Abstract
Genome-wide association studies (GWAS) have transformed our understanding of susceptibility to multiple myeloma (MM), but much of the heritability remains unexplained. We report a new GWAS, a meta-analysis with previous GWAS and a replication series, totalling 9974 MM cases and 247,556 controls of European ancestry. Collectively, these data provide evidence for six new MM risk loci, bringing the total number to 23. Integration of information from gene expression, epigenetic profiling and in situ Hi-C data for the 23 risk loci implicate disruption of developmental transcriptional regulators as a basis of MM susceptibility, compatible with altered B-cell differentiation as a key mechanism. Dysregulation of autophagy/apoptosis and cell cycle signalling feature as recurrently perturbed pathways. Our findings provide further insight into the biological basis of MM.
Multiple myeloma is a cancer of the plasma cells, and the complete aetiology of the disease is still unclear. Here the authors perform an additional GWAS analysis followed by a meta-analysis with existing GWAS and replication genotyping and identify 6 novel risk loci and utilise gene expression, epigenetic profiling and in situ Hi-C data to further our understanding of MM susceptibility.
Introduction
Multiple myeloma (MM) is a malignancy of plasma cells primarily located within the bone marrow. Although no lifestyle or environmental exposures have been consistently linked to an increased risk of MM, the two- to four-fold increased risk observed in relatives of MM patients provides support for inherited genetic predisposition1. Our understanding of MM susceptibility has recently been informed by genome-wide association studies (GWAS), which have so far identified 17 independent risk loci for MM2–5, with an additional locus being subtype-specific for t(11;14) translocation MM6. Much of the heritable risk of MM, however, remains unexplained and statistical modelling indicates that further common risk variants remain to be discovered7.
To gain a more comprehensive insight into MM aetiology, we performed a new GWAS followed by a meta-analysis with existing GWAS and replication genotyping (totalling 9974 cases and 247,556 controls). Here, we report the identification of six new MM susceptibility loci as well as refined risk estimates for the previously reported loci. In addition, we have investigated the possible gene regulatory mechanisms underlying the associations seen at all 23 GWAS risk loci by analysing in situ promoter Capture Hi-C (CHi-C) in MM cells to characterise chromatin interactions between predisposition single-nucleotide polymorphism (SNPs) and target genes, integrating these data with chromatin immunoprecipitation-sequencing (ChIP-seq) data generated in house and a range of publicly available genomics data. Finally, we have quantified the contribution of both new and previously discovered loci to the heritable risk of MM and implemented a likelihood-based approach to estimate sample sizes required to explain 80% of the heritability.
Results
Association analysis
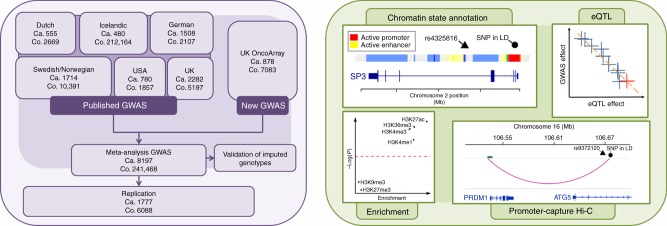
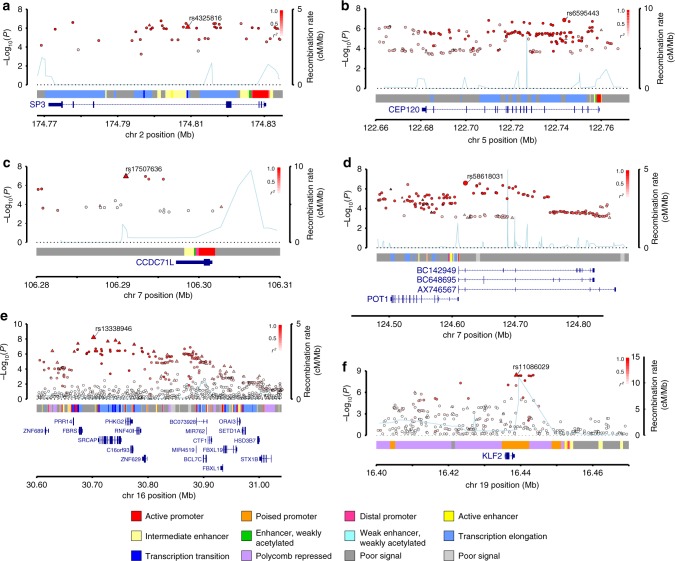
