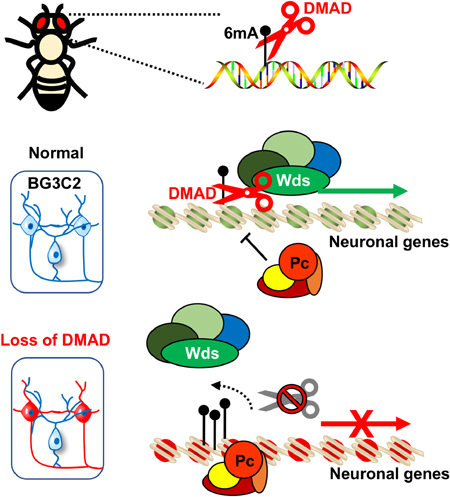
SUMMARY
A Ten-Eleven Translocation (TET) ortholog exists as a DNA N6-Methyladenine (6mA) demethylase (DMAD) in Drosophila. However, the molecular roles of 6mA and DMAD remain unexplored. Through genome-wide 6mA and transcriptome profiling in Drosophila brains and neuronal cells, we found that 6mA may epigenetically regulate a group of genes involved in neurodevelopment and neuronal functions. Mechanistically, DMAD interacts with Trithorax-related complex protein Wds to maintain active transcription by dynamically demethylating intragenic 6mA. Accumulation of 6mA by depleting DMAD coordinates with Polycomb proteins and contributes to transcriptional repression of these genes. Our findings suggest that active 6mA demethylation by DMAD plays essential roles in fly CNS by orchestrating through added epigenetic mechanisms.
eTOC Blurb
A DNA N6-Methyladenine (6mA) demethylase (DMAD) was found in Drosophila to actively remove 6mA. Yao et al demonstrate that DMAD depletion in neurons leads to impaired neurodevelopment accompanied by 6mA accumulation. DMAD and 6mA epigenetically modulate a group of neuronal genes by coordinating with the Trithorax and Polycomb histone modifiers.
INTRODUCTION
Cytosine methylation at the 5-carbon position (5-methylcytosine; 5mC) is a critical repressive epigenetic mark in the mammalian genome (Bird, 2002; Ma et al., 2010; Schubeler, 2015; Spivakov and Fisher, 2007). 5mC is generally viewed as a stable and irreversible covalent modification to DNA; however, the fact that ten-eleven translocation (TET) proteins can oxidize 5mC to 5-hydroxymethylcytosine (5hmC) and downstream derivatives gives us a new perspective on the plasticity of 5mC-dependent regulatory processes (Zhang et al., 2012). Cytosine modifications exist in bacteria, archaea, viruses, fungi, vertebrates, and plants, but their presence and functions remain controversial in model organisms such as Drosophila (Lyko et al., 2000; Zhang et al., 2015b).
Surprisingly, a TET ortholog with unknown function exists in the fly genome (Dunwell et al., 2013). Recent studies demonstrated that this Drosophila TET ortholog could demethylate the DNA modification N6-Methyladenine (6mA) in eukaryotes (Zhang et al., 2015b), a prevalent DNA modification previously only found in bacteria. 6mA was also recently found in algae, worms, fungi and mammals (Fu et al., 2015; Greer et al., 2015; Koziol et al., 2016; Mondo et al., 2017; Wu et al., 2016; Zhang et al., 2015b). Although this Drosophila TET ortholog was identified as a 6mA demethylase (DMAD) (Zhang et al., 2015b), the precise molecular functions of 6mA and DMAD in Drosophila genome remain unknown.
RESULTS
DMAD depletion in Drosophila brain results in brain developmental defects accompanied by 6mA accumulation
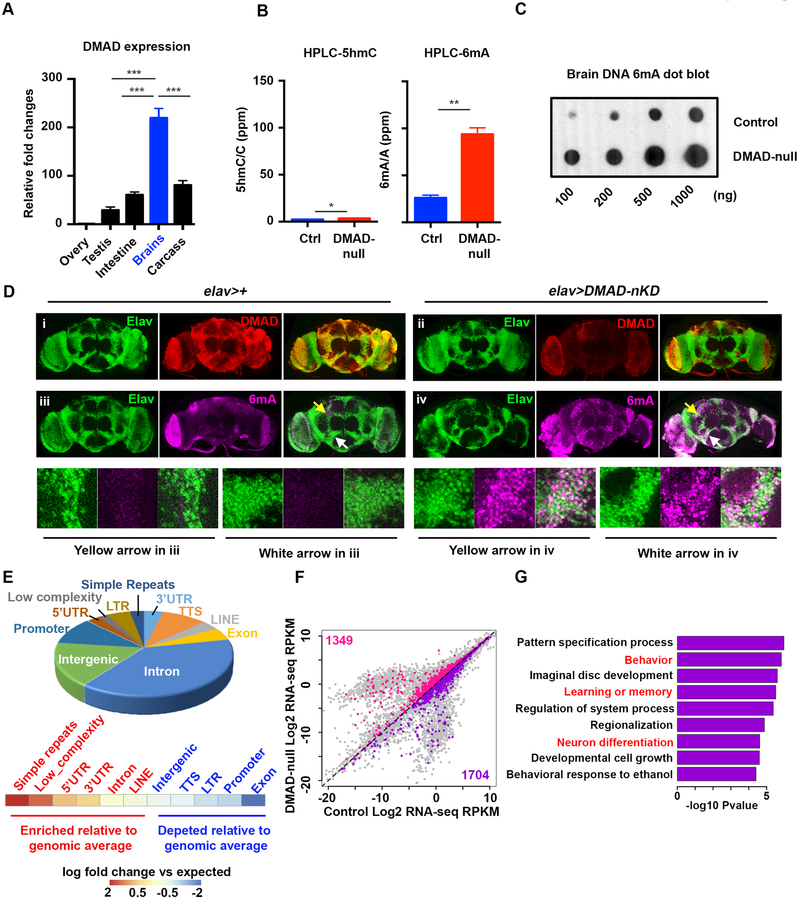
Gene expression analyses across adult fly tissues show that DMAD is highly expressed in fly brains (Figure 1A). Previous work showed that DMAD is essential for development since only a small fraction of DMAD-null mutants can survive through the pupa stage, although these mutants die within 3 days post-eclosion (Zhang et al., 2015b). To understand the role DMAD plays in fly brain function, we generated multiple transgenic lines, P{UASp-artimiR-DMAD}, which carry artificial miRNAs targeting DMAD. Consistent with DMAD-null flies (Zhang et al., 2015b), ubiquitous expression of artificial miRNAs against DMAD using the driver Tubulin-GAL4 (DMAD-KD) resulted in developmental defects, as most flies died pre-eclosion (Figure S1A). The mRNA of DMAD was effectively depleted in two DMAD-KD lines (Figure S1B). Neuronal-specific DMAD knockdown (DMAD-nKD) under the control of the pan-neuronal driver Elav-GAL4 resulted in mushroom body (MB) abnormalities in adult flies. MBs are one of the best-characterized brain regions involved in learning and memory in flies and are composed of axon-like fiber structures forming Fas II-positive neuronal lobes (the a-, b- and g-lobes) (Figure S1C) (Heisenberg, 2003). In DMAD-nKD flies, the b-lobes, which normally end at the midline cleft, often crossed the midline. Also, DMAD-nKD flies also bore accessory MB phenotypes, such as missing or misdirected a- and b-lobes, as well as truncated or overbranched lobes (Figure S1C). These data suggest that DMAD may contribute to neuronal morphology, development, and function in the fly brain.
Figure 1. DMAD demethylates intragenic 6mA in Drosophila brains.
(A) qRT-PCR across fly tissues revealed high levels of DMAD expression in heads compared to other somatic tissues (n=3).(B) High resolution HPLC quantification of 5hmC and 6mA in Drosophila brains in the presence (Ctrl) and absence of DMAD (DMAD-null). 5hmC/total C or 6mA/total A are shown as percentage per million nucleotide (ppm). There was a ~3.6-fold increase in 6mA with DMAD depletion while 5hmC was undetectable (n=2).(C) Dot blots using an antibody specific for 6mA confirmed the accumulation of 6mA in DMAD-null fly brains.(D) Confocal images of adult brains stained with anti-Elav (Green), anti-DMAD (Red) and anti-6mA (Purple) in the background of elav-Gal4 alone (i & iii) or in combination with UAS-DMAD miRNA (DMAD-nKD) (ii & iv). Enlarged views of arrowed area in iii and iv are shown below. Compared to control brains (elav-Gal4 alone), DMAD knockdown significantly increased nuclear 6mA levels in elav-expressing cells.(E) Genomic annotation of gain-of-6mA regions in DMAD-null fly brains revealed their intragenic characteristics, A heatmap shows the enrichment of each genomic feature versus expected values.(F) Plot of the global transcriptome in control and DMAD-null fly brains obtained from RNA-seq (n=2). Genes bearing gain-of-6mA regions in their gene bodies were highlighted (up-regulated genes in pink and down-regulated genes in purple).(G) Gene Ontology analysis was performed on a subset of downregulated genes in Figure 1F (purple). Log2 fold change [(KD: WT) < −0.5] was applied as the threshold cut-off. Several biological processes involved in neurodevelopment and neuronal functions were enriched and are highlighted in red. Data are represented as mean ° SEM.
