ABSTRACT
Despite the opposite roles of Tbet and Foxp3 in the immune system as well as in tumour biology, recent studies have demonstrated the presence of of CD4+ T cells, expressing both, Tbet and Foxp3. Although Tbet+Foxp3+ T cells are currently a subject of intense research, less is known about their biological function especially in cancer. Here we found a considerable accumulation of Tbet+Foxp3+CD4+ T cells, mediated by the immunosuppressive cytokine TGFβ in the lungs of tumour bearing mice. This is in line with previous studies, demonstrating the important role of TGFβ for the immunopathogenesis of cancer. By gathering results both in murine model and in human disease, we demonstrate that, the conversion of IFNγ producing anti-tumoral T-bet+Th1 CD4+ T cells into immunosuppressive Tbet and Foxp3-PD1 co-expressing regulatory cells could represent an additional important mechanism of TGFβ-mediated blockade of anti-tumour immunity.
KEYWORDS: Tbet, Foxp3, PD1, NSCLC, Immunotherapy
Introduction
Lung cancer represents the leading cause of cancer deaths worldwide, being responsible for more than one million deaths every year.1,2 Recent studies suggest that immunotherapy could be a promising approach to improve the prognosis for this disease.3,4 The recognition and elimination of tumour cells by the immune system requires the presence of Tbet (Tbx21), a transcriptional regulator that induces the differentiation of Th1 cells and promotes the production of cytolytic effector molecules by CD8+ T cells and NK cells.4-8 Accordingly, Tbet-deficiency leads to significantly increased lung tumour growth in mice.3 Tbet is an immune cell specific member of the T-box protein family, which is encoded by the Tbx21 gene and is expressed by different kinds of immune cells that contribute to tumour rejection.4-6 The significance of Tbet-mediated effects for efficient anti-tumour immune responses is supported by a previous study, demonstrating that Tbet-deficiency leads to significantly increased tumour development in a murine model of lung cancer.3 Furthermore, we recently demonstrated that reduced mRNA levels of TBX21 in the lungs of patients with non-small cell lung cancer are associated with increased tumour diameters, pointing out that Tbet-dependent immune responses are not only of high relevance for the limitation of tumour growth in a murine model, but also in human lung cancer.9
Despite of the general ability of the immune system to recognize and eliminate malignancies, tumour cells have evolved numerous strategies to escape immune-mediated eradication.10,11 One of the main mechanisms, which tumour cells are thought to use for this purpose is the induction and the recruitment of regulatory T cells (Tregs).11,12 Regulatory T cells play an important role for the maintenance of immunological homeostasis. They hamper overshooting pro-inflammatory responses and inhibit immune reactions against innocuous environmental antigens or self-antigens. However, they are also suggested to block tumour cell directed immune responses.13,14 In accordance with this assumption, an increased amount of regulatory T cells could be detected in the peripheral blood of patients with different kinds of cancer, including lung carcinoma. In addition to that, an increased ratio of regulatory to effector T cells at the tumour site correlates with poor prognosis for the patients.15–17 Moreover, various murine model studies confirmed the tumour promoting role of regulatory T cells.13,18 A characteristic feature of regulatory T cells is the expression of the transcription factor Foxp3 that is required for the definition of the Treg cell phenotype as well as for their suppressive function.19-21 In spite of the opposite roles of Tbet and Foxp3 in the immune system as well as in tumour biology, recent studies have revealed the existence of CD4+ T cells, which express both, Tbet and Foxp3.22-25 Although Tbet+Foxp3+ T cells are currently a subject of intense research, less is known about the biological function of these cells as well as their role in diseases. In the present study we focused on the role of Tbet and Foxp3 co-expressing CD4+ T cells in a murine model of lung carcinoma, in which we observed a considerable accumulation of this cell subtype and finally provided evidences of this cell lineage expansion and immunosuppressive properties in human cells derived from lung tissues after surgery from patients affected by NSCLC.
Results
Accumulation of Foxp3 co-expressing Tbet+CD4+ T-cells in lung cancer
In this study we analyzed CD4+ T cell responses in a murine model of lung cancer. For this purpose we induced lung tumour growth in mice via intravenous injection of luciferase expressing LL/2-luc-M38 lung carcinoma cells (Fig 1a). Here we found that tumor progression is associated with an increase of Foxp-3+CD25+ expressing CD4+ T cells in the lung and a reduced percentage of them in the spleen, whereas their numbers in the lymph nodes remain unchanged before and 15 days after the tumour induction (Fig. 1a). Th1 mediated immune responses play a major role for tumour cell rejection.26 Therefore, we next analyzed Th1 associated factors, expressed by CD4+ T-cells in naïve as well as tumour- bearing mice at different time points after lung carcinoma induction. Interestingly, we found that more than 40% of all Tbet+ CD4+ Tcells in the lungs of tumor bearing mice co-expressed Foxp3 and CD25 (Fig. 1b) indicating a loss of anti-tumour effector function of T-bet+ T cells (Fig. 1b). Similarly to Foxp-3+ T cells, we observed a significant increase of T-bet-expressing CD4+ T cells, infiltrating the lungs of tumour-bearing mice simultaneously to their decrease in the spleen (Fig. 1c) and an expansion of Foxp-3 co-expression along with T-bet in tumour infiltrating CD4+ T cells by using different gating strategies (Fig. 1d,e). We further observed a significant expansion of Tbet+Foxp3+ T cells during the tumour development, exclusively in the lung, as well as a positive correlation between the percentage of these double positive cells and the lung tumor growth (Fig. 1f, g). These data support the notion that anti-tumour T-bet+ CD4+ T cells are targeted and silenced by the tumour cells.
Figure 1.
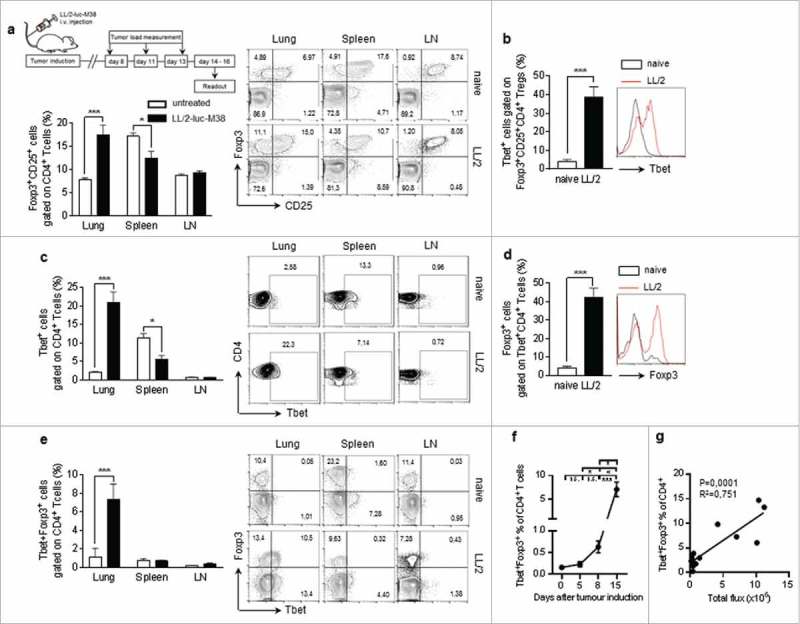
Lung cancer is associated with Foxp3 induction in Tbet+ CD4+ T-cells a.Experimental procedure: Lung tumor induction in mice occured via intravenous injection of LL/2 M38 tumor cells. Tumor growth was measured at day 8, 11 and 13 after tumor induction by an in vivo bioluminescence imaging system detecting luciferase activity. The experiment ended at day 14 to 16. b-d. Accumulation of Tbet+Foxp3+CD4+ T cells in lung tumor bearing mice: b. Increased percentage of Tbet+ cells gated on Foxp3+CD25+CD4+ T cells in the lung of tumor-bearing as compared to naïve control mice. c. Increased percentage of Tbet+ cells gated on CD4+ T cells in the lung of tumor-bearing as compared to naïve control mice, whereas no induction was observed in the spleen or the lymph nodes (LN). Flow cytometry analysis of Tbet+ expressing CD4+ T cells in whole cell suspensions from the lungs, the spleens and the lymph nodes of naïve (untreated) as well as tumour bearing (LL/2-luc-M38) WT mice after the i.v. injection of LL/2-luc-M38 lung carcinoma cells (N-lung = 4–11; N-spleen = 4–9; N lymph nodes = 4–13). (d-g) Increased percentage of Tbet+Foxp3+CD4+ T cells in the lung of tumor-bearing mice increased with progressing lung tumor growth. d Flow cytometry analysis of Foxp3 expressing cells gated on Tbet+CD4+ T cells in the lungs of naïve and tumour-bearing WT mice (N = 13–14). e Flow cytometry analysis of Tbet and Foxp3 ;co-expressing cells gated on CD4+ T cells in the lungs, the spleens and the lymph nodes of naïve (day 0) as well as tumour bearing WT mice (Nlung = 4–11; Nspleen = 4–9; Nlymph nodes = 4–13). f.Percentage of Tbet and Foxp3 co-expressing cells gated on CD4+ T cells in the lungs of WT mice at day 0 to 15 after the tumour induction assessed via flow cytometry (N = 4–11). g Correlation between the percentage of Tbet and Foxp3 co-expressing cells gated on CD4+ T cells in the lungs of tumor bearing WT mice and the tumor load, measured as the total photon flux per second (N = 12). Bar charts indicate mean values +/- s.e.m. using student´s two-tailed t-test *P = 0.05; **P = 0.01; ***P = 0.001.
