Abstract
Objective: Controlled medical terminologies (CMTs) have been recognized as important tools in a variety of medical informatics applications, ranging from patient-record systems to decision-support systems. Controlled medical terminologies are typically organized in semantic network structures consisting of tens to hundreds of thousands of concepts. This overwhelming size and complexity can be a serious barrier to their maintenance and widespread utilization. The authors propose the use of object-oriented databases to address the problems posed by the extensive scope and high complexity of most CMTs for maintenance personnel and general users alike.
Design: The authors present a methodology that allows an existing CMT, modeled as a semantic network, to be represented as an equivalent object-oriented database. Such a representation is called an object-oriented health care terminology repository (OOHTR).
Results: The major benefit of an OOHTR is its schema, which provides an important layer of structural abstraction. Using the high-level view of a CMT afforded by the schema, one can gain insight into the CMT's overarching organization and begin to better comprehend it. The authors' methodology is applied to the Medical Entities Dictionary (MED), a large CMT developed at Columbia-Presbyterian Medical Center. Examples of how the OOHTR schema facilitated updating, correcting, and improving the design of the MED are presented.
Conclusion: The OOHTR schema can serve as an important abstraction mechanism for enhancing comprehension of a large CMT, and thus promotes its usability.
Controlled medical terminologies (CMTs) are collections of medical concepts that consolidate aspects of medical knowledge.1, 2, 3, 4 Large CMTs have been emerging as important resources for use in medical informatics applications, such as hospital departmental systems, patient record systems, expert systems, and medical information systems.5 Examples of CMTs include terminologies—such as MeSH, CPM93, CPT98, SNOMED, and ICD-9-CM—that are integrated into the UMLS6, 7, 8, 9, 10, 11 (a complex collection of terms, concepts, and relationships retrieved and integrated from a variety of existing medical information sources), GALEN's Core Model12 (expressed in GRAIL13), and the Columbia-Presbyterian Medical Entities Dictionary (MED).5, 14
Acceptance of these CMTs and others has been slow, however, partly because of their wide scope and high complexity; CMTs typically comprise tens of thousands to hundreds of thousands of interconnected concepts. The size and complexity of CMTs make them hard to comprehend and maintain. In this paper, we address some of the problems of terminology comprehension by presenting a methodology for representing a CMT, modeled using the semantic network paradigm,15, 16, 17 as an object-oriented database.18, 19, 20 We refer to such a representation as an object-oriented health care terminology repository (OOHTR).21, 22 One of the most important components of the OOHTR is its schema, which provides an abstraction layer through which the CMT can be viewed and studied. This compact presentation of the CMT helps shed light on its overarching structure.
We use the MED as our test bed. Studies have shown that users of the MED at Columbia-Presbyterian Medical Center have trouble navigating through its constituent semantic network to find desired concepts.23 The complexity of the MED also presents challenges to its maintenance personnel, who often find it difficult to add concepts or create links without a clear understanding of the underlying terminology structural model. Others approaching the MED have encountered similar difficulties.24 In this paper, we demonstrate how the schema of the OOHTR was used to uncover some conceptual errors and inconsistencies in the MED—some that had been introduced initially and others that had crept in over time. These discoveries led directly to improvements in the MED's design.
A short version of this work appeared previously in the Proceedings of the 1996 AMIA Annual Fall Symposium.25 In that paper we gave an overview of our modeling approach and showed some of the MED improvements derived from the OOHTR schema. In this paper, we give a complete description of the modeling theory and the OOHTR schema for the MED. We also use additional examples to demonstrate the benefits of the object-oriented database representation of a CMT.
The paper is organized as follows: In the next section, we give an overview of the structural characteristics of CMTs, like the MED, that are modeled as semantic networks. The third section describes our methodology for modeling a CMT as an object-oriented database and presents the results of applying the methodology to the MED to produce an OOHTR. Finally, in the fourth section, we discuss how the OOHTR's schema improved the MED design by exposing errors that were subsequently corrected. In the fifth section we compare our approach with approaches based on the use of description logics. Conclusions appear in the last section.
Semantic Network CMTs
A CMT that is amenable to our methodology must have the structure of a semantic network. Such a CMT is a collection of medical concepts, each of which consists of properties that are either attributes (holding literal data values) or relationships (storing references to other concepts). One attribute needed in each concept contains the concept's associated term (or textual denotation).26 In the MED this attribute is called name. Another attribute, called synonyms in the MED, needs to hold additional denotations for a concept. As an example of a relationship in the MED, is-measured-by connects the concept Chemical to the concept Chemistry Test.*
The CMT's concepts must be organized into a concept subsumption hierarchy—i.e., a directed acyclic graph (DAG) composed of concepts (nodes) and IS-A links, each connecting a concept to its superconcept. The IS-A links provide the means for the inheritance of attributes and relationships, and they support subsumption-based reasoning. A concept may have more than one parent in the hierarchy—for example, in the MED, Chemical IS-A Measurable Substance and Chemical IS-A Etiologic Agent. We assume that all CMTs are singly rooted. The MED's IS-A hierarchy is rooted overall at the concept Medical Entity.
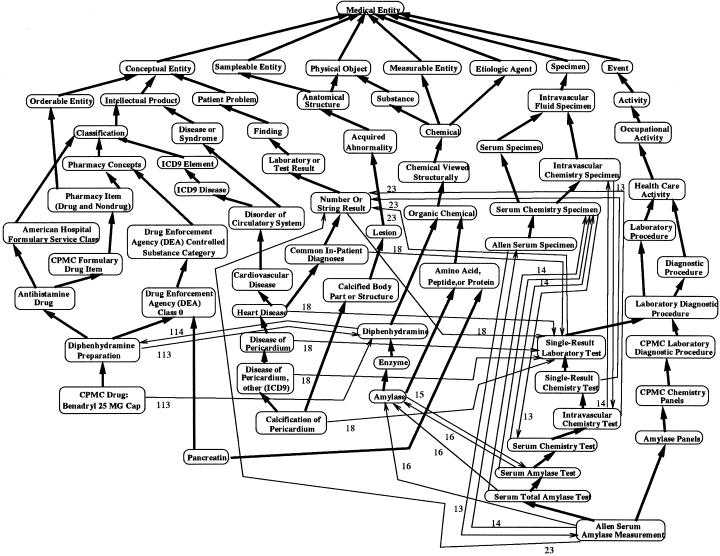
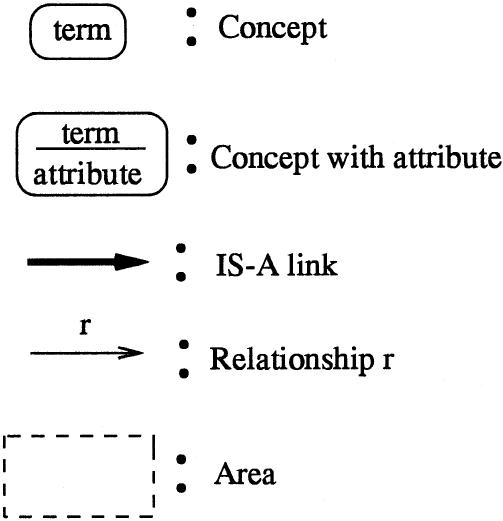
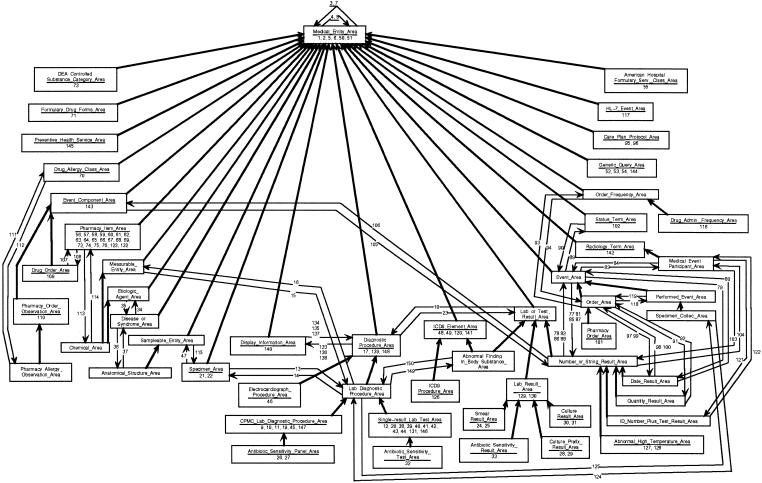
Controlled medical terminologies tend to be large and complex in scope. At the time of our research, the MED comprised about 43,000 concepts, which were connected by more than 71,000 (nonhierarchic) relationships. The IS-A links totaled more than 61,000.† ▶ shows a small portion of the MED (68 concepts, or about 0.16 percent of the entire MED). ▶ presents the notational conventions used for semantic networks.