DMAD possesses enzymatic activity as a DNA N6-Methyladenine (6mA) demethylase in fly ovaries (Zhang et al., 2015b). To confirm this enzymatic activity in fly brains, we applied the highly sensitive ultra-performance liquid chromatography tandem mass spectrometer (UHPLCMS/MS) to quantify 6mA and 5hmC abundance in control and DMAD-null fly brains (Schubeler, 2015; Zhang et al., 2015b). The DMAD protein level was abolished entirely in DMAD-null fly brains (Figure S1D). Consistent with previous findings, a shallow level of 5hmC was detectable in control fly brains (Dunwell et al., 2013; Raddatz et al., 2013), and DMAD depletion had minimal effects on 5hmC abundance (average 5hmC/C ppm increased from 2.29 to 3.44). In contrast to 5hmC, we detected higher levels of 6mA than 5hmC in control fly brains (average 26.0 6mA/A ppm). Importantly, 6mA levels significantly increased 4-fold in DMAD-null flies relative to controls (Figure 1B and Table S1). We confirmed this finding with dot blots using a 6mA-specific antibody (Figure 1C). To further confirm that DMAD is a 6mA demethylase in fly neuronal cells, we performed 6mA immunostaining in control and DMAD-nKD flies. Neuronal-specific DMAD knockdown using Elav-Gal4 resulted in 6mA accumulation explicitly in Elav-expressing neuronal cells (Figure 1D). Also, in vitro, 6mA demethylation assays using double-stranded synthetic oligonucleotide substrates indicated that recombinant DMAD demethylates 6mA (Figure S1E and S1F). These findings provide strong support for the role of DMAD as 6mA demethylase in fly brains. One recent study suggested that DMAD can generate 5-hydroxymethylcytosine in RNA molecules (5hmrC), and DMAD-deficient S2 cells showed decreased 5hmrC (Delatte et al., 2016). To test this, we applied UHPLC-MS/MS to precisely quantify the 5hmrC levels in control, DMAD-null fly brains, and DMAD-knockdown S2 cells. Extremely low levels of 5hmrC (0.2 ppm in fly brains and 2 ppm in S2 cells versus 25–100 ppm 6mA) were detected (Figure S1G), indicating 6mA is a major substrate for DMAD in Drosophila genome. No significant change in RNA 5mC was observed (Figure S1H). Furthermore, we tested the 6mA levels in an AlkB neuronal knockdown, which is a homolog of mammalian 6mA putative demethylase Alkbh1 (Wu et al., 2016). No substantial 6mA accumulation was found in the absence of AlkB (Figure S1I), suggesting DMAD could be the key 6mA demethylase in Drosophila. Since the key residues responsible for DMAD’s demethylation activities were defined (Zhang et al., 2015b), we generated the recombinant DMAD catalytic domain (DMADCD) and its catalytically dead mutations (DMAD-CD-mut) to test their direct demethylation activity in vitro. The DMAD-CD has comparable demethylation activities with bacterial 6mA demethylase ALKB. In agreement with previous observations, the DMAD-CD-mutant displayed drastic and significant reduced enzymatic activity for 6mA demethylation (Figure S1J).
To understand the molecular functions of active 6mA demethylation, we generated genome-wide 6mA maps from control and DMAD-null fly brains. We immunoprecipitated 6mA-containing DNA isolated from ~1,000 dissected fly brains and used high-throughput sequencing to identify genomic loci enriched for 6mA. The specificity and reproducibility of 6mA immunoprecipitation were ensured using competitive elution with excess 6mA. Overall, the number of binned genomic regions containing high 6mA reads was larger in DMAD-null brains (Figure S1K). We identified 5340 high confidence gain-of-6mA regions in DMAD-null mutants relative to control flies. Thus, these regions represent active 6mA demethylation loci in wild-type brains (Figure S1L and Table S1). Genomic annotation of gain-of-6mA regions revealed that a large percentage of 6mA increases occurred in intragenic regions, with enrichment in introns and untranslated regions (UTRs) compared to expected values. Consistent with previous observations (Zhang et al., 2015b), gain-of-6mA regions were also enriched for several classes of repetitive elements relative to expected values (Figure 1E),.
To investigate the connection between dynamic 6mA demethylation and global transcriptome changes, we performed RNA-seq on control and DMAD-null fly brains (Table S1). 1704 downregulated, and 1349 upregulated genes bearing significant accumulation of 6mA on their gene bodies upon DMAD depletion were found (Figure 1F, purple and pink, respectively). Gene Ontology (GO) analysis of these downregulated genes showed enrichment in several key ontology terms reflecting DMAD-null fly phenotypes such as behavior, learning and memory, and neuronal differentiation (Figure 1G). In contrast, upregulated genes were enriched in general development-related functions (Figure S1M). To further investigate the potential regulatory mechanisms underlying 6mA-mediated gene regulation, we searched for common motifs within gain-of-6mA regions and predicted possible binding factors based on these motifs. Intriguingly, the 6mA motif “AGAAGGAG” in fly brains was previously found in C. elegans (Greer et al., 2015), potentially suggesting a conserved mechanism of 6mA regulation across species. Several known DNA-binding proteins, such as transcription factors Aef1 and Adf1, were predicted to bind to these regions (Figure S1N). Interestingly, both Aef1 and Adf1 define Polycomb response elements (PRE) by interacting with Polycomb PRC1 component Pc (Orsi et al., 2014). These findings inspired us to explore the possible interplay between 6mA and histone modifiers.
6mA is associated with the binding of Polycomb protein in Drosophila neuronal cells
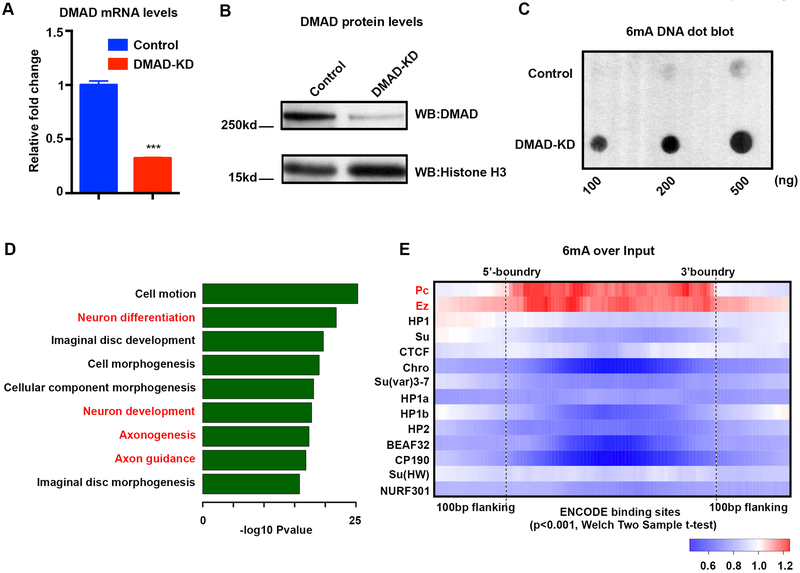
To gain in-depth mechanistic insight into 6mA dynamics and gene regulation, we assessed the effects of reduced DMAD expression in a fly neuronal cell line, BG3C2 (Ui et al., 1994), which was extensively analyzed in the modENCODE project. DMAD was knocked down (KD) using double-stranded RNA (dsRNAs), and successful KD was confirmed by qPCR and western blots (Figure 2A and 2B). Consistent with our data in fly brains, DMAD knockdown in BG3C2 neuronal cell line also led to an overall increase in 6mA levels (Figure 2C). A total of 6093 gain-of-6mA regions were identified in BG3C2 cells from replicates of control and DMAD-KD cells (Figure S2A and Table S2). In parallel, RNA-seq analyses occurred for control and DMAD-KD BG3C2 cells (Table S2). Like our findings in the brain, gain-of-6mA regions were associated with intragenic regions such as introns and repetitive elements relative to expected values (Figure S2B). Also, GO analysis of downregulated genes containing intragenic gain-of-6mA regions showed preferential enrichment for neuronal functions (Figure 2D). Conversely, upregulated genes having gain-of-6mA regions were enriched for general developmental pathways (Figure S2C). Furthermore, we found 42.5% and 21.2% of neuronally expressed Long interspersed elements (LINEs), and Long terminal repeats (LTRs) overlapped with gain-of-6mA regions (Figure S2D), consistent with the previous report that 6mA could potentially impact transposon expression (Zhang et al., 2015b). To confirm the 6mA-IP seq data, we performed restriction enzyme digests to validate 6mA differential loci found by BG3C2 6mA-IP (Methods). As expected, the qPCR results were consistent with our 6mA-IP that these loci contain methylation on the adenines and 6mA accumulated upon DMAD-KD (Figure S2E).
Figure 2. 6mA is enriched at Polycomb-binding sites in neuronal cells.
(A) qRT-PCR validated a 70% reduction in DMAD mRNA levels after double-strand siRNA knockdown in BG3C2 cells.(B) Western blot using a DMAD-specific antibody confirmed effective DMAD knockdown in BG3C2 cells.(C) Dot blots using a 6mA-specific antibody confirmed 6mA accumulation in the absence of DMAD in BG3C2 cells.(D) GO analysis showed specific enrichment for neurodevelopment and neuronal functions from downregulated genes carrying intragenic BG3C2 gain-of-6mA regions.(E) Average fold change in 6mA mapped reads versus non-enriched input DNA was calculated for various binned ChIP-chip regions of epigenetic regulators available from the modENCODE database. Average fold change is plotted in Heatmap view. Red (fold change >1) indicates enrichment over input while blue (fold change <1) indicates depletion. 6mA was explicitly enriched at Polycomb protein binding sites. Enrichment and depletion were significant with p-value < 0.001, Welch Two sample t-tests. Data are represented as mean ° SEM.