Increased IL-10 but not IFN-gamma production in the supernatants of total lung cells isolated from lung tumour-bearing mice
We next asked whether the number of T-bet+Foxp3- T cells is increased in tumour and would compensate for the fact that an increased fraction of the cells produce less Th1 cytokines. To this aim we analyzed the number of T-bet+Foxp3- T cells gated on lung CD4+ T cells and found them significantly induced in the lung of mice bearing-tumour (Fig 2a, b). However, by looking at the IFN-gamma production from total lung cells from tumour-bearing mice after anti-CD3 and anti-CD28 antibodies challenge, we found a small, not significant, induction of total lung IFN-gamma in mice bearing-tumour (Fig 2c). Overall the total amount of IFN-gamma in the lung of mice bearing is very low. There has been the suggestion that Tbet and Foxp3 co-expressing cells might be Tregs that specifically suppress Th1 responses. The fact that there is an increased number of Tbet+Foxp3- cells but no increase of Th1 cytokines, may support this assumption.
Figure 2.

Increased IL-10 but not IFN-gamma production in the supernatants of total lung cells isolated from lung tumour-bearing mice a. Representative FACS analysis of the increased percentage of Tbet+Foxp3- lung CD4+ T cells in the lung of tumor- bearing as compared to naïve control mice. b. FACS analysis of T-bet+/Foxp3- cells gated on lung CD4+ T cells in the lung of mice bearing tumour (N = 4,5; p = 0.0118). c. IFN-gamma, p = 0.128) and d.IL-10 (p = 0.0001) production from total lung cells from mice bearing-tumour after anti CD3 and CD28 antibodies challenge (N = 10 per group). Bar charts indicate mean values +/- s.e.m. using student´s two-tailed t-test *P = 0.05; **P = 0.01; ***P = 0.001.
By contrast, we found a strong significant induction in IL-10 in the supernatants of total lung cells from tumour-bearing mice (Fig 2d). In conclusion, there is not a compensation of IFN-gamma production in T-bet+Foxp3+ T cells in the lung of tumour-bearing mice, rather T cells in the lung of tumour bearing mice release the immunosuppressive cytokine IL-10.
The role of TGFβ for Foxp3 induction in Tbet+CD4+ T-cells
Previous studies suggest that Tbet+Foxp3+CD4+ T cells can develop from Foxp3+ regulatory T cells through IFNγ and STAT1-mediated conversion.22 Moreover, we reasoned that the tumour-microenvironment is generally characterized by high levels of anti-inflammatory cytokines, such as TGFβ. Different kinds of tumour cells, including lung carcinoma cells, are able to produce TGFβ.27,28 Moreover, it is well known that TGFβ induces the expression of Foxp3 in naïve T cells.29,30 We therefore asked, if the expansion of Tbet+Foxp3+ cells in the lungs of tumour-bearing mice could be attributed to a TGFβ-dependent induction of Foxp3 expression in T-bet+ Th1 cells. This hypothesis is supported by our finding that Tbet+CD4+ T cells express higher levels of TGFβRII than Tbet−CD4+ T-cells in the lungs of tumour-bearing mice (Fig. 3a). To investigate the ability of TGFβ to induce Foxp3 expression in Tbet+CD4+ T cells, we cultured CD4+CD62 L+ splenocytes from naïve WT mice under Th1 polarizing conditions for 5 days. These fully differentiated Th1 cells were incubated in the presence or absence of TGFβ for additional 2 days. Here we observed a significant up-regulation of Foxp3 in TGFβ-treated Th1 cells, supporting the notion that TGFβ could induce Tbet+Foxp3+CD4+ T cell development (Fig. 3b-d). Notably, we also observed that TGF-beta given before Th1 differentiation was more efficient in inducing Foxp-3+T-bet+ CD4+ T cells than at day 5 when Th1 were already differentiated. This property of TGF-beta was further confirmed by the finding that the numbers of Foxp3+Tbet+ double positive CD4+T cells was strongly reduced in tumour-bearing hCD2-ΔkTβRII mice, expressing a truncated form of the TGFβRII on their T-cells as compared to wild-type mice (Fig. 3e).
Figure 3.
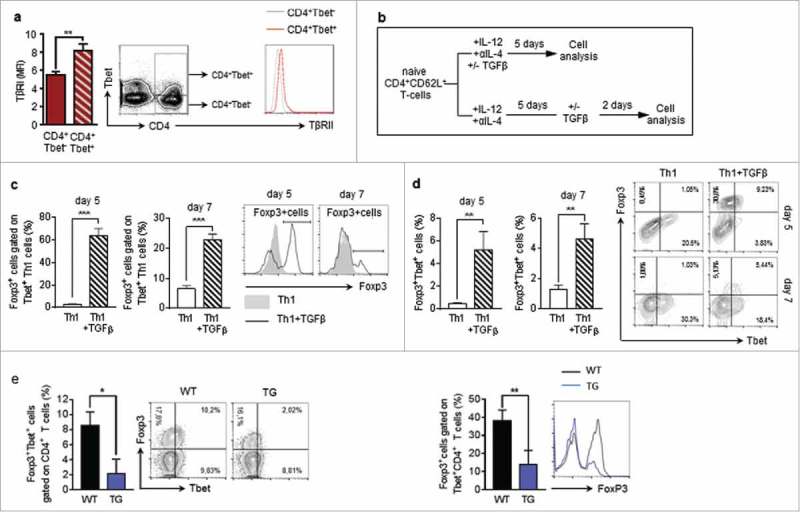
TGFβ induces Foxp3 expression in Tbet+CD4+ T-cells a Flow cytometry analysis of TβRII expression on Tbet positive and Tbet negative CD4+ T cells in the lungs of tumour-bearing WT mice (N = 16). b Experimental design of Flow cytometry analysis of Foxp3 induction in CD4+CD62L+ T cells, isolated from the spleens of naïve WT mice, cultured under Th1 polarizing conditions for 5 days and subsequently cultured in the presence or absence of TGFβ for another 2 days (N = 3). c,d Flow cytometry analysis of Foxp3 expressing cells gated on Tbet+CD4+ T cells as well as Tbet+Foxp3+ (e) Foxp3+Tbet+ cells gated on CD4+ T cells in the lungs of tumour-bearing WT and hCD2-ΔkTβRII mice (N = 9–13).