Figure 1.
Sample content from the Columbia-Presbyterian Medical Entities Dictionary.
Figure 2.
Key to symbols used in figures of semantic networks.
▶ contains the six concepts CPMC Drug: Benadryl 25MG Cap, Pancreatin, Calcification of Pericardium, Amylase, Allen Serum Specimen, and Allen Serum Amylase Measurement, along with most of their ancestors in the IS-A hierarchy and some of the relationships between the respective concepts. Included are concepts for laboratory tests, medications, and diagnoses. For brevity, some details have been omitted, including additional children of the ancestor concepts, all attributes, and some relationships. The names of relationships have been written as numeric codes, whose meanings can be found in ▶.
Table 1.
Names of Properties Shown in Figures 1 and 4
| 1 umls-code |
| 2 name |
| 3 has-subconcept |
| 4 has-superconcept |
| 5 synonyms |
| 6 print-name |
| 7 has-parts |
| 8 part-of |
| 9 cpmc-lab-proc-code |
| 10 service-code |
| 11 cpmc-unit-names |
| 12 cpmc-lab-test-names |
| 13 specimen-of |
| 14 specimen |
| 15 measured-by |
| 16 substance-measured |
| 17 units |
| 18 result-of-tests |
| 19 cpmc-lab-proc-name |
| 20 cpmc-lab-test-code |
| 21 cpmc-lab-spec-code |
| 22 cpmc-lab-spec-name |
| 23 result-type |
| 24 cpmc-smear-code |
| 25 cpmc-smear-name |
| 26 cpmc-panel-code |
| 27 cpmc-panel-name |
| 28 cpmc-prefix-code |
| 29 cpmc-prefix-name |
| 30 cpmc-result-code |
| 31 cpmc-result-name |
| 32 cpmc-sensitivity-name |
| 33 cpmc-sensitivity-result-name |
| 34 etiology |
| 35 causes-diseases |
| 36 site |
| 37 site-of-diseases |
| 38 normal-value |
| 39 low-normal-value |
| 40 high-normal-value |
| 41 male-low-normal-value |
| 42 male-high-normal-value |
| 43 female-low-normal-value |
| 44 female-high-normal-value |
| 45 normal-ranges-text |
| 46 cpmc-ecg-name |
| 47 substance-sampled |
| 48 icd9-code |
| 49 icd9-entry-code |
| 50 main-mesh |
| 51 supplementary-mesh |
| 52 question-type |
| 53 english-question |
| 54 brs-question |
| 55 ahfs-class-code |
| 56 dose-strength-units |
| 57 dose-strength-number |
| 58 formulary-name |
| 59 short-formulary-name |
| 60 formulary-code |
| 61 drug-trade-name |
| 62 drug-generic-name |
| 63 drug-manufacturer |
| 64 drug-rx-vs-otc |
| 65 drug-form-code |
| 66 drug-floor-stock |
| 67 drug-route |
| 68 drug-in-formulary |
| 69 drug-volume |
| 70 allergy-class-code |
| 71 drug-description |
| 72 drug-category |
| 73 dea-code |
| 74 drug-specifier |
| 75 drug-generic-code |
| 76 drug-interaction-codes |
| 77 event-id |
| 78 event-id-of |
| 79 event-date |
| 80 event-date-of |
| 81 event-patient-id |
| 82 event-patient-id-of |
| 83 event-participant |
| 84 participant-of |
| 85 event-organization |
| 86 event-organization-of |
| 87 event-location |
| 88 event-location-of |
| 89 event-status |
| 90 status-of |
| 91 order-quantity |
| 92 order-quantity-of |
| 93 order-frequency |
| 94 order-frequency-of |
| 95 protocol-name |
| 96 protocol-short-name |
| 97 order-start-date |
| 98 order-start-date-of |
| 99 order-stop-date |
| 100 order-stop-date-of |
| 101 pharmacy-order-code |
| 102 status-code |
| 103 participant-id |
| 104 participant-id-of |
| 105 order-value |
| 106 order-value-of |
| 107 ordered-drug |
| 108 ordered-in |
| 109 drug-role-code |
| 110 pharmacy-observation-code |
| 111 observed-allergy |
| 112 allergy-observed-in |
| 113 pharmaceutic-component |
| 114 pharmaceutic-component-of |
| 115 sampled-by |
| 116 admin-frequency-abbrev |
| 117 h17-event-code |
| 118 event-object |
| 119 object-of-event |
| 120 old-icd9-code |
| 121 participant-name |
| 122 participant-name-of |
| 123 drug-id |
| 124 collected-for |
| 125 collected-by |
| 126 cpt4-code |
| 127 lower-limit-for-input |
| 128 upper-limit-for-input |
| 129 lab-message-code |
| 130 lab-message-text |
| 131 cpmc-long-test-name |
| 132 drug-alert-code |
| 133 has-default-displays |
| 134 default-display-for |
| 135 displays-elements-of |
| 136 elements-displayed-by |
| 137 has-display-parameters |
| 138 is-display-parameter-of |
| 139 has-test-display-class-name |
| 140 display-parameter-order |
| 141 icd9-name |
| 142 cpmc-radiology-code |
| 143 event-component-display-name |
| 144 query-fillers |
| 145 preventive-health-name |
| 146 lab-alt-test-name |
| 147 lab-alt-proc-name |
| 148 has-proc-display-class-name |
| 149 defined-by-test |
| 150 defines-abnormal-finding |
As discussed in Cimino et al.,5 the content of a CMT should satisfy the following seven basic requirements:
Domain completeness: There should be no numeric limitation on the size of any of the CMT's dimensions (e.g., no limit on the depth of the IS-A hierarchy).
Synonymy: Concepts can be recognized by multiple names.
Nonvagueness: Each concept must have a well-formed meaning.
Nonredundancy: No two concepts may have the same meaning.
Nonambiguity: Each concept may have no more than one meaning.
Multiple classification: Concepts may have more than one superconcept in the IS-A hierarchy.
Consistency of views: A concept should appear the same (and have the same properties and children) no matter how the concept is arrived at in the hierarchy.
We also assume that a CMT satisfies the following rule regarding the introduction of properties.
Rule 1: A given property x (whether it be an attribute or a relationship) can be introduced at only one concept in the CMT.
This requirement is not limiting, because if several concepts need to introduce the property x, then an “artificial” parent of them can be added to accommodate the unique introduction.27 The MED, to which we apply our methodology, satisfies Rule 1.
The OOHTR Schema
Initial OOHTR Schema
The strategy that we chose for modeling a CMT as an OOHTR utilizes special concepts as the basis for the definitions of object classes in the schema. In fact, it produces an abstraction of the underlying pattern in which properties are introduced into the CMT.
In general, the purpose of an object class in an object-oriented database schema is to define abstractly a collection of properties for a group of objects (or instances) that exhibit those exact properties and have a common semantics. In a CMT, some concepts function in an analogous role: each introduces (defines) attributes and relationships that are exhibited by all its children and descendants in the IS-A hierarchy (because of the inheritance mechanism). We call such concepts property-introducing concepts.‡ As discussed later, very few concepts in a CMT are property-introducing. Almost all of them inherit all their properties.
A property-introducing concept also plays the role of the most general conceptual entity among its descendants. In this way, it captures the overarching semantics of the descendants.
Because of these facts, it is sensible to construct object classes with respect to all the property-introducing concepts appearing in the CMT. Toward that end, we define the notion of area to be a set containing one property-introducing concept plus all that concept's descendants that have the same properties. Notice that some descendants can have more properties than the property-introducing concept; in such cases, the descendants do not belong to the area. The property-introducing concept of an area is called the area's root, since it is the area's highest concept in the IS-A hierarchy (i.e., the property-introducing concept's parents are not in the same area because they lack the properties it introduces). An area will also be named by its property-introducing concept. An area with property-introducing concept A will be called “Area A” or “A Area.”
