Abstract
Atrial fibrillation (AF) affects over 33 million individuals worldwide1 and has a complex heritability.2 We conducted the largest meta-analysis of genome-wide association studies for AF to date, consisting of over half a million individuals including 65,446 with AF. In total, we identified 97 loci significantly associated with AF including 67 of which were novel in a combined-ancestry analysis, and 3 in a European specific analysis. We sought to identify AF-associated genes at the GWAS loci by performing RNA-sequencing and expression quantitative trait loci (eQTL) analyses in 101 left atrial samples, the most relevant tissue for AF. We also performed transcriptome-wide analyses that identified 57 AF-associated genes, 42 of which overlap with GWAS loci. The identified loci implicate genes enriched within cardiac developmental, electrophysiological, contractile and structural pathways. These results extend our understanding of the biological pathways underlying AF and may facilitate the development of therapeutics for AF.
Atrial fibrillation (AF) is the most common heart rhythm disorder, and is a leading cause of heart failure and stroke.3 Prior genome-wide association studies (GWAS) have identified at least 30 loci associated with AF.4–9 We conducted a large-scale analysis with over half a million participants, including 65,446 with AF, from more than 50 studies. Our AF sample was composed of 84.2% European, 12.5% Japanese, 2% African American, and 1.3% Brazilian and Hispanic populations (Supplementary Table 1). We used the Haplotype Reference Consortium (HRC) reference panel to impute variants from SNP array data for 75% of the samples (Figure 1). In the remainder, we included HRC overlapping variants from 1000 Genomes imputed data, or from a combined reference panel. We analyzed 8,328,530 common variants (minor allele frequency (MAF) >5%), 2,884,670 low frequency variants (1%> MAF ≥5%), and 936,779 rare variants (MAF ≤1%).
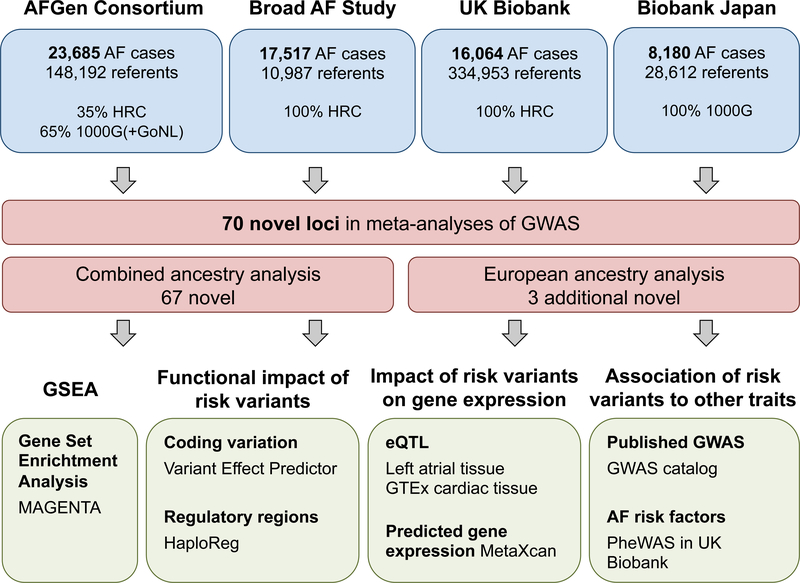
Figure 1. Study and analysis flowchart.
Top, overview of the participating studies, number of AF cases and referents, and the percent of samples imputed with each reference panel. Middle, summary of the primary analyses and the newly discovered loci for AF. Bottom, overview of the secondary analyses to evaluate AF risk variants and loci.
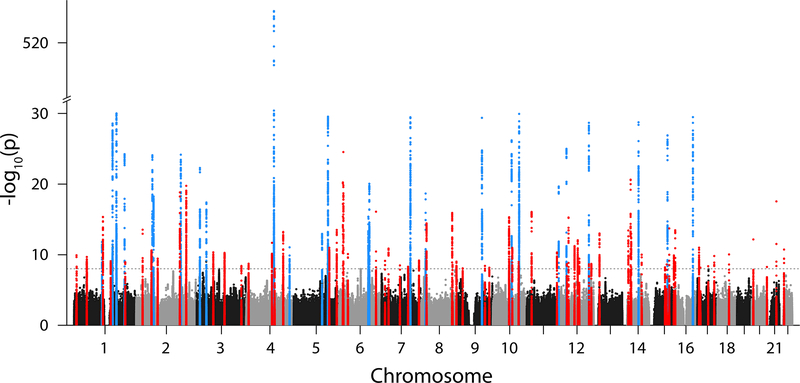
The combined-ancestry meta-analysis revealed 94 AF-associated loci, 67 of which were novel at genome-wide significance (P-value (P) < 1×10−8). This conservative threshold accounts for testing independent variants with MAF ≥0.1% using a Bonferroni correction, while use of a more commonly utilized threshold of 5×10−8 resulted in the identification of an additional 10 loci (Supplementary Table 2). The majority of sentinel variants (n=92) were common (MAF >5%), with relative risks ranging from 1.04 to 1.55. Two low frequency sentinel variants were identified within the genes C1orf185 and UBE4B (Figure 2, Table 1, Supplementary Table 3, Supplementary Figure 1).
Figure 2. Manhattan plot of combined-ancestry meta-analysis.
The plot shows 67 novel (red) and 27 known (blue) genetic loci associated with AF at a significance level of P < 1×10−8 (dashed line), for the combined-ancestry meta-analysis (n=588,190). The significance level accounts for multiple testing of independent variants with MAF ≥0.1% using a Bonferroni correction. P-values (two-sided) were derived from a meta-analysis using a fixed effects model with an inverse-variance weighted approach. The y-axis has a break between –log10(P) of 30 and 510 to emphasize the novel loci
Table 1.
