Abstract
We examined specimens from 111 HIV-infected participants virally suppressed on ART for a minimum of 5 years who had donated serial peripheral blood mononuclear cell (PBMC) specimens to the University of Washington / Fred Hutch Center for AIDS Research (CFAR) Specimen Repository. We determined the HIV proviral copy number per million PBMCs, corrected for CD4 count, in 477 specimens collected after a minimum of 5-years of follow up and up to 15.5 years of clinical viral suppression. Generalized estimating equation regression was used to examine the association between the reservoir size and time, age at study entry, antiretroviral (ARV) regimen, and risk factors for HIV acquisition.
We found that: 1) the inter-participant baseline HIV DNA level varied widely between 0.01 and 4.8 pol-copies per microgram genomic DNA and per CD4 cell number / micoliter; 2) the HIV DNA level declined with time (half-life was estimated at 12 years, 95% confidence interval of 6.2-240 years); 3) the HIV DNA level was lower for those who achieved viral suppression at a younger age; and 4) the HIV DNA level was not affected by the specific antiretroviral regimen used to achieve and maintain suppression.
Background
The HIV reservoir is comprised of cells that contain integrated HIV DNA (proviruses) capable of producing new virions, and is the impediment to HIV cure. The majority of replication-competent HIV proviruses reside in a population of resting CD4 T-cells that can be either central memory, transitional memory, effector memory or other subtypes 1. In persons on antiretroviral therapy (ART) the reservoir can be maintained by several mechanisms; long-lived cells contribute to the reservoir by persisting for years while cells with shorter life spans can either die or proliferate leading to contraction or expansion of the reservoir, respectively2. The reservoir could also be maintained by new infection of uninfected target cells with rounds of complete viral replication 3.
The size of the reservoir varies between individuals4,5 and is difficult to quantify as all currently available methods of measurement are imperfect. Quantitative viral outgrowth (QVOA) from highly purified CD4 T-cells is the gold-standard method, but is expensive, time consuming, and likely underestimates the reservoir size since not all intact and full-length integrated proviruses can be induced to make new infectious virions 6. Quantitative PCR (qPCR) based methods detect integrated proviral sequences, but qPCR methods cannot distinguish between replication competent proviruses (the true reservoir) and other incomplete HIV DNA fragments or mutated proviruses that are replication incompetent and, thus, likely overestimate the reservoir size by orders of magnitude. Nevertheless, studies that used both techniques (QVOA and qPCR) on the same sample have demonstrated the measurements are proportionate to one another7, correlate with the actual reservoir size, and can be used to measure the change in reservoir over time.
Studies using QVOA estimated the HIV reservoir half-life to be as short as 6.3 months to as long as 96 months1,3,4,6,8. The broad range of observed half-lives is likely explained by differences in assay methods, patient characteristics, degree of viral suppression, duration of observation, as well as the small sample size. In some studies, the reservoir half-life was longer (the decay rate slower) for participants who experienced intermittent ‘blips’ of plasma viremia as compared to those without blips 3,4.
To address limitations of prior studies of HIV reservoir decay (low sample sizes, the unclear effect of long-term viral suppression, and relatively short periods of observation) we investigated the clinical correlates with PBMC HIV DNA levels and decay in a large cohort of 111 HIV patients who had contributed serial peripheral blood mononuclear cell (PBMC) specimens to the University of Washington / Fred Hutch Center for AIDS Research (CFAR) Specimen Repository after at least five years of viral suppression before the first measurement of reservoir size.
Methods
Participants, specimens and clinical data:
University of Washington / Fred Hutch CFAR maintains a cohort of HIV-infected individuals who receive their care at the UW HIV outpatient clinics and have agreed to contribute their clinical data to the UW HIV Information System (UWHIS) 9. Many of these patients have also joined the UW HIV Specimen Repository and serially donated blood for HIV research. For this study, we identified chronically HIV infected participants in the cohort who had continuously taken suppressive antiretroviral therapy (ART) for at least 5 years with evidence for continuous viral suppression, defined as having HIV plasma viral loads (HIV VL) that were not detectable or consistently < 40 copies per mL; patients with viral load ‘blips’ in this five-year, pre-measurement period were excluded from the study. Figure S1 shows the frequency of HIV VL measurement during this five-year period; demonstrating a median of 99 days between observations with nearly all individuals having at least annual testing. Clinical and demographic data were obtained from the UWHIS including ART history and laboratory tests (HIV RNA plasma viral load, CD4 and CD8 T-cell counts). All patients were chronically infected. All patients provided informed consent before participating.
