCC serum and the normal people serum was calculated with the equation 2−ΔΔCT(ΔΔCT = ΔCTHCC−ΔCTnormal).17, 18, 19
Statistical analysis
SPSS version 22.0 (IBM, Chicago, IL) and GraphPad Prism 6 (GraphPad Software, USA) were used for statistical analyses. Qualitative variables were expressed as mean ± standard deviation. Therefore, the significant differences between the different research groups were determined by t-test and one-way analysis of variance (ANOVA) test. The difference was statistically significant at P value < 0.05.
Results
The expression level of miRNA-4463 was up-regulated in HCC
In order to clarify the difference of the expression level of miRNA-4463 and miRNA-6087 in HCC and control group, we had collected the HCC and healthy people blood, and detected the expression level of miRNA-4463 of each serum by using the RT-PCR. The results illustrated that the expression level of miR-4463 in HCC was indeed higher than the control group, with the mean (7.40 ± 1.33) in HCC and the mean (3.65 ± 0.79) in non-HCC (Fig. 1A). And the difference was statistically significant (P < 0.05). At the same time, we had made analysis of miRNA-6087 in the same way, and the results showed the there was no difference in the HCC and control group (Fig. 1B).
Fig. 1.
The expression levels of microRNA-4463 in HCC and in healthy people. A shows that the expression level of miRNA-4463 was significant different (*P < 0.05) between HCC and normal group. Besides the expression of miRNA-6087 in HCC and control group was no different.
There had no correlation between the expression level of miR-4463 and the clinical features in HCC
The results showed that the expression level of miRNA-4463 was up-regulated in HCC. We next wonder whether the clinical features had the correlation with the expression level of miRNA-4463. So we make comparisons of the expression level of miRNA-4463 in HCC, which we set groups at gender, cell types, staging of the disease, the size of the tumor, ALT, AST, TBIL and HBsAg status (Table 1). The final results indicated that there were no significant correlation of miRNA-4463 with clinical characteristics such as gender, cell types, the stages of the disease, the size of the tumor, ALT, AST, TBIL and HBV status (P > 0.05).
Table 1.
Expression and clinical correlation analyses in HCC.
| n | % | Mean ± SD | P-Value | |
|---|---|---|---|---|
| Sex | ||||
| Female | 12 | 27 | 7.82 ± 1.34 | 0.22 |
| Male | 33 | 73 | 7.25 ± 1.31 | |
| Size of tumor | ||||
| ≤5 cm | 15 | 33 | 7.34 ± 1.34 | 0.82 |
| >5 cm | 30 | 67 | 7.44 ± 1.34 | |
| Staging* | ||||
| I, II | 9 | 20 | 7.54 ± 1.15 | 0.71 |
| III, IV | 36 | 80 | 7.37 ± 1.38 | |
| Cell Types | ||||
| Hepatocellular | 30 | 67 | 7.05 ± 1.02 | 0.29 |
| Cholangiocarcinoma | 8 | 18 | 8.05 ± 1.92 | |
| Mixed | 7 | 15 | 7.32 ± 1.18 | |
| ALT | ||||
| ≥65IU/L | 19 | 42 | 7.41 ± 1.22 | 0.53 |
| <65IU/L | 26 | 58 | 7.40 ± 1.43 | |
| AST | ||||
| ≥65IU/L | 14 | 31 | 7.58 ± 1.15 | 0.97 |
| <65IU/L | 31 | 69 | 7.33 ± 1.41 | |
| TBIL | ||||
| ≥25 umol/L | 16 | 36 | 7.45 ± 1.29 | 0.87 |
| <25 umol/L | 29 | 64 | 7.38 ± 1.36 | |
| HBsAg status | ||||
| HBsAg+ | 19 | 42 | 7.17 ± 1.12 | 0.30 |
| HBsAg− | 26 | 58 | 7.57 ± 1.46 | |
However, the expression level of miRNA-4463 was related with AFP levels. Since the level of AFP helps to diagnose HCC, we tried to found out whether there was a difference in the group of AFP higher than 400 μg/l and in the group of AFP lower than 400 μg/l (Table 2). It showed that the expression level of miRNA-4463 had a significant difference (P < 0.05). But the expression level of miRNA-4463 with AFP level is negative correlation. The group of AFP lower than 400 μg/l, has a higher expression level of miRNA-4463. Furthermore, we had made a comparison between the group of AFP <400 μg/l and the control group for the purpose of to diagnose the patients with HCC in AFP level lower than 400 μg/l (Table 3). The data indicates that the expression level of miRNA-4463 is higher in the group of AFP level lower than 400 μg/l compare to the control group. And the statistical significance is calculated (P < 0.05). For the low sensitivity of AFP to diagnose HCC, detecting the expression level of miRNA-4463 will help a lot to make diagnosis of HCC, even those patients have the AFP level lower than 400 μg/l.