Novel loci in combined-ancestry meta-analysis
| Rsid | Ch r |
hg19 | Risk/Re f Allele |
RA F [%] |
RR | 95% Cl | PMETA | Nearest Gene(s)* | Func | imp Qua I |
I2HET | PHET |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs18758553 0 | 1 | 10167425 | A/G | 0.5 | 1.55 | 1.36–1.77 | 1.18×10−10 | UBE4B | missense | 0.81 | 0.0 | 1.000 |
| rs880315 | 1 | 10796866 | C/T | 37.4 | 1.04 | 1.03–1.06 | 5.04×10−09 | CASZ1 | intronic | 0.97 | 40.7 | 0.150 |
| rs14651872 6 | 1 | 51535039 | A/G | 2.6 | 1.18 | 1.12–1.24 | 2.05×10−10 | C1orf185 | intronic | 0.96 | 0.0 | 1.000 |
| rs4484922 | 1 | 116310818 | G/C | 68.3 | 1.07 | 1.05–1.08 | 4.57×10−16 | CASQ2 | intronic | 0.98 | 0.0 | 0.689 |
| rs79187193 | 1 | 147255831 | G/A | 94.8 | 1.12 | 1.08–1.16 | 8.07×10−10 | GJA5 | upstream | 0.97 | 39.8 | 0.190 |
| rs4951261 | 1 | 205717823 | C/A | 38.2 | 1.05 | 1.03–1.06 | 1.17×10−09 | NUCKS1 | intronic | 0.99 | 0.0 | 0.788 |
| rs6546620 | 2 | 26159940 | C/T | 75.3 | 1.07 | 1.05–1.09 | 2.96×10−14 | KIF3C | intronic | 0.95 | 33.0 | 0.201 |
| rs6742276 | 2 | 61768745 | A/G | 61.2 | 1.05 | 1.03–1.06 | 2.42×10−11 | XP01 | upstream | 0.99 | 0.0 | 0.731 |
| rs72926475 | 2 | 86594487 | G/A | 87.0 | 1.07 | 1.05–1.10 | 3.49×10−10 | REEP1,KDM3A | intergenic | 0.97 | 38.7 | 0.180 |
| rs56181519 | 2 | 175555714 | C/T | 74.0 | 1.08 | 1.06–1.10 | 1.52×10−19 | WIPF1 ,CHRNA1 | intergenic | 0.94 | 0.0 | 0.519 |
| rs295114 | 2 | 201195602 | C/T | 59.7 | 1.07 | 1.05–1.09 | 1.76×10−20 | SPATS2L | intronic | 1.00 | 21.9 | 0.275 |
| rs2306272 | 3 | 66434643 | C/T | 31.8 | 1.05 | 1.04–1.07 | 4.54×10−11 | LRIG1 | missense | 0.99 | 30.6 | 0.218 |
| rs17490701 | 3 | 111587879 | G/A | 85.7 | 1.07 | 1.05–1.10 | 5.43×10−11 | PHLDB2 | intronic | 0.97 | 46.8 | 0.111 |
| rs4855075 | 3 | 179170494 | T/C | 14.3 | 1.06 | 1.04–1.08 | 4.00×10−09 | GNB4 | upstream | 0.95 | 10.1 | 0.348 |
| rs3822259 | 4 | 10118745 | T/G | 67.9 | 1.05 | 1.03–1.06 | 1.93×10−09 | WDR1 | upstream | 0.96 | 0.0 | 0.922 |
| rs3960788 | 4 | 103915618 | C/T | 42.4 | 1.05 | 1.04–1.07 | 2.09×10−12 | SLC9B1 | intronic | 0.98 | 35.7 | 0.183 |
| rs55754224 | 4 | 114428714 | T/C | 25.0 | 1.05 | 1.03–1.07 | 9.25×10−09 | CAMK2D | intronic | 0.99 | 0.0 | 0.511 |
| rs10213171 | 4 | 148937537 | G/C | 8.2 | 1.11 | 1.08–1.14 | 6.09×10−14 | ARHGAP10 | intronic | 0.96 | 0.0 | 0.584 |
| rs174048 | 5 | 142650404 | C/T | 15.7 | 1.07 | 1.05–1.09 | 1.05×10−11 | ARHGAP26,NR3C1 | intergenic | 0.99 | 0.0 | 0.852 |
| rs6882776 | 5 | 172664163 | G/A | 67.2 | 1.06 | 1.05–1.08 | 3.18×10−14 | NKX2–5 | upstream | 0.95 | 0.0 | 0.858 |
| rs73366713 | 6 | 16415751 | G/A | 86.2 | 1.11 | 1.09–1.14 | 5.80×10−21 | ATXN1 | intronic | 0.94 | 0.0 | 0.879 |
| rs34969716 | 6 | 18210109 | A/G | 31.1 | 1.09 | 1.07–1.11 | 2.91×10−25 | KDM1B | intronic | 0.80 | 19.5 | 0.290 |
| rs3176326 | 6 | 36647289 | G/A | 80.4 | 1.06 | 1.04–1.08 | 7.95×10−11 | CDKN1A | intronic | 0.95 | 0.0 | 0.450 |
| rs11798485 3 | 6 | 149399100 | T/G | 8.9 | 1.12 | 1.09–1.15 | 8.38×10−17 | UST | downstream | 0.83 | 56.5 | 0.100 |
| rs55734480 | 7 | 14372009 | A/G | 26.6 | 1.05 | 1.03–1.07 | 7.34×10−10 | DGKB | intronic | 0.94 | 0.0 | 0.441 |
| rs6462078 | 7 | 28413187 | A/C | 74.7 | 1.06 | 1.04–1.08 | 1.35×10−11 | CREB5 | intronic | 0.98 | 22.2 | 0.278 |
| rs74910854 | 7 | 74110705 | G/A | 6.9 | 1.10 | 1.07–1.13 | 3.36×10−09 | GTF2I | intronic | 0.74 | 24.4 | 0.265 |
| rs62483627 | 7 | 106856002 | A/G | 23.5 | 1.05 | 1.03–1.07 | 5.17×10−09 | COG5 | intronic | 0.98 | 15.1 | 0.318 |
| rs7789146 | 7 | 150661409 | G/A | 80.3 | 1.06 | 1.04–1.08 | 6.51×10−10 | KCNH2 | intronic | 0.96 | 66.0 | 0.019 |
| rs7846485 | 8 | 21803735 | C/A | 86.8 | 1.09 | 1.07–1.12 | 3.71×10−15 | XP07 | intronic | 0.99 | 0.0 | 0.676 |
| rs62521286 | 8 | 124551975 | G/A | 6.7 | 1.13 | 1.10–1.16 | 1.24×10−16 | FBX032 | intronic | 0.96 | 0.0 | 0.678 |
| rs35006907 | 8 | 125859817 | A/C | 32.9 | 1.05 | 1.03–1.06 | 2.76×10−09 | MTSS1, LINC0096 4 | regulatory reg. | 0.97 | 0.0 | 0.542 |
| rs6993266 | 8 | 141762659 | A/G | 53.8 | 1.05 | 1.03–1.06 | 9.73×10−10 | PTK2 | intronic | 0.99 | 5.7 | 0.374 |
| rs4977397 | 9 | 20235004 | A/G | 57.0 | 1.04 | 1.03–1.06 | 8.60×10−09 | SLC24A2,MLLT3 | intergenic | 0.95 | 38.3 | 0.166 |
| rs4743034 | 9 | 109632353 | A/G | 23.4 | 1.05 | 1.03–1.07 | 3.98×10−09 | ZNF462 | intronic | 1.00 | 0.0 | 0.963 |
| rs10760361 | 9 | 127178266 | G/T | 64.7 | 1.04 | 1.03–1.06 | 7.03×10−09 | PSMB7 | upstream | 0.97 | 0.0 | 0.680 |
| rs7919685 | 10 | 65315800 | G/T | 53.3 | 1.06 | 1.04–1.07 | 5.00×10−16 | REEP3 | intronic | 1.00 | 49.2 | 0.097 |
| rs11001667 | 10 | 77935345 | G/A | 22.2 | 1.06 | 1.05–1.08 | 1.06×10−11 | C10orf11 | intronic | 0.98 | 26.8 | 0.243 |
| rs1044258 | 10 | 103605714 | T/C | 66.2 | 1.05 | 1.03–1.06 | 1.07×10−09 | C10orf76 | 3’ UTR | 0.98 | 14.0 | 0.325 |
| rs1822273 | 11 | 20010513 | G/A | 27.1 | 1.07 | 1.05–1.09 | 8.99×10−17 | NAV2 | intronic | 0.98 | 0.0 | 0.764 |
| rs949078 | 11 | 121629007 | C/T | 27.1 | 1.05 | 1.04–1.07 | 4.77×10−11 | sorli,mirioohG | intergenic | 0.97 | 0.0 | 0.600 |
| rs11381953 7 | 12 | 26348429 | C/G | 74.3 | 1.05 | 1.03–1.07 | 2.23×10−09 | SSPN | upstream | 0.98 | 0.0 | 0.597 |
| rs12809354 | 12 | 32978437 | C/T | 14.7 | 1.08 | 1.06–1.11 | 5.48×10−16 | PKP2 | intronic | 0.97 | 31.5 | 0.211 |
| rs7978685 | 12 | 57103154 | T/C | 27.9 | 1.06 | 1.04–1.07 | 5.99×10−12 | NACA | downstream | 0.98 | 2.4 | 0.393 |
| rs35349325 | 12 | 70097464 | T/C | 54.1 | 1.05 | 1.04–1.07 | 9.04×10−13 | BEST3 | upstream | 0.96 | 0.0 | 0.863 |
| rs11180703 | 12 | 76223817 | G/A | 56.0 | 1.05 | 1.03–1.06 | 3.58×10−10 | KRR1,PHLDA1 | intergenic | 0.97 | 0.0 | 0.482 |
| rs12810346 | 12 | 115091017 | T/C | 14.9 | 1.07 | 1.05–1.09 | 2.34×10−09 | TBX5-AS1, TBX3 | intergenic | 0.84 | 0.0 | 0.428 |
| rs12298484 | 12 | 124418674 | C/T | 67.4 | 1.05 | 1.03–1.06 | 2.05×10−09 | DNAH10 | intronic | 1.00 | 0.0 | 0.973 |
| rs9580438 | 13 | 23373406 | C/T | 32.5 | 1.06 | 1.04–1.07 | 1.01×10−13 | LINC00540,BASP1P1 | intergenic | 0.98 | 0.0 | 0.485 |
| rs28631169 | 14 | 23888183 | T/C | 19.9 | 1.07 | 1.05–1.09 | 3.80×10−14 | MYH7 | intronic | 0.97 | 14.5 | 0.319 |
| rs2145587 | 14 | 32981484 | A/G | 28.1 | 1.08 | 1.06–1.10 | 2.32×10−21 | AKAP6 | intronic | 0.94 | 0.0 | 0.888 |
| rs73241997 | 14 | 35173775 | T/C | 16.4 | 1.07 | 1.05–1.10 | 1.10×10−13 | SNX6, CFL2 | intergenic | 0.98 | 62.2 | 0.032 |