Increased survival of hCD2 ΔkTβRII as compared to wt lung tumour-bearing mice
To demonstrate that Foxp-3-T-bet double positive cells are not able to limit the tumour, we analyzed the lung tumour development in the absence of TGF-beta signaling in T cells. In preliminary analysis we found that our model of lung tumour is characterized by induction of tumour infiltrating CD4+ T cells carrying the TGF-beta RII (Fig 4a). Moreover, interrupted TGFβ signaling in T cells leads to decreased tumour load (Fig. 4b) and increased survival (Fig 4c) accompanied by induced anti-tumoral immune responses as shown by the observation that CD4+ T cells isolated from the lung of TGFβRII transgenic mice produce increased levels of IFNγ, TNFα and IL-2 upon in vitro stimulation with αCD3 and αCD28 antibodies as compared to those isolated from WT littermates (Fig 4d-g).
Figure 4.

Increased survival of hCD2 ΔkTβRII as compared to wt lung tumour bearing mice a. Flow cytometry analysis of the percentage of TGFβRII expressing CD4+ T cells in the lungs of naive and tumor-bearing wild-type (WT) mice (n(naive) = 10, n(LL/2) = 11)b. Tumour load analysis at day 8, 11 and 13 after intravenous injection of LL/2-luc-M38 lung carcinoma cells in WT and hCD2 ΔkTβRII mice via the measurement of photone flux per second (N day8 = 10-12; N day11 = 14–16; N day13 = 14–16). c. Percentage of survival after the intravenous injection of L1C2 tumour cells into WT and hCD2 ΔkTβRII mice. d. Flow cytometry analysis of Tbet and IFNγ co-expressing cells gated on CD4+ T cells in in the lungs of tumour-bearing WT and hCD2 ΔkTβRII mice (N = 4–9). e-g ELISA analysis of IFNγ (e), TNFα (f) and IL-2 (g) levels in the supernatants of CD4+ T cells, isolated from the lungs of tumour-bearing WT and hCD2-ΔkTβRII mice and cultured for 40 h in the presence of αCD3 and αCD28 antibodies (NIFNγ = 10–12; NTNFα = 9–12; NIL-2 = 9–16). N values are given per group. Bar charts indicate mean values +/- s.e.m. using student´s two-tailed t-test *P = 0.05; **P = 0.01; ***P = 0.001.
The impact of Tbet and Foxp3 co-expression on the T cell phenotype
Tbet+Foxp3+CD4+ T cells are characterized by the co-expression of two transcription factors that are responsible for the establishment of opposite T cell phenotypes. In fact, Tbet is the master regulator of Th1 cells, whereas Foxp3 controls the development and the function of regulatory T cells, while it inhibits the expression of some Tbet-induced factors, as for example IFNγ and IL-2.13,18,31 This leads to the question which T cell phenotype and function is induced in the presence of both, Tbet and Foxp3. In order to clarify whether the Tbet+Foxp3+CD4+ T cells found in the lungs of tumour-bearing mice exhibit regulatory or rather pro-inflammatory properties, we analyzed the expression of typical Treg- and Th1- associated markers. We found that Tbet and Foxp3 double positive CD4+ T cells show a higher expression of typical Treg-factors, such as ICOS, GITR, CD103, CTLA4 and IL-10 as compared to CD4+ T cells, which express either Tbet or Foxp3. Interestingly, a higher percentage of Tbet+Foxp3+CD4+ T cells is positive for the inhibitory receptor PD-1 as compared to Foxp3+CD4+ T-cells without Tbet, although there is no difference compared to Tbet+CD4+ T cells without Foxp3 (Fig. 5a, c).
Figure 5.
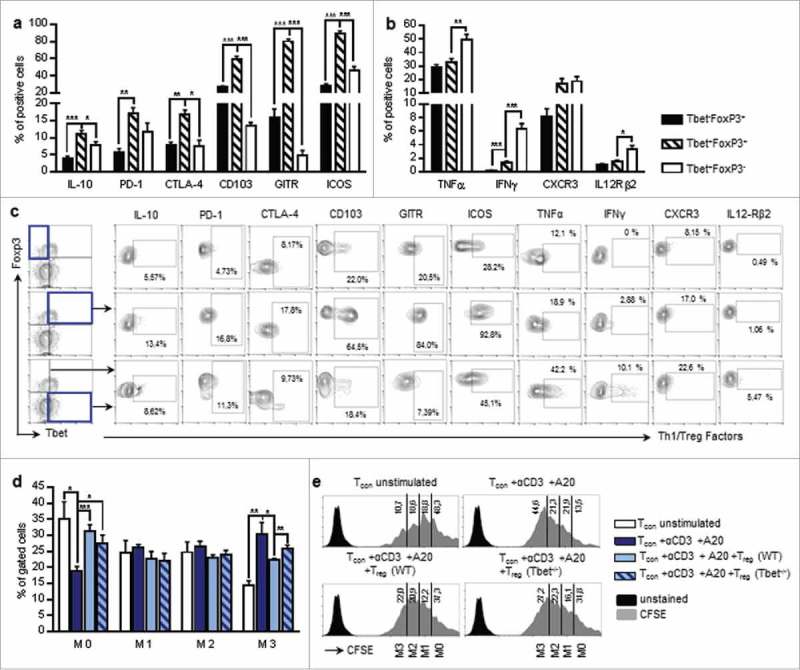
Characterization of Tbet and Foxp3 double positive T-cells. a-c Flow cytometry analysis of Treg-associated factors (IL-10, PD-1, CTLA-4, CD103, GITR, ICOS). (a) and Th1-associated factors (IFNγ, TNFα, CXCR3, IL-12Rβ2). (b) in the lungs of tumour-bearing WT mice as a percentage of Tbet+Foxp3−, Tbet+Foxp3+ or Tbet−Foxp3+ cells, respectively (NIL-10 = 5; NPD-1 = 3; NCTLA-4 = 3; NCD103 = 3; NGITR = 3; NICOS = 9; NTNFα = 7; NIFNγ = 9; NCXCR3 = 3; N IL12Rβ2 = 6). (c) Specific gating strategy for e is depicted. d-e Treg in vitro suppression assay. CD4+CD25+ cells (Tregs) were isolated from the lungs of tumour-bearing WT and Tbet−/− mice and co-cultured with CFSE stained CD4+CD25− (Tcon) cells from the lungs of tumour bearing WT mice for 96 h in the presence of αCD3 antibodies and A20 cells for T cell stimulation. The proliferation of Tcon cells was determined by flow cytometry, based on the measurement of CFSE dilution. d The percentage of not proliferating cells (M0) as well as cells, which have divided 1–3 times (M1–3) is shown for unstimulated Tcon cells, stimulated Tcon cells without Treg cells as well as stimulated Tcon cells after the co-culture with WT or Tbet−/− Treg cells, respectively (N = 6-8). e Histograms with gating strategy for (d) are depicted. N values are given per group. Bar charts indicate mean values +/- s.e.m. using student´s two-tailed t-test *P = 0.05; **P = 0.01; ***P = 0.001.
The subsequent analysis of Th1-associated markers revealed that only a small proportion of Tbet+Foxp3+CD4+ T-cells express IL-12Rβ2 in contrast to CD4+ T-cells expressing T-bet alone. Accordingly, Tbet+Foxp3+ T-cells produce less IFN-γ and TNFα as compared to Tbet+Foxp3− cells. Nevertheless, the expression of these Th1-hallmark factors is significantly higher in cells, co-expressing T-bet and Foxp3, as compared to Foxp3+ cells that don´t express T-bet. Unlike the other analyzed Th1-markers, CXCR3 expression does not differ between the Tbet+Foxp3+CD4+ and the Foxp3 negative Tbet+CD4+ T cell population (Fig. 5b, c). This is in line with previous studies, which indicate that the presence of Tbet in Foxp3+CD4+ Treg cells is required for CXCR3 induction and thus the migration of Foxp3+ Treg cells to the site of Th1 inflammation.6,23 In previous studies, Tbet+Foxp3+CD4+ T cells are being described as a specific kind of regulatory T cells.22,23,32 In accordance with this our results indicate that, despite the presence of Tbet, Tbet+Foxp3+CD4+ T-cells are characterized by regulatory, rather than pro-inflammatory properties.