Measurement of reservoir size via quantitative PCR (qPCR):
One aliquot of five million PBMCs from each of 477 specimen donation times from all 111 patients was tested for HIV proviral DNA by qPCR. Cryopreserved PBMC specimens were thawed viably and extra chromosomal nucleic acid was removed using the Qiagen QIAprep Miniprep Kit. The remaining genomic DNA was extracted using the Qiagen QIAamp DNA mini kit. HIV DNA was amplified by PCR using primers targeting both gag (SK431 and HXB2) and pol (Abbott proprietary) regions, quantified using gag mix probes (HXB2 and AR-8) on the ABI 7900 and Abbott proprietary pol probe on the Abbott m2000, normalized for genomic DNA content based on the DNA concentration (NanoDrop) and corrected for the closest CD4 count. CD4 and reservoir measurements were on the same day for 198 of the 477 samples (41.5%); the CD4 measurements were within 30 days of reservoir measurements for 320 of the samples (67.1%); and the measurements were within 90 days of each other for 444 of the reservoir measurements (93.1%).
Statistical analysis:
The HIV DNA level assessed by qPCR was first normalized to the total PBMC count of the sample, and then to the nearest absolute CD4 count (cells per microliter) for the patient. The log10-transformed normalized qPCR value with zero values replaced with half of the minimum detected value before log-transformation defined the log-HIV DNA level, the dependent variable for subsequent analysis. We used linear mixed effects modeling, accounting for repeated measures on the same individual by including a random effects term to test for associations between clinical factors and HIV DNA level at baseline and the rate of change in level over time.
Using the estimated coefficient for time in years in the linear mixed effects model, we calculated the half-life of the HIV DNA level by the formula:
The statsmodel10 package (version 0.6.1) of Python was used to implement the strategy, with the pandas11 package (version 0.19.2) of Python used to normalize the data.
We examined whether there was a difference in the HIV DNA decay rate (half-life) among patients for whom all plasma viral loads (pVLs) remained undetectable versus those with detectable-but-not-quantifiable pVLs (i.e., a positive pVL that was below the level of assay quantification, i.e. < 40 copies per microliter for most of the assays), as well as those with detectable and quantifiable pVLs (referred to as “viral blips”). We used linear mixed effects modeling with a per-participant intercept as a random effect to correct for multiple comparisons.
Results
Study Cohort.
We identified 111 individuals meeting criteria for analysis; demographic and clinical features are shown in Table 1. All study participants received combinations of a nucleotide or nucleoside reverse transcriptase inhibitors. A majority (86%) of participants also received a protease inhibitor and approximately one-half, (47; 42.3%) received an integrase inhibitor.
Table 1:
Study Participant Characteristics
| Patient characteristics (n = 111) | |
| Age (median and range) | 48 (31 - 66) years |
| Male / Female / Male-to-Female | 93 / 15 / 3 |
| Race (Caucasian / Black / Asian or Pacific Islander / multiple) | 82 / 17 / 11 / 1 |
| Antiretroviral exposure (all exposed to nRTI) | |
| Protease inhibitors | 96 |
| Non-nucleoside RTI | 89 |
| Integrase inhibitors | 47 |
| Median Years Clinically Suppressed at T0 | 8 years |
| CD4 T-cell count at T0 (median and range) | 554 (83 – 1260) |
| Study details | |
| Median follow-up period after T0 (range) | 1.4 (0 – 8.5) years |
| Median number of HIV proviral DNA measurements (range) | 3 (1 - 23) |
Quantitative PCR of gag and pol for reservoir size and decay assessment.
Study participants contributed a median of 3 (range 1-23) blood donations to the repository, over a median of 1.4 years (range 0-8.5; Fig S2), resulting in 477 specimens that were tested for HIV proviral DNA by quantitative PCR (qPCR).
The HIV DNA levels as determined by qPCR for the HIV pol gene were consistent with the estimates for the HIV gag gene (Spearman’s R2 of 0.60), as depicted in Figure 1. However, there were 68 (14.1%) samples for which only the pol gene amplified but the gag gene did not and 3 (0.63%) samples for which the reverse was true. For all subsequent analysis, we estimated HIV DNA levels based on the pol gene qPCR normalized to both total PBMC genomic DNA in the sample, and to the most recent absolute CD4 count (in cells per mcL) which were then log (base 10) transformed.
Figure 1: HIV DNA size as determined by quantitative PCR against the HIV gag or pol gene.