Table 2.
Expression and the analysis of the correlation of AFP in HCC.
| AFP | n | % | Mean ± SD | P-Value |
|---|---|---|---|---|
| >400 | 23 | 51 | 6.53 ± 0.95 | P < 0.05 |
| <400 | 22 | 49 | 7.84 ± 1.28 |
Table 3.
Analyse between the group of AFP<400 ug/L and the control group.
| n | Range | Mean ± SD | P-Value | |
|---|---|---|---|---|
| Normal people | 45 | 2.21–5.61 | 3.65 ± 0.79 | P < 0.05 |
| AFP<400 ug/L | 30 | 5.31–9.98 | 7.84 ± 1.28 |
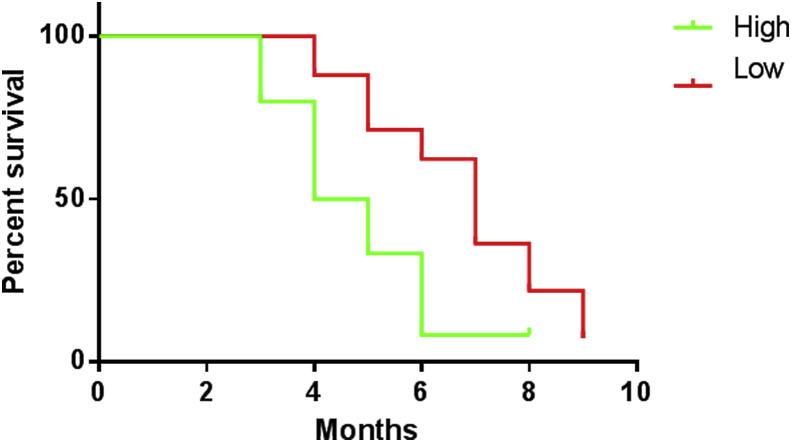
We used the median expression level of all 45 patients as the cut-off point.19 And patients were divided into two groups with low or high expression level of miRNA-4463 (Fig. 2). Kaplan–Meier analysis showed that patients with high level of miRNA-4463 had a shorter survival time (P < 0.05). The mean survive time of high group is (4.55 ± 0.64 months), which is shorter than the other group (mean 6.28 ± 1.56 months).
Fig. 2.
The survival times of HCC patients. Red line means the patients has a longer survival time in the lower expression level of miRNA-4463 (<7.40), with the mean time (6.28 ± 1.56 months). The green line shows that patients has a shorter survival time in the group, of which the miRNA-4463 (>7.40) expression level is higher, with the mean time (4.55 ± 0.64 months). And the difference is statistically significant (P < 0.05).
Serum miRNA-4463 could be a new marker for diagnose and prognostic evaluation in HCC
In this study, the results suggested that the serum miRNA-4463 in HCC was much higher than the control group certainly. And the disorder of miRNA-4463 happened at the every stages in HCC, including the early-stage. So miRNA-4463 can be detected as a method for early diagnostic of HCC, and it could be a new marker in the diagnosis of HCC. We found that the 95% confidence interval (95%CI) of the HCC is 7.01–7.79. It illustrates that people who gets the expression level of miR-4463 in this area can be highly thought to diagnose HCC. For the patients who have the AFP level lower than 400 ug/l, the expression level of miRNA-4463 is high mostly, and will help to diagnose HCC.