To directly link 6mA to epigenetic modulators, we used ChIP-chip data available for BG3C2 cells from modENCODE (Ho et al., 2014) to investigate 6mA enrichment at epigenetic regulator binding sites. We calculated 6mA reads over non-enriched input at the epigenetic regulator binding sites, finding that 6mA is explicitly enriched at Polycomb protein binding sites, including Polycomb complex components Pc, dRING, Psc, and Ez, but not other epigenetic regulators such as HP1, Su(var)3–7 and CTCF (Figure 2E and S2F). Moreover, knockdown of DMAD led to further 6mA accumulation primarily on Polycomb-binding sites (Figure S2G). Therefore, 6mA could potentially work cooperatively with Polycomb proteins to mediate epigenetic regulation of gene expression.
DMAD coordinates with Trithorax-related protein Wds to regulate gene expression by maintaining 6mA homeostasis at a group of neuronal genes
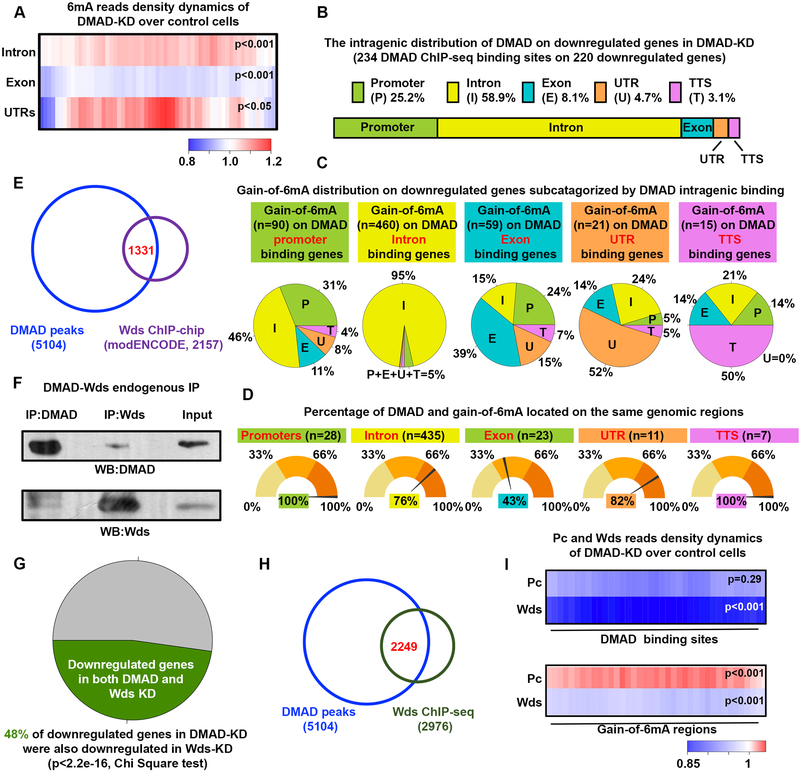
To investigate the regulatory roles of DMAD in gene expression, we performed ChIP-seq to map genome-wide DMAD binding sites in BG3C2 cells (Table S3). Genome-wide annotation of DMAD binding sites revealed its general intragenic enrichment, such as exons (Figure S3A). Interestingly, 6mA accumulated on introns and UTRs upon DMAD-KD (Figure 3A), consistent with genomic annotations of gain-of-6mA regions (Fig. 1E). We focused next on the DMAD-bound genes bearing accumulation of 6mA in their gene bodies in the DMAD-KD cells as they could be modulated by DMAD through 6mA demethylation. These genes were further separated into downregulated and upregulated genes (Figure 3B and S3B). Interestingly, instead of more exon enrichment of general DMAD binding sites, DMAD showed more enrichment in introns of these genes, coinciding with 6mA accumulation in introns (Figure 1E and S2B). These genes were subcategorized based on the DMAD intragenic binding regions (Figure 3C and S3C). In general, DMAD binding sites and 6mA accumulation upon DMAD-KD were associated with the same genomic regions. For instance, a remarkable 95% and 98% of intronic 6mA accumulation were found in genes with DMAD binding to their introns, showing DMAD influenced these genes through their intronic 6mA demethylation (Figure 3C and S3C). Interestingly, 76% of introns in downregulated genes harbored gain-of-6mA regions and DMAD binding sites in the exact intronic regions, further supporting that DMAD could demethylate 6mA in cis (Figure 3D). Similar observations were also found in the upregulated genes, although the percentage of DMAD and gain-of-6mA binding to the same introns showed a lesser extent (Figure S3D). Given that DMAD depletion downregulated a group of critical neuronal genes, we hypothesized that DMAD may coordinate with transcriptional activators to maintain active expression profiles. Using the modENCODE ChIP-chip dataset, we found that binding sites of Wds (Will die slowly), a protein component of the Trithorax-related complex that is responsible for gene activation (Mohan et al., 2011), substantially overlapped with DMAD binding sites (Figure 3E). This overlap appeared specific, as the binding sites of many other transcription factors available in ChIP-chip datasets did not overlap with DMAD (Figure S3E). Wds is the Drosophila ortholog of the mammalian WDR5, and has been shown to modulate active transcription with other Trithorax components through histone modifications (Herz et al., 2012; Mohan et al., 2011). Substantial overlap between Wds and DMAD binding sites suggested that Wds and DMAD may form a complex in vivo. To test this, we performed co-immunoprecipitation (co-IP) experiments and observed a biochemical association between DMAD and Wds in BG3C2 cells (Figure 3F). Altered genes in DMAD-KD BG3C2 cells significantly overlapped with differentially expressed genes due to the Wds knockdown (Figure 3G and S3F). Interestingly, knockdown of Wds did not affect the global 6mA level, suggesting the DMAD target recognition and demethylation are independent of Wds (Figure S3G and 3H). Co-IP experiments using Wds and DMAD deletion constructs were performed to map their interaction domains. WD40 domains of Wds were critical for interaction with DMAD, as the complete removal of WD-40 domains abolished its binding to DMAD (Figure S3I). On the other hand, the C-terminal DMAD (aa 1657–2860) bearing the Tet-JBP catalytic domain, but not the DMAD N-terminal region (aa 1–1657) with the CXXC DNA-binding domain, was responsible for association with Wds (Figure S3J). Additional fine mapping revealed that DMAD 1657–2666, including the 6mA catalytic domain (aa 1796–2666), bind to Wds (Figure S3K). These data together suggest that the DMAD interaction with Wds is coupled with its 6mA demethylation activities.
Figure 3. DMAD demethylates intragenic 6mA in cis and coordinates with Trithorax-related protein Wds to regulate gene expression.
(A) Average fold changes in 6mA reads between DMAD-KD and control were calculated for DMAD-occupied gene introns, exons, and untranslated regions. Enrichment or depletion of 6mA on these genomic regions were significant (p<0.001 or p<0.05, Welch Two sample t-tests) (B) Intragenic distributions of DMAD on DMAD-bound downregulated genes bearing accumulation of 6mA upon DMAD knockdown was demonstrated proportionally. DMAD showed strong intronic enrichment on these genes. (C) The genes in (B) were further subcategorized based on the DMAD intragenic association. Gain-of-6mA distributions on each subset of genes were calculated, and the percentages shown in the pie chart. (D) The percentage of genes with both DMAD and gain-of-6mA binding to the same genomic feature were calculated.(E) Venn diagram shows substantial and significant overlap between DMAD and Wds ChIP-chip data (Binomial tests, p<0.001).(F) Co-immunoprecipitation experiments indicated a physical interaction between DMAD and Wds. (G) Downregulated genes in the DMAD-KD cells showed significant overlapping with downregulated genes in the absence of Wds. Chi-square tests were performed. RNA-seq were performed in triplicates. RPKM fold changes <−0.1 were included.(H) Substantial overlap between DMAD and Wds ChIPseq peaks suggested their functional coordination in regulating gene expression.(I) Average fold change in Pc and Wds reads of DMAD-KD over control were calculated for both DMAD-binding sites and gain-of-6mA regions to explore Pc and Wds dynamics in these regions. Heatmap demonstrated a general decrease in both Pc and Wds at DMAD binding sites when DMAD is depleted. A specific and significant increase in Pc binding on gain-of-6mA regions with DMAD depletion was found. Welch Two sample t-tests, p-values were indicated. Data are represented as mean ° SEM.
Since ChIP-chip assays in general have lower resolution than ChIP-seq, we performed Wds ChIP-seq in control and DMAD-KD BG3C2 cells (Table S3). 2249 of 2976 Wds peaks overlapped with DMAD binding sites, actively supporting their cooperation in transcriptional control (Figure 3H). We further analyzed ChIP-seq data to understand the dynamic changes of Pc (Table S3) and Wds on DMAD binding sites and gain-of-6mA regions. We computed the normalized Pc, and Wds ChIP-seq reads ratios at DMAD binding sites using DMAD-KD over control cells. Wds, but not Pc, showed a significant reduction in the absence of DMAD (Figure 3I, blue indicates KD reads < WT reads, p<0.001). Interestingly, substantial, and significant increases in Pc binding were found on gain-of-6mA regions, suggesting potential crosstalk between 6mA and Polycomb proteins (Figure 3I, red indicates KD reads > WT reads, p<0.001). Using micrococcal nuclease (MNase) digestion coupled with high-throughput sequencing, we investigated the dynamic changes of nucleosome positioning on intragenic DMAD-binding sites and gain-of-6mA regions when DMAD was depleted. DMAD-binding sites showed no significant changes in nucleosome occupancy when DMAD was knocked down (Figure S3L and S3N, p=0.4436). In contrast, gain-of-6mA regions displayed significantly higher nucleosome occupancy in DMAD-KD cells (Figure S3M and S3O, p<0.0001), possibly due to Polycomb recruitment for chromatin remodeling (Orsi et al., 2014). Our findings suggest that DMAD binds to specific sets of genes to modulate intragenic 6mA levels and coordinate with histone modifiers, thereby regulating gene expression.