| rs10873299 | 14 | 77426711 | A/G | 38.4 | 1.05 | 1.03–1.07 | 9.62×10−11 | LRRC 74, IRF2BPL | intergenic | 0.96 | 4.4 | 0.381 |
| rs62011291 | 15 | 63800013 | G/A | 22.9 | 1.05 | 1.04–1.07 | 6.14×10−09 | USP3 | intronic | 0.96 | 0.0 | 0.727 |
| rs12591736 | 15 | 70454139 | G/A | 82.0 | 1.06 | 1.04–1.08 | 2.47×10−09 | TLE3,UACA | intergenic | 0.92 | 0.0 | 0.966 |
| rs12908004 | 15 | 80676925 | G/A | 15.9 | 1.08 | 1.06–1.10 | 1.95×10−14 | LINC00927,ARNT2 | intronic | 0.96 | 57.4 | 0.052 |
| rs12908437 | 15 | 99287375 | T/C | 39.2 | 1.05 | 1.03–1.06 | 1.25×10−10 | IGF1R | intronic | 0.98 | 0.0 | 0.818 |
| rs2286466 | 16 | 2014283 | G/A | 80.9 | 1.07 | 1.05–1.09 | 3.53×10−14 | RPS2 | synonymous | 0.92 | 0.0 | 0.882 |
| rs8073937 | 17 | 7435040 | G/A | 36.6 | 1.05 | 1.04–1.07 | 1.02×10−11 | POLR2A, TNFSF1 2 | intergenic | 0.96 | 12.3 | 0.335 |
| rs72811294 | 17 | 12618680 | G/C | 88.7 | 1.07 | 1.05–1.09 | 6.87×10−09 | MYOCD | intronic | 0.95 | 32.3 | 0.206 |
| rs242557 | 17 | 44019712 | G/A | 61.3 | 1.04 | 1.03–1.06 | 4.35×10−09 | MAPT | intronic | 0.94 | 62.1 | 0.032 |
| rs7219869 | 17 | 68337185 | G/C | 43.9 | 1.05 | 1.03–1.06 | 1.49×10−10 | KCNJ2,CASC17 | intergenic | 0.99 | 16.1 | 0.312 |
| rs9953366 | 18 | 46474192 | C/T | 65.5 | 1.05 | 1.04–1.07 | 9.03×10−11 | SMAD7 | intronic | 0.93 | 0.0 | 0.565 |
| rs2145274 | 20 | 6572014 | A/C | 91.3 | 1.11 | 1.08–1.14 | 6.97×10−13 | CASC20,BMP2 | regulatory reg. | 0.96 | 19.0 | 0.295 |
| rs7269123 | 20 | 61157939 | C/T | 58.5 | 1.05 | 1.03–1.06 | 5.59×10−09 | C20orf166 | intronic | 0.85 | 68.7 | 0.012 |
| rs2834618 | 21 | 36119111 | T/G | 89.8 | 1.12 | 1.09–1.14 | 2.93×10−18 | LOC100506385 | intronic | 0.93 | 21.6 | 0.277 |
| rs465276 | 22 | 18600583 | G/A | 61.5 | 1.05 | 1.04–1.07 | 1.84×10−11 | TUBA8 | intronic | 0.90 | 0.0 | 0.654 |
Sentinel variants at novel genetic loci associated with AF at a significance level of P < 1×10-8, for the combined-ancestry meta-analysis (n=588,190). The significance level accounts for multiple testing of independent variants with MAF >0.1% using a Bonferroni correction. PMETA (two-sided) was derived from a meta-analysis using a fixed effects model with an inverse-variance weighted approach. PHET was derived from a Cochran’s Q-test (two-sided) for heterogeneity. Abbreviations, Chr, chromosome, Cl, confidence interval, Func, functional consequence (most severe consequence by variant effect predictor), HET, heterogeneity, I2, l-square, impQual, average imputation quality, META, metaanalysis, P, P-value, RAF, risk allele frequency, reg, region, Ref, reference, RR, relative risk.
Reported is either the gene that overlaps with the sentinel variant, or the nearest gene(s) up- and downstream of the sentinel variant (separated by comma).
We then conducted a gene set enrichment analysis with the results from the combined-ancestry meta-analysis using MAGENTA. We identified 55 enriched gene sets or pathways that largely fall into cardiac developmental, electrophysiological, and cardiomyocyte contractile or structural functional groups (Supplementary Table 4). In total, 48 of the 67 novel loci contain one or more genes within 500kb of the sentinel variant that were part of an enriched gene set or pathway (Supplementary Figure 2).
Next, we performed ancestry-specific meta-analyses. Among individuals of European ancestry, we identified 3 additional loci associated with AF, each of which had a sub-threshold association (P < 1×10−6) in the combined-ancestry meta-analysis. These loci were located close to or within the genes CDK6, EPHA3, and GOSR2 (Supplementary Table 5, Supplementary Figure 3-4). The region most significantly associated with AF in Europeans, Japanese, and African Americans (Supplementary Figure 5–6) was on chromosome 4q25, upstream of the gene PITX2 (Supplementary Figure 7). We did not observe significant heterogeneity of effect estimates across ancestries for most associations, suggesting that top genetic susceptibility signals for AF have a relatively constant effect across ancestries (Table 1, Supplementary Table 3, Supplementary Figure 8). The proportion of heritability explained by the loci from the European ancestry analysis was 42%, compared to previously reported 25%10 (Supplementary Table 6).
In conditional and joint analyses of the European ancestry results, we found 11 loci with multiple, independent AF-associated signals. At a locus centered on a cluster of sodium channel genes, we identified 3 regions that independently associate with AF within SCN10A, SCN5A and a third signal between both genes. At the previously described TBX5 locus,8 we detected a novel independent signal close to TBX3. Pairwise linkage disequilibrium (LD) estimates between the independent variants at both loci were extremely low (r2 <0.03; Supplementary Table 7).
For 13 AF loci, the sentinel variant or a proxy (r2 >0.6) was a missense variant. A missense variant (rs11057401) in CCDC92 was predicted to be damaging by 4 of 5 in silico prediction algorithms (Supplementary Table 8); and was previously associated with coronary artery disease.11 Since most AF-associated variants reside in non-coding regions we sought to determine if the sentinel variants or their proxies (r2 >0.6) fell within regulatory regions in heart tissues based on chromatin states from the Roadmap Epigenomics Consortium. At 64 out of 67 novel loci, variants were located within regulatory elements (Supplementary Table 9); AF-associated loci were also significantly enriched within regulatory elements (Supplementary Figure 9).
We then sought to link risk variants to candidate genes by assessing their effect on gene expression levels. First, since AF often arises from the pulmonary veins and left atrium (LA), we performed RNA sequencing, genotyping, and eQTL analyses in 101 human left atrial samples without structural heart disease from the Myocardial Applied Genomics Network repository. Second, we identified eQTLs from right atrial (RA) and left ventricular (LV) cardiac tissue from the Genotype Tissue Expression (GTEx) project. Finally, we performed a transcriptome-wide analysis using the MetaXcan12 method, which infers the association between genetically predicted gene expression and disease risk.