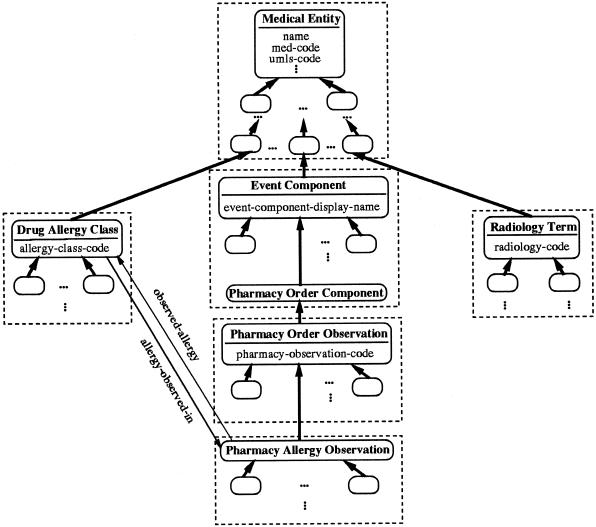
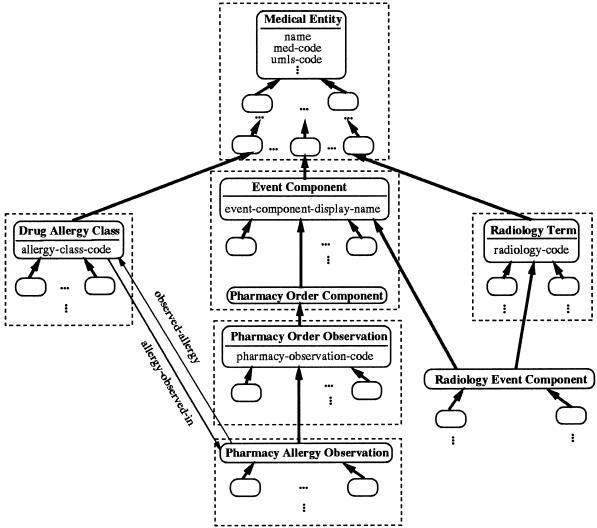
To illustrate the notations of property-introducing concept and area, ▶ shows an excerpt from the MED. The figure contains six property-introducing concepts: Medical Entity, Drug Allergy Class, Event Component, Radiology Term, Pharmacy Order Observation, and Pharmacy Allergy Observation. It also contains the concept Pharmacy Order Component, which is not property-introducing. Other concepts are left unlabeled. The concept Medical Entity, the root of the entire MED, introduces the attributes name, med-code, and umls-code (among others). The concept Drug Allergy Class introduces the attribute allergy-class-code and the relationship allergy-observed-in directed to the concept Pharmacy Allergy Observation. The concepts Event Component, Radiology Term, and Pharmacy Order Observation each introduce a single attribute.
Figure 3.
Six areas of the Medical Entities Dictionary (see key at ▶).
Finally, Pharmacy Allergy Observation introduces the relationship observed-allergy, the converse of allergy-observed-in.
Each of the six areas in ▶ is enclosed in a large, dashed rectangle. The area rooted at concept Medical Entity extends down to, but excludes, concepts Drug Allergy Class, Event Component, and Radiology Term. Those three concepts are roots of areas of their own. Examples of some of the Drug Allergy Class Area's concepts, not shown in the figure, are Glucocorticoids, Codeine, Morphine, Barbiturates, Tetracyclines, and Phenothiazines. The Event Component Area extends down to include the concept Pharmacy Order Component, which is the parent of Pharmacy Order Observation, the root of the Pharmacy Order Observation Area. The last area is rooted at Pharmacy Allergy Observation.
Once the property-introducing concepts and their respective areas have been identified, object classes can be created to represent them. Such a class serves the dual purposes of defining the properties for an area and holding the area's concepts—all of which have identical structure and semantic similarity—as its instances. For this reason, we refer to a class in the OOHTR schema as an area class.
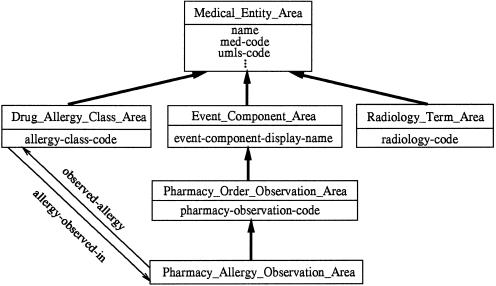
To be more precise, for each area in the CMT we define an object class whose instances will be exactly the area's concepts, including its root. The class's name is formed by concatenating the name of the area's root and “_Area.” The properties defined by an area class are identical to those introduced by the area's root in the CMT. So, for example, the Medical Entity Area would have the corresponding class Medical_Entity_Area, which would define the properties name, med-code, and umls-code, among others.
Notice that the root of an area exhibits all the properties that it itself introduces, plus the properties that it inherits from its parent(s). The area's other concepts, of course, also have these same properties. To reflect this situation, we utilize the standard subclass inheritance of object-oriented database schemas. A given area class A_Area, corresponding to Area A, is made a subclass of each area class that contains a parent of the root of Area A. As we discussed above, a concept may have more than one parent, and the subclass hierarchy induced by this process is therefore not necessarily a tree—that is, an area class can have multiple area classes as superclasses. In addition to the properties that it defines intrinsically, an area class has all the properties of its superclasses through inheritance.
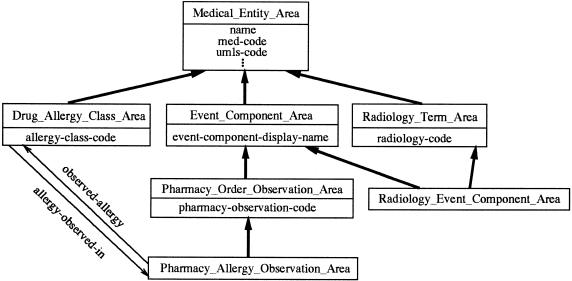
▶ shows the schema corresponding to the six areas in ▶; the key to symbols used there is shown in ▶. Notice that the schema is represented using our OOdini-2 graphic notation.28 ▶ shows the notation used for object-oriented database schemas. With OOdini-2, a class is represented as a rectangle and a relationship as a labeled arrow. A subclass relationship is drawn as a bold arrow directed upward from the subclass to the superclass. An attribute is listed inside its class rectangle beneath the class's name.
Figure 4.
Area classes corresponding to the areas of the Medical Entities Dictionary shown in ▶.
Figure 5.
Key to symbols used in figures of object-oriented database schemas.
We can see that there are six object classes, one for each area in ▶. The classes have the properties introduced by the corresponding roots. For example, the class Medical_Entity_Area has the attributes name, med-code, umls-code, etc. As another example, the class Pharmacy_Allergy_Observation_Area has the relationship observed-allergy directed to the class Drug_Allergy_Class_Area, and it is a subclass of Pharmacy_Order_Observation_Area, from which it inherits the attributes name, med-code, umls-code (and so on), event-component-display-name, and pharmacy-observation-code.
The overall OOHTR schema produced by this mapping turns out to be very compact in its number of classes, particularly when one compares that number to the MED's tens of thousands of concepts. The compactness results from the fact that the total number of distinct properties in the MED is only 150. This implies that there are at most 150 property-introducing concepts of the 43,000 concepts in the entire terminology. So, it can be seen that most concepts in the MED are not property-introducing. Because some concepts introduce multiple properties, the number of property-introducing concepts is actually just 53. This process thus identifies 53 areas for the MED's 43,000 concepts, and the OOHTR schema consists of only 53 area classes.
▶ presents the entire OOHTR schema obtained via the mapping described above. To save space, only numeric codes of attributes and relationships are shown. For example, attribute “9” is lab-procedure-code; relationship “18” is result-of-tests. ▶ gives the codes and corresponding names for all attributes and relationships. The area class Medical_Entity_Area that corresponds to the MED's overall root Medical Entity becomes the top class in the OOHTR schema's class hierarchy. As we mentioned, an area class can have more than one superclass. This is demonstrated by the class Chemical_Area, which has the superclasses Measurable_Entity_Area and Etiologic_Agent_Area.
Figure 6.
Schema derived from the areas of the Medical Entities Dictionary (see key at ▶).