We conducted a new GWAS using the OncoArray platform8 (878 MM cases and 7083 controls from the UK), followed by a meta-analysis with six published MM GWAS data sets (totalling 7319 cases and 234,385 controls) (Fig. 1, Supplementary Tables 1–3)2–5. To increase genomic resolution, we imputed data to >10 million SNPs. Quantile–quantile (Q–Q) plots for SNPs with minor allele frequency (MAF) >1% after imputation did not show evidence of substantive over-dispersion for the OncoArray GWAS (λ = 1.03, λ1000 = 1.02, Supplementary Fig. 1). We derived joint odds ratios (ORs) under a fixed-effects model for each SNP with MAF >1%. Finally, we sought validation of nine SNPs associated at P < 1 × 10−6 in the meta-analysis, which did not map to known MM risk loci and displayed a consistent OR across all GWAS data sets, by genotyping an additional 1777 cases and 6088 controls from three independent series (Germany, Denmark and Sweden). After meta-analysis of the new and pre-existing GWAS data sets and replication series, we identified genome-wide significant associations (i.e. P < 5 × 10−8)9 for six new loci at 2q31.1, 5q23.2, 7q22.3, 7q31.33, 16p11.2 and 19p13.11 (Table 1, Supplementary Table 4 and 5, Fig. 2). Additionally, borderline associations were identified at two loci with P values of 5.93 × 10−8 (6p25.3) and 9.90 × 10−8 (7q21.11), which have corresponding Bayesian false-discovery probabilities (BFDP)10 of 4% and 6%, respectively (Supplementary Table 4 and 5). We found no evidence for significant interactions between any of the 23 risk loci. Finally, we found no evidence to support the existence of the putative risk locus at 2p12.3 (rs1214346), previously proposed by Erickson et al.11 (GWAS meta-analysis P value = 0.32).
Fig. 1.
GWAS study design. Details of the new and existing GWAS samples, including recruitment centres or trials and quality control, are provided in Supplementary Tables 1 and 2. Trials or centres from which replication samples were recruited are detailed in Supplementary Table 3. Ca.: cases, Co.: controls, eQTL: expression quantitative trait loci, SNP: single-nucleotide polymorphism, LD: linkage disequilibrium
Table 1.
Summary of genotyping results for all 23 risk SNPs
| OncoArray | Previous data | Replication | Combined meta | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Locus | Pos. (b37) | Risk Allele | RAF | OR | P trend | OR | P trend | OR | P trend | OR | P meta | I 2 |
| rs7577599 | 2p23.3 | 25613146 | T | 0.81 | 1.22 | 2.63×10−3 | 1.24 | 1.24×10−16 | – | – | 1.23 | 1.29×10−18 | 0 |
| rs4325816 | 2q31.1 | 174808899 | T | 0.77 | 1.16 | 1.23×10 − 2 | 1.11 | 1.30×10 − 5 | 1.16 | 3.00×10 − 3 | 1.12 | 7.37×10 − 9 | 9 |
| rs6599192 | 3p22.1 | 41992408 | G | 0.16 | 1.24 | 1.35×10−3 | 1.26 | 8.75×10−18 | – | – | 1.26 | 4.96×10−20 | 0 |
| rs10936600 | 3q26.2 | 169514585 | A | 0.75 | 1.18 | 5.12×10−3 | 1.20 | 5.94×10−15 | – | – | 1.20 | 1.20×10−16 | 0 |
| rs1423269 | 5q15 | 95255724 | A | 0.75 | 1.09 | 0.125 | 1.17 | 1.57×10−11 | – | – | 1.16 | 8.30×10−12 | 23 |
| rs6595443 | 5q23.2 | 122743325 | T | 0.43 | 1.14 | 9.87×10 − 3 | 1.10 | 4.69×10 − 6 | 1.10 | 0.022 | 1.11 | 1.20×10 − 8 | 0 |
| rs34229995 | 6p22.3 | 15244018 | G | 0.02 | 1.05 | 0.781 | 1.40 | 1.76×10−8 | – | – | 1.36 | 5.60×10−8 | 0 |
| rs3132535 | 6p21.3 | 31116526 | A | 0.29 | 1.26 | 2.67×10−5 | 1.20 | 2.97×10−17 | – | – | 1.21 | 6.00×10−21 | 0 |
| rs9372120 | 6q21 | 106667535 | G | 0.21 | 1.18 | 7.74×10−3 | 1.20 | 8.72×10−14 | – | – | 1.19 | 2.40×10−15 | 0 |
| rs4487645 | 7p15.3 | 21938240 | C | 0.65 | 1.23 | 1.06×10−4 | 1.24 | 5.30×10−25 | – | – | 1.24 | 2.80×10−28 | 0 |