The results show that patients with high expression level of miRNA-4463 seem to live a shorter time. And the patients are likely to live a longer time with low miRNA-4463 expression. So the expression level of miRNA-4463 may evaluate prognosis.
Discussion
Hepatocellular carcinoma is always associated with a poor prognosis. Owing to the hiding of clinical symptoms, patients may ignore the ultrasound, CT and other imaging examinations, which may cause a delay in diagnosis, finally to the advanced-stage of HCC.20, 21, 22 At present, the diagnosis of HCC is mainly carried out by serum AFP and imaging examination, but the specificity and sensitivity of these diagnostic tests remain deficient for detecting HCC at early stage.23, 24 At the same time, due to the appearance of chemo-resistance and the toxic side effects, intra-hepatic and distant metastases lead to a high cancer recurrence rate and short survival time even with the advanced therapeutic strategies.25, 26, 27 Invasion and metastasis are still problems which need to be solved urgently. If early diagnosis possible, especially when the only nodule or the nodules' size smaller than 2 cm, then the treatment effect may be much better, and the 5-year survival rate will be greatly extended. So we need an effective method to improve the early diagnosis and the results of treatment of HCC. Alpha-fetoprotein (AFP) acts as the most common bio-marker of HCC, but the accuracy of the diagnosis was limited.28, 29 The up-regulating of AFP is inconspicuous at the early stage of HCC, even without any rise. Besides, the AFP also increased in benign liver diseases such as viral hepatitis.30, 31 Detecting AFP has variable effectiveness for diagnosis HCC.14, 32, 33, 34
However, for miRNAs, the expression in body tissues or fluids is earlier and more stable.35, 36, 37 Studies showed that miRNAs are to be resistant to extreme pH, high temperature, and even repeated freezing–thawing cycles. What's more, the expression of miRNAs can be found in HCC tissues, cells and serum.15, 38, 39, 40 A growing number of studies have indicated that miRNAs had played an important role in many biological processes, including the occurrence, development, proliferation and metastasis of many diseases.41, 42, 43, 44, 45, 46 Studies showed that miR-122a is a rich liver-specific miRNA, which down-regulated in about 70% HCC.47, 48 Chen WX et al. found the expression of miR-630 had a relationship of metastasis and TNM. It showed that patients with the up-regulated expression of miR-630 met the longer survival time and lower recurrence rate than those who with the down-regulated expression. The research indicated that the miR-630 could inhibit the metastasis of HCC, which can be used as marker of prognosis.49 Another study suggested that the ectopic over-expression of miR-379-5P could promote the invasion and metastasis of HCC in vivo or in vitro, through the miR-379-5P/FAK/AKT signaling pathway as well as the EMT process.50 Tomimaru et al. detected the expression level of miR-21 was significantly higher in HCC than in no-HCC.51 At the same time, miR-21 had been confirmed to be regarded as a marker for early diagnosis of liver cancer.52 Although there are a considerable number of studies on miRNAs in HCC, and it also shows the relationship between the diagnosis, metastasis and prognosis of HCC, but there is no report about the expression of miR-4463.