In another paper,21 we present a program called the OOHTR Generator, which automatically generates the object-oriented database schema for a given CMT. That program was used to build the MED's OOHTR schema shown in ▶. It has also been applied to the InterMED.21
The MED's IS-A hierarchy served as the basis for the mapping into the OOHTR schema. In fact, the mapping constituted the identification of the property-introducing concepts and a “collapsing” of the inheritance paths between them. Thus, the OOHTR schema can be seen as an abstraction of the property definitions and accompanying inheritance that occur in the MED. We call this kind of schema a network abstraction schema. To preserve the actual IS-A connections between concepts from within the source CMT, a pair of converse relationships has-superconcept and has-subconcept is added to the area class Medical_Entity_Area. Because of this, the two properties are exhibited by all concepts in the OOHTR. The relationships are used to connect a given concept to its parents and children, respectively. If the concept A IS-A B in the CMT, then the object representing A in the OOHTR refers to the object denoting B via has-superconcept. Conversely, B relates to A through has-subconcept.
Extended OOHTR Schema
One complication in this mapping arises because of the multiple inheritance that occurs in the CMT's IS-A hierarchy. (Recall that it is a DAG, not a tree.) The problem is illustrated for the MED in ▶, which expands ▶ to include the concept Radiology Event Component (and some of its descendants). Notice that Radiology Event Component is not a property-introducing concept. It is, however, a child of two property-introducing concepts, Event Component and Radiology Term, and inherits its properties from both of them. The latter point gives rise to the difficulty. Since Radiology Event Component inherits from Event Component, it has a different set of properties than its parent Radiology Term and is therefore not in the Radiology Term Area. Likewise, it is not in the Event Component Area, either. In fact, Radiology Event Component does not reside in any area! As such, it currently has no representation within the OOHTR. The same is true of its descendants.
Figure 7.
Expanded version of the six areas shown in ▶ (see key at ▶).
Our solution is to introduce a new kind of area to include concepts like Radiology Event Component. In general, such a concept is characterized by the fact that its property set differs from the property sets of all property-introducing concepts in the CMT. While such a concept does not introduce any new properties of its own, it does lie at the juncture of “independent” inheritance paths and uniquely collects groups of properties. For this reason, we call such a concept an intersection concept. Notice that we preclude a concept from being an intersection concept if it has an intersection concept ancestor with the same set of properties. For example, in ▶, Radiology Event Component's two children—Radiology Report Event Component and Radiology Service Modifier—are not intersection concepts.
We now define a new kind of area (called an intersection area) to be a set containing one or more intersection concepts having the same set of properties and all their descendants with the same properties. The intersection concepts residing in an intersection area are called the roots of the area because they are the area's highest concepts in the IS-A hierarchy (i.e., their parents do not belong to the area). An example of an intersection area is the one containing the root Radiology Event Component, its two children Radiology Report Event Component and Radiology Service Modifier, and an additional 79 descendants. This intersection area contains just one root; however, an intersection area can be multirooted. As an example, the three concepts Antihistamine Drugs, Anti-Infective Agents, and Autonomic Drugs are children of the two property-introducing concepts AHFS Service Class and Formulary Drug Item. Hence, all three are intersection concepts, have the same set of properties, and root the same intersection area. In fact, there are 28 other intersection concepts that also have this property set, and therefore this particular intersection area has a total of 31 roots.
As with the areas rooted at property-introducing concepts, a separate class is created in the OOHTR schema for each intersection area. This new kind of class is referred to as an intersection (area) class. The concepts in the intersection area become instances of this intersection class, which, interestingly, does not define any new properties (just like its root[s]). Instead, it gets its properties entirely via object-oriented subclass inheritance. The subclass relationships for an intersection class are determined by the parentage of its root(s) in an analogous manner to that for ordinary area classes. Another interesting point is that an intersection class must have at least two superclasses in the schema's subclass hierarchy. Hence, the presence of intersection concepts in the CMT implies multiple inheritance within the OOHTR schema.
If an intersection area has a unique root, then its corresponding intersection area class is naturally denoted using the root's name (concatenated with “_Area”). Otherwise, one of the roots—say, the one appearing first in some search of the CMT—is arbitrarily selected as the name of the intersection class. In ▶, we show the classes for the areas appearing in ▶. The only addition to the schema from ▶ is the class Radiology_Event_Component_Area, representing the intersection area rooted at the concept Radiology Event Component. Notice that it is a subclass of both Event_Component_Area and Radiology_Term_Area.
Figure 8.
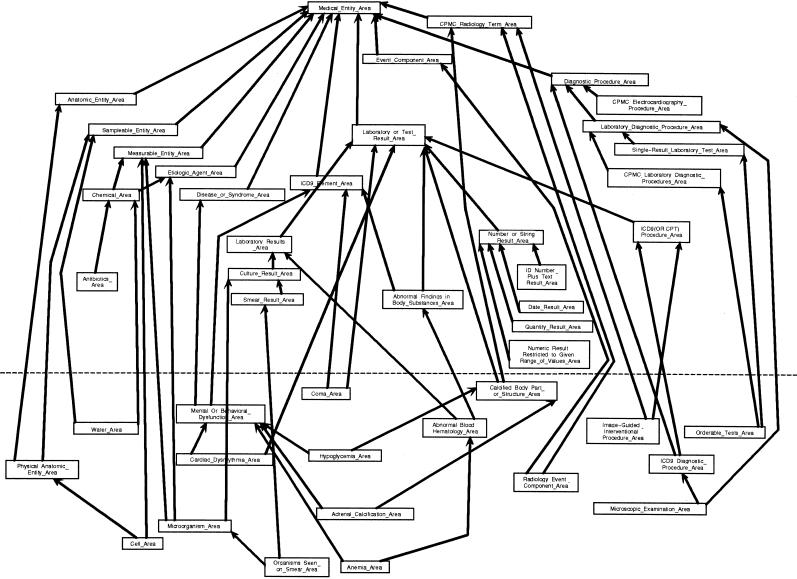
The entire OOHTR schema for the MED comprises 90 area classes, 37 of which are intersection classes, and 131 subclass relationships. Of the 37 intersection classes, 22 contain a single root and 15 are multirooted. Even though the schema is large, one should bear in mind that it abstracts a CMT of 43,000 concepts—a network 632 times the size of the excerpt of 68 concepts shown in ▶. Each class contains, on average, about 477 concepts.
In ▶, we show a large portion of the OOHTR schema's subclass hierarchy, with attributes and relationships omitted by applying “information thinning.” 29, 30 The figure contains about half the property-introducing classes and the intersection classes. The area classes above the dashed line represent areas rooted at property-introducing concepts. Those below the line are intersection classes.
Figure 9.
Excerpt from the schema for the object-oriented health care terminology repository (see key at ▶).
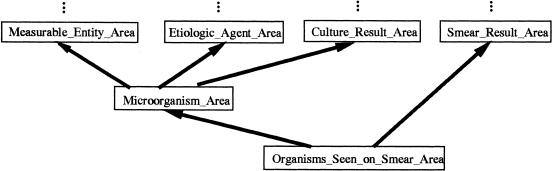
Let us point out that intersection concepts may lie at the juncture of three or more inheritance paths. The intersection class for such an area will be a subclass of at least three other classes. An example is Microorganism_Area, which is a subclass of Measurable_Substance_Area, Etiologic_Agent_Area, and Culture_Result_Area (▶). It is also possible for an intersection class to be a subclass of another intersection class. For example, in ▶, Anemia_Area is a subclass of the intersection class Abnormal_Blood_Hematology_Area.
CMT Improvement Based on the OOHTR Schema View
The development of specialized views, such as network abstraction schemas, is of more than theoretic interest. The maintenance of a CMT like the MED at Columbia—Presbyterian Medical Center is a complex and difficult task. The challenges faced by maintenance personnel include updating the CMT (see Robinson et al.,31 for example), adding terms and relationships,32 and in general developing a change model for CMTS.33 Furthermore, proper maintenance should include improving a CMT's organization and uncovering and correcting inconsistencies and errors in its content. All these require an understanding of the CMT's underlying structure. However, providing users of terminologies with comprehensible, comprehensive views remains difficult. This is true for terminology administrators as well as for those who would build applications or knowledge bases with respect to the CMTs.