| rs17507636 | 7q22.3 | 106291118 | C | 0.74 | 1.12 | 5.71×10 − 2 | 1.12 | 5.54×10 − 7 | 1.10 | 0.036 | 1.12 | 9.20×10 − 9 | 50 |
| rs58618031 | 7q31.33 | 124583896 | T | 0.72 | 1.17 | 7.61×10 − 3 | 1.11 | 4.70×10 − 6 | 1.10 | 0.061 | 1.12 | 2.73×10 − 8 | 0 |
| rs7781265 | 7q36.1 | 150950940 | A | 0.12 | 1.33 | 3.23×10−4 | 1.20 | 1.82×10−7 | – | – | 1.22 | 4.82×10−10 | 49 |
| rs1948915 | 8q24.21 | 128222421 | C | 0.32 | 1.19 | 1.68×10−3 | 1.14 | 3.14×10−10 | – | – | 1.15 | 2.53×10−12 | 26 |
| rs2811710 | 9p21.3 | 21991923 | C | 0.63 | 1.13 | 1.76×10−2 | 1.14 | 6.50×10−10 | – | – | 1.14 | 3.64×10−11 | 0 |
| rs2790457 | 10p12.1 | 28856819 | G | 0.73 | 1.09 | 0.124 | 1.12 | 8.44×10−7 | – | – | 1.11 | 2.66×10−6 | 0 |
| rs13338946 | 16p11.2 | 30700858 | C | 0.26 | 1.17 | 7.90×10 − 3 | 1.12 | 2.22×10 − 7 | 1.26 | 2.5×10 − 7 | 1.15 | 1.02×10 − 13 | 26 |
| rs7193541 | 16q23.1 | 74664743 | T | 0.58 | 1.14 | 9.01×10−3 | 1.12 | 1.14×10−8 | – | – | 1.12 | 3.68×10−10 | 34 |
| rs34562254 | 17p11.2 | 16842991 | A | 0.10 | 1.32 | 7.63×10−4 | 1.30 | 3.63×10−17 | – | – | 1.30 | 1.18×10−19 | 29 |
| rs11086029 | 19p13.11 | 16438661 | T | 0.24 | 1.26 | 1.02×10 − 4 | 1.12 | 1.69×10 − 6 | 1.15 | 5.00×10 − 3 | 1.14 | 6.79×10 − 11 | 42 |
| rs6066835 | 20q13.13 | 47355009 | C | 0.08 | 1.13 | 0.162 | 1.24 | 1.16×10−9 | – | – | 1.23 | 6.58×10−10 | 38 |
| rs138747 | 22q13.1 | 35700488 | A | 0.66 | – | – | 1.21 | 2.58×10−8 | – | – | 1.21 | 2.58×10−8 | 0 |
| rs139402 | 22q13.1 | 39546145 | C | 0.44 | 1.11 | 4.146×10−2 | 1.23 | 4.98×10−26 | – | – | 1.22 | 3.84×10−26 | 56 |
Fig. 2.
Regional plots of the six new risk loci. Regional plots of loci a 2q31.1, b 5q23.2, c 7q22.3, d 7q31.33, e 16p11.2 and f 19p13.11. Plots show results of the meta-analysis for both genotyped (triangles) and imputed (circles) single-nucleotide polymorphisms (SNPs) and recombination rates. −log10(P) (y axes) of the SNPs are shown according to their chromosomal positions (x axes). The sentinel SNP in each combined analysis is shown as a large circle or triangle and is labelled by its rsID. The colour intensity of each symbol reflects the extent of LD with the top SNP, white (r2 = 0) through to dark red (r2 = 1.0). Genetic recombination rates, estimated using 1000 Genomes Project samples, are shown with a light blue line. Physical positions are based on NCBI build 37 of the human genome. Also shown are the relative positions of genes and transcripts mapping to the region of association. Genes have been redrawn to show their relative positions; therefore maps are not to physical scale. The middle track represents the chromatin-state segmentation track (ChromHMM) for KMS11
Risk SNPs and myeloma phenotype
We did not find any association between sex or age at diagnosis and the 23 MM risk SNPs using case-only analysis (Supplementary Table 6 and 7). Aside from previously reported relationships between the risk loci at 11q13.3 and 5q15 with t(11;14) MM6 and hyperdiploid MM12, respectively, we found no evidence for subtype-specific associations (Supplementary Table 8-11) or an impact on MM-specific survival (Supplementary Table 12). A failure to demonstrate additional relationships may, however, be reflective of limited study power. Collectively, these data suggest that the risk variants are likely to have generic effects on MM development.