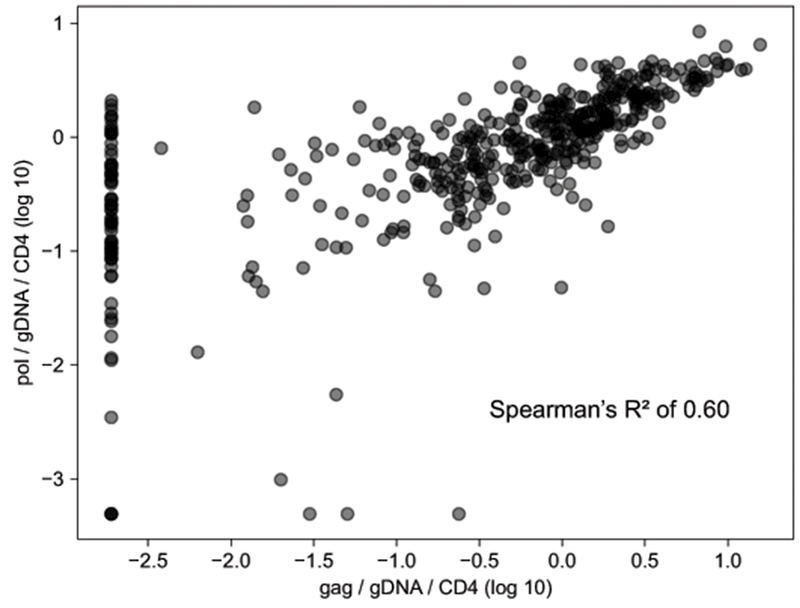
Each circle represents one sample. The x-axis is the reservoir size as determined by gag qPCR, the y-axis as determined by pol qPCR. There were 68 samples for which the pol gene amplified by the gag gene did not.
PBMC HIV DNA level correlates:
We used linear mixed / random effects modeling to test the associations between the HIV DNA levels and demographic or clinical characteristics of a patient, and to calculate an HIV DNA level half-life while accounting for repeated sampling of individuals.
Table 2 shows the results of the multivariate regression. HIV DNA levels (at all observed time points, correcting for time) were significantly associated with the age (as a continuous variable) of the patient when clinical suppression was achieved. Age did not correlate with the pre-ART peak observed viral load (Fig S3; Spearman R2 = 0.001) or CD4 nadir (Fig S4; Spearman R2 = 0.003). We did not find an association between the HIV DNA levels and the risk factor for HIV acquisition, specific ARV class used (analyzed by “ever exposed” and “months on particular ARV classes”), gender or race.
Table 2:
Associations with HIV Proviral DNA Level and Estimation of the Adjusted Decay Rate as per linear mixed effects modeling.
| Estimate | p | |
|---|---|---|
| HIV Risk Factor | ||
| Heterosexual Contact | 0.283 | 0.261 |
| MSM | 0.356 | 0.120 |
| IDU | 0.120 | 0.344 |
| ARV Exposure (Before) | ||
| nNRTI | 0.069 | 0.565 |
| Protease Inhibitor | 0.193 | 0.110 |
| Integrase Inhibitor | 0.087 | 0.707 |
| Demographics | ||
| Age (after 5 years of clinical suppression) | 0.016 | 0.023 |
| Male (Biological gender) | 0.163 | 0.400 |
| White Race | −0.191 | 0.125 |
| Time | ||
| Time (years) after 5 years of clinical suppression | −0.026 | 0.023 |
Time was included as a covariate in this model, corresponding to time in years since the first observation of reservoir size for each individual T0. We found that HIV DNA levels were significantly associated with time since study entry. The regression coefficient for time estimated a decay rate of −0.26 (pol/gDNA/CD4 log 10 over time in years).
PBMC HIV DNA decay:
Figure 2 shows the HIV DNA levels and change over time for the entire study cohort with the median and 95% confidence interval (CI) adjusted decay rate as determined by the linear mixed effects model. By this method, the half-life of the HIV DNA content for all study participants was 11.7 years (95% CI of 6.3 to 86 years).
Figure 2: Change in HIV DNA over time.