We observed eQTLs to one or more genes at 17 novel loci. Of the 10 eQTLs detected in LA tissue 8 were also detected in RA or LV, with consistent directionality. For example, we observed that rs4484922 was an eQTL for CASQ2 in LA tissue only. Although we detected more AF loci with eQTLs in the RA or LV data, for many of these (n=8) the results pointed to multiple genes per locus (Supplementary Table 10–12). LA eQTL studies may facilitate the prioritization of candidate genes, but are currently limited by sample size.
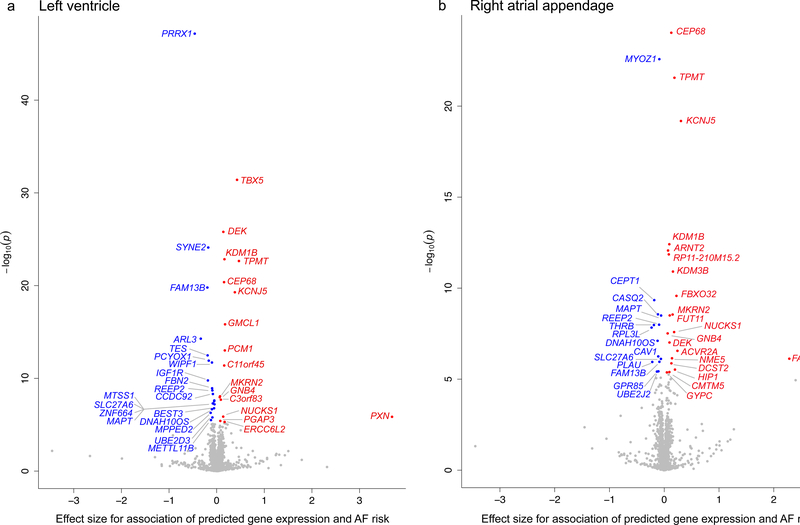
For the transcriptome-wide analyses we used GTEx human atrial and ventricular expression data as a reference. We identified 57 genes significantly associated with AF. Of these, 42 genes were located at AF loci, whereas the remaining 15 were >500kb from an AF sentinel variant (Supplementary Table 13, Figure 3). The probable candidate genes at each locus are summarized in Supplementary Table 12. For example, at the locus with lead variant rs4484922 we observed results from all downstream analyses pointing towards the nearest gene CASQ2, at rs12908437 towards the gene IGFR1, and at rs113819537 towards the gene SSPN. However, for many loci the evaluation of candidate genes remains challenging.
Figure 3. Volcano plot of transcriptome-wide analysis from human heart tissues.
The plots show the results from the transcriptome-wide analysis based on left ventricle (a, n=190) and right atrial appendage (b, n=159) tissue from GTEx, calculated with the MetaXcan method based on the combined-ancestry summary level results (n=588,190). Each plotted point represents the association results for an individual gene. The x-axis shows the effect size for associations of predicted gene expression and AF risk for each tested gene. The y-axis shows the –log10(P) for the associations per gene. Genes with positive effect (red) showed an association of increased predicted gene expression with AF risk. Genes with negative effect (blue) showed an association of decreased predicted gene expression with AF risk. The highlighted genes are significant after Bonferroni correction for all tested genes and tissues with a P-value < 5.36×10-6. The result for one gene for right atrial appendage (b) is not shown (SNX4, Effect = 6.94, P = 0.2).
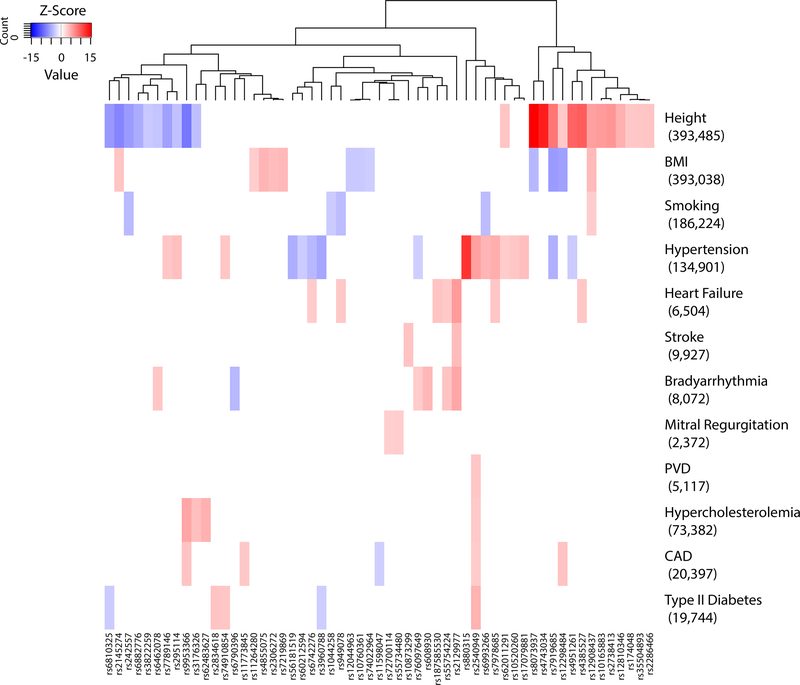
We then sought to assess the pleiotropic effects of the identified AF risk variants. First, we queried the NHGRI-EBI GWAS Catalog to detect associations to other phenotypes (Supplementary Table 14). Second, using the UK Biobank,13 we performed a phenome-wide association study (pheWAS) for 12 AF risk factors (Supplementary Table 15). As illustrated in Figure 4, distinct clusters of variants were associated with AF as well as height, BMI, and hypertension. For example, we observed a pleiotropic effect at rs880315 (CASZ1) for blood pressure14 and hypertension14, that was also observed in the UK Biobank (association with hypertension, P = 2.56×10−34).
Figure 4. Cross-trait associations of AF risk variants with AF risk factors in the UK Biobank.
The heatmap shows associations of novel and known sentinel variants at AF risk loci from the combined-ancestry meta-analysis. Shown are variants and phenotypes with significant associations after correcting for 12 phenotypes via Bonferroni with P < 4.17×10-3. P-values (two-sided) were derived from linear and logistic regression models. Listed next to each trait is the number of cases for binary traits or total sample size for quantitative traits. Hierarchical clustering was performed on a variant level using the complete linkage method based on Euclidian distance. Coloring represents Z-scores for each respective trait or disease, oriented toward the AF risk allele. Red indicates an increase in the trait or disease risk while blue indicates a decrease in the trait or disease risk. Abbreviations, BMI, body-mass index, CAD, coronary artery disease, PVD, pulmonary vascular disease.
In sum, we identified a total of 97 distinct AF loci from 65,446 AF cases and more than 522,000 referents. In recent pre-publication results, Nielsen et al., reported 111 loci from 60,620 AF cases and more than 970,000 referents,15 including more than 18,000 AF cases from our prior report.8 We therefore performed a preliminary meta-analysis for the top loci in nonoverlapping participants from these two large efforts with a resulting total of over 93,000 AF cases and more than 1 million referents. In aggregate, we identified at least 134 distinct AFassociated loci (Supplementary Table 16).
Four major themes emerge from the identified AF loci. First, two AF loci contain genes that are primary targets for current antiarrhythmic medications used to treat AF. The SCN5A gene encodes a sodium channel in the heart, the target of sodium-channel-blockers such as flecainide and propafenone. Similarly, KCNH2 encodes the alpha subunit of the potassium channel complex, the target of potassium-channel-inhibiting medications such as amiodarone, sotalol, and dofetilide. SCN5A and KCNH2 have previously been implicated in AF through GWAS,8 candidate gene analysis16 and family-based studies.17,18
Second, transcriptional regulation appears to be a key feature of AF etiology. TBX3 and the adjacent gene TBX5 encode transcription factors, that have been shown to regulate the development of the cardiac conduction system.19 Similarly, the NKX2–5 encodes a transcription factor, that is an early cue for cardiac development and has been associated with congenital heart disease20 and heart rate21 (Supplementary Table 14). Further, reduced function of the transcription factor encoded by PITX2 has been associated with AF, shortening of the left atrial action potential, and with modulation of sodium channel blocker therapy in the adult left atrium.22–24 A transcriptional co-regulatory network governed by transcription factors encoded by TBX5 and PITX2 has been shown to be critical for atrial development.25
Third, the transcriptome-wide analyses revealed a number of compelling findings. Decreased expression of PRRX1 associated with AF, a result consistent with findings where reduction of PRRX1 in zebrafish and stem cell-derived cardiomyocytes was associated with action potential shortening.26 Further, increased expression of TBX5 and KCNJ5 was associated with AF, a finding consistent with gain-of-function mutations in TBX5 reported in a family with Holt-Oram syndrome and a high penetrance of AF.27 Similarly, KCNJ5 encodes a potassium channel that underlies a component of the IKAch current, a channel that is upregulated in AF. Thus, prior studies support both the role of PRRX1, TBX5, and KCNJ5 in AF and the observed directionality.