MiRNA-4463 and miRNA-6087 were chose by high-throughput screened, and we made the gene chip to make sure the difference of HCC and normal blood, which the expression level has significant difference between the hepatocellular carcinoma patients and normal human (data no shown). And the blood of HCC patients without any treatments and the blood of control group were collected for this study. Finally we detected the expression level of miR-4463 and miR-6087 in HCC patients and control group by qRT-PCR. And to ensure the accuracy and credibility of the experimental results, we had repeated the experiment three times. We had compared the expression level of serum in HCC and the normal serum. We had found that the miR-4463 was also expressed in normal serum, but is extremely lower than the HCC. So we could infer that the miR-4463 in serum comes from tumor cells. And we will do further explore in future study. Compared with the control group, the expression of miR-4463 was significantly higher in HCC, and the difference was statistically significant (P < 0.05), but there was no difference in sex, pathological type, the stages of the disease, the size of the tumor, ALT, AST, TBIL and HBsAg status (P > 0.05). So we have drawn a conclusion that miR-4463 is up-regulated in HCC no matter of the stages, sexes, cell types, size of tumor, ALT, AST, TBIL and HBsAg status. The disorder of expression of miRNA-4463 can be detected in HCC, if only the patients were diagnosed as HCC. In a word, the miRNA-4463 is highly thought to be a new marker to diagnose HCC including in the early-stage. Besides, there was a significant difference of different level of AFP in HCC (P < 0.05). In this study, we had the higher expression level of miRNA-4463 in the group of lower-AFP (P < 0.05). For the low specificity and sensitivity of AFP, the detection of miRNA-4463 will contribute more to the diagnosis of HCC. In this study, people who have the expression level between 7.01 and 7.79 will highly be diagnosed of HCC. So serum miRNA-4463 may serve as a new potential marker in the diagnosis of HCC. But it still need more study to make sure. Besides, we had made the follow-up investigation to the all patients. The results show that the patients who had a higher level of miRNA-4463 were likely to have a shorter survival time. So serum miRNA-4463 also may be a new marker in the prognostic evaluation. But due to the limit of test time and the sample amount, we couldn't know whether the expression level of miR-4463 after any treatment reduces or rises. And we will do more research in future. We need to do more research about how to set the diagnosis standard to a certain value, and evaluate the survival time before and after treatment. Thereby more eloquent testimony is needed to their accuracy of miRNA-4463, to help to make diagnose of HCC, and even to help to predict the prognosis of HCC. There only two articles had done study about miR-4463 in the research of cancer field. Ding had found that the expression of miR-4463 were significant up-regulated in polycystic ovary syndrome, but the biological function remains unclear.53 But the expression of miR-4463 was down-regulated in arteriosclerosis obliterans in He's research. And they had found that the miR-4463 is participant in regulating the cell migration and polarity via targeting AMOT gene.54
With the widely application of gene chip technology and RT-PCR reaction technique, the importance of microRNAs in disease may gradually clear, and the detection of the expression level of microRNAs for diagnosis may be gradually applied in clinic, finally it may become a new marker for diagnosis of tumors. In our study, the expression level of miR-4463 in patients with HCC is significantly higher than non-HCC, and the patients with higher level of miRNA-4463 have a shorter survival time. It indicated that the miRNA-4463 plays an important role in the prognosis of HCC. Serum miRNA-4463 provided new ideas to the diagnosis of HCC. In the future, we may evaluate the therapeutic effect by detecting the expression of miRNA-4463 before and after treatment. And the detecting of miRNA-4463 may bring a new method for the prognosis evaluation, and lay the foundation for the new molecular targeted therapy.
Conflicts of interest
None.
Footnotes
Grantsponsor: National Nature Science Foundation of China, Grant number: 81272960; Key Research Program from Science and Technology Department of Hunan Province China, Grant number: 2013WK2010 and 2014SK2015; Key Research Program from Ministry of human Resources and Social Security of the People's Republic of China, Grant number: (2016)176; The fund of Tianqing liver disease research, Grant number: (TQGB20140155).
Peer review under responsibility of Chongqing Medical University.
Contributor Information
Meiling Wen, Email: 1115910801@qq.com.
Jianhong Zuo, Email: 632138414@qq.com.