At present, few commercial tools are suitable to support this maintenance work. Most such tools and environments aid developers in construction of CMTs and provide support for managing and enhancing terminologies. Also, they facilitate distributed-development tasks. For example, Rocha et al.34 described the Voser project for designing a CMT server. MEME II (Metathesaurus Enhancement and Maintenance Environment, version II), described by Suarez-Munist et al.,35 is a tool to support UMLS Metathesaurus maintenance and enhancement. It allows remote enhancements to a terminology to be incorporated locally, and local enhancements to be shared remotely. In a study by Mays et al.,36 K-Rep, a knowledge representation system based on description logic, is used to model CMTs. This approach increases semantic consistency and inferential capability. Gálapagos is a configuration management and conflict resolution environment built on top of K-Rep.37 It provides support for handling the inevitable conflicts generated by concurrent development of enhancements to a terminology. A proof-of-concept of Gálapagos is shown using an example in a report by Campbell et al.37 The use of semantic-based methods for managing concurrent terminology development to avoid disadvantages of traditional lock-based approaches common in database systems is presented by Campbell.38
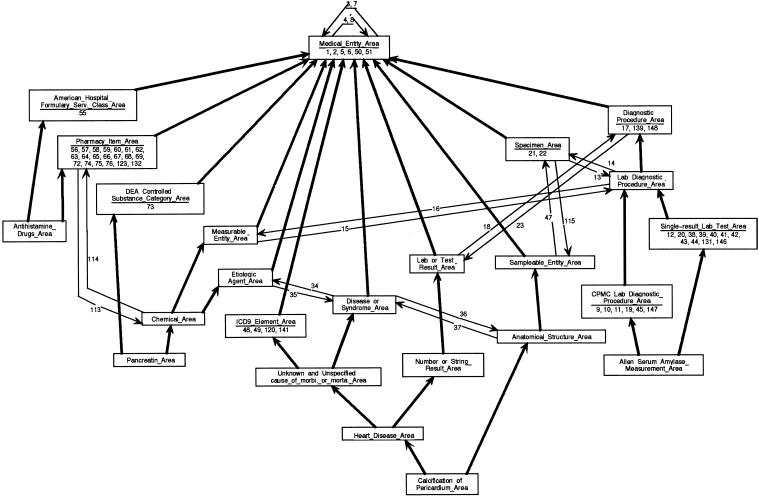
In our approach, an existing CMT is partitioned by using the object-oriented database representation. This partitioning supplies an abstract view of the CMT, which helps the user understand the CMT. As we mentioned in the previous section, the OOHTR schema is very compact compared with the overall size of the MED—90 area classes for 43,000 concepts. In ▶, we present another portion of the schema, which comprises 24 area classes, corresponding to the 68 concepts of the MED shown in ▶. These 24 classes amount to 26 percent of the whole OOHTR schema and represent not only the 68 concepts shown in ▶ but also an additional 27,900 concepts, or 65 percent of the entire MED. Compared with the complicated network shown in ▶ and, even more, compared with a semantic network of about 28,000 concepts, the schema in ▶ is much simpler and easier to understand. Even so, it still completely and correctly captures the structure of a significant portion of the MED.
Figure 10.
Partial schema for the object-oriented health care terminology repository, showing the area classes that account for ▶ (see key at ▶).
In the following sections, we discuss how the OOHTR schema facilitated various improvements of the MED design.25 Specifically, we present examples of support for updating the MED, improving its general design, and correcting errors.
Support for Updating the CMT
The MED comprises more than 43,000 concepts, with 88 different kinds of attributes, 62 different kinds of relationships (divided into 31 pairs of reciprocal relationships), 61,000 IS-A links, and 71,000 nonhierarchic links. Therefore, understanding the “big picture” of the MED is difficult. When new concepts are to be added, or when someone needs to find appropriate concepts in the MED, any lack of understanding becomes immediately apparent. The situation is often worsened because those people who maintain and use the MED may not be the same people who modeled a particular aspect originally.
Some ability to provide users with a manageable, high-level view of a CMT like the MED is needed to support user orientation. The OOHTR's network abstraction schema affords such a view. By reducing the MED hierarchy about 500-fold, one can quickly see what the important areas (as represented by area classes in the schema) are and what attributes and relationships they exhibit. Someone looking to add a new concept to the MED can easily traverse these areas. During that traversal, the user can review the areas' properties to determine the appropriate area for the new concept. For example, a user faced with the task of adding a new laboratory panel to the MED can traverse the 90 class schema to find which area should contain the concept to be added; then the person can switch to traversing the concepts inside the area. Such traversal is easier and faster than a traversal of the whole terminology hierarchy of 43,000 concepts. This is analogous to commuting on the highways until reaching the vicinity of the destination and then taking an exit and continuing the ride on the local roads to the destination.
In our example (see ▶), we start in the Medical_Entity_Area and move to the Diagnostic_Procedure_Area and then to the Lab_Diagnostic_Procedure_Area. This area has two children, Single_Result_Lab_Test_Area and CPMC_Lab_Diagnostic_Procedure_Area. Scanning the attributes of these two candidate areas reveals that the latter has an attribute lab_procedure_code (encoded in ▶ as “9”). Since the concept to be entered is known by the user to have such an attribute, this area is clearly the appropriate one to choose. It has a child, Antibiotic_Sensitivity_Panel_Area, which is obviously not relevant to the new concept. We therefore switch to traversing the concepts within the CPMC_Lab_Diagnostic_Procedure_Area to find the proper position in the hierarchy where this new concept should be added. In this example, we need only traverse five areas to find the appropriate position for the new concept. Compared with a traversal of the CMT's hierarchy of concepts, the schema traversal is more efficient for such an update. Thus, the OOHTR schema can be seen to provide a valuable gestalt of the MED complexity, an understanding of which is needed to support updates.
Improving the CMT Organizational Structure
The MED content has grown steadily, averaging 500 additional concepts per month over the past ten years. Much of this growth has been the result of work by a variety of individuals, at times using automated mechanisms for adding concepts. When several people share the task of maintaining a content domain, and each has a slightly different organizational philosophy (e.g., “lumpers” versus “splitters”), it is easy for concepts to be characterized differently depending on who added them. The network abstraction schema provides a way for different people to share the same high-level view of the MED and to identify differences in their personal views. It also makes the MED's overall organization simpler to follow for all parties.
For example, the laboratory system at Columbia-Presbyterian Medical Center has concepts for individual laboratory tests (like Serum Glucose Test) and other concepts for orderable collections of tests (such as CHEM-7, a panel of seven individual tests). These concepts are all represented in the MED with attributes appropriate to each (e.g., tests have units of measurement and normal ranges, while panels have codes used for billing). The concepts are linked to each other via relationships, e.g., Tests are part-of Panels, and Tests measure Measurable Substances. Users of the MED are often confused about the differences between tests and panels (the latter are also called “procedures” by some and “batteries” by others.23, 24 This confusion is exacerbated by the fact that individual tests can be ordered separately and can therefore take on the characteristics of both tests and panels.