Fourth, many of the novel loci implicate genes that underlie Mendelian forms of arrhythmia syndromes. Mutations in CASQ2 lead to catecholaminergic polymorphic ventricular tachycardia.28,29 Pathogenic variants in PKP2 impair cardiomyocyte communication and structural integrity, and are a common cause of arrhythmogenic right ventricular cardiomyopathy.30,31 Mutations in GJA5, KCNH2, SCN5A, KCNJ2, MYH7, NKX2–5, have been mapped in a variety of inherited arrhythmia, cardiomyopathy, or conduction system diseases.32 Our observations highlight the pleiotropy of variation in genes specifying cardiac conduction, morphology, and function, and underscore the complex, polygenic nature of AF.
In conclusion, we conducted the largest AF meta-analysis to date and report a more than three-fold increase in the number of loci associated with this common arrhythmia. Our results lay the groundwork for functional evaluations of genes implicated by AF risk loci. Our findings also broaden our understanding of biological pathways involved in AF and may facilitate the development of therapeutics for AF.
Online Methods
Samples
Participants from more than 50 studies were included in this analysis. Participants were collected from both case-control studies for atrial fibrillation (AF) and population based studies. The majority of studies were part of the Atrial Fibrillation Genetics (AFGen) consortium and the Broad AF Study (Broad AF). Additional summary level results from the UK Biobank (UKBB) and the Biobank Japan (BBJ) were included (Figure 1). Cases include participants with paroxysmal or permanent atrial fibrillation, or atrial flutter, and referents were free of these diagnoses. Adjudication of atrial fibrillation for each study is described in the Supplementary Notes. Ascertainment of AF in the UK Biobank includes samples with one or more of the following codes 1) Non-cancer illness code, self-reported (1471, 1483), 2) Operation code (1524), 3) Diagnoses – main/secondary ICD10 (I48, I48.0–4, I48.9), 4) Underlying (primary/secondary) cause of death: ICD10 (I48, I48.0–4, I48.9) 5) Diagnoses – main/secondary ICD9 (4273), 6) Operative procedures – main/secondary OPCS (K57.1, K62.1–4).8,10,33 Baseline characteristics for each study are reported in Supplementary Table 17. We analyzed: 55,114 cases and 482,295 referents of European ancestry, 1,307 cases and 7,660 referents of African American ancestry, 8,180 cases and 28,612 referents of Japanese ancestry, 568 cases and 1,096 referents from Brazil and 277 cases and 3,081 referents of Hispanic ethnicity. Samples from the UK Biobank, the Broad AF Study, and the following studies from the AFGen consortium: SiGN, EGCUT, PHB and the Vanderbilt Atrial Fibrillation Registry, were previously not included in primary AF GWAS discovery analyses. There is minimal sample overlap from the studies MGH AF, BBJ and AFLMU between this and previous analyses. Ethics approval for participation was obtained individually by each study. All relevant ethical regulations were followed for this work. Written informed consent was obtained from all study participants.
The Institutional Review Board (IRB) at Massachusetts General Hospital reviewed and approved the overall study.
Genotyping and Genotype Calling
Samples within the Broad AF Study were genotyped at the Broad Institute using the Infinium PsychArray-24 v1.2 Bead Chip. They were genotyped in 19 batches, grouped by origin of the samples and with a balanced case control mix on each array. Common variants (≥1% MAF) were called with GenomeStudio v1.6.2.2 and Birdseed v1.33,34 while rare variants (<1% MAF) were called with zCall.35 Batch specific quality control (QC) was performed on each call-set including >95% sample call rate, Hardy-Weinberg-Equilibrium (HWE) P > 1×10−6 and variant call-rate >97%. For common variants, a consensus merge was performed between the call-sets from GenomeStudio and Birdseed. For each genotype only concordant calls between the two algorithms were kept. The common variants from the consensus call were then combined with the rare variants calls from the zCall algorithm. Samples from all batches were joined prior to performing pre-imputation QC steps. Detailed procedures for genotyping and genotype calling for the SiGN study,36 the UK Biobank,37,38 and the Biobank Japan9 are described elsewhere. Details on genotyping and calling for all participating studies are listed in Supplementary Table 18.
Imputation
Pre-imputation QC filtering of samples and variants was conducted based on recommended guidelines as described in Supplementary Table 19. QC steps were performed by each study and are described in Supplementary Table 18. Most studies with European ancestry samples performed imputation with the HRC reference v1.139 panel on the Michigan Imputation Server v1.0.1.40 Studies without available HRC imputation were included based on imputation to the 1000 Genomes Phase 1 integrated v3 panel (March 2012).41 Participants of the SiGN study were imputed to a combined reference panel consisting of 1000 Genomes phase 1 plus Genome of the Netherlands.42 Studies from Brazil were imputed with the HRC reference v1.1 panel. Studies of Japanese ancestry or Hispanic ethnicity were imputed to the 1000G Phase 1 integrated v3 panel (March 2012). Studies of African American ancestry were imputed to the HRC reference v1.1 panel or the 1000G Phase 1 integrated v3 panel (March 2012). Studies were advised to use the HRC preparation and checking tool (http://www.well.ox.ac.uk/~wrayner/tools/) prior to imputation. Prephasing and imputation methods for each study are described in Supplementary Table 18.
Primary statistical analyses
Genome-wide association testing on autosomal chromosomes was performed using an additive genetic effect model based on genotype probabilities. Each ancestry group was analyzed separately for each study. For the Broad AF Study, the primary statistical analysis was performed jointly on unrelated individuals, excluding one of each pair for related samples with PI_HAT >0.2 as calculated in PLINK v1.90.43,44 Samples with sex mismatches and sample call rate <97% were excluded. Ancestry groups were defined with ADMIXTURE45 based on genotyped, independent, and high quality variants, using the supervised method with 1000Genomes phase 1 v3 samples as reference. A cutoff of 80% European ancestry was used to define the European subset and a cutoff of 60% African ancestry was used to define the African American subset. A Brazilian cohort within the Broad AF Study was analyzed separately. Principal components were calculated within each ancestry group with the smartpca program from EIGENSOFT v6.1.146. For the UK Biobank, a European subset was selected within samples with self-reported white race (British, Irish, or other) and similar genetic ancestry. Genetic similarity was defined with the aberrant47 package in R based on principal components, following the same method as described for the UK Biobank.38 We excluded samples with sex mismatches, outliers in heterozygosity and missing rates, samples that carry sex chromosome configurations other than XX or XY, and samples that were excluded from the kinship inference procedure as flagged in the UK Biobank QC file. We further removed one sample for each pair of third degree or closer relatives (kinship coefficient >0.0442), preferentially keeping samples with AF case status. Primary analyses for all other studies were performed at the study sites and the summary level data of the results were provided. Prevalent cases were analyzed in a logistic regression model and most incident cases were analyzed in a Cox proportional hazards model. Studies with both prevalent and incident cases analyzed these either separately using a logistic regression model or Cox proportional hazards model respectively, or jointly in a logistic regression model. The following tools were used for primary GWAS: ProbABEL,48 SNPTEST,49 FAST,50 mach2dat (http://www.sph.umich.edu/csg/yli), R,51 EPACTS (http://genome.sph.umich.edu/wiki/EPACTS), Hail (https://github.com/hail-is/hail) and PLINK44 (Supplementary Table 18). Summary level results were filtered, keeping variants with imputation quality >0.3 and MAF * imputation quality * N events ≥10. Post-analysis QC steps of summary level results included a check of allele frequencies, inspection of Manhattan-plots, QQ-plots, PZ-plots, and the distribution of effect estimates and standard errors, calculation of genomic inflation (λGC), and consistent directionality for known AF risk variants.5
Meta-analyses
Summary level results were meta-analyzed jointly with METAL (released on 2011–03-25) using a fixed effects model with inverse-variance weighted approach, correcting for genomic control.52 Separate meta-analyses were conducted for each ancestry. The results for the Japanese9 and Hispanic8 specific analyses have previously been reported and therefore their ancestry-specific results are not shown. Variants were included if they were present in at least two studies and showed an average MAF ≥0.1%. To correct for multiple testing, a genome-wide significance threshold of P < 1×10−8 was applied for each analysis. This threshold is based on a naive Bonferroni correction for independent variants with MAF ≥0.1%, using an LD threshold of r2 <0.8 to estimate the number of independent variants based on European ancestry LD.53 As these meta-analyses are based on effect estimates and standard errors from both logistic regression and Cox proportional hazards regression, we report variant effects as relative risk, calculated as the exponential of effect estimates. For sentinel variants reaching genome-wide significance in the combined ancestry meta-analysis, we assessed if effect estimates were homogeneous across ancestries by calculating an I2 statistic54 across ancestry specific meta-analyses. We account for multiple testing across 94 variants using a Bonferroni correction, resulting in a significance threshold of P < 5.32×10−4 for the heterogeneity test.