The role of Tbet expression in regulatory T cells has been investigated in numerous studies. On the one hand, it has been demonstrated that Tbet-deficient Treg cells are characterized by reduced survival as well as an inability to inhibit Th1-mediated inflammation after the adoptive transfer into Scurfy mice.23 On the other hand, there are a number of studies indicating that Tbet-deficiency does not affect the suppressive function of regulatory T cells.6,33,34 Here we demonstrate that a higher percentage of Tbet+Foxp3+CD4+ T cells co-express regulatory markers as compared to CD4+ T cells, expressing Foxp3 alone (Fig. 5a, c). To analyze their regulatory function, we performed an in vitro suppression assay with tumour-derived WT and Tbet−/− regulatory T cells. In order to specifically test the capacity of WT and Tbet−/− Treg cells to suppress tumour infiltrating effector cell, we used CD4+CD25− conventional T cells (Tcon), isolated from the lungs of tumour bearing WT mice as responder cells. Indeed, we observed a reduced ability of tumour derived Tbet−/− Treg cells to suppress the cell proliferation of Tcon cells (Fig. 5d, e). This is in line with some previous studies, indicating that the expression of Tbet in regulatory T cells could be required for the specific suppression of Th1-mediated immune responses that are known to be important for successful anti-tumour immunity.23
Induction of Foxp-3 and TGF-beta in the lung of human subjects affected by NSCLC.
We next performed translational analysis in human lung samples obtained after surgery because of lung cancer (NSCLC).
In this cohort of patients we recently described and reported clinical characteristics,35,36 we found an induction of FOXP3 and TGF-beta1 in the tumoural region as compared to their control tumour-free lung region (Fig 6a-c). Moreover, we found a direct positive correlation between TGF-beta and Foxp3 (Fig 6d), confirming the murine data on the Foxp-3 inducing properties of TGF-beta 1.
Figure 6.
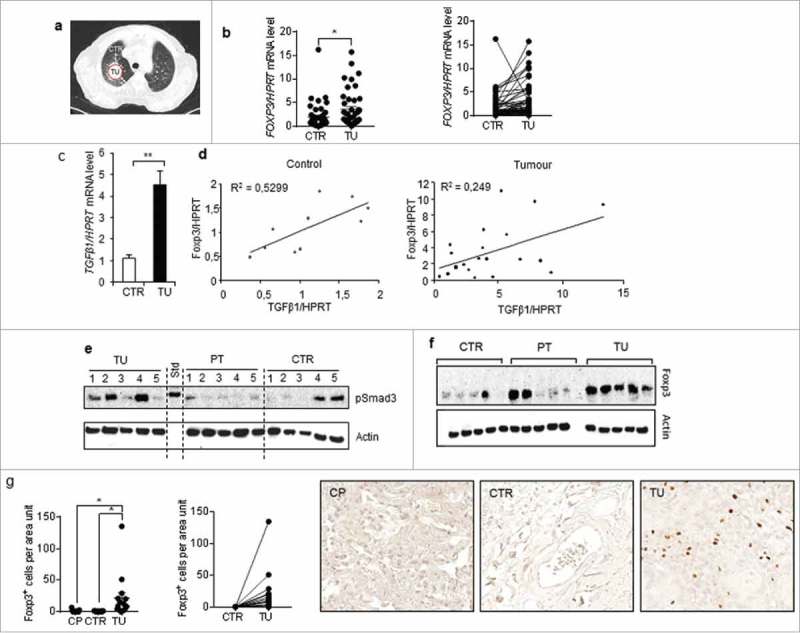
Foxp3 correlated with TGF-beta 1 expression in the lung tissue samples from patients with non-small cell lung cancer. a-d.Comparison between Foxp-3 and TGFbeta1 mRNA expression inthe tumoral lung region (TU), taken from the solid tumour, and the tumour free control region (CTR), which is at least 5 cm away from the solid tumour.d. Correlation between Foxp-3 and TGF-beta mRNA levels in those lung tissues. e and f. Western blot analysis performed with proteins isolated from the lung of patients as previously described and incubated with anti pSMAD3 and anti Foxp-3 antibodies.52g. Immunohistochemistry performed with anti Foxp-3 antibodies in lung tissue arrays from our cohort of patients with NSCLC and control lung tissue array obtained from healthy subjects and quantified as recently described.36
To add functional studies, we analyzed activated SMAD3 (pSMAD3) expression downstream of TGF-beta Receptor (Fig 6e). Consistent with TGF-beta induction in the tumoural region here pSMAD3 was found induced along with Foxp-3 at the protein level (Fig 6e, f).
Similarly, at the cellular level, by Immunohistochemistry, Foxp-3 + T cells were found induced in the tumoural region of these patients (Fig 6g).
Identification of Foxp3+T-bet+ T cells in human lung carcinoma samples
We next isolated immediately after surgery, T cells infiltrating the lung tumour and analyzed the presence of Foxp-3+T-bet+T cells in their tumoural versus the control region (Fig 7). Consistent with the murine data, we found an induction of Foxp-3+T-bet+ CD4+ T cells in the tumoural region as compared to the control region of these patients (Fig 7a-f). Moreover, in the tumoural region of patients with NSCLC, 50% of the T-bet positive cells expressed IFN-gamma whereas when Foxp-3 was co-expressed with T-bet this percentage dropped down to the 8% similar to Foxp-3+T-bet negative CD4+ T regulatory cells (Fig 7g, h).
Figure 7.

Percentage of Foxp3+Tbet+CD4+ T cells suppressed IFN-gamma production in lung cells isolated from lung tissue samples obtained after lung surgery in patients with non-small cell lung cancer. FACS analysis and comparison between the cells isolated from the tumoral lung region (TU), taken from the solid tumour, and the tumour free control region (CTR), which is at least 5 cm away from the solid tumour (N = 8).
Discussion
Upon antigen binding, naïve CD4+ T cells can differentiate into distinct subtypes, such as Th1, Th2, Th17 but also peripheral regulatory T cells (iTregs). The development of each T cell subtype can be induced by a specific cytokine milieu and is characterized by the expression of lineage specific hallmark transcription factors, such as Tbet, Gata3, RorγT or Foxp3. Previous studies have shown that the expression of these master transcription factors not only promotes the differentiation of the respective T cell phenotype, but also inhibits alternative differentiation pathways.6,25,37-40 Moreover, it has been assumed that the differentiation of naïve T cells is irreversible. However, recent studies have proved the existence of CD4+ T cells that co-express two or more key transcription factors of different T cell subtypes.22–25 For instance, it has been discovered that Th1 inducing conditions are associated with the development of CD4+ T cells, co-expressing the Treg transcription factor Foxp3 and the Th1 hallmark factor Tbet. Thus, Tbet+Foxp3+ T cells can be induced in a murine model in vivo via treatment with an agonistic αCD40 antibody.23 In the present study we found that the induction of lung carcinoma growth in a murine model is associated with increased amounts of Tbet expressing CD4+ T cells in the lungs. About 40 % of these Tbet+CD4+ T cells in the tumour bearing lungs co-expressed Foxp3. The analysis of Tbet+Foxp3+CD4+ T cells regarding the expression of Treg and Th1 associated markers leads to the conclusion that, despite the presence of Tbet, this T cell subtype is characterized by regulatory, rather than pro-inflammatory properties. The results of previous studies indicate that the expression of Tbet in regulatory T cells could be required for the expansion of these immunosuppressive cells which limit Th1-mediated immune responses, known to be important for successful anti-tumour immunity.23 In accordance with this, we demonstrated that tumour infiltrating Tbet deficient Treg cells have a reduced ability to suppress the proliferation of responder CD4+ T cells, derived from the tumour site.