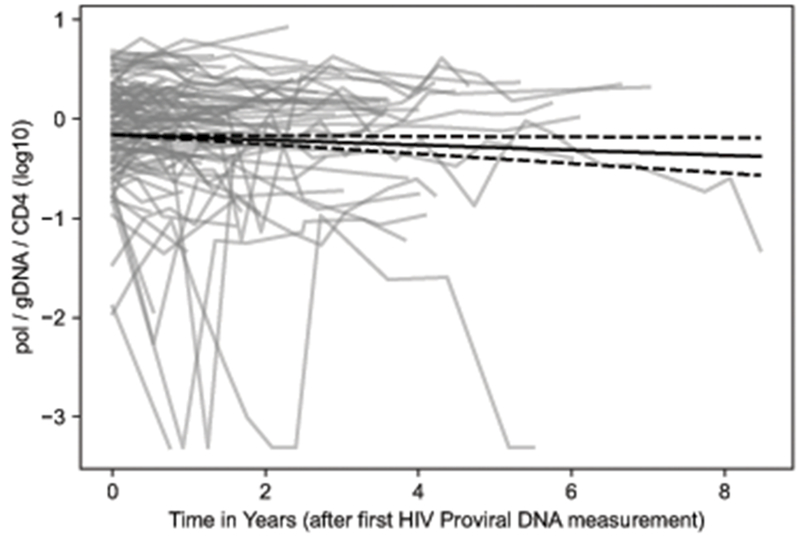
Each grey line represents one participant’s reservoir size over time. The x-axis is time in years starting from the individual’s first sample. The y-axis represents reservoir size as estimated by qPCR for the HIV pol gene, normalized to total genomic DNA (gDNA) and then absolute CD4 count (cells per mm3), finally log transformed (base 10). The solid black line is the corrected decay rate as estimated by our linear mixed effect model, with dashed black lines for the estimated 95% confidence interval of the decay rate.
Results of the subgroup analysis among patients for whom all plasma viral loads (pVLs) remained undetectable versus those with detectable-but-not-quantifiable pVLs (i.e., a positive pVL that was below the level of assay quantification, i.e. < 40 copies per microliter for most of the assays) and those with detectable and quantifiable pVLs (referred to as “viral blips”) are shown in Table 3. We discovered a (not statistically significant) trend towards shorter HIV DNA level half-lives in 21 patients (19.6%) who remained consistently undetectable at 87 months, compared to the 82 patients (76.6%) with at least one episode of detectable-but-not-quantifiable of 145 months, or 4 people (3.7%) with blips of quantifiable viral RNA in the plasma at a half-life of 264 months.
Table 3:
Decay Rate Stratified by Serum HIV Viral Load
| Category | # Pts | # Reservoir assays | Decay rate (slope) | Half-life (mo) | |
|---|---|---|---|---|---|
| Mean | 95% CI | Mean | |||
| Entire cohort | 111 | 477 | −0.026 | −0.048 to −0.004 | 140 |
| Patients with: | |||||
| Undetectable | 21 | 59 | −0.042 | −0.088 to −0.004 | 87 |
| Detectable | 82 | 400 | −0.025 | −0.049 to −0.000 | 145 |
| Quantifiable | 4 | 14 | −0.014 | −0.076 to 0.049 | 264 |
The subset of patients without blips had the same HIV DNA level (1.3 copies of pol / CD4) as those with detectable-but-not-quantifiable plasma RNA (1.3 copes of pol / CD4). Surprisingly, for those with blips (quantifiable plasma RNA) there was a trend towards higher HIV DNA level (0.6 copies of pol / CD4).
Discussion
This is one of the largest observational cohort studies of PBMC HIV DNA level kinetics to date, with 477 separate measurements from 111 participants on suppressive ART for a median of 8 years. Among patients with consistently undetectable pVLs throughout the observation period, the half-life was estimated to be 87 months, which is consistent with other studies of chronically virally suppressed HIV patients. Most patients in our study (77%) had at least brief stretches of detectable-but-not-quantifiable serum HIV virus, for whom the half-life (144 months) was similar to the cohort as a whole (140 months), and longer than the typical HIV reservoir half-life previously reported 1,3,4,6,8.
Our study used quantitative DNA PCR for the pol gene as a surrogate for HIV persistance. We paired quantitative PCR measurements of the gag gene, but found in a subset of samples gag did not amplify when pol was detectable (often at high levels). We hypothesize that mutations in the gag gene (perhaps better tolerated than mutations in pol) prevented proviral amplification with gag-specific primers in these instances. Sequence analyses of the gag and pol from discordant samples are needed to test this hypothesis.