Broad AF LD reference and proxies
A linkage disequilibrium (LD) reference file was created including 26,796 European ancestry individuals from the Broad AF study. The LD reference was based on HRC imputed genotypes. Monomorphic variants and variants with imputation quality <0.1 were removed prior to conversion to hard calls. A genotype probability (GP) threshold filter of GP >0.8 was applied during hard call conversion. For multi-allelic sites the more common alleles were kept. Variants were included in the final reference file if the variant call rate was >70%.
We identified proxies of sentinel variants as variants in LD of r2 >0.6 based on the Broad AF LD reference file, using PLINK v1.90.43,44
Meta-analysis of provisional loci
We meta-analyzed 111 variants from externally reported15 provisional loci within predominantly non-overlapping samples from the Broad AF Study, BBJ, EGCUT, PHB, SiGN and the Vanderbilt AF Registry with METAL (released on 2011–03-25).52 The predominantly nonoverlapping samples included a total of 32,957 AF cases and 83,546 referents, with minimal overlap from the studies MGH AF, BBJ and AFLMU. We subsequently meta-analyzed these results with the reported provisional results with METAL using a fixed effects model with inverse-variance weighted approach. We analyzed a total of 93,577 AF cases and 1,053,762 referents. We compared our discovery results with the provisional loci using the same significance cutoff of P < 5×10−8 for both results. Overlapping loci were identified, if the reported sentinel variants were located within 500kb of each other. For overlapping loci with differing sentinel variants we calculated the LD between the sentinel variants, based on the Broad AF LD reference panel of European ancestry.
Variant consequence on protein coding sequence
The most severe consequence for variants was identified with the Ensembl Variant Effect Predictor version 89.7 using RefSeq as gene reference and the option “pick” to identify one consequence per variant with the default pick order.55 We queried sentinel variants and their proxies to identify tagged variants with HIGH and MODERATE impact including the following consequences: “transcript_ablation”, “splice_acceptor_variant”, “splice_donor_variant”, “stop_gained”, “frameshift_variant”, “stop_lost”, “start_lost”, “transcript_amplification”, “inframe_insertion”, “inframe_deletion”, “missense_variant” and “protein_altering_variant”. We evaluated each identified consequence on the protein coding sequence with in silico prediction tools to assess potentially damaging effects. The evaluation included MutationTaster56 (disease causing automatic or disease causing), SIFT57 (damaging), LRT58 (deleterious), Polyphen259 prediction based on HumDiv and HumVar (probably damaging or possibly damaging).
Chromatin states
Chromatin state annotation.
We identified chromatin states for sentinel variants and their proxies from the Roadmap Epigenomics Consortium 25-state model (2015)60 using HaploReg v4.61 We looked for chromatin states occurring in any included tissues as well as chromatin states occurring in heart tissue. Heart tissues include E065: Aorta, E083: Fetal Heart, E095: Left Ventricle, E104: Right Atrium and E105: Right Ventricle.
Regulatory region enrichment.
1,000 sets of control loci were generated by matching SNPs to sentinel variants from the AF combined-ancestry analysis, with the SNPSnap62 tool. We used the European 1000 Genomes Phase 3 population to match via minor allele frequency, gene density, distance to nearest gene and LD buddies using r2 >0.6 as LD cutoff and otherwise default settings. We excluded input SNPs and HLA SNPs from the matched SNPs. Loci were defined as SNPs and their proxies with r2 >0.6 based on LD from the European 1000 Genomes Phase 3 population. We identified SNPs in regulatory regions across all tissues and in cardiac tissues (E065, E095, E104, E105) based on the Roadmap Epigenomics Consortium 25-state model (2015)60 using HaploReg v4.61 Regulatory regions included the following states: 2_PromU, 3_PromD1, 4_PromD2, 9_TxReg, 10_TxEnh5, 11_TxEnh3, 12_TxEnhW, 13_EnhA1, 14_EnhA2, 15_EnhAF, 16_EnhW1, 17_EnhW2, 18_EnhAc, 19_DNase, 22_PromP and 23_PromBiv. We calculated the percent overlap of each annotation per locus, defined as number of SNPs per locus that fall in regulatory regions divided by total number of SNPs per locus. Statistical significance was calculated with a permutation test from the perm package in R.63
Expression quantitative trait loci (eQTL)
Variants identified from GWAS were assessed for overlap with eQTLs from two sources: 1) Left atrial (LA) tissue from the Myocardial Applied Genomics Network (MAGNet) repository. We performed RNA sequencing (RNA-seq) on 101 left atrial tissue samples from the MAGNet repository (http://www.med.upenn.edu/magnet/) on the Illumina HiSeq 4000 platform at the Broad Institute Genomic Services. Left atrial tissue was obtained at the time of cardiac transplantation from normal donors with no evidence of structural heart disease. All left atrial samples were from individuals of European ancestry. A summary of the clinical characteristics for these samples is shown in Supplementary Table 20. Reads were aligned to the reference genome by STAR v2.4.1a64 and assigned to genes based on the GENCODE gene annotation.65 Gene expression was measured in fragments per kilobase of transcript per million mapped reads (FPKM) and subsequently quantile-normalized and adjusted for age, sex, and the first 10 principal components. Genotyping was performed on the Illumina OmniExpressExome-8v1 array and imputed to the HRC reference panel. Principal components were calculated with the smartpca program from EIGENSOFT v6.1.146 and European ancestry was confirmed by assessing principal components in the samples combined with 1000 Genomes European samples.41 Associations between gene expression and genotypes were tested in a linear regression model with QTLtools v1.0,66 in order to detect cis-eQTLs, defined as eQTLs within 1MB of the transcription start site of a gene. To account for multiple testing, an empirical false discovery rate (FDR) was used to identify significant eQTLs with a FDR <5%. 2) Genotype-Tissue Expression (GTEx) project.67 We queried the GTEx version 6p database for cis-eQTLs with significant associations to gene expression levels in the two available heart tissues: left ventricle and right atrial appendage.68
Association between predicted gene expression and risk of atrial fibrillation
To investigate transcriptome-wide associations between predicted gene expression and AF disease risk, we employed the method MetaXcan v0.3.5.12 MetaXcan extends the previous method PrediXcan69 to predict the association between gene expression and a phenotype of interest, using summary association statistics. Gene expression prediction models were generated from eQTL datasets using Elastic-Net to identify the most predictive set of SNPs. Only models that significantly predict gene expression in the reference eQTL dataset (FDR <0.05) were considered. Pre-computed MetaXcan models for the two available heart tissues (left ventricle and right atrial appendage) in the genotype-tissue expression project version 6p (GTEx)68 were used to predict the association between gene expression and risk of AF. Summary level statistics from the combined ancestry meta-analysis were used as input. 4859 genes were tested for left ventricle and 4467 genes were tested for right atrial appendage. Bonferroni correction was applied to account for the number of genes tested across both tissues, resulting in a significance threshold of P < 5.36×10−6, calculated as 0.05/(4859 + 4467).