Contribution of risk SNPs to heritability
Using linkage disequilibrium adjusted kinships (LDAK)13, the heritability of MM ascribable to all common variation was 15.6% (±4.7); collectively the previously identified and new risk loci account for 15.7% of the GWAS heritability (13.6% and 2.1%, respectively). To assess the collective impact of all identified risk SNPs, we constructed polygenic risk scores (PRS) considering the combined effect of all risk SNPs modelled under a log-normal relative risk distribution14. Using this approach, an individual in the top 1% of genetic risk has a threefold increased risk of MM when compared to an individual with median genetic risk (Supplementary Fig. 2). We observed an enrichment of risk variants among familial MM compared with both sporadic MM cases and population-based controls comparable to that expected in the absence of a strong monogenic predisposition (respective P values 0.027 and 1.60 × 10−5; Supplementary Fig. 3). Undoubtedly, the identification of further risk loci through the analysis of larger GWAS are likely to improve the performance of any PRS model. To estimate the sample size required to explain a greater proportion of the GWAS heritability, we implemented a likelihood-based approach using association statistics in combination with LD information to model the effect-size distribution15,16. The effect-size distributions for susceptibility SNPs were best modelled using the three-component model (mixture of two normal distributions) (Supplementary Fig. 4). Under this model, to identify SNPs explaining 80% of the GWAS heritability is likely to require sample sizes in excess of 50,000 (Supplementary Fig. 5).
Functional annotation and biological inference of risk loci
To the extent that they have been studied, many GWAS risk SNPs localise to non-coding regions and influence gene regulation17. To investigate the functional role of previously reported and new MM risk SNPs, we performed a global analysis of SNP associations using ChIP-seq data generated on the MM cell line KMS11 and publicly accessible naive B-cell Blueprint Epigenome Project data18. We found enrichment of MM SNPs in regions of active chromatin, as indicated by the presence of H3K27ac, H3K4Me3 and H3K4Me1 marks (Supplementary Fig. 6). We also observed an enrichment of relevant B-cell transcription factor-(TF) binding sites using ENCODE GM12878 lymphoblastoid cell line data (Supplementary Fig. 7). Collectively these data support the tenet that the MM predisposition loci influence risk through effects on cis-regulatory networks involved in transcriptional initiation and enhancement.
Since genomic spatial proximity and chromatin looping interactions are key to the regulation of gene expression, we interrogated physical interactions at respective genomic regions in KMS11 and naive B-cells using CHi-C data19. We also sought to gain insight into the possible biological mechanisms for associations by performing an expression quantitative trait locus (eQTL) analysis using mRNA expression data on CD138-purified MM plasma cells; specifically, we used Summary data-based Mendelian Randomisation (SMR) analysis20 to test for pleiotropy between GWAS signal and cis-eQTL for genes within 1 Mb of the sentinel SNP to identify a causal relationship. We additionally annotated risk loci with variants mapping to binding motifs of B-cell-specific TFs. Finally, we catalogued direct promoter variants and non-synonymous coding mutations for genes within risk loci (Supplementary Data 1 and Fig. 1).
Although preliminary, and requiring functional validation, our analysis delineates four potential candidate disease mechanisms across the 23 MM risk loci (Supplementary Data 1). Firstly, four of the risk loci contain candidate genes linked to regulation of cell cycle and genomic instability, as evidenced by Hi-C looping interactions in KMS11 cells to MTAP (at 9p21.3) and eQTL effects for CEP120 (at 5q23.2). CEP120 is required for microtubule assembly and elongation with overexpression of CEP120 leading to uncontrolled centriole elongation21. rs58618031 (7q31.33) maps 5′ of POT1, the protection of telomeres 1 gene. POT1 is part of the shelterin complex that functions to protect telomeres and maintain chromosomal stability22,23. While mutated POT1 is not a feature of MM, it is commonly observed in B-cell chronic lymphocytic leukaemia24–26