It has been shown before that Tbet+Foxp3+ T cells can be the result of the conversion of Foxp3+ T cells into Tbet expressing Treg cells in an IFNγ and STAT1 dependent manner.22 Moreover, our results indicate that this cell type can also develop from Tbet+CD4+Th1 cells due to the action of the immune suppressive cytokine TGFβ. The connection between the immunosuppressive function of TGFβ and cancer has been demonstrated in several previous studies. For instance, it has been shown that TGFβ could be responsible for increased numbers of immature dendritic cells at the tumour site, and thus for insufficient presentation of tumour antigen, leading to T cell anergy.41,42 Moreover, it has been shown before, that the interruption of TGFβ signaling in T cells is associated with an improved ability to fight tumour cells in a murine model for melanoma as well as thymoma. Furthermore, it has been demonstrated that TGFβ promotes tumour growth through the inhibition of CD8+ T cell production of effector molecules, such as IFNγ, Perforin and Granzyme B.43,44 Our results indicate that the conversion of IFNγ producing anti-tumoral Th1 cells into immunosuppressive Tbet and Foxp3 co-expressing regulatory T cells could represent an additional mechanism of TGFβ-mediated blockade of anti-tumour immunity. Moreover, we found that TGF-beta given in naïve cells during Th1 development induces a higher number of T-bet+Foxp-3+ T cells as compared to its action on mature Th1 cells. These Th1 cells expressing Foxp-3 and PD1 and other immune-suppressive markers could promote tumour growth. Th1 cells expressing-Foxp-3-PD1 are generated by the tumour cells via TGF-beta and are then targeted by the lung tumour cells expressing PDL1, thus evading the immune responses.35,36,45 Although further analysis in this direction are needed, altogether we conclude that the accumulation of Tbet+Foxp3+CD4+ T cells at the tumour site could contribute to the immunopathogenesis of lung cancer. Thus targeting these cells could be a good avenue to improve current immune-therapy against lung cancer. In our translational studies in human samples we could demonstrate the increased number of these cells in the tumoural region of patients with non small cell lung cancer. Functionally, we show a direct correlation between TGF-beta and Foxp-3 in the lungof patients with NSCLC. These data indicate the presence of immunosuppressive cells induced by TGF-beta which directly correlate with T-bet in the tumoural region of patients with NSCLC. Further studies in this direction are underway in our laboratories. Thus, these data support an increase suppressive function of T-bet+Foxp-3+T cells in NSCLC that is relevant for anti tumour immunotherapies.
Experimental procedures
Human subjects and study population
This study was performed at the University of Erlangen in Germany and was approved by the ethics review board of the University of Erlangen (Re-No: 56_12B; DRKS-ID: DRKS00005376). Patients, who were suffering from NSCLC, underwent surgery and gave their approval to being enrolled in this study. Patients' confidentiality was maintained. The diagnosis of lung cancer was based on pathological confirmation. The histological types of lung cancer were classified according to the classification of the World Health Organization (WHO), formulated in 2004. The staging of lung cancer was based on the Cancer TNM Staging Manual, formulated by the International Association for the Study of Lung Cancer (IASLC) in 2010. Tissue samples were taken from the tumoural area (TU: solid tumour tissue), the peri-tumoural area (PT: up to 2 cm away from the solid tumour) as well as from the tumour free control area (CTR: at least 5 ;cm away from the solid tumour) of the surgically removed lung material. Most of the material and methods related to the human studies where recently reported in details.3,35,36,45,46
Total cells isolation from human lung samples
Samples were cut into very small pieces using scalpels (1-3 mm2). The shredded material was then subjected to collagenase/DNase digestion (20 ml solution per 1 g of material) and incubated over night at 37°C under constant shaking (300 r.p.m). The digestion solution contained 4.8 µg/ml collagenase (Liberase TM Research Grade REF 05 401 119 001, Roche Diagnostics, Mannheim, Germany) and 15 µg/ml DNase (Roche Diagnostics, Mannheim, Germany), dissolved in PBS. In order to get a single cell suspension the digested material was sieved through a cell strainer (70 µm). The cell suspension was centrifuged (10 min, 1000 rpm, 4°C) and the supernatant was discarded. The cells were resuspended in 10 ml of hypotonic solution (Ammonium-Chloride-Potassium buffer (ACK)) and immediately centrifuged again (5 min, 1000 rpm, 4°C) to quickly remove the ACK lysis buffer. This was followed by cell resuspension in 10 ml of PBS+EDTA+1% Penicilin/Streptomycin and centrifugation (10 min, 1000 r.p.m., 4°C). Cells were finally resuspended in PBS+ EDTA+ 1% Penicilin/Streptomycin+ 5% FCS.
Mice
Balb/c wild-type (WT), CD2-ΔkTFβRII and Tbet−/− were bred and maintained under specific pathogen-free conditions in our animal facility (Center of Molecular Inflammation and Cancer Research, Erlangen). CD2-ΔkTFβRII and Tbet−/− mice were previously described.3,47 All experiments were performed according to institutional animal care guidelines under our ethical grant 54–2532.1-36/13.1.
Cell Lines
The LL/2-luc-M38 cell line (Bioware cell line, Caliper LifeScience, Waltham, Massachussets, USA) was cultured in DMEM medium (Gibco, Invitrogen, Darmstadt, Germany), supplemented with 10 % of fetal calf serum (FCS) (PAA Laboratories, Cölbe, Germany) and 1% of the antibiotics penicillin and streptavidin (Pen/Strep) (PAA Laboratories, Cölbe, Germany) at 37°C and 5% of CO2.
L1C2 lung adenocarcinoma cells were cultured in RPMI medium (Gibco, Invitrogen, Darmstadt, Germany) enriched with 10% FCS (Biofluids, Rockville, MD) and 1% of the antibiotics penicillin and streptavidin (Pen/Strep) (PAA Laboratories, Cölbe, Germany) at 37°C and 5% of CO2.
A20 B cell lymphoma cells were generously provided by PD. Dr. S. Wirts (Medical Clinic I, Friedrich –Alexander University Erlangen-Nürnberg). The cells were cultured in ISCOVE medium (Lonza, Köln, Germany), supplemented with 5 % of FCS (PAA Laboratories, Cölbe, Germany), 1 % of Pen/Strep (PAA Laboratories, Cölbe, Germany), 1 % of Pyruvate (PAA Laboratories, Cölbe, Germany) and 0,1 % of β-Mercaptoethanol (Roth, Karlsruhe, Germany) at 37°C and 5% of CO2.
Murine model for lung adenocarcinoma and tumour load analysis
The Lewis lung carcinoma model was induced via injection of 5 × 105 LL/2-luc-M38 cells, resuspended in 200 µl of DMEM medium (Gibco, Invitrogen, Darmstadt, Germany) without supplements, into the tail vein of 6–8 week old female mice. Mice were sacrificed at day 5 to 16 after tumour cell injection. Tumour load was analyzed in vivo at day 8, 11 and 13 after injection of LL/2-luc-M38 cells. For that purpose mice were injected with 150 µl of Luciferin solution (15 mg/ml) intraperitoneally (i.p.) (Promega, Madison, Wisconsin, USA). After 10 min of incubation, mice were anaesthetized using isoflurane and luminescence was measured by the Ivis Imaging System, as previously described (Bioware cell line, Caliper LifeScience, Waltham, Massachussets, USA).3 Briefly, the analysis was done in a logarithmic scale mode. For quantification of the tumour load the total flux (photons per second) was determined.
For the induction of L1C2 derived lung tumours, 2 × 105 cells, resuspended in 200 µl of RPMI (Gibco, Invitrogen, Darmstadt, Germany) without supplements were injected into the tail vein of the mice.