Conditional and joint analyses
Conditional and joint analyses70 of GWAS summary statistics were performed with Genomewide Complex Trait Analysis (GCTA v1.25.2)71 using a stepwise selection procedure to identify independently-associated variants on each chromosome. We used the Broad AF LD reference file for LD calculations.
Gene set enrichment analysis (GSEA)
A Meta-Analysis Gene-set Enrichment of variaNT Associations (MAGENTA) v2.472 was performed with a combined gene set input database (GO_PANTHER_INGENUITY_KEGG_REACTOME_BIOCARTA) based on publicly available data. The analysis was conducted using the summary level results from the combined ancestry meta-analysis. 4045 gene sets were included and multiple testing was corrected via false discovery rate (FDR). Gene sets were manually assigned to one or more of the following functional groups: developmental, electrophysiological, contractile/structural, and other. Genes within 500 kilobases of a sentinel variant were identified based on the longest spanning transcribed region in the RefSeq gene reference. For each gene set, genes close to significant loci were listed. The selected genes were assigned to one or more functional groups based on their affiliation to gene sets. Functional groups from gene sets with a single label were preferentially assigned.
Association with other phenotypes
To determine if the sentinel AF risk variants had associations with other phenotypes, two sources of data were used:
1). GWAS catalog.
We queried the NHGRI-EBI Catalog of published genome-wide association studies73,74 (accessed 2017–08-31) to detect associations of AF risk variants with other phenotypes.
2). UK Biobank phenome-wide association study (PheWAS).
A PheWAS was conducted in the UK Biobank in European ancestry individuals. Ancestry definition and sample QC exclusions were performed in the same manner as for the primary statistical analysis, as described above. We further removed one sample for each pair of second degree or closer relatives (kinship coefficient >0.0884), preferentially keeping the sample with case status or non-missing phenotype. We included the following phenotypes: height, body mass index (BMI), smoking, hypertension, heart failure, stroke, mitral regurgitation, bradyarrhythmia, peripheral vascular disease (PVD), hypercholesterolemia, coronary artery disease (CAD), and type II diabetes. Phenotype definitions are shown in Supplementary Table 21. Number of samples analyzed, as well as case and referent counts for each phenotype are listed in Supplementary Table 22. Binary phenotypes were analyzed with a logistic regression model and quantitative phenotypes with a linear regression model using imputed genotype dosages in PLINK 2.00.44 As covariates we included sex, age at first visit, genotyping array, and the first 10 principal components.
Proportion of heritability explained
We calculated SNP-heritability (h2g) of AF-associated loci with the REML algorithm in BOLT-LMM v2.275 in 120,286 unrelated samples of European ancestry from a subset of the UK Biobank dataset comprising a prior interim release as previously described in separate work from our group.10 We defined loci based on a 1MB (+/− 500kb) window around 84 sentinel variants from the European ancestry meta-analysis. We transformed the h2g estimates into liability scale (AF prevalence = 2.45% in UK Biobank). We then calculated the proportion of h2g explained at AF loci by dividing the h2g estimate of AF-associated loci by the total h2g for AF, that was based on 811,488 LD-pruned and hard-called common variants (MAF ≥1%).10
Life Sciences Reporting Summary
Further information on experimental esign is available in the Life Sciences Reporting Summary.
Data Availability and Accession Code Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request. The results of this study are available on the Cardiovascular Disease Knowledge Portal (http://www.broadcvdi.org/). The left atrial RNAsequencing data can be accessed via dbGaP under the accession number phs001539.
Supplementary Material
Acknowledgements
A full list of acknowledgments appears in the Supplementary Note.
Footnotes
Competing financial interests
Dr. Ellinor is the PI on a grant from Bayer to the Broad Institute focused on the genetics and therapeutics of atrial fibrillation. Dr. Psaty serves on the DSMB of a clinical trial funded by Zoll LifeCor and on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. Dr. Kirchhof receives research support from European Union, British Heart Foundation, Leducq Foundation, Medical Research Council (UK), and German Centre for Cardiovascular Research, from several drug and device companies active in atrial fibrillation, and has received honoraria from several such companies. Dr. Kirchhof is also listed as inventor on two patents held by University of Birmingham (Atrial Fibrillation Therapy WO 2015140571, Markers for Atrial Fibrillation WO 2016012783). Dr. Leineweber is an employee of Bayer. The genotyping of participants in the Broad AF Study and the expression analysis of left atrial tissue samples were supported by a grant from Bayer to the Broad Institute. Dr. Nazarian is a consultant to Biosense Webster, Siemens, and Cardiosolv. Dr. Nazarian also receives research grants from NIH/NHLBI, Siemens, Biosense Webster, and Imricor. S. Kathiresan has received grant support from Bayer and Amarin; holds equity in San Therapeutics and Catabasis; and has received personal fees for participation in scientific advisory boards for Catabasis, Regeneron Genetics Center, Merck, Celera, Genomics PLC, Corvidia Therapeutics, Novo Ventures. S. Kathiresan also received personal fees from consulting services from Novartis, AstraZeneca, Alnylam, Eli Lilly Company, Leerink Partners, Merck, Noble Insights, Bayer, Ionis Pharmaceuticals, Novo Ventures, Haug Partners LLC. Genetic Modifiers Newco, Inc. Dr. Lubitz receives sponsored research support from Bristol Myers Squibb, Bayer, Biotronik, and Boehringer Ingelheim, and has consulted for St. Jude Medical / Abbott and Quest Diagnostics. The remaining authors have no disclosures.