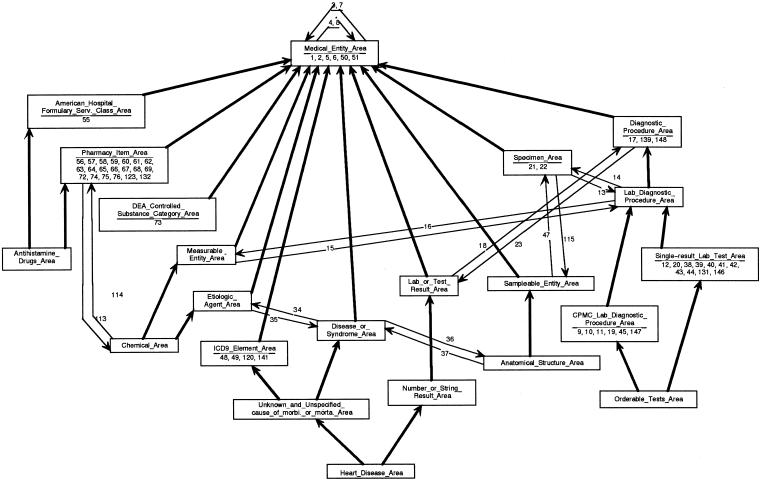
In the schema, the tests belong to the class Single_Result_Lab_Test_Area and the panels are contained in the class CPMC_Lab_Diagnostic_Procedure_Area. The schema grouped the tests that have the properties of panels into the intersection class that is a subclass of Single_Result_Lab_Test_Area and CPMC_Lab_Diagnostic_Procedure_Area (see ▶). In the case where an intersection class has a unique root, that concept's name is chosen to name the area class. Otherwise, one of the roots is arbitrarily chosen as the name. In our example, the intersection area is indeed multirooted and is named the Allen_Serum_Amylase_Measurement_Area. When the intersection area classes were displayed, it was realized that an implicit, natural grouping of tests with panel properties exists. The MED could be simplified by making this group explicit. However, no single concept in the MED was the parent of these particular tests. Thus, a new concept was created called Orderable Tests, as a child of both Single-Result Lab Test and CPMC Lab Diagnostic Procedure. All the tests in Allen_Serum_Amylase_Measurement_Area (such as Allen Serum Amylase Measurement Test) were then linked to Orderable Tests as its children. When the schema was redrawn (▶), the Allen_Serum_Amylase_Measurement_Area took on the new name Orderable_Tests_Area, since that concept was now the single root of the area. Having such an intersection class in the schema as a a subclass of its parent classes, Single-Result_Lab_Test_Area and CPMC_Lab_Diagnostic_Procedure_Area, helps clear up for the user the confusion about tests, panels, and orderable tests. Interestingly, soon after the Orderable Tests concept was added to the MED, New York State required that Columbia—Presbyterian Medical Center make explicit to its physicians and computer systems how the previously “bundled” tests could be ordered and reported individually. Thanks to the Orderable_Tests_Area class, the transition was relatively painless and completely transparent to CPMC's information systems.
Figure 11.
From the above, we derived a general rule for dealing with multirooted intersection areas in the MED and the OOHTR schema. Instead of picking an arbitrary concept to name the area class, we create in the MED a new general parent concept to summarize all the concepts in the area. This new area root is then used in the OOHTR to name the area class.
Finding Inconsistencies and Errors in the CMT
Given the ambiguities that often occur in medical terminology, it is likely that the MED contains a concept with a name that has multiple meanings—contradicting the nonambiguity condition required for the MED, which was described earlier. Since the inception of the MED model,14 it was thought that such ambiguity could be detected through automated means. The intersection areas have provided the basis for such a method.
As an example, it can be seen in ▶ that Calcification_of_the_Pericardium_Area contains all concepts that are both heart diseases and anatomic structures (40 in all). Until the MED was viewed from this perspective, no one realized that the same concepts were listed as both diseases and anatomic structures! This is an example of ambiguity, as the concept Calcification of the Pericardium (and its descendants) has one meaning as a body part and another meaning as a heart disease. This is not consistent with the original design of the MED, in which a disease can be linked to body parts as the site of the disease but can not itself be a body part. Thus, one or the other of the parent-child links had to be removed from the MED. On closer inspection of the Calcification_of_the_Pericardium_Area, we found that there are many such “Calcification of the X” concepts in the MED, all of which are included as descendants of Calcification of Body Part. This concept is a child of Body Part, and both are in the Anatomical_Structure_Area.
Calcification of Body Part has 40 children (and three grandchildren) that are also classified as diseases, while other children are not. The discovery of this intersection class led directly to a study of these 40 concepts and their reclassification as either body parts or diseases, as deemed appropriate by external domain experts. So, for example, the link between Calcification of the Pericardium and Heart Disease was removed. This caused Calcification of the Pericardium to be in a single area, namely, Anatomical_Structure_Area. Therefore, it no longer defined an intersection area of its own. When the schema was re-created (▶), there was no longer any intersection area class that was a subclass of both Heart_Disease_Area and Anatomical_Structure_Area.
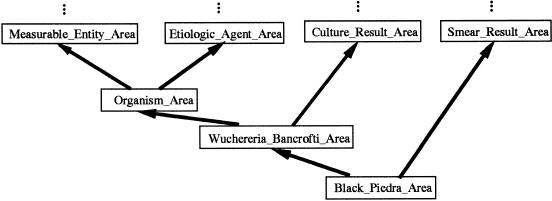
Let us look at another example of ambiguity. In the schema shown in ▶, Black_Piedra_Area is an intersection area class that has two superclasses, Smear_Result_Area and Wuchereria_Bancrofti_Area. Wuchereria_Bancrofti_Area itself is an intersection area class, which contains diseases caused by organisms. That means all concepts in Black_Piedra_Area are classified as smear results and as diseases caused by organisms. After viewing the schema, the designer decided to disambiguate this situation by letting the concept Black Piedra refer only to the organism and not the disease caused by the organism (that disease now being called Black Piedra Infection). So, as an organism Black Piedra is classified under the concept Microorganism.
Figure 12.
Partial schema for the object-oriented health care terminology repository, detecting the ambiguity of “Black Piedra” (see key at ▶).
There is a class of things that are seen under a microscope on microbiologic laboratory tests; they are called “smear results” because the specimens are “smeared” on the slides, stained, and examined. Not all smear results are organisms, and not all microorganisms (e.g., viruses) are seen on smears. Thus, the concept Organisms Seen on Smear was created. The concept Black Piedra became a child of Organisms Seen on Smear in the MED and sits in the intersection area class Organisms_Seen_on_Smear_Area, which is the subclass of Microorganism_Area and Smear_Result_Area in the schema (▶).
Figure 13.
The process of adding concepts to the MED was done by various experts in different fields, sometimes using automatic mechanisms to integrate concepts from a variety of sources. As a result, inconsistencies and outright errors have, not surprisingly, crept into the MED. An example of an error discovered through the use of the schema was the Pancreatin_Area intersection area class (▶). In the MED, it had been decided that medications (such as those classified by their Drug Enforcement Agency controlled substance category) would have chemicals as their pharmaceutic components, but medications themselves would not be chemicals. The OOHTR schema (▶) clearly shows that Pancreatin_Area violates this rule. On closer inspection, it was found that the concept Pancreatin Preparations was properly classified as a medication and that it was linked appropriately to the concept Pancreatin. However, the concept Pancreatin was classified not only as a chemical (which allowed it to have the pharmaceutic-component-of relationship to Pancreatin Preparations) but also as a medication (as shown in ▶). Once this error was seen, it was corrected easily by removing the IS-A link between Pancreatin and DEA Class 0. Since Pancreatin was the only concept in the MED to have attributes of both chemicals and medications, Pancreatin_Area had only one concept prior to the correction. After the correction, the intersection area class no longer existed, since the concept Pancreatin was now included in Chemical_Area (▶).
Object-oriented Databases versus Description Logics
A number of approaches to medical terminologies are based on the use of description logics. Description logics are close descendants of KL-ONE,39, 40, 41 which is itself a descendant of Quillian's original semantic network.42 Quite a number of KL-ONE descendants exist, which makes this probably the largest and most successful family of implemented knowledge representation systems. Two excellent overviews are provided by Sowa43 and Lehman,44 while some of the family members are described in a number of papers.40, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55
Several other semantic networks that do not belong to the family of description logics exist, such as those described by Shapiro and Rapaport56 and by Winston.57
Our own choice of using object-oriented databases instead of description logics is based on two kinds of reasons, which may be summarized under the labels of abstraction ability and commercial viability. Concerning abstraction ability, databases naturally come with two layers of representation, the schema layer and the data layer. As explained earlier, the schema is, by orders of magnitude, smaller than the data. This makes the schema a valuable orientation aid to users, maintainers, and newly hired or newly trained developers of a medical terminology.
In traditional database applications, the object-oriented database schema is developed first, and then the database is populated by instances of the schema classes. It is the strength of our approach that we are deriving a schema after the fact from terminology data that already exists. Thus, we are supplying a road map for information that was not designed with a schema in mind and that is, therefore, naturally harder to understand. We note that the abstraction supplied by a schema is different in nature from the abstraction supplied by the top-level classes of a description logic network, because the schema registers “significant structural changes” in the terminology, no matter at what level they occur. This is not the case for description logic networks, because looking at the top levels gives just that, the top levels. Furthermore, intersections of top-level concepts at lower levels are not reflected at the top levels, whereas they are reflected in our schema approach.