Flow cytometry analyses (FACS)
Flow cytometry analyses were performed with 1 × 106 total cells per sample and staining. The cells were washed with PBS, followed by incubation (30 min., 4°C, in the dark) with the respective master mix of antibodies against surface proteins, solved in PBS (ad 80 µl). For FACS analyses of surface markers alone, the cells were washed with PBS and resuspended in PBS + EDTA (Lonza, Köln, Germany). No fixation and permeabilization was performed in that case. For FACS analysis of intracellular molecules, the cells were fixed and permeabilized with Fixation and Permeabilization solutions according to manufacturer´s protocol (eBioscience, Frankfurt, Germany) and incubated (30 min, 4°C, in the dark) with antibodies against intracellular proteins, solved in Permeabilization buffer (eBioscience, Frankfurt, Germany) (ad 80 µl). After a final washing step with Permeabilization buffer (eBioscience, Frankfurt, Germany) the cells were resuspended in PBS + EDTA (Lonza, Köln, Germany). For the analysis of cytokine expression by flow cytometry, whole lung cells were cultured with 5 µg/ml plate bound anti-mouse CD3 antibody (BD Biosciences, Heidelberg, Germany) and 2 µg/ml soluble anti-mouse CD28 antibody (BioLegend, Fell, Germany) at 37 °C and 5 % of CO2 for 24 h and incubated with Phobol-12-myristate 13-acetate (PMA) (Sigma-Aldrich Chemie, Munich, Germany), Ionomycin (Sigma-Aldrich Chemie, Munich, Germany) and the protein transport inhibitor Golgi Stop (BD Biosciences, Heidelberg, Germany) for 4 h. The subsequent flow cytometry measurements were performed via FACS Calibur and analyzed using Cell Quest Pro version 4.02 (BD Biosciences, Heidelberg, Germany). The following antibodies were used during this study: anti-mouse CD4 FITC (BD Biosciences, Heidelberg, Germany), anti-mouse CD4 PerCP (BD Biosciences, Heidelberg, Germany), anti-mouse CD4 PE (BD Biosciences, Heidelberg, Germany), anti-mouse CD4 APC (BD Biosciences, Heidelberg, Germany), anti-human CD4 FITC (eBioscience, San Diego, USA), anti-human/mouse T-bet PE (BD Biosciences, Heidelberg, Germany), anti-human/mouse Foxp3 APC (Miltenyi Biotec, Bergisch-Gladbach, Germany), anti-mouse TβRII PerCP (R&D Systems, Wiesbaden, Germany), anti-mouse IFNγ FITC (BD Biosciences, Heidelberg, Germany), anti-mouse TNF-α FITC (BD Biosciences, Heidelberg, Germany), anti-mouse CXCR3 FITC (eBioscience, Frankfurt, Germany), anti-mouse IL12Rβ2 Alexa488 (R&D Systems, Wiesbaden, Germany), anti-mouse IL-10 FITC (BD Biosciences, Heidelberg, Germany), anti-mouse PD-1 PE (BD Biosciences, Heidelberg, Germany), anti-mouse CTLA4 PE (BD Biosciences, Heidelberg, Germany), anti-mouse CD103 FITC (Miltenyi Biotec, Bergisch-Gladbach, Germany), anti-mouse GITR FITC (eBioscience, Frankfurt, Germany), anti-mouse ICOS FITC (eBioscience, Frankfurt, Germany), anti-mouse CD25 PerCP (BD Biosciences, Heidelberg, Germany) antibodies.
Isolation of murine lung CD4+ T-cells
Single cell suspension from murine lungs were prepared as previously described.48 Isolation of CD4+ T-cells from total lung cells was performed using anti-mouse CD4 Microbeads (Miltenyi Biotec, Bergisch-Gladbach, Germany) by positive selection according to manufacturer´s protocol. CD4+ T-cells were cultured in RPMI medium (Gibco, Invitrogen, Darmstadt, Germany), supplemented with 10% FCS (PAA Laboratories, Cölbe, Germany) and 1% Pen/Strep (PAA Laboratories, Cölbe, Germany), at 37 °C and 5 % CO2, in the presence of 5 µg/ml plate bound anti-mouse CD3 antibody (BD Biosciences, Heidelberg, Germany) and 2 µg/ml soluble anti-mouse CD28 antibody (BD Biosciences, Heidelberg, Germany). Supernatants and cells were harvested after 40 h of cell culture for further analyses.
Isolation of naïve CD4+CD62 L+ T cells
CD4+CD62 L+ naïve T cells were isolated from splenic whole cell suspensions via magnetic activated cell sorting (MACS) using the mouse CD4+CD62 L+ T cell isolation Kit II (Miltenyi Biotec, Bergisch-Gladbach, Germany) according to manufacturer´s instructions. The isolated cells were used for in vitro T cell differentiation assays, as described below.
In vitro Th1 cell differentiation and TGFβ treatment
In order to analyze T cell differentiation, CD4+CD62 L+ T cell were cultured in RPMI medium (Gibco, Invitrogen, Darmstadt, Germany), supplemented with 10 % FCS (PAA Laboratories, Cölbe, Germany) and 1 % Pen/Strep (PAA Laboratories, Cölbe, Germany), at 37°C and 5 % CO2 in the presence of 5 µg/ml plate bound anti-mouse CD3 antibody (BD Biosciences, Heidelberg, Germany) and 2 µg/ml soluble anti-mouse CD28 antibody (BD Biosciences, Heidelberg, Germany).
For Th1 differentiation, the cells were additionally treated with 100 ng/ml IL-2 (PeproTech, Hamburg; Germany), 5 µg/ml anti-IL-4 antibody (11B11 anti-IL-4 mAb, produced by hybridoma cells, kindly provided by Prof. Dr. med. E. Schmitt, Mainz, Germany) and 12 ng/ml IL-12 (PeproTech, Hamburg; Germany). After 3 days in culture the cells were split, transferred to a new culture plate and restimulated with the Th1-polarizing cocktail as described above as well as with 5 µg/ml plate bound anti-mouse CD3 antibody (BD Biosciences, Heidelberg, Germany) and 2 µg/ml soluble anti-mouse CD28 antibody (BD Biosciences, Heidelberg, Germany) for another 2 days. To test the influence of TGFβ on Th1 differentiated cells, 30 ng/ml of recombinant TGFβ (PeproTech, Hamburg; Germany) was added to the cell culture after the Th1 induction (at day 5) and incubated for additional 2 days. At day 7 the cells were harvested for flow cytometry analysis.
Enzyme-linked immunosorbent assay (ELISA)
Sandwich ELISA sets were used for the detection of IL-2 (BD Biosciences, Heidelberg, Germany), TNF-α (BD Biosciences, Heidelberg, Germany), IL-10 and IFN-γ (BD Biosciences, Heidelberg, Germany) in the cell supernatants, according to manufacturer´s protocols.
Isolation of lung infiltrating CD4+CD25+ and CD4+CD25− T cells
Isolation of CD4+ T-cells from a single cell suspension of murine lungs was performed via positive selection using anti-mouse CD4 Dynabeads (Life Technologies, Darmstadt, Germany) according to manufacturer´s protocol. Dynabeads were removed from the isolated CD4+ T cells by mouse CD4 Detachabeads (Life Technologies, Darmstadt, Germany) follwing the manufacturer´s instructions. CD25+CD4+ T cells were separated from the CD25−CD4+ fraction by magnetic sorting of the bead-free CD4+ T cells using the mouse CD25 Microbead Kit (Miltenyi Biotec, Bergisch-Gladbach, Germany) as described in the manufacturer´s protocol as well as in previous studies.49 CD4+CD25+ and CD4+CD25− T cells were used in an in vitro suppression assay, described below.
In vitro Treg-suppression assay
CD4+CD25− T cells, isolated from the lungs of tumour-bearing wild-type mice served as responder cells and were labeled with 2,5 µM of CFSE using the Cell Trace CFSE Cell Proliferation kit (Invitrogen, Karlsruhe, Germany) according to manufacturer´s instructions. Labeled CD4+CD25− responder cells (1 × 105) were co-cultured with CD4+CD25+ Treg-cells (1 × 105), isolated from tumour-bearing wild-type or Tbet−/− mice. To stimulate the cell proliferation the culture also included mitomycin-treated A20 cells (1 × 104) and 5 µg/ml of soluble anti-mouse CD3 antibody (BD Biosciences, Heidelberg, Germany). The cells were cultured in RPMI medium, supplemented with 10 % FCS and 1 % Pen/Strep for 96 h at 37 °C and 5 % CO2. Afterwards, the cells were harvested and the CFSE dilution in CD4+CD25− responder cells was measured using the FACS Calibur and analyzed using Cell Quest Pro version 4.02 (BD Biosciences, Heidelberg, Germany).
Statistical evaluation
Statistical analysis excluding correlations were done with the Student's t- test (*p < 0,05;**p < 0,01; ***p < 0,001) and presented as mean values±SEM. Correlations were examined by importing data, in XY-tables of GraphPad Prism 7 software, diagram it with linear regression curve and perform the two tailed Pearson correlation analysis to get the r and p value (*p < 0,05;**p < 0,01;***p < 0,001).