6mA dynamic regulation by DMAD coordinates with Trithorax and Polycomb-mediated epigenetic mechanisms
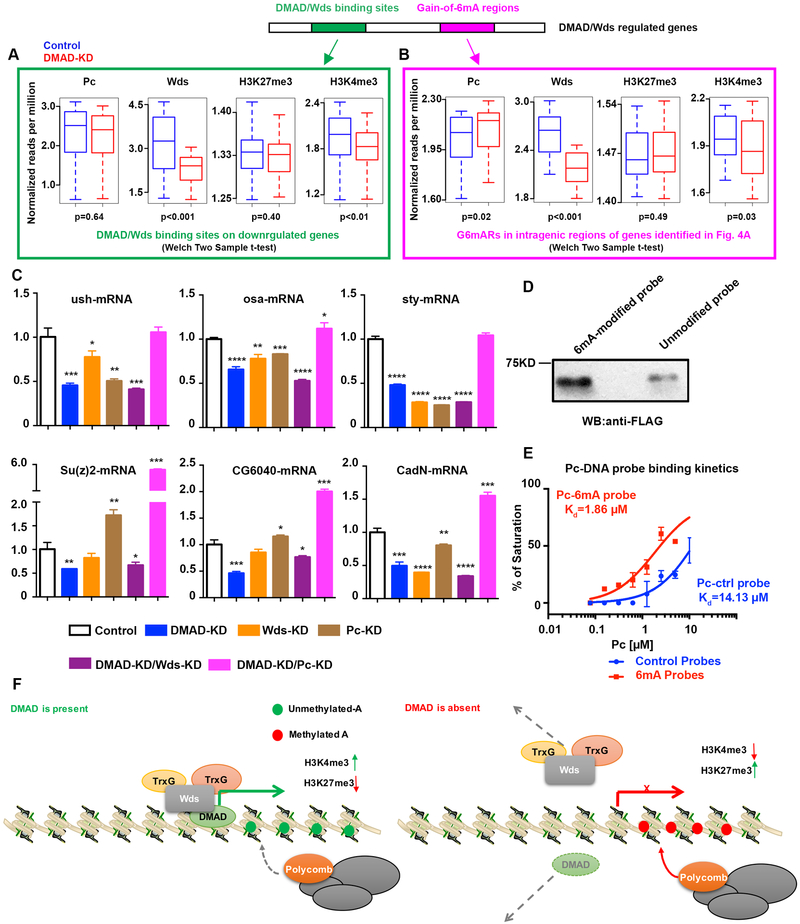
To further define DMAD-mediated epigenetic regulatory mechanisms, we focused on two sets of regions located in downregulated genes that have vital neuronal functions: 1. Intragenic regions commonly bound by DMAD/Wds identified by ChIP-seq (Figure 4A, green), and 2. Gain-of-6mA regions in intragenic regions of genes identified in Figure 4A that are also commonly bound by DMAD/Wds (Figure 4B, pink). We examined the dynamic changes of Pc and Wds binding as well as signature histone modifications, such as H3K27me3 and H3K4me3, at these loci in both control and DMAD-depleted cells. Intragenic regions bound by DMAD/Wds showed a slight but not significant reduction in Pc and H3K27me3 upon DMAD depletion. However, DMAD loss led to a substantial and significant reduction in Wds and the active histone marker H3K4me3 (Figure 4A). These data suggest that interplay between DMAD and Wds at these loci maintains transcriptional activation. We also noted that Wds was solely enriched at downregulated genes compared to non-enriched input (Figure S3P and S3Q). These data show that Wds, not Pc, is responsible for maintaining active expression of downregulated genes by interacting with DMAD. Genetic interaction between DMAD and Wds was further examined in the context of MB development. Either control (elav>+) or pan-neuronal knockdown of Wds alone (elav>WdsRNAi) does not affect α lobe development. Pan-neuronal knockdown of DMAD (elav>DMADRNAi) causes α lobe defect that is significantly enhanced by the Wds knockdown (elav> WdsRNAi + DMADRNAi) (Figure S3R). These results further confirmed functional interplay between DMAD and Wds and suggested DMAD preceded and coordinated with Wds (Figure S3R). It also suggested that DMAD could partner with other Trithorax proteins in fly neurons. In agreement with this, Trithorax-related (Trr), a major H3K4 monomethyltransferase on enhancers, shared substantial overlap with DMAD (Herz et al., 2012) (Figure S3S).
Figure 4. DMAD and 6mA coordinate with Trithorax and Polycomb.
(A) Average normalized reads (per million) dynamics in Pc, Wds, H3K27me3, and H3K4me3 at DMAD/Wds binding sites in genes downregulated in the absence of DMAD compared to the control are shown. Welch Two sample t-tests, p-values were indicated.(B) Average normalized reads (per million) dynamics in Pc, Wds, H3K27me3 and H3K4me3 at gain-of-6mA regions in intragenic regions of genes identified in Figure 4A that were bound by DMAD/Wds. Welch Two sample t-tests, p-values were indicated.(C) Loci from Figure 4A and 4B were further tested by qPCR for expression changes in the absence of DMAD, Wds, Pc, or combined depletion of DMAD and Pc, as well as DMAD and Wds. t-tests were performed. *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001.(D) In vitro 6mA-Pc binding assays were performed to confirm direct correlation between 6mA and Pc.(E) Pc binding kinetics to control and 6mA-modified probes showed that Pc displayed stronger binding to 6mA-modified DNA probes, as measured by fluorescence polarization assays. (F) DMAD binds to a group of genes involved in neurodevelopment and neuronal functions. These genes are directly targeted by the Trithorax protein Wds to maintain an active transcription profile. Additionally, DMAD actively demethylates intragenic 6mA. In the absence of DMAD,Wds binding is reduced at these loci, and accumulation of intragenic 6mA recruits Polycomb proteins. Data are presented as mean ± SEM.
Analysis of the second set of regions, which contain intragenic regions with increased 6mA upon DMAD-KD, show that Pc binding was concomitantly and significantly enhanced (Figure 4B). This observation supports the notion that accumulation of intragenic 6mA due to DMAD loss could facilitate Polycomb recruitment to secure transcriptional repression at these loci. In the absence of DMAD, these data suggest a dynamic switch between Trithorax and Polycomb proteins at these downregulated genes.
We also investigated upregulated genes bearing DMAD/Wds binding and gain-of-6mA regions. The DMAD-binding sites on these genes displayed a more sophisticated modulating network, showed by the co-occupancy of Wds and Pc on these regions (Figure S3Q, S4A, and S4B). Importantly, Gene Ontology analyses showed a specific enrichment of neurodevelopment and neuronal functional terms of these downregulated genes bound by DMAD, compared to general terms of upregulated genes (Figure S4C and S4D). This evidence implies that these downregulated genes could be the primary and direct targets of the DMAD-Wds complex related to neuronal development and functions.
To confirm the direct targets of DMAD and Wds, we independently knocked down DMAD, Wds, and Pc in BG3C2 cells. We also generated Pc/DMAD or Wds/DMAD double knockdown cell lines (Figure S4E and S4F). Several loci shown in Figure 4A and 4B were tested for expression changes with these knockdown conditions. Similar genes were downregulated with either Wds knockdown or DMAD knockdown, supporting that Wds interacts and coordinates with DMAD to transcriptionally activate these genes. DMAD and Wds double knockdown showed synergistic effects on some loci. On the other hand, simultaneous depletion of DMAD and Pc sustained or stimulated gene expression, suggesting that Pc contributes to transcription repression (Figure 4C and S4F). However, we found the knockdown of Pc itself resulted in a significant reduction of Wds (Figure S4E). The reduction in Wds might represent interesting crosstalk between TrxG and PcG (Schuettengruber et al., 2007). Considering this finding, we found that Pc knockdown resulted in reduced expression of some loci, possibly due to indirect effects of regulating Wds or other factor expression.
To obtain direct evidence that Pc preferentially binds to the 6mA mark, we synthesized DNA oligos with the gain-of-6mA/Pc common consensus sequence. The adenines in the consensuses sequence were either methylated or unmodified as a negative control. Control and 6mA-containing probes were used for in vitro binding with recombinant FLAG-Pc produced from baculovirus. We found that Pc preferentially binds to 6mA-modified probes over controls in vitro (Figure 4D). By using a Fluorescence-based DNA-binding assay (Hashimoto et al., 2014), we calculated the binding kinetics. FLAG-Pc bound to probes in a dose-dependent manner, and 6mA-modified probes showed stronger binding kinetics (Kd) to Pc relative to control (Figure 4E).
We generated three Pc deletion constructs covering the chromodomain (Pc aa1–86), the linker region (Pc aa 75–228), and the cbox domain (Pc aa222–390) and tested their binding affinities to control and 6mA-modified DNA oligos (Figure S4G and S4H) to define the 6mA-binding domain of Pc. Only the C-terminal Pc deletion (aa 222–390) bound to DNA probes and displayeda preference for 6mA-modified DNA probes, although with lower affinity than full-length Pc (Figure S4I). These data provide further evidence that 6mA has crosstalk with Polycomb proteins.