The commercial viability of object-oriented databases seems to be better than that of description logics. A number of vendors deliver “full service object-oriented database systems.” Those systems incorporate basic database features such as persistence and multiuser access. Some of them go far beyond these features, including, for example, versioning and schema evolution. Documentation and help lines are standard for most of these systems. We mention as the most widely advertised products ORION/ITASCA, GemStone, ONTOS, ObjectStore, VERSANT, Jasmine, and O2. While description logics have recently become more available as commercial products, the balance still tilts toward object-oriented database systems. We mention tools for the maintenance of description logic-based medical terminologies from Lexical Technologies § and Ontyx. ∥ Some description logics—e.g., K-Rep36 have been extended to include persistence and other features of object-oriented databases. There are also freely available prototypes, both of object-oriented databases (e.g., ODE58) and description logics (e.g., LOOM59¶).
Naturally, the description logic approaches are superior to object-oriented databases in what we might label reasoning-based support. They make better use of inheritance than object-oriented databases, as their notion of inheritance is based on structure and values, while object-oriented database inheritance is purely structural. The classification algorithm of description logics is an outstanding achievement that has not been duplicated in object-oriented databases. However, a reasoning layer can be added on top of an object-oriented database representation.
However, the nature of description logics themselves imposes severe limitations on their abilities in those areas that are considered their strengths. Specifically, as Brachman and Levesque discussed in their fundamental paper on the tradeoff between representation and reasoning,60 “... subsumption of descriptions in FL [first-order logic] is intractable....” The only way to make the appropriate algorithms computable in polynomial time is to severely limit the power of the representation language. Second, description logics thrive in areas where the Aristotelian view of categorization, by necessary and sufficient conditions, is most applicable. When the number of natural kinds (“primitive concepts” in KL-ONE61) that cannot be defined that way increases, then classification algorithms lose some of their usefulness. In an area like medicine, many terms are highly subjective (“pain” comes to mind) and therefore cannot be defined by necessary and sufficient conditions.
Even in the face of these limitations, description logics are valuable and interesting experimental, scientific, and commercial vehicles. Thus, we consider object-oriented databases for medical terminologies as complementary to description logics.
Conclusions
The job of maintaining a CMT can be daunting because of the typical CMT's large size and extensive scope. Among the tasks that need to be performed by maintenance personnel are updating the CMT with new concepts, reorganizing its design to enhance usability, and correcting mistakes that can arise from various sources. To handle these chores, a person must have a solid understanding of the overall structure of the CMT and its content.
Toward that end, we have proposed the use of the object-oriented database paradigm for the representation of CMTs. We have introduced the notion of an object-oriented health care terminology repository (OOHTR)—that is, a CMT represented in the form of an object-oriented database. An OOHTR is derived from an underlying CMT through a partitioning process based on the pattern in which properties are introduced and distributed among the CMT's constituent concepts. In that context, we defined the notions of property-introducing concept and intersection concept. From these emerged the basic unit of the partitioning process called an area, a collection of concepts that have the same set of properties.
The partitioning process yields an object-oriented database schema that captures the CMTs overall structure. The benefit of the schema is that it provides an extra level of abstraction and summarization for the CMT. After applying our methodology to the MED to produce an OOHTR, we demonstrated how the view afforded by the schema facilitated a variety of improvements. In general, the OOHTR schema can serve as an important mechanism for enhancing comprehension of a large CMT by users and maintainers alike.
Acknowledgments
The authors thank Jim Cimino for his important feedback on earlier drafts of this paper, for sharing the MED with them, helping them understand it, and helping them interpret results of the experiments with the MED. They also thank Boris Harmeyer and Venkatesh Jayaraman for their help in drawing some of the figures. Finally, they thank Eric Mays of Ontyx, Inc. for providing access to the current version of the MED.
Appendix
Glossary
Property-introducing concept: A concept that introduces (defines) attributes or relationships that are exhibited by all its children and descendants in the IS-A hierarchy.
Property-introducing area: A set containing one property-introducing concept plus all that concept's descendants that have the same properties.
Intersection concept: A concept that does not introduce any new properties of its own and that has multiple superconcepts. The set of properties of an intersection concept differs from the set of properties of each of its superconcepts.
Intersection area: A set containing one or more intersection concepts having the same set of properties and all their descendants with the same properties.
This work was supported in part by a cooperative agreement between the Advanced Technology Program of the National Institute of Standards and Technology, under HIIT contract 70NANB5H1011, and the Healthcare Open Systems and Trials consortium.
Footnotes
Throughout the paper, terms appear in boldface type. Property names appear in italics and are written in lowercase letters only.
This was the 1996 version; the MED has since grown to more than 59,000 concepts.
See the glossary in the appendix for a collection of the most important technical terms introduced in this paper.
Information available at http://www.lexical.com.
Information available at http://www.ontyx.com.
Information available at http://www.isi.edu/isd/LOOM.
References
- 1.Cimino JJ. Coding systems in health care. Methods Inf Med. 1996. ;35:273-84. [PubMed] [Google Scholar]
- 2.Cimino JJ. Vocabulary and health care information technology: state of the art. J Am Soc Inf Sci. 1995. ;46(10):777-82. [Google Scholar]
- 3.Campbell KE, Oliver DE, Spackman KA, Shortliffe EH. Representing thoughts, words, and things in the Unified Medical Language System. J Am Med Inform Assoc. 1998. ;5(5):421-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell KE, Oliver DE, Shortliffe EH. The Unified Medical Language System: toward a collaborative approach for solving terminologic problems. J Am Med Inform Assoc . 1998. ;5(1):12-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cimino JJ, Clayton PD, Hripcsak G, Johnson SB. Knowledge-based approaches to the maintenance of a large controlled medical terminology. J Am Med Inform Assoc . 1994. ;1(1):35-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Humphreys BL, Lindberg DAB, Schoolman HM, Barnett GO. The Unified Medical Language System: an informatics research collaboration. J Am Med Inform Assoc. 1998. ;5(1):1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindberg DAB, Humphreys BL, McCray AT. The Unified Medical Language System. Methods Inf Med. 1993. ;32:281-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tuttle MS, Nelson SJ. The role of the UMLS in “storing” and “sharing” across systems. Int J Biomed Comput. 1994. ;34:207-37. [DOI] [PubMed] [Google Scholar]
- 9.Humphreys BL, Lindberg DAB. Building the Unified Medical Language System. Proc 13th Annu Symp Comput Appl Med Care. 1989. ;475-80.
- 10.Côté RA (ed). Systematized Nomenclature of Medicine, 2nd ed. Skokie, Ill.: College of American Pathologists, 1979; updated 1982. .
- 11.U.S. National Center for Health Statistics: International Classification of Diseases: 9th revision, with clinical modifications (ICD-9-CM). Washington, D.C.: Health Care Financing Administration, 1989. .
- 12.Rector A. Coordinating taxonomies: key to re-usable concept representations. Artif Intell Med. 1995. :17-28.
- 13.Rector A, Bechhofer S, Goble C, Horrocks I, Nowlan W, Solomon W. The GRAIL concept modelling language for medical terminology. Artif Intell Med. 1997. ;9:139-71. [DOI] [PubMed] [Google Scholar]
- 14.Cimino JJ, Hripcsak G, Johnson SB, Clayton PD. Designing an introspective, multipurpose, controlled medical vocabulary. Proc 13th Annu Symp Comput Appl Med Care. 1989. ;513-7.
- 15.Lehmann F. Semantic networks. In: Lehmann F (ed). Semantic Networks in Artificial Intelligence. Tarrytown, N.Y.: Pergamon Press, 1992. :1-50.
- 16.Sowa JF. Principles of Semantic Networks, Explorations in the Representation of Knowledge. San Mateo, Calif.: Morgan Kaufmann, 1991. .
- 17.Woods WA. What's in a link: foundations for semantic networks. In: Brachman RJ, Levesque HJ (eds). “Readings in Knowledge Representation.” San Mateo, Calif.: Morgan Kaufmann, 1985. :218-41.
- 18.Bertino E, Martino L. Object-oriented Database Systems, Concepts and Architectures. Menlo Park, Calif.: Addison-Wesley, 1993. .
- 19.Kim W, Lochovsky FH (eds). Object-oriented Concepts, Databases, and Applications. New York: ACM Press, 1989. .