Supplementary Material
Funding Statement
This work was supported by an ELAN 16-05-24-1-Andreev Grant awarded to Katerina Kachler, by the IZKF grant A59 and by the grant B12/SFB643 both awarded to Susetta Finotto and by the Molecular Pneumology department at the University FAU Erlangen-Nürnberg.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank the whole team of the Molecular Pneumology department at the University of Erlangen for their technical support. We also thank Kinga Soó Becker from the University of Mainz, providing us with the results on the survival of hCD2-ΔkTβRII as compared to WT mice upon tumor induction. This work was supported by an ELAN16-05-24-1-Andreev Grant awarded to Katerina Kachler, by the IZKF grant A59 and by the grant B12 in the SFB643 both awarded to Susetta Finotto and by the Molecular Pneumology department at the University FAU Erlangen-Nürnberg.
Author contributions
S.F. supervised the research and helped with data analysis as well as with writing and the revision of the manuscript. K.K designed and performed the experiments presented in this study, analysed the data in the paper and wrote the manuscript. D.I.T. contributed to cell isolation from lung tissue obtained after surgery from patients. H.S. and D.I.T. contributed to selection of patients, performed surgery and collected the clinical data. The authors declare no competing financial interests. Correspondence and requests for materials should be addressed to S.F. (susetta.finotto@uk-erlangen.de).
References
- 1.van Klaveren R. J. Lung cancer screening with low dose computed tomography: where do we stand today? Eur J Cancer. 2009;45(Suppl 1):375–6. doi: 10.1016/S0959-8049(09)70054-1. PMID:19775636 [DOI] [PubMed] [Google Scholar]
- 2.Rogers T. K., Hamilton W., Tod A., Neal R. Response to: What characteristics of primary care and patients are associated with early death in patients with lung cancer in the UK? Thorax. 2015;70:184. doi: 10.1136/thoraxjnl-2014-206514. PMID:25535292 [DOI] [PubMed] [Google Scholar]
- 3.Reppert S. Boross I, Koslowski M, Ö Türeci, Koch S, Lehr HA, Finotto S. A role for T-bet-mediated tumour immune surveillance in anti-IL-17 ;A treatment of lung cancer. Nature communications. 2011;2:600. doi: 10.1038/ncomms1609. PMID:22186896 [DOI] [PubMed] [Google Scholar]
- 4.Werneck M. B., Lugo-Villarino G., Hwang E. S., Cantor H., Glimcher L. H. T-bet plays a key role in NK-mediated control of melanoma metastatic disease. Journal of immunology. 2008;180:8004–10. doi: 10.4049/jimmunol.180.12.8004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cruz-Guilloty F, Pipkin ME, Djuretic IM, Levanon D, Lotem J, Lichtenheld MG, Groner Y, Rao A.. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. The Journal of experimental medicine. 2009;206:51–59. doi: 10.1084/jem.20081242. PMID:19139168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lazarevic V., Glimcher L. H. T-bet in disease. Nat Immunol. 2011;12:597–606. doi: 10.1038/ni.2059. PMID:21685955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szabo S. J. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–69. doi: 10.1016/S0092-8674(00)80702-3. PMID:10761931 [DOI] [PubMed] [Google Scholar]
- 8.Townsend M. J, Weinmann AS, Matsuda JL, Salomon R, Farnham PJ, Biron CA, Gapin L, Glimcher LH. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20:477–94. doi: 10.1016/S1074-7613(04)00076-7. PMID:15084276 [DOI] [PubMed] [Google Scholar]
- 9.Andreev K. Impaired T-bet-pSTAT1alpha and perforin-mediated immune responses in the tumoral region of lung adenocarcinoma. British journal of cancer. 2015:113:902–13. doi: 10.1038/bjc.2015.255. PMID:26348446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perales M. A, Blachere NE, Engelhorn ME, Ferrone CR, Gold JS, Gregor PD, Noffz G, Wolchok JD, Houghton AN. Strategies to overcome immune ignorance and tolerance. Seminars in Cancer Biology. 2002;12:63–71. doi: 10.1006/scbi.2001.0397. PMID:11926414 [DOI] [PubMed] [Google Scholar]
- 11.Motz G. T., Coukos G.. Deciphering and Reversing Tumor Immune Suppression. Immunity. 2013;39:61–73. doi: 10.1016/j.immuni.2013.07.005. PMID:23890064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curiel T. J, Coukos G., Zou L, Alvarez X, Cheng P, Mottram P, Evdemon-Hogan M, Conejo-Garcia JR, Zhang L, Burow M, et al.. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nature medicine. 2004;10:942–9. doi: 10.1038/nm1093. PMID:15322536 [DOI] [PubMed] [Google Scholar]
- 13.Sakaguchi S. Regulatory T cells in the past and for the future. European journal of immunology. 2008;38:901–3. doi: 10.1002/eji.200890012. PMID:18395855 [DOI] [PubMed] [Google Scholar]
- 14.Berendt M. J., North R. J. T-Cell-Mediated Suppression of Anti-Tumor Immunity – Explanation for Progressive Growth of an Immunogenic Tumor. Journal of Experimental Medicine. 1980;151:69–80. doi: 10.1084/jem.151.1.69. PMID:6444236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Byrne W. L., Mills K. H. G., Lederer J. A, O'Sullivan G. C. Targeting Regulatory T Cells in Cancer. Cancer research. 2011;71:6915–20. doi: 10.1158/0008-5472.CAN-11-1156. PMID:22068034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woo E. Y, Chu CS, Goletz TJ, Schlienger K, Yeh H, Coukos G, Rubin SC, Kaiser LR, June CH. Regulatory CD4(+)CD25(+) T cells in tumors from patients with early-stage non-small cell lung cancer and late-stage ovarian cancer. Cancer research. 2001;61:4766–72. PMID:11406550 [PubMed] [Google Scholar]
- 17.Li H, Zhao H, Yu J, Su Y, Cao S, An X, Ren X. Increased prevalence of regulatory T cells in the lung cancer microenvironment: a role of thymic stromal lymphopoietin. Cancer Immunology Immunotherapy. 2011;60:1587–96. doi: 10.1007/s00262-011-1059-6. PMID:21681373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimizu K, Nakata M, Hirami Y, Yukawa T, Maeda A, Tanemoto K. Tumor-Infiltrating Foxp3+Regulatory T Cells are Correlated with Cyclooxygenase-2 Expression and are Associated with Recurrence in Resected Non-small Cell Lung Cancer. Journal of Thoracic Oncology. 2010;5:585–90. doi: 10.1097/JTO.0b013e3181d60fd7. PMID:20234320 [DOI] [PubMed] [Google Scholar]
- 19.Khattri R., Cox T., Yasayko S. A, Ramsdell F. An essential role for Scurfin in CD4(+)CD25(+) T regulatory cells. Nature Immunology. 2003;4:337–42. doi: 10.1038/ni909. PMID:12612581 [DOI] [PubMed] [Google Scholar]
- 20.Fontenot J. D., Gavin M. A, Rudensky A. Y. Foxp3 programs the development and function of CD4(+)CD25(+) regulatory T cells. Nature Immunology. 2003;4:330–6. doi: 10.1038/ni904. PMID:12612578 [DOI] [PubMed] [Google Scholar]
- 21.Hori S., Nomura T., Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–61. doi: 10.1126/science.1079490. PMID:12522256 [DOI] [PubMed] [Google Scholar]
- 22.Koch M. A, Thomas KR, Perdue NR, Smigiel KS, Srivastava S, Campbell DJ. T-bet(+) Treg cells undergo abortive Th1 cell differentiation due to impaired expression of IL-12 receptor beta2. Immunity. 2012;37:501–10. doi: 10.1016/j.immuni.2012.05.031. PMID:22960221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koch M. A, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol. 2009;10:595–602. doi: 10.1038/ni.1731. PMID:19412181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu F., Sharma S., Edwards J., Feigenbaum L., Zhu J. Dynamic expression of transcription factors T-bet and GATA-3 by regulatory T cells maintains immunotolerance. Nat Immunol. 2015;16:197–206. doi: 10.1038/ni.3053. PMID:25501630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou L., Chong M. M., Littman D. R. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30:646–55. doi: 10.1016/j.immuni.2009.05.001. PMID:19464987 [DOI] [PubMed] [Google Scholar]