DISCUSSION
Cytosine modifications are rare if present at all in many model organisms including worms (C. elegans) and insects (D. melanogaster). Speculation exists that 6mA could serve as a viable DNA modification in these organisms to epigenetically modulate transcription. Crosstalk between 6mA and H3K4me2 in C. elegans (Greer et al., 2015), as well as communication between cytosine methylation and H3K9me3 (Du et al., 2015), has been documented. Our data identify the molecular mechanisms of 6mA and its demethylase DMAD in gene regulation. Our findings suggest that 6mA plays epigenetic roles in regulating a group of genes involved in Drosophila neurodevelopment and neuronal functions. DMAD coordinates with Trithorax-related protein Wds to maintain active transcription by removing intragenic 6mA. Depletion of DMAD results in a reduction of Wds and accumulation of 6mA. This 6mA accumulation recruits Polycomb Pc to implement transcriptional repression on these loci. Also, our data link DNA modification to histone modifications in insect cells (Figure 4F). It is worth noting that, like 5mC in mammals, the specific epigenetic role(s) of 6mA, either active or repressive, could depend on its co-factors and varies in different species, tissues, and cell types.
STAR★METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER | |||||
|---|---|---|---|---|---|---|---|
| Antibodies | |||||||
| Anti-N6-methyladenosine antibody | Synaptic Systems | 202003 | |||||
| Anti-H3K4me3 | Abeam | ab8580 | |||||
| Anti-H3K27me3 | Abeam | ab6002 | |||||
| Anti-Pc antibody (dN-19) | Santa Cruz | sc-15814 | |||||
| Anti-Wds | Novus Biologicals | 40630002 | |||||
| Anti-DMAD | Chen Lab | (Zhang et al., 2015b) | |||||
| Anti-Fasciclin II | DSHB | 1D4 | |||||
| Anti-HA(16B12) | Covance | MMS-101P | |||||
| Anti-Myc(9E10) | Thermo Fisher | MA1-980 | |||||
| Rabbit TrueBlot anti-Rabbit IgG HRP | Rockland | 18-8816-33 | |||||
| Mouse TrueBlot anti-Mouse IgG HRP | Rockland | 18-8817-30 | |||||
| Alexa Fluor® 488 Anti-Rabbit IgG | Jackson ImmunoResearch | 711-545-152 | |||||
| Cy™3 Anti-RabbitlgG (H+L) | Jackson ImmunoResearch | 711-165-152 | |||||
| Anti-FLAG® M2 Magnetic Beads | Sigma | M8823 | |||||
| Anti-HA-Agarose | Sigma | A2095 | |||||
| EZview™ Red Anti-c-Myc Affinity Gel | Sigma | E6654 | |||||
| Dynabeads™ Protein G | Thermo Fisher | 10003D | |||||
| Chemicals, Peptides, and Recombinant Proteins | |||||||
| Recombinant Pc full length | This study | Pc full length | |||||
| Recombinant Pc (aa 1–86) | This study | Pc(aal-86) | |||||
| Recombinant Pc (aa 75–228) | This study | Pc (aa 75–228) | |||||
| Recombinant Pc (aa 222–390) | This study | Pc (aa 222–390) | |||||
| DMAD catalytic domain | This study | N/A | |||||
| Agencourt AMPure XP beads | Beckman Coulter | A63880 | |||||
| Dynabeads™ MyOne™ Streptavidin Cl | Thermo Fisher | 65001 | |||||
| Cellfectin II Reagent | Thermo Fisher | 10362100 | |||||
| Dpnl I | New England Biolabs | R0176S | |||||
| Calf intestinal phosphatase | New England Biolabs | M0290S | |||||
| Phosphodiesterase I from Crotalus adamanteus venom | Sigma | P3243 | |||||
| Insulin solution human | Sigma | 19278 | |||||
| Sodium Ascorbate | Sigma | A4034 | |||||
| Alpha-ketoglutaric acid | Sigma | K3752 | |||||
| Lithium chloride | Sigma | L4408 | |||||
| Bacterial and Virus Strains | |||||||
| E. coli (DH5a)-One Shot Top1O | Thermo Fisher | C404003 | |||||
| Baculovirus | LakePhama | N/A | |||||
| Critical Commercial Assays | |||||||
| Qubit™ dsDNA HS Assay Kit | Thermo Fisher | Q32854 | |||||
| High Sensitivity DNA Analysis Kits | Agilent | 5067-4627 | |||||
| NEBNext DNA Library Prep Reagent Set | New England Biolabs | E6000 | |||||
| HiScnbe™ T7 High Yield RNA Synthesis Kit | New England Biolabs | E2040S | |||||
| TruSeq™ RNA Sample Prep Kit V2 | Ilium ina | RS-122-2001 | |||||
| Pierce BCA Protein Assay Kit | Thermo Fisher | 23225 | |||||
| Q5® Site-Directed Mutagenesis Kit | New England Biolabs | E0554S | |||||
| Pierce™ Biotin 3’ End DNA Labeling Kit | Thermo Fisher | 89818 | |||||
| Experimental Models: Cell lines | |||||||
| BG3C2 cells | The Drosophila Genomics Resource Center (DGRC) | (Ui et al., 1994) | |||||
| SF9 cells | Lab of Xiaodong Cheng | N/A | |||||
| Experimental Models: Organisms/Strains | |||||||
| DMAD1 | Chen Lab | (Zhang et al., 2015b) | |||||
| DMAD2 | Chen Lab | (Zhang et al., 2015b) | |||||
| DMAD RNAi | This study | N/A | |||||
| Wds RNAi | Bloomington stock center | 60399 | |||||
| Recombinant DNA | |||||||
| pET-Pc-1-390 | Harte Lab | (Tie et al., 2016) | |||||
| pET-Pc-1-86 | Harte Lab | (Tie et al., 2016) | |||||
| pET- Pc-75-390 | Harte Lab | (Tie et al., 2016) | |||||
| pET- Pc-75-228 | Harte Lab | (Tie et al., 2016) | |||||
| pFastBac FPC | Addgene | Plasmid#1927 | |||||
| pFastBac-Flag-Pc-1-86 | This study | N/A | |||||
| pFastBac-Flag-Pc-75-228 | This study | N/A | |||||
| pFastBac-Flag-Pc-222-390 | This study | N/A | |||||
| pAc5.1-HA-Wds full length | This study | N/A | |||||
| pAc5.1-HA-Wds-l-61 | This study | N/A | |||||
| pAc5.1-HA-Wds-l-187 | This study | N/A | |||||
| pAc5.1-HA-Wds-l-262 | This study | N/A | |||||
| pAc5.1-myc- DMAD full length | This study | N/A | |||||
| pAc5.1 -myc- DMAD-1 −1657 | This study | N/A | |||||
| pAc5.1 -myc-DMAD-1657-2860 | This study | N/A | |||||
| pAc5.1-myc-1658-1796 | This study | N/A | |||||
| pAc5.1-myc-1797-2666 | This study | N/A | |||||
| pAc5.1-myc-2667-2860 | This study | N/A | |||||
| Oligonucleotides | |||||||
| artmiR-DMAD-l -s: 5 ‘-ctagcagtCGATGTACTAGAATGGCTGGAtagttatattcaagcataTGCAGCCATTGTAGTACATCGgcg-3’ | IDT | N/A | |||||
| artmiR-DMAD-2-s: 5’-ctagcagtCGCCTATGATCCCTATCAGTAtagttatattcaagcataTTCTGATAGGCATCATAGGCGgcg-3’ | IDT | N/A | |||||
| artmiR-DMAD-2-as: 5’-aattcgcCGCCTATGATGCCTATCAGAAtatgcttgaatataactaTACTGATAGGGATCATAGGCGactg-3’ | IDT | N/A | |||||
| 6mA-modified probe for Pc binding assay: GAT CGA TCG ACA /iN6Me-dA/CA /iN6Me-dA/CA /iN6Me-dA/CA /iN6Me-dA/CA /iN6Me-dA/CA /iN6Me-dA/CA /iN6Me-dA/CA /iN6Me-dA/GA TCG ATC GA | IDT | N/A | |||||
| Reverse complemented probe: TCG ATC GAT CTG TGT GTG TGT GTG TGT CGA TCG ATC | IDT | N/A | |||||
| Software and Algorithms | |||||||
| Bowtie vl.1.2 | (Langmead et al., 2009) | ||||||
| MACS 1.4.2 | (Zhang et al., 2008) | ||||||
| Ngsplot 2.61 | (Shen et al., 2014) | ||||||
| HOMER v4.9.1 | (Heinz et al., 2010) | ||||||
| TopHat v2.0.13 | (Trapnell et al., 2012) | ||||||
| Cuffdiff v2.2.1 | (Trapnell et al., 2012) | ||||||
| Bedtools v2.17.0 | (Quinlan and Hall, 2010). | ||||||
| R/Bioconductor package DSS 3.7 | (Wu et al., 2013) | ||||||
| Cis Genome v2.0 | (Ji et al., 2006) | ||||||
| TEToolkit vl.5.1 | (Jin et al., 2015) | ||||||
| dynamic analysis of nucleosome position and occupancy by sequencing (DANPOS) v2 | (Chen et al., 2013) | ||||||
| Deposited Data | |||||||
| All sequencing data | GSE67855 | ||||||
| Raw images | Mendeley Data | ||||||
| DOI: 10.17632/wy3jshg29 m.3 | |||||||
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and classifiers should be directed to and will be fulfilled by Bing Yao (bing.yao@emory.edu) or Peng Jin (peng.jin@emory.edu). There are no restrictions for use of the materials disclosed.