Only age at the start of study correlated with reservoir size. This may be due to age at entry being confounded by or being a surrogate for a longer time before HIV diagnosis and longer time without virologic control that, in turn, led to higher PBMC HIV DNA levels. Unfortunately, we do not know the dates of HIV infection, and therefore the timing of ART relative to infection, for these patients. However, we found no clear relationship between age at study entry and peak plasma viral load or CD4 nadir prior to the study observation period, suggesting that the relationship of age to PBMC HIV DNA level is not simply mediated by plasma virus zenith or disease stage (see supplemental figures S3 and S4).
Our results demonstrate a longer HIV persistence than previously reported, which may be due to: 1) differences in the reservoir composition as estimated by qPCR as compared to QVOA; 2) non-log-linear kinetics of PBMC HIV DNA decay resulting in a slowing of the half-life for participants with a longer time interval of viral suppression; 3) the larger size of our cohort better captured the HIV DNA decay kinetics compared to other studies with fewer participants; and 4) the possibility that limited viral replication due to incomplete adherence led to reseeding of the HIV DNA pool.
We have used quantitative PCR to measure integrated HIV proviral DNA as a surrogate for HIV reservoir size, or HIV proviral persistence. The QVOA is the gold standard for measuring the inducible, infectious HIV reservoir. We note prior studies 7,8 found a linear relationship between reservoir size as estimated by qPCR and QVOA. Estimates of HIV persistence by quantitative PCR are orders of magnitude greater than that estimated by QVOA, since qPCR can detect defective provirus that is not replication competent. However, changes in the PBMC HIV DNA level over time (the decay rate or half-life) should be similar whether measured by qPCR or QVOA, provided the death and proliferation rates of cells with replication-competent or defective provirus are similar. However, the relative persistence of cells with defective or replication-competent proviruses remains controversial; one study reported an inverse correlation between proviral DNA clone size and the probability of reactivation suggesting cells with replication competent provirus are targeted for destruction over proliferation 12. In contrast, a more recent publication using ex vivo CD4+ T-cells from HIV infected, ART-suppressed persons, demonstrated CD8+ T-cell killing of cells with defective provirus with no effect on cells harboring replication-competent provirus 13. Our results are similar to that of Siliciano et al. 4 and showed a trend toward a longer HIV persistence in those patients with detectable viral RNA in plasma, which likely represents expansion of the PBMC HIV DNA pool due to cell proliferation 3 given that all patients were maintained on suppressive ART throughout the period of observation. Additional studies that examine proviral integration site analysis are needed to prove the concept of clonal expansion and to determine whether the ratio of cells with defective and replication competent provirus changes over time.
Supplementary Material
Acknowledgements:
None
Funding:
ACTG Laboratory Center (UM1-AI-106701); CFAR (P30-AI-027757); HVTN HIV Diagnostic Laboratory (UM-AI-068618)
Footnotes
Conflicts of Interest:
The authors have no conflicts of interest to disclose.
A portion of this work was previously presented as an oral abstract (#953) at ID Week 2016 (New Orleans, LA, USA)
References
- 1.Finzi D et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med 5, 512–517 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Chomont N et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat. Med 15, 893–900 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramratnam B et al. The decay of the latent reservoir of replication-competent HIV-1 is inversely correlated with the extent of residual viral replication during prolonged anti-retroviral therapy. Nat. Med 6, 82–85 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Siliciano JD et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med 9, 727–728 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Cockerham LR et al. CD4+ and CD8+ T Cell Activation Are Associated with HIV DNA in Resting CD4+ T Cells. PLoS ONE 9, e110731 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eriksson S et al. Comparative Analysis of Measures of Viral Reservoirs in HIV-1 Eradication Studies. PLoS Pathog 9, e1003174 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiselinova M et al. Integrated and Total HIV-1 DNA Predict Ex Vivo Viral Outgrowth. PLOS Pathog 12, e1005472 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crooks AM et al. Precise Quantitation of the Latent HIV-1 Reservoir: Implications for Eradication Strategies. J. Infect. Dis 212, 1361–1365 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kitahata MM et al. Electronic human immunodeficiency virus (HIV) clinical reminder system improves adherence to practice guidelines among the University of Washington HIV Study Cohort. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am 36, 803–811 (2003). [DOI] [PubMed] [Google Scholar]
- 10.statsmodels.
- 11.Python Data Analysis Library.
- 12.Lorenzi JCC et al. Paired quantitative and qualitative assessment of the replication-competent HIV-1 reservoir and comparison with integrated proviral DNA. Proc. Natl. Acad. Sci 113, E7908–E7916 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang S-H et al. Latent HIV reservoirs exhibit inherent resistance to elimination by CD8+ T cells. J. Clin. Invest 128, 876–889 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