- 26.Andreev K., Graser A., Maier A., Mousset S., Finotto S. Therapeutical measures to control airway tolerance in asthma and lung cancer. Frontiers in immunology. 2012;3:216. doi: 10.3389/fimmu.2012.00216. PMID:22855687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fischer J. R, Darjes H, Lahm H, Schindel M, Drings P, Krammer PH. Constitutive secretion of bioactive transforming growth factor beta 1 by small cell lung cancer cell lines. Eur J Cancer. 1994;30A:2125–9. doi: 10.1016/0959-8049(94)00364-B. [DOI] [PubMed] [Google Scholar]
- 28.Sargent E. R, Gomella LG, Wade TP, Ewing MW, Kasid A, Linehan WM. Expression of mRNA for transforming growth factors-alpha and -beta and secretion of transforming growth factor-beta by renal cell carcinoma cell lines. Cancer Commun. 1989;1:317–22. doi: 10.3727/095535489820874904. PMID:2488683 [DOI] [PubMed] [Google Scholar]
- 29.Chen W Jin W, Hardegen N, Lei KJ, Li L, Marinos N, McGrady G, Wahl SM. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. The Journal of experimental medicine. 2003;198:1875–86. doi: 10.1084/jem.20030152. PMID:14676299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fantini M, C Becker C, Monteleone G, Pallone F, Galle PR, Neurath MF. Cutting edge: TGF-beta induces a regulatory phenotype in CD4+CD25- T cells through Foxp3 induction and down-regulation of Smad7. Journal of immunology. 2004;172:5149–53. doi: 10.4049/jimmunol.172.9.5149. [DOI] [PubMed] [Google Scholar]
- 31.Lazarevic V., Glimcher L. H., Lord G. M. T-bet: A bridge between innate and adaptive immunity. Nature Reviews Immunology. 2013;13:777–89. doi: 10.1038/nri3536. PMID:24113868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hall A. O, Beiting DP, Tato C, John B, Oldenhove G, Lombana CG, Pritchard GH, Silver JS, Bouladoux N, Stumhofer JS, et al.. The Cytokines Interleukin 27 and Interferon-gamma Promote Distinct Treg Cell Populations Required to Limit Infection-Induced Pathology. Immunity. 2012;37:511–23. doi: 10.1016/j.immuni.2012.06.014. PMID:22981537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finotto S, Hausding M, Doganci A, Maxeiner JH, Lehr HA, Luft C, Galle PR, Glimcher LH. Asthmatic changes in mice lacking T-bet are mediated by IL-13. International immunology. 2005;17:993–1007. doi: 10.1093/intimm/dxh281. PMID:16000330 [DOI] [PubMed] [Google Scholar]
- 34.Neurath M. F, Weigmann B, Finotto S, Glickman J, Nieuwenhuis E, Iijima H, Mizoguchi A, Mizoguchi E, Mudter J, Galle PR. The transcription factor T-bet regulates mucosal T cell activation in experimental colitis and Crohn's disease. The Journal of experimental medicine. 2002;195:1129–43. doi: 10.1084/jem.20011956. PMID:11994418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kachler K, Bailer M., Heim L, Schumacher F, Reichel M, Holzinger CD, Trump S, Mittler S, Monti J, Trufa DI, et al.. Enhanced Acid Sphingomyelinase Activity Drives Immune Evasion and Tumor Growth in Non-Small Cell Lung Carcinoma. Cancer Res. 2017;77(21):5963–5976. doi: 10.1158/0008-5472.CAN-16-3313. PMID:28883000 [DOI] [PubMed] [Google Scholar]
- 36.Vahl J. M, Friedrich J, Mittler S, Trump S, Heim L, Kachler K, Balabko L, Fuhrich N, Geppert CI, Trufa DI, et al.. Interleukin-10-regulated tumour tolerance in non-small cell lung cancer. British journal of cancer. 2017;117(11):1644–1655. doi: 10.1038/bjc.2017.336. PMID:29016555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lazarevic V, Chen X, Shim JH, Hwang ES, Jang E, Bolm AN, Oukka M, Kuchroo VK, Glimcher LH. T-bet represses T(H)17 differentiation by preventing Runx1-mediated activation of the gene encoding RORgammat. Nat Immunol. 2011;12:96–104. doi: 10.1038/ni.1969. PMID:21151104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang F., Meng G., Strober W. Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat Immunol. 2008;9:1297–306. doi: 10.1038/ni.1663. PMID:18849990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng W., Flavell R. A. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 ;T cells. Cell. 1997;89:587–96. doi: 10.1016/S0092-8674(00)80240-8. PMID:9160750 [DOI] [PubMed] [Google Scholar]
- 40.Zhu J, Jankovic D, Oler AJ, Wei G, Sharma S, Hu G, Guo L, Yagi R, Yamane H, Punkosdy G. The transcription factor T-bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses. Immunity. 2012;37:660–73. doi: 10.1016/j.immuni.2012.09.007. PMID:23041064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geissmann F, Revy P, Regnault A, Lepelletier Y, Dy M, Brousse N, Amigorena S, Hermine O, Durandy A. TGF-beta 1 prevents the noncognate maturation of human dendritic Langerhans cells. Journal of immunology. 1999;162:4567–75. [PubMed] [Google Scholar]
- 42.Siegel P. M., Massague J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nature Reviews Cancer. 2003;3:807–20. doi: 10.1038/nrc1208. PMID:14557817 [DOI] [PubMed] [Google Scholar]
- 43.Massague J. TGF beta in cancer. Cell. 2008;134:215–30. doi: 10.1016/j.cell.2008.07.001. PMID:18662538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas D. A., Massague J. TGF-beta directly targets cytotoxic T cell functions during tumor evasion of immune sur-veillance. Cancer cell. 2005;8:369–80. doi: 10.1016/j.ccr.2005.10.012. PMID:16286245 [DOI] [PubMed] [Google Scholar]
- 45.Eisenhut F Heim L, Trump S, Mittler S, Sopel N, Andreev K, Ferrazzi F, Ekici AB, Rieker R, Springel R, et al.. FAM13 A is associated with non-small cell lung cancer (NSCLC) progression and controls tumor cell proliferation and survival. Oncoimmunology. 2017;6:e1256526. doi: 10.1080/2162402X.2016.1256526. PMID:28197372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Balabko L, Andreev K, Burmann N, Schubert M, Mathews M, Trufa DI, Reppert S, Rau T, Schicht M, Sirbu H, et al.. Increased expression of the Th17-IL-6R/pSTAT3/BATF/RorgammaT-axis in the tumoural region of adenocarcinoma as compared to squamous cell carcinoma of the lung. Scientific reports. 2014;4:7396. doi: 10.1038/srep07396. PMID:25491772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schramm C, Protschka M, Köhler HH, Podlech J, Reddehase MJ, Schirmacher P, Galle PR, Lohse AW, Blessing M. Impairment of TGF-beta signaling in T cells increases susceptibility to experimental autoimmune hepatitis in mice. Am J Physiol Gastrointest Liver Physiol. 2003;284:G525–535. doi: 10.1152/ajpgi.00286.2002. PMID:12466145 [DOI] [PubMed] [Google Scholar]
- 48.Sauer K. A., Scholtes P., Karwot R., Finotto S.. Isolation of CD4+ T cells from murine lungs: a method to analyze ongoing immune responses in the lung. Nature protocols. 2006;1:2870–5. doi: 10.1038/nprot.2006.435. PMID:17406546 [DOI] [PubMed] [Google Scholar]
- 49.Doganci A, Eigenbrod T, Krug N, De Sanctis GT, Hausding M, Erpenbeck VJ, el-B Haddad, Lehr HA, Schmitt E, Bopp T, et al.. The IL-6R alpha chain controls lung CD4+CD25+ Treg development and function during allergic airway inflammation in vivo. The Journal of clinical investigation. 2005;115:313–25. doi: 10.1172/JCI200522433. PMID:15668741 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