METHOD DETAILS
DMAD-null and DMAD-KD fly lines
DMAD-null flies were described previously (Zhang et al., 2015b). DMAD transgenic RNAi lines, P{uasp-2×artmiR-DMAD}, in which two sets of 71 nt oligos containing a small hairpin sequence targeting the DMAD coding region under the control of the uasp promoter, were generated according to the method described previously (Zhang et al., 2015b). The sequences of these oligos are below.
artmiR-DMAD-1-s: 5’-
ctagcagtCGATGTACTAGAATGGCTGGAtagttatattcaagcataTGCAGCCATTGTAGTACATCG
gcg-3’
artmiR-DMAD-1-as:
5’-
aattcgcCGATGTACTACAATGGCTGCAtatgcttgaatataactaTCCAGCCATTCTAGTACATCGa
ctg-3’
artmiR-DMAD-2-s:
5’-
ctagcagtCGCCTATGATCCCTATCAGTAtagttatattcaagcataTTCTGATAGGCATCATAGGCGgcg
−3’
artmiR-DMAD-2-as:
5’-
aattcgcCGCCTATGATGCCTATCAGAAtatgcttgaatataactaTACTGATAGGGATCATAGGCGactg
−3’
Immunostaining
Fly brains were dissected in calcium-free 1x PBS and fixed in 4% paraformaldehyde for 1 hour on ice. Fixed tissues were permeabilized with 0.3% PTX (1x PBS + 0.3% Triton X-100) at room temperature for 30 minutes and then blocked in 0.1% PTX (1x PBS + 0.1% Triton X-100) + 5% Normal Goat Serum for 30 more minutes. Tissues were then incubated with anti-DMAD (1:200, Rb) or anti-6mA (1:500, Rb) in PBTG (PBST + 5% normal goat serum) overnight at 4ºC. Anti-Fasciclin II monoclonal antibody (DHSB, 1D4) was used to stain the Drosophila mushroom body. The next day, tissues were washed 3 times with 1x PBST for 5 minutes and incubated with secondary antibody for 1 hour at room temperature. The tissues were washed 3 times with 1x PBST and mounted onto slides with Vectashield to prevent photobleaching. Samples were kept at −20ºC until observed under confocal microscopy.
Cell culture and RNAi
BG3C2 cells were cultured as described previously (Ui et al., 1994). For double stranded RNA (dsRNA) knockdown, a 580bp dsRNA fragment targeting DMAD coding region was generated via T7 promoter-driven in vitro transcription using an RT-PCR product as template. The following primers were used for RT-PCR: forward −5’gaaatCTCGAGtaatacgactcactatagggCGGAGCCAGTAGTTTTCAGC3’and reverse −5’gaaatGAATTCtaatacgactcactatagggCATGGGGTTGATCTTCTCGT3’. The in vitro transcription was performed using HiScribe T7 High Yield RNA Synthesis Kit from NEB following manufacturer’s instructions. dsRNA products were purified by two-step extractions with phenol: chloroform: isoamyl alcohol =25:24:1, followed by chloroform alone. The extracted dsRNA was precipitated with absolute ethanol. The dsRNA was incubated with BG3C2 cells that had been grown in Shields and Sang M3 insect medium under FBS starvation for at least 24 hours. After 24-hour incubation with dsRNA, FBS was added to the culture media to a final concentration of 3%. Fresh media containing dsRNA was replaced every 48 hours and the total incubation time was 144 hours to ensure effective knockdown.Drosophila S2 cells were cultured as described previously for co-immunoprecipitation experiments (Zhang et al., 2015a).
Isolation of genomic DNA
Lysis buffer containing 100mM Tris-HCl (pH 8.5), 5mM EDTA (pH 8.0), 0.2% SDS, 200mM NaCl, and 20–25 l of protease K (20mg/ml) was added to homogenized tissues or cells, mixed well, and incubated at 55°C overnight. After the overnight digestion, the lysates were brought to room temperature, and incubated with 5 l of RNase A solution (20mg/ml) for at least 1 hour at room temperature. DNA was extracted by adding equal volume of phenol: chloroform: isoamyl alcohol at a ratio of 25:24:1 and centrifuged at 13,000 rpm for 10 minutes. Supernatant was transferred to clean tubes. Equal volume of isopropanol was then added to the supernatant and mixed well at room temperature to precipitate DNA. Once flocky DNA was visible, the precipitate was transferred to a new tube containing 1ml of 70% ethanol to wash. DNA was then collected by centrifugation and air dried.
UHPLC-MRM-MS/MS analysis
Genomic DNA was enzymatically digested into single nucleosides with a mixture of DNaseI, calf intestinal phosphatase, and snake venom phosphodiesterase I at 37°C for 12 h. After the enzymes were removed by ultrafiltration, the digested DNA was subjected to UHPLC-MS/MS analysis. HPLC fractionation of Drosophila m6dA and UHPLC-QTOF-MS/MS analysis were performed as described previously (Zhang et al., 2015b).
Dot blot
Dot blot was performed as described previously (Szulwach et al., 2011) using 6mA rabbit polyclonal antibody. DNA samples were subjected to extensive RNase treatments before loading on Nitrocellulose membrane.
6mA Immunoprecipitation
Genomic DNA was sonicated to 200–300 bp fragments for 6mA enrichment using rabbit polyclonal antibody (Synaptic Systems) at 1:100 in 1x IP buffer containing 100mM Tris-HCl (pH7.4), 150mM NaCl and 0.05% Triton X-100. The DNA-antibody incubation was conducted on a rotating platform at 4°C overnight. Dynabeads Protein G (Novex by Life Technologies, REF 10009D 30mg/ml) were added the next day for additional 2 hours at 4°C. The beads were then washed six times at room temperature with 1x ice-cold IP buffer. After the wash, the immunoprecipitated DNA fragments were eluted by adding IP buffer containing 2.6mM 6mA for competitive elution. The elutions were repeated for three times at room temperature. The eluted DNA fragments were precipitated by isopropanol.
Chromatin Immunoprecipitation (ChIP)
BG3C2 cells (5–10 × 106) in the presence or absence of DMAD were fixed in 1% formaldehyde for 10 minutes at room temperature with gentle shaking, then 0.125 M final concentration of Glycine was added for additional 5-minute incubation to stop the fixation. Fixed cells were lysed on ice for 10 minutes in a NP-40 lysis buffer (10 mM Hepes/pH7.9, 0.5% NP-40, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT and protease inhibitor cocktail) to release nuclei. After centrifugation at 4000 rpm for 5 minutes, the nuclear pellets were further lysed by sonication on ice in a nuclear lysis buffer (20 mM Hepes/pH7.9, 25% glycerol, 0.5% NP-40, 0.5% Triton X-100, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA and protease inhibitor cocktail), then centrifuged at 13,000 rpm for 10 minutes at 4°C. The supernatant was diluted with 2 volumes of dilution buffer (0.01% SDS, 1% Triton X-100, 1.2 mM EDTA, 167 mM NaCl, 16.7 mM Tris-HCl/pH8.0 and protease inhibitor cocktail). Immunoprecipitation was performed with desired antibodies for 6 hours to overnight at 4ºC. After antibody incubation, 20 l salmon sperm blocked DNA/protein G agarose (Upstate) were added and incubated for additional 1 hour. Precipitates were sequentially washed with TSE I (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl/pH 8.0), TSE II (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 500 mM NaCl, 20 mM Tris-HCl/pH 8.0), TSE III (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl/pH 8.0), and then washed twice with TE buffer. The ChIP’ed DNA was eluted by elution buffer containing 1% SDS and 0.1 M NaHCO3. NaCl at final concentration of 0.2M was added to the elution along with 1 g RNaseA for reverse-crosslinking at 65 ºC for at least 6 hours. DNA fragments were purified using the PCR Purification Kit (Qiagen). ChIP’ed DNA was subjected to library preparation.
Co-Immunoprecipitation
BG3C2 or S2 cells were lysed for 10 minutes on ice with lysis buffer containing 25 mM Tris-HCl (pH7.5), 2mM MgCl2, 5mM DTT, 0.5%Triton-X100, and 300mM NaCl in the presence of DNase and RNase. The cell lysate was sonicated before centrifugation at maximum speed for 10 min; the supernatant was incubated with antibodies overnight at 4ºC. After antibody incubation, 20 l salmon sperm blocked DNA/protein G agarose beads (Upstate) were added and incubated for additional 1 hr. Beads were then washed 3 times by lysis buffer. The immunoprecipitated proteins were eluted with 2x Laemmli Sample Buffer (Bio-Rad) followed by western blots.