References
- 1.Chugh SS et al. Worldwide epidemiology of atrial fibrillation: a Global Burden of Disease 2010 Study. Circulation 129, 837–47 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lubitz SA et al. Association between familial atrial fibrillation and risk of new-onset atrial fibrillation. JAMA 304, 2263–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.January CT et al. 2014 AHA/ACC/HRS Guideline for the Management of Patients With Atrial Fibrillation: Executive Summary. J. Am. Coll. Cardiol. 64, (2014). [DOI] [PubMed] [Google Scholar]
- 4.Benjamin EJ et al. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat. Genet. 41, 879–81 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellinor PT et al. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat. Genet. 44, 670–5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sinner MF et al. Integrating genetic, transcriptional, and functional analyses to identify 5 novel genes for atrial fibrillation. Circulation 130, 1225–35 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellinor PT et al. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat. Genet. 42, 240–4 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christophersen IE et al. Large-scale analyses of common and rare variants identify 12 new loci associated with atrial fibrillation. Nat. Genet. 49, 946–952 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Low S-K et al. Identification of six new genetic loci associated with atrial fibrillation in the Japanese population. Nat. Genet. 49, 953–958 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Weng L-C et al. Heritability of Atrial Fibrillation. Circ. Cardiovasc. Genet. 10, e001838 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klarin D et al. Genetic analysis in UK Biobank links insulin resistance and transendothelial migration pathways to coronary artery disease. Nat. Genet. 49, 1392–1397 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barbeira A et al. Integrating tissue specific mechanisms into GWAS summary results. bioRxiv (2017). at <http://biorxiv.org/content/early/2017/05/21/045260.abstract> [Google Scholar]
- 13.Sudlow C et al. UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. 12, e1001779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu X et al. Genome-wide association study in Chinese identifies novel loci for blood pressure and hypertension. Hum. Mol. Genet. 24, 865–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nielsen JB et al. Genome-wide association study of 1 million people identifies 111 loci for atrial fibrillation. bioRxiv 242149 (2018). doi: 10.1101/242149. [DOI] [Google Scholar]
- 16.Sinner MF et al. The non-synonymous coding IKr-channel variant KCNH2-K897T is associated with atrial fibrillation: results from a systematic candidate gene-based analysis of KCNH2 (HERG). Eur. Heart J. 29, 907–914 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Olson TM et al. Sodium channel mutations and susceptibility to heart failure and atrial fibrillation. JAMA 293, 447–54 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McNair WP et al. SCN5A Mutation Associated With Dilated Cardiomyopathy, Conduction Disorder, and Arrhythmia. Circulation 110, 2163–2167 (2004). [DOI] [PubMed] [Google Scholar]
- 19.van Weerd JH et al. A large permissive regulatory domain exclusively controls Tbx3 expression in the cardiac conduction system. Circ. Res. 115, 432–41 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Schott JJ et al. Congenital heart disease caused by mutations in the transcription factor NKX2–5. Science 281, 108–11 (1998). [DOI] [PubMed] [Google Scholar]
- 21.den Hoed M et al. Identification of heart rate-associated loci and their effects on cardiac conduction and rhythm disorders. Nat. Genet. 45, 621–31 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirchhof P et al. PITX2c is Expressed in the Adult Left Atrium, and Reducing Pitx2c Expression Promotes Atrial Fibrillation Inducibility and Complex Changes in Gene Expression. Circ. Cardiovasc. Genet. 4, 123–133 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Wang J et al. Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc. Natl. Acad. Sci. U. S. A. 107, 9753–8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Syeda F et al. PITX2 Modulates Atrial Membrane Potential and the Antiarrhythmic Effects of Sodium-Channel Blockers. J. Am. Coll. Cardiol. 68, 1881–1894 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nadadur RD et al. Pitx2 modulates a Tbx5-dependent gene regulatory network to maintain atrial rhythm. Sci. Transl. Med. 8, 354ra115 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tucker NR et al. Diminished PRRX1 Expression Is Associated With Increased Risk of Atrial Fibrillation and Shortening of the Cardiac Action Potential. Circ. Cardiovasc. Genet. 10, e001902 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Postma AV et al. A gain-of-function TBX5 mutation is associated with atypical Holt-Oram syndrome and paroxysmal atrial fibrillation. Circ. Res. 102, 1433–42 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Lahat H et al. A missense mutation in a highly conserved region of CASQ2 is associated with autosomal recessive catecholamine-induced polymorphic ventricular tachycardia in Bedouin families from Israel. Am. J. Hum. Genet. 69, 1378–84 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lahat H et al. Autosomal recessive catecholamine- or exercise-induced polymorphic ventricular tachycardia: clinical features and assignment of the disease gene to chromosome 1p13–21. Circulation 103, 2822–7 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Corrado D, Link MS & Calkins H Arrhythmogenic Right Ventricular Cardiomyopathy. N. Engl. J. Med. 376, 61–72 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Gerull B et al. Mutations in the desmosomal protein plakophilin-2 are common in arrhythmogenic right ventricular cardiomyopathy. Nat. Genet. 36, 1162–1164 (2004). [DOI] [PubMed] [Google Scholar]
- 32.HRS/EHRA Expert Consensus Statement on the State of Genetic Testing for the Channelopathies and Cardiomyopathies: This document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA). Hear. Rhythm 8, 1308–1339 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Weng L-C et al. Genetic Predisposition, Clinical Risk Factor Burden, and Lifetime Risk of Atrial Fibrillation. Circulation CIRCULATIONAHA.117031431 (2017). doi: 10.1161/CIRCULATIONAHA.117.031431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korn JM et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat. Genet. 40, 1253–1260 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goldstein JI et al. zCall: a rare variant caller for array-based genotyping. Bioinformatics 28, 2543–2545 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pulit SL et al. Loci associated with ischaemic stroke and its subtypes (SiGN): a genome-wide association study. Lancet Neurol. 15, 174–184 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Genotyping and quality control of UK Biobank, a large-scale, extensively phenotyped prospective resource. at <http://www.ukbiobank.ac.uk/wpcontent/uploads/2014/04/UKBiobank_genotyping_QC_documentation-web.pdf>
- 38.Bycroft C et al. Genome-wide genetic data on ~500,000 UK Biobank participants. 166298 (2017). doi: 10.1101/166298 [DOI]
- 39.HRC Consortium et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 48, 1279–1283 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Das S et al. Next-generation genotype imputation service and methods. Nat. Genet. 48, 1284–1287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Auton A et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Francioli LC et al. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat. Genet. 46, 818–825 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Shaun P & Christopher C PLINK v1.90b3.32.
- 44.Chang CC et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alexander DH, Novembre J & Lange K Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–64 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Price AL et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Bellenguez C, Strange A, Freeman C, Donnelly P & Spencer CCA A robust clustering algorithm for identifying problematic samples in genome-wide association studies. Bioinformatics 28, 134–135 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aulchenko YS, Struchalin MV & van Duijn CM ProbABEL package for genomewide association analysis of imputed data. BMC Bioinformatics 11, 134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marchini J, Howie B, Myers S, McVean G & Donnelly P A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 39, 906–913 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Chanda P, Huang H, Arking DE & Bader JS Fast Association Tests for Genes with FAST. PLoS One 8, e68585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.R Core Team. R: A Language and Environment for Statistical Computing. (2015). at <http://www.r-project.org/>
- 52.Willer CJ, Li Y & Abecasis GR METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–1 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fadista J, Manning AK, Florez JC & Groop L The (in)famous GWAS P-value threshold revisited and updated for low-frequency variants. Eur. J. Hum. Genet. 24, 1202–1205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Higgins JPT, Thompson SG, Deeks JJ & Altman DG Measuring inconsistency in meta-analyses. BMJ 327, 557–60 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McLaren W et al. The Ensembl Variant Effect Predictor. Genome Biol. 17, 122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schwarz JM, Rödelsperger C, Schuelke M & Seelow D MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods 7, 575–576 (2010). [DOI] [PubMed] [Google Scholar]
- 57.Kumar P, Henikoff S & Ng PC Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4, 1073–1081 (2009). [DOI] [PubMed] [Google Scholar]
- 58.Chun S & Fay JC Identification of deleterious mutations within three human genomes. Genome Res. 19, 1553–61 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adzhubei IA et al. A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ernst J & Kellis M Large-scale imputation of epigenomic datasets for systematic annotation of diverse human tissues. Nat. Biotechnol. 33, 364–376 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ward LD & Kellis M HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930–4 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pers TH, Timshel P & Hirschhorn JN SNPsnap: a Web-based tool for identification and annotation of matched SNPs. Bioinformatics 31, 418–20 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fay MP & Shaw PA Exact and Asymptotic Weighted Logrank Tests for Interval Censored Data: The interval R Package. J. Stat. Softw. 36, 1–34 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harrow J et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Delaneau O et al. A complete tool set for molecular QTL discovery and analysis. Nat. Commun. 8, 15452 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.The GTEx Consortium et al. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 348, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aguet F et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gamazon ER et al. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 47, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang J et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 44, 369–75, S1–3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang J, Lee SH, Goddard ME & Visscher PM GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Segrè AV et al. Common Inherited Variation in Mitochondrial Genes Is Not Enriched for Associations with Type 2 Diabetes or Related Glycemic Traits. PLoS Genet. 6, e1001058 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Welter D et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 42, D1001–6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Burdett T et al. The NHGRI-EBI Catalog of published genome-wide association studies. at <www.ebi.ac.uk/gwas> [DOI] [PMC free article] [PubMed]
- 75.Loh P-R et al. Contrasting genetic architectures of schizophrenia and other complex diseases using fast variance-components analysis. Nat. Genet. 47, 1385–1392 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request. The results of this study are available on the Cardiovascular Disease Knowledge Portal (http://www.broadcvdi.org/). The left atrial RNAsequencing data can be accessed via dbGaP under the accession number phs001539.