References
- 1.Gokuladhas K., Jayakumar S., Rajan B. Exploring the potential role of chemopreventive agent, hesperetin conjugated pegylated gold nanoparticles in diethylnitrosamine-induced hepatocellular carcinoma in male wistar albino rats. Indian J Clin Biochem. 2016;31:171–184. doi: 10.1007/s12291-015-0520-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afonso M.B., Rodrigues P.M., Simão A.L. Circulating microRNAs as potential biomarkers in non-alcoholic fatty liver disease and hepatocellular carcinoma. J Clin Med. 2016;5:1–20. doi: 10.3390/jcm5030030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu J.X., Chen Q., Yu Y.Q. Upregulation of colonic and hepatic tumor overexpressed gene is significantly associated with the unfavorable prognosis marker of human Hepatocellular Carcinoma. Am J Cancer Res. 2016;6:690–700. [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y.Q. The expression and potential clinical value of microRNA in hepatocellular carcinoma. J Clin Exp Med. 2015;14:27–29. [Google Scholar]
- 5.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;11:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 6.Siegel R.L., Sahar L., Portier K.M. Cancer death rates in US congressional districts. CA Cancer J Clin. 2015;65:339–444. doi: 10.3322/caac.21292. [DOI] [PubMed] [Google Scholar]
- 7.He S., Hu X.W., Wang D. Accuracy of microRNAs for the diagnosis of hepatocellular carcinoma: a systematic review and meta-analysis. Clin Res Hepatol Gastroenterol. 2016;16:30026–30032. doi: 10.1016/j.clinre.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Skipper M. Cancer genomics: a panoramic view of cancer. Nat Rev Genet. 2013;11:750–759. doi: 10.1038/nrg3602. [DOI] [PubMed] [Google Scholar]
- 9.Ghosh A., Datta S., Dasgupta D. Hepatic miR-126 is a potential plasma biomarker for detection of hepatitis B virus infected Hepatocellular Carcinoma. Int J Cancer. 2016;138:2732–2744. doi: 10.1002/ijc.29999. [DOI] [PubMed] [Google Scholar]
- 10.Garzon R., Calin G.A., Croce C.M. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 11.Saus E., Brunet-Vega A., Iraola-Guzman S. Long non-coding RNAs as potential novel prognostic biomarkers in colorectal cancer. Front Genet. 2016;54:1–14. doi: 10.3389/fgene.2016.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park H.K., Jo W., Choi H.J. Time-course changes in the expression of miR-122,-155, and -21 as markers of liver cell damage, inflammation, and regeneration in acetaminophen-induced liver injury in rats. J Vet Sci. 2016;17:45–51. doi: 10.4142/jvs.2016.17.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okamoto K., Koda M., Okamoto T. A series of microRNA in the chromosome 14q32.2 maternally impritnted region related to progression of non-alcoholic fatty liver disease in a mouse model. PLoS One. 2016;11:1–19. doi: 10.1371/journal.pone.0154676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He S., Zhang D.C., Wei C. MicroRNAs as biomarkers for hepatocellular carcinoma diagnosis and prognosis. Clin Res Hepatol Gastroenterol. 2015;39:426–434. doi: 10.1016/j.clinre.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 15.Morishita A., Masaki T. MicroRNAs in hepatocellular carcinoma. Hepatol Res. 2015;45:128–141. doi: 10.1111/hepr.12386. [DOI] [PubMed] [Google Scholar]
- 16.Zhu R.X., Seto W.K., Lai C.L. Epidemiology of hepatocellular carcinoma in the asia-pacific region. Gut Liver. 2016;10:331–339. doi: 10.5009/gnl15257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farinati F., Vitale A., Spolverato G. Development and validation of a new prognostic system for patients with hepatocellular carcinoma. PLos Med. 2016;13:1–18. doi: 10.1371/journal.pmed.1002006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qi F., Zhang Y., Yang Y. Low serum miR-19a expression as a novel poor prognostic indicator in multiple myeloma. Int J Cancer. 2015;136:1835–1844. doi: 10.1002/ijc.29199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zuo Jianhong, Wen Meiling, Lei Mingsheng. MiR-210 links hypoxia with cell proliferation regulation in human laryngocarcinoma cancer. J Cell Biochem. 2015;116:1039–1049. doi: 10.1002/jcb.25059. [DOI] [PubMed] [Google Scholar]
- 20.Zhang L.P., Xu L.T., Wang P. Serum miR-128-2 serves as a prognostic marker for patients with hepatocellular carcinoma. PLos One. 2015;10:1–12. doi: 10.1371/journal.pone.0117274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takahashi K., Yan I.K., Kogure T. Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human Hepatocellular Cancer. FEBS Open Bio. 2014;21:458–467. doi: 10.1016/j.fob.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blachier M., Leleu H., Peck-Radosavljevic M. The burden of liver disease in Europe: a review of available epidemiological data. J Hepatol. 2013;58:593–608. doi: 10.1016/j.jhep.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 23.Jelic S., Sotiropoulos G.C., GroupEGW Hepatocellular carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. AnnOncol. 2010;21:59–64. doi: 10.1093/annonc/mdq166. [DOI] [PubMed] [Google Scholar]
- 24.Liu J., Wei X., Wu Y. Giganteaside D induces ROS-mediated apoptosis in human Hepatocellular Carcinoma cells through the MAPK pathway. Cell Oncol. 2016;6:1–10. doi: 10.1007/s13402-016-0273-9. [DOI] [PubMed] [Google Scholar]
- 25.Yuen M.F., Cheng C.C., Lauder I.J. Early detection of hepatocellular carcinoma increases the chance of treatment: Hong Kong experience. Hepatology. 2000;31:330–335. doi: 10.1002/hep.510310211. [DOI] [PubMed] [Google Scholar]
- 26.Fukuda S., Itamoto T., Nakahara H. Clinicopathologic features and prognostic factors of resected solitary small-sized hepatocellular carcinoma. Hepatol Gastroenterol. 2005;52:1163–1167. [PubMed] [Google Scholar]
- 27.Bruix J., Gores G.J., Mazzaferro V. Hepatocellular carcinoma: clinical frontiers and perspectives. Gut. 2014;63:844–855. doi: 10.1136/gutjnl-2013-306627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han L.L., Lv Y., Guo H. Implications of biomarkers in human hepatocellular carcinoma pathogenesis and therapy. World J Gastroenterol. 2014;20:10249–10261. doi: 10.3748/wjg.v20.i30.10249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu L., Zhao Y., Jia J. The prognostic value of alpha-fetoprotein response for advanced-stage hepatocellular carcinoma treated with sorafenib combined with transarterial chemoembolization. Sci Rep. 2016;6:1–9. doi: 10.1038/srep19851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rapaccini G.L., Pompili M., Caturelli E. Hepatocellular carcinomas <2 cm in diameter complicating cirrhosis: ultrasound and clinical features in 153 consecutive patients. Liver Int. 2004;24:124–130. doi: 10.1111/j.1478-3231.2004.0903.x. [DOI] [PubMed] [Google Scholar]
- 31.Debruyne E.N., Delanghe J.R. Diagnosing and monitoring hepatocellular carcinoma with alpha-fetoprotein: new aspects and applications. Clin Chim Acta. 2008;395:19–26. doi: 10.1016/j.cca.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 32.Yao L. The expression and significance of microRNA in primary hepatocellular carcinoma. Pract J Cancer. 2016;31:165–168. [Google Scholar]
- 33.Songlin A., Weiqi R., Liming W. Analysis of clinicopathological features and prognosis between alpha-fetoprotein negative and positive hepatocellular carcinoma patients after R0 radical hepatectomy. Chin J Oncol. 2015;37:308–311. [PubMed] [Google Scholar]
- 34.Seleem M.I., Abdelraouf A., Gerges S.S. Laparoscopic assisted per-frequency ablation (Laprfa)as a new modality for treatment of HCC in cirrhotic liver. J Egypt Soc Parasitol. 2015;45:451–456. doi: 10.12816/0017599. [DOI] [PubMed] [Google Scholar]
- 35.Arroyo J.D., Chevillet J.R., Kroh E.M. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U. S. A. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vickers K.C., Palmisano B.T., Shoucri B.M. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang K., Zhang S., Weber J. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38:7248–7259. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Hees S., Michielsen P., Vanwolleghem T. Circulating predictive and diagnostic biomarkers for hepatitis B virus-associated hepatocellular carcinoma. World J Gastroenterol. 2016;37:8271–8282. doi: 10.3748/wjg.v22.i37.8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilad S., Meiri E., Yogev Y. Serum microRNAs are promising novel biomarkers. PLoS One. 2008;3:3148.1–3148.7. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kasinski A., Slack F. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dykxhoor D. MicroRNAs and metastasis: little RNAs go a long way. Cancer Res. 2010;70:6401–6406. doi: 10.1158/0008-5472.CAN-10-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kong Y., Ferland-McCollough D., Jackson T. microRNAs in cancer management. Lancet Oncol. 2012;6:249–258. doi: 10.1016/S1470-2045(12)70073-6. [DOI] [PubMed] [Google Scholar]
- 43.Croce C. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hua H.W., Jiang F., Huang Q. MicroRNA-153 promotes Wnt/beta-catenin activation in hepatocellular carcinoma through suppression of WWOX. Oncotarget. 2015;6:3840–3847. doi: 10.18632/oncotarget.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan S.Y., Chen M.M., Li G.M. MiR-32 induces cell proliferation, migration, and invasion in hepatocellular carcinoma by targeting PTEN. Tumour Biol. 2015;36:4747–4755. doi: 10.1007/s13277-015-3124-9. [DOI] [PubMed] [Google Scholar]
- 46.Schutte K., Schulz C., Link A. Current biomarkers for hepatocellular carcinoma: surveillance, diagnosis and prediction of prognosis. World J Hepatol. 2015;7:139–149. doi: 10.4254/wjh.v7.i2.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kutay H., Bai S., Datta J. Down-regulation of miR-122 in the rodent and human Hepatocellular Carcinoma. J Cell Biochem. 2006;99:671–678. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Diao S., Zhang J.F., Wang H. Protemic identification of microRNA-122a target proteins in Hepatocellular Carcinoma. Proteomics. 2010;20:3723–3731. doi: 10.1002/pmic.201000050. [DOI] [PubMed] [Google Scholar]
- 49.Chen W.X., Zhang Z.G., Ding Z.Y. MicroRNA-630 suppresses tumor metastasis through the TGF-β-miR-630-Slug signaling pathway and correlates inversely with poor prognosis in Hepatocellular Carcinoma. Oncotarget. 2016;25:1–13. doi: 10.18632/oncotarget.8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen J.S., Li H.S., Huang J.Q. MicroRNA-379-5P inhibits tumor invasion and metastasis by targeting FAK/AKT signaling in Hepatocellular Carcinoma. Cancer Lett. 2016;375:73–83. doi: 10.1016/j.canlet.2016.02.043. [DOI] [PubMed] [Google Scholar]
- 51.Tomimaru Y., Eguchi H., Nagano H. Circulating microRNA-21 as a novel biomarker for Hepatocellular Carcinoma. J Hepatal. 2012;56:167–175. doi: 10.1016/j.jhep.2011.04.026. [DOI] [PubMed] [Google Scholar]
- 52.Liao Q., Han P., Huang Y. Potential role of circulating microRNA-21 for hepatocellular carcinoma diagnosis: A meta-analysis. PLoS One. 2015;6:1–11. doi: 10.1371/journal.pone.0130677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ding C.F., Chen W.Q., Zhu Y.T. Circulating microRNAs in patients with polycystic ovary syndrome. Hum Fertil (Camb) 2015;18:22–29. doi: 10.3109/14647273.2014.956811. [DOI] [PubMed] [Google Scholar]
- 54.He X.M., Zheng Y.Q., Liu S.Z. Altered plasma MicroRNAs as novel biomarkers for arteriosclerosis obliterans. J Atheroscler Thromb. 2016;23:196–206. doi: 10.5551/jat.30775. [DOI] [PubMed] [Google Scholar]