Baculovirus infection and FLAG-tagged Pc purification
SF9 cells were a gift from Dr. Xiaodong Cheng’s lab in the Department of Biochemistry at the Emory University School of Medicine. SF9 cells were cultured in Sf-900™ III SFM media from Thermo Fisher Scientific, and infected with Baculovirus generated with pFastBac-FPC (full length Pc) with FLAG tag obtained from Addgene. Baculovirus were produced by LakePharma. 72-hour post infection, SF9 cells were collected and lysed in lysis buffer containing 25mM Tris-Cl (pH 7.5), 300mM NaCl, 0.5% Triton-X100, and protease inhibitor tablet (Roche) for 30 minutes on ice followed by sonication at 1.5V, 0–3W for 20 seconds. Samples with 300 l volume in each tube were sonicated 4 times. Lysate was centrifuged at 13,000 rpm for 10 minutes and supernatant was transferred to a new tube for IP purification. The supernatant was incubated with FLAG-M2 dynabeads (Sigma) and rotated at 4°C for 3 hours. Beads were then washed 5 times with 1x lysis buffer. To elute the FLAG-Pc, beads were incubated with the elution buffer containing 3X FLAG peptide at 100ug/ml for 2 hours at 4°C with gentle rotation. FLAG-Pc were tested by western blots.
Probe labeling and annealing
To compare Pc binding efficiency to 6mA DNA and unmodified DNA, two oligos were designed based on gain-of-6mA regions and the consensus sequence recognized by Pc (−5’-GAT CGA TCG A-CA CAC ACA CAC ACA CA-G ATC GAT CGA-3’). The forward and reverse oligos were separately labeled using the Biotin 3’ End DNA Labeling Kit (Catalog number: 89818) from Pierce™Fisher Scientific per manufacturer’s instruction. Labeled oligos were precipitated with 0.1 volume of 3M NaAc (pH 5.2) and 3 volumes of absolute ethanol. Precipitated oligos were dissolved in 50 l of nuclease free water and annealed into double stranded oligos.
Pc and DNA probe binding assays
dsDNA probes with either unmodified A or 6mA were independently mobilized on Dynabeads Streptavidin MyOne C1 (Life Technologies) with 1x W/B buffer containing 25mM Tris-Cl (pH7.5), 1mM EDTA, and 1M NaCl at room temperature for 30 minutes with gentle rotation. The beads were then immobilized with probes and blocked with Blocker™ BSA (10x), Thermo Fisher Scientific (Catalog number: 37520) in TBS for 30 minutes at room temperature. Beads were washed twice with 1x lysis buffer. A range of purified FLAG-Pc protein concentrations (0.01 M to 1 M) was added to blocked beads and incubated for 1 hour at room temperature with gentle rotation. The beads were then washed extensively with 1x lysis buffer 5 times. FLAG-Pc was eluted in 2x Laemmli Sample Buffer (Bio-Rad) at 100°C for 10 minutes and loaded onto an SDS-polyacrylamide gel for Western blot to detect binding efficiency.
Generation of deletion/truncation constructs of DMAD, Wds and Pc
Site-directed mutagenesis kit (NEB) was used to generate series of deletion constructs from full length cDNA from DMAD, Wds and Pc. All constructs were subjected to Sanger sequencing to confirm the correct insertion. DMAD and Wds constructs were cloned into pAc 5.1 vector and expressed in Drosophila S2 cells for co-immunoprecipitation experiments. Pc deletion constructs were cloned into pFASTBAC plasmid for Baculovirus production (LakePharma).
Fluorescence-based Pc-6mA binding assays
The 6-carboxy-fluorescein (FAM)-labelled control or 6mA-modified DNA probes were annealed with their reverse complementary strand to form double-stranded DNA oligos. Various concentrations (0.01–1 M) of Pc full length or C-terminal truncation recombinant proteins were incubated with 2 nM DNA oligos for 15 minutes at room temperature in nicking buffer (10mM Tris-Cl pH8.0, 1mM EDTA, 0.1% BSA). Fluorescence polarization measurements were carried out at 25°C on a Synergy 4 microplate reader (BioTek). Curves were fit individually using GraphPad Prism 7.0 software (GraphPad Software, Inc.). Binding constants (Kd) were calculated described previously (Hashimoto et al., 2014).
In vitro 6mA demethylation assay
Double stranded control and 6mA-modified DNA oligos were mixed with DMAD C-terminal catalytic domain (aa 1657–2860) in reaction buffer containing 50mM Hepes (pH8.0), 2mM ascorbate, 1mM a-KG and 150 mM Fe at room temperature for 3 hours. The reactions were stopped by proteinase K digestion at 50°C for 2 hours. The purified DNA samples were subjected to 6mA dot blots and ImageJ quantification.
DpnI digestion and qPCR
DpnI digestion and qPCR was conducted as previously described (Luo et al., 2016). Briefly, restriction enzyme digestion was performed by treating 1 ug of genomic DNA with 5 uL of 5 U/uL DpnI restriction enzyme (NEB) at 37°C for 1 hour. The digested DNA and non-digested DNA (5 ng) were subjected to qPCR using FastStart SYBR Green Master kit. The restriction enzyme digestion method takes advantage of the 6mA-sensitive restriction enzyme DpnI that preferentially cleaves methylated adenine at GATC/CATC/GATG sites. Equal amounts of DpnI-digested DNA and undigested control DNA were subjected to qPCR analyses with primers targeting 6mA dynamic regions identified by 6mA-IP. The percentage of 6mA in either control or DMAD-KD can be assessed by qPCR amplification and normalized to undigested DNA control (digested/undigested). Loci with lower 6mA modification in controls would hinder DpnI digestion, resulting in higher PCR fold changes than DMAD-KD samples.
Library Preparation and High-throughput Sequencing
Enriched DNA from 6mA-IP and ChIP were subjected to library construction using the NEBNext ChIP-Seq Library Prep Reagent Set from Illumina according the manufacturer’s protocol. Briefly, 25 ng of input genomic DNA or experimental enriched DNA were utilized for each library construction. DNA fragments (150–300 bp) were selected by AMPure XP Beads (Beckman Coulter) after adapter ligation. An Agilent 2100 BioAnalyzer was used to quantify amplified DNA and qPCR was applied to accurately quantify library concentration. 20 pM diluted libraries were used for sequencing. 50-cycle single-end sequencings were performed using Illumina HISeq 2000. Image processing and sequence extraction were done using the standard Illumina Pipeline. RNA-seq libraries were generated from duplicated samples per condition using the Illumina TruSeq RNA Sample Preparation Kit v2 following manufacturer’s protocol. RNA-seq libraries were sequenced as 50-cycle pair-end runs using Illumina HISeq 2000.
Bioinformatics analyses
Bioinformatics analyses for ChIP-Seq and 6mA-IP-seq were conducted as described previously (Szulwach et al., 2011; Yao et al., 2014). Briefly, FASTQ sequence files were aligned to the dm3 reference genome using Bowtie v1.1.2 (Langmead et al., 2009). Peaks were identified by Model-based Analysis of ChIP-Seq (MACS) software (Zhang et al., 2008). Ngsplot software was used to calculate and plot unique 6mA and ChIP-Seq mapped reads various genomic regions and generated heatmaps (Shen et al., 2014). Annotation and motif analysis were performed using the HOMER (Heinz et al., 2010) suite. RNA-seq reads were aligned using Tophat v2.0.8 (Trapnell et al., 2012) and differential RPKM expression values were extracted using cuffdiff v2.2.1 (Trapnell et al., 2012). Genomic interval overlapping analyses were performed using Bedtools (Quinlan and Hall, 2010). Gene Ontology analyses were performed by The Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 (Huang da et al., 2009) and Gene Ontology Consortium (Ashburner et al., 2000). Gain-of-6mA regions in BG3C2 cells were identified by published computational algorithm (R/Bioconductor package DSS) that implements a series of differential methylation detection algorithms based on the dispersion shrinkage method followed by Wald statistical test to rigorously interrogate the 6mA differential regions between replicated samples of control and DMAD-KD group (Feng et al., 2014). Top-ranked gain-of-6mA regions were further intersected with 6mA peaks identified by MACS with FDR<0.05. modENCODE ChIP-chip regions were generated by CisGenome (Ji et al., 2006). Transposon expression was analyzed by TEtranscripts (Jin et al., 2015). Gain-of-6mA regions in brains were identified by bedtools (Quinlan and Hall, 2010). MNase-seq data were analyzed using dynamic analysis of nucleosome position and occupancy by sequencing (DANPOS) (Chen et al., 2013).
Sample size and Statistics
Fly brain RNA-seq experiments were performed using two control and DMAD-null pooled samples as biological replicates. Each pooled sample contained 500–1000 fly brains per sample per genotype. Fly brains were dissected from heads to avoid 6mA contamination from bacteria. BG3C2 RNA-seq samples were performed in triplicate from control and DMAD-KD cells. Brain RNA-seq were performed in duplicates. Differential expression analyses were performed by cuffdiff (Trapnell et al., 2012). 6mA-IP in control and DMAD-null brains were performed using biological replicates from 1000 pooled fly brains per sample per genotype. 6mA-IP experiments in BG3C2 cells were performed in duplicate using control and DMAD-KD cells. Control and DMAD-KD replicate 1 was sequenced twice serving as a technical replicate. DMAD ChIP-Seq experiments in BG3C2 cells were performed in triplicate. All replicated samples from each condition were merged for downstream bioinformatic analyses. Pearson’s Chi-squared tests with Yates’ continuity correction and Welch Two Sample t-tests were performed in R computational environment (http://www.r-project.org/). Student’s t-tests were performed in Graphpad Prism (http://www.graphpad.com/scientific-software/prism/).
DATA AND SOFTWARE AVAILABILITY
Original imaging data have been deposited to Mendeley Data and are available at https://data.mendeley.com/datasets/wy3jshg29m/draft?a=374e997d-4c76-49e0-9d08-d9e763dd55b1
Supplementary Material
Table S1, Related to Figure 1. HPLC quantification of 5hmC and 6mA in Drosophila brains; Active 6mA demethylation loci in Drosophila brains; Gene expression analyses of control and DMAD-null fly brains
Table S2, Related to Figure 2. Active 6mA demethylation loci in fly neuronal cells, BG3C2; Gene expression analyses of control and DMAD-KD BG3C2 cells.
Table S4, Related to STAR methods. Primer sequences
Highlights.
DMAD is critical in Drosophila brain by actively demethylating 6mA
DMAD interacts with Trithorax-related protein Wds to regulate gene expression
6mA correlates with the binding of Polycomb proteins in Drosophila genome
DMAD and 6mA coordinates with Trithorax and Polycomb proteins for gene regulation
ACKNOWLEDGMENTS
We would like to thank S. Warren, K. Garber and D. Cook for critical reading of the manuscript. This work was supported in part by National Institutes of Health (NS051630, NS079625, NS097206, NS091859, MH102690, AG052476 and HG008935 to P.J.), March of Dimes (6-FY13-121 to P.J.), and the Emory Genetics Discovery Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes four f gures and can be found with this article online at
DECLARATION OF INTERESTS
The authors declare no conflict of interest.
REFERENCES
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. (2000).Gene ontology: tool for the unification of biology.The Gene Ontology Consortium. Nature genetics 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A (2002).DNA methylation patterns and epigenetic memory.Genes Dev 16, 6–21. [DOI] [PubMed] [Google Scholar]
- Chen K, Xi Y, Pan X, Li Z, Kaestner K, Tyler J, Dent S, He X, and Li W (2013).DANPOS: dynamic analysis of nucleosome position and occupancy by sequencing.Genome Res 23, 341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatte B, Wang F, Ngoc LV, Collignon E, Bonvin E, Deplus R, Calonne E, Hassabi B, Putmans P, Awe S, et al. (2016).RNA biochemistry. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine.Science 351, 282–285. [DOI] [PubMed] [Google Scholar]
- Du J, Johnson LM, Jacobsen SE, and Patel DJ (2015).DNA methylation pathways and their crosstalk with histone methylation.Nature reviews. Molecular cell biology 16, 519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunwell TL, McGuffin LJ, Dunwell JM, and Pfeifer GP (2013).The mysterious presence of a 5-methylcytosine oxidase in the Drosophila genome: possible explanations.Cell cycle 12, 3357–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Conneely KN, and Wu H (2014).A Bayesian hierarchical model to detect differentially methylated loci from single nucleotide resolution sequencing data.Nucleic acids research 42, e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Luo GZ, Chen K, Deng X, Yu M, Han D, Hao Z, Liu J, Lu X, Dore LC, et al. (2015).N6-methyldeoxyadenosine marks active transcription start sites in Chlamydomonas.Cell 161, 879–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Blanco MA, Gu L, Sendinc E, Liu J, Aristizabal-Corrales D, Hsu CH, Aravind L, He C, and Shi Y (2015).DNA Methylation on N6-Adenine in C.elegans. Cell 161, 868–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Olanrewaju YO, Zheng Y, Wilson GG, Zhang X, and Cheng X (2014).Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.Genes & development 28, 2304–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010).Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities.Mol Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisenberg M (2003).Mushroom body memoir: from maps to models.Nat Rev Neurosci 4, 266–275. [DOI] [PubMed] [Google Scholar]
- Herz HM, Mohan M, Garruss AS, Liang K, Takahashi YH, Mickey K, Voets O, Verrijzer CP, and Shilatifard A (2012).Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4.Genes & development 26, 2604–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JW, Jung YL, Liu T, Alver BH, Lee S, Ikegami K, Sohn KA, Minoda A, Tolstorukov MY, Appert A, et al. (2014).Comparative analysis of metazoan chromatin organization.Nature 512, 449–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, and Lempicki RA (2009).Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources.Nature protocols 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Ji H, Vokes SA, and Wong WH (2006).A comparative analysis of genome-wide chromatin immunoprecipitation data for mammalian transcription factors.Nucleic acids research 34, e146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Tam OH, Paniagua E, and Hammell M (2015).TEtranscripts: a package for including transposable elements in differential expression analysis of RNA-seq datasets.Bioinformatics 31, 3593–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziol MJ, Bradshaw CR, Allen GE, Costa AS, Frezza C, and Gurdon JB (2016).Identification of methylated deoxyadenosines in vertebrates reveals diversity in DNA modifications.Nature structural & molecular biology 23, 24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, and Salzberg SL (2009).Ultrafast and memory-efficient alignment of short DNA sequences to the human genome.Genome biology 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo GZ, Wang F, Weng X, Chen K, Hao Z, Yu M, Deng X, Liu J, and He C (2016).Characterization of eukaryotic DNA N(6)-methyladenine by a highly sensitive restriction enzyme-assisted sequencing.Nature communications 7, 11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyko F, Ramsahoye BH, and Jaenisch R (2000).DNA methylation in Drosophila melanogaster.Nature 408, 538–540. [DOI] [PubMed] [Google Scholar]
- Ma DK, Marchetto MC, Guo JU, Ming GL, Gage FH, and Song H (2010).Epigenetic choreographers of neurogenesis in the adult mammalian brain.Nature neuroscience 13, 1338–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M, Herz HM, Smith ER, Zhang Y, Jackson J, Washburn MP, Florens L, Eissenberg JC, and Shilatifard A (2011).The COMPASS family of H3K4 methylases in Drosophila.Molecular and cellular biology 31, 4310–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondo SJ, Dannebaum RO, Kuo RC, Louie KB, Bewick AJ, LaButti K, Haridas S, Kuo A, Salamov A, Ahrendt SR, et al. (2017).Widespread adenine N6-methylation of active genes in fungi.Nature genetics 49, 964–968. [DOI] [PubMed] [Google Scholar]
- Orsi GA, Kasinathan S, Hughes KT, Saminadin-Peter S, Henikoff S, and Ahmad K (2014).High-resolution mapping defines the cooperative architecture of Polycomb response elements.Genome Res 24, 809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010).BEDTools: a flexible suite of utilities for comparing genomic features.Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raddatz G, Guzzardo PM, Olova N, Fantappie MR, Rampp M, Schaefer M, Reik W, Hannon GJ, and Lyko F (2013).Dnmt2-dependent methylomes lack defined DNA methylation patterns.Proc Natl Acad Sci U S A 110, 8627–8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler D (2015).Function and information content of DNA methylation.Nature 517, 321–326. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, and Cavalli G (2007).Genome regulation by polycomb and trithorax proteins.Cell 128, 735–745. [DOI] [PubMed] [Google Scholar]
- Shen L, Shao N, Liu X, and Nestler E (2014).ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases.BMC genomics 15, 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivakov M, and Fisher AG (2007).Epigenetic signatures of stem-cell identity.Nature reviews. Genetics 8, 263–271. [DOI] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, Irier H, Upadhyay AK, Gearing M, Levey AI, et al. (2011).5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging.Nature neuroscience 14, 1607–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie F, Banerjee R, Fu C, Stratton CA, Fang M, and Harte PJ (2016).Polycomb inhibits histone acetylation by CBP by binding directly to its catalytic domain.Proceedings of the National Academy of Sciences of the United States of America 113, E744–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, and Pachter L (2012).Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks.Nature protocols 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ui K, Nishihara S, Sakuma M, Togashi S, Ueda R, Miyata Y, and Miyake T (1994).Newly established cell lines from Drosophila larval CNS express neural specific characteristics.In Vitro Cell Dev Biol Anim 30A, 209–216. [DOI] [PubMed] [Google Scholar]
- Wu H, Wang C, and Wu Z (2013).A new shrinkage estimator for dispersion improves differential expression detection in RNA-seq data.Biostatistics 14, 232–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TP, Wang T, Seetin MG, Lai Y, Zhu S, Lin K, Liu Y, Byrum SD, Mackintosh SG, Zhong M, et al. (2016).DNA methylation on N(6)-adenine in mammalian embryonic stem cells.Nature 532, 329–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao B, Lin L, Street RC, Zalewski ZA, Galloway JN, Wu H, Nelson DL, and Jin P (2014).Genome-wide alteration of 5-hydroxymethylcytosine in a mouse model of fragile X-associated tremor/ataxia syndrome.Human molecular genetics 23, 1095–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Robinson BS, Xu W, Yang L, Yao B, Zhao H, Byun PK, Jin P, Veraksa A, and Moberg KH (2015a).The ecdysone receptor coactivator Taiman links Yorkie to transcriptional control of germline stem cell factors in somatic tissue.Developmental cell 34, 168–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Huang H, Liu D, Cheng Y, Liu X, Zhang W, Yin R, Zhang D, Zhang P, Liu J, et al. (2015b).N6-methyladenine DNA modification in Drosophila.Cell 161, 893–906. [DOI] [PubMed] [Google Scholar]
- Zhang L, Lu X, Lu J, Liang H, Dai Q, Xu GL, Luo C, Jiang H, and He C (2012).Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA.Nature chemical biology 8, 328–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008).Model-based analysis of ChIP-Seq (MACS).Genome biology 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1, Related to Figure 1. HPLC quantification of 5hmC and 6mA in Drosophila brains; Active 6mA demethylation loci in Drosophila brains; Gene expression analyses of control and DMAD-null fly brains
Table S2, Related to Figure 2. Active 6mA demethylation loci in fly neuronal cells, BG3C2; Gene expression analyses of control and DMAD-KD BG3C2 cells.
Table S4, Related to STAR methods. Primer sequences