- 20.Zdonik SB, Maier D (eds). Readings in Object-oriented Database Systems. San Mateo, Calif.: Morgan Kaufmann Publishers, 1990. .
- 21.Liu L, Halper M, Geller J, Perl Y. Controlled vocabularies in OODBs: modeling issues and implementation. Distributed and Parallel Databases. 1999. ;7(1):37-65. [Google Scholar]
- 22.Liu L, Halper M, Gu H, Geller J, Perl Y. Modeling a vocabulary in an object-oriented database. In: Barker K, Özsu MT (eds). CIKM-96. Proc 5th International Conference on Information and Knowledge Management. 1996. :179-88.
- 23.Hripcsak G, Allen B, Cimino JJ, Lee R. Access to data: comparing AccessMed to Query by review. J Am Med Inform Assoc. 1996. ;3(4):288-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kannry J, Wright L, Shifman M, Silverstein S, Miller PL. Portability issues for a structured clinical vocabulary: mapping from Yale to the Columbia Medical Entities Dictionary. J Am Med Inform Assoc. 1996. ;3:66-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu H, Cimino JJ, Halper M, Geller J, Perl Y. Utilizing OODB schema modeling for vocabulary management. Proc AMIA Annu Fall Symp. 1996. ;274-8. [PMC free article] [PubMed]
- 26.Fischer DH. Consistency rules and triggers for multilingual terminology. Proc TKE93 Terminology and Knowledge Engineering. 1993. :333-42.
- 27.Cimino JJ. Personal communication, 1997. .
- 28.Halper M, Geller J, Perl Y, Neuhold EJ. A graphical schema representation for object-oriented databases. In: Cooper R (ed). Interfaces to Database Systems . London, England: Springer-Verlag, 1993. :282-307.
- 29.Perl Y, Geller J, Gu H. Identifying a forest hierarchy in an OODB specialization hierarchy satisfying disciplined modeling. Proc 1st IFCIS Int Conf Cooperative Inf Syst (CoopIS96). 1996. :182-95.
- 30.Gu H, Perl Y, Geller J, Halper M, Singh M. A methodology for partitioning a vocabulary hierarchy into trees. Artif Intelli Med. 1999. ;15(1):77-98. [DOI] [PubMed] [Google Scholar]
- 31.Robinson D, Comp D, Schulz E, Brown P, Price C. Updating the read codes: user-interactive maintenance of a dynamic clinical vocabulary. J Am Med Inform Assoc. 1997. ;4(6):465-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tuttle MS, Sherertz DD, Erlbaum MS, et al. Adding your terms and relationships to the UMLS metathesaurus. Proc 15th Annu Symp Comput Appl Med Care. 1991. :219-23. [PMC free article] [PubMed]
- 33.Oliver DE, Shahar Y. Development of a change model for a controlled medical vocabulary. Proc AMIA Annu Fall Symp. 1997. :605-9. [PMC free article] [PubMed]
- 34.Rocha RA, Huff SM, Haug PJ, Warner HR. Designing a controlled medical vocabulary server: the Voser project. Comput Biomed Res. 1994. ;27:472-507. [DOI] [PubMed] [Google Scholar]
- 35.Suarez-Munist ON, Tuttle MS, Olson NE, Erlbaum MS, Sheretz DD, Lipow SS, et al. MEME II supports the cooperative management of terminology. Proc AMIA Annu Fall Symp . 1996. :84-8. [PMC free article] [PubMed]
- 36.Mays E, Weida R, Dionne R, et al. Scalable and expressive medical terminologies. Proc AMIA Annu Fall Symp. 1996. :259-63. [PMC free article] [PubMed]
- 37.Campbell KE, Cohn SP, Chute CG, Rennels G, Shortliffe EH. Gálapagos: computer-based support for evolution of a convergent medical terminology. Proc AMIA Annu Fall Symp. 1996:269-73. [PMC free article] [PubMed]
- 38.Campbell KE. Distributed Development of a Logical-based Controlled Medical Terminology. [PhD thesis] Stanford, Calif.: Stanford University, 1997. Thesis no. CS-TR-97-1596.
- 39.Woods WA. What's in a link? Foundations for semantic networks. In: Bobrow DG, Collins AM (eds). Representation and Understanding. New York: Academic Press, 1975. :35-82.
- 40.Brachman RJ. On the epistemological status of semantic networks. In: Findler N (ed). Associative Networks. New York: Academic Press, 1979:3-50.
- 41.Brachman RJ, Schmolze J. An overview of the KL-ONE knowledge representation system. Cognitive Sci. 1985. ;9(2):171-216. [Google Scholar]
- 42.Quillian MR. Semantic memory. In: Minsky ML (ed). Semantic Information Processing. Cambridge, Mass.: The MIT Press, 1968. ;227-70.
- 43.Sowa JF (ed). Principles of Semantic Networks. San Mateo, Calif.: Morgan Kaufmann, 1991.
- 44.Lehman F (ed). Semantic Networks in Artificial Intelligence. Oxford, England: Pergamon Press, 1992. .
- 45.Borgida A, Brachman RJ, McGuinness DL, Resnick LA. CLASSIC: a structural data model for objects. Proc ACM SIGMOD Int Conf Manage Data (SIGMOD). 1989. ;18(2):58-67. [Google Scholar]
- 46.Brachman RJ, Fikes RE, Levesque HJ. KRYPTON: a functional approach to knowledge representation. IEEE Comput. 1983. ;16(10):67-73. [Google Scholar]
- 47.MacGregor RM. A deductive pattern matcher. Proc 7th Nat Conf Artif Intell. 1988. :403-8.
- 48.Nebel B, Luck K. Issues of integration and balancing in hybrid knowledge. In: Morik K (ed). German Workshop on Artificial Intelligence. 1987. :114-23.
- 49.Vilain M. The restricted language architecture of a hybrid representation system. In: Proc 9th Int Joint Conf Artif Intell. 1985:547-51.
- 50.Woods WA. Knowledge representation: what's important about it? In: Cercone N, McCalla G (eds). The Knowledge Frontier. New York: Springer-Verlag, 1987:44-79.
- 51.Baader F, Hollunder B. KRIS: knowledge representation and inference system. SIGART Bull. 1991. ;2(3):8-14. [Google Scholar]
- 52.Bayer S, Vilain M. The relation-based knowledge representation of King Kong. SIGART Bull. 1991. ;2(3):15-21. [Google Scholar]
- 53.Kobsa A. First experience with the SB-ONE knowledge representation workbench in natural-language applications. SIGART Bull. 1991. ;2(3):70-76. [Google Scholar]
- 54.Patel-Schneider P, McGuiness DL, Brachman RJ, Resnick LA, Borgida A. The CLASSIC knowledge representation system: guiding principles and implementation rationale. SIGART Bull. 1991. ;2(3):108-113. [Google Scholar]
- 55.Bergmann FW, Quantz JJ. Parallel propagation in the description-logic system FLEX. In: Geller J, Kitano H, Suttner CB (eds). Parallel Processing for Artificial Intelligence 3 . New York: North-Holland, 1997;181-207.
- 56.Shapiro SC, Rapaport WJ. SNePS considered as a fully intensional propositional semantic network. In: Cercone N, McCalla G (eds). The Knowledge Frontier. New York: Springer-Verlag, 1987. ;262-315.
- 57.Winston PH. Learning structural descriptions from examples. In: Brachman RJ, Levesque HJ (eds). Readings in Knowledge Representation. Los Altos, Calif: Morgan Kaufmann, 1985. :141-68.
- 58.Agrawal R, Gehani NH. ODE (object database and environment): the language and data model. Proc ACM SIGMOD Int Conf Manage Data. 1989. :36-45.
- 59.MacGregor RM. The evolving technology of classification-based knowledge representation systems. In: Lehman F (ed). Semantic Networks in Artificial Intelligence. Oxford, England: Pergamon Press, 1992. :385-400.
- 60.Brachman RJ, Levesque HJ. The tractability of subsumption in frame-based description languages. Proc American Association for Artificial Intelligence. 1984. :34-37.
- 61.Woods WA, Schmolze JG. The kl-one family. In: Lehman F (ed). Semantic Networks in Artificial Intelligence . Oxford, England: Pergamon Press, 1992:133-77.
- 62.Lehmann F (ed). Semantic Networks in Artificial Intelligence. Tarrytown, N.Y.: Pergamon Press, 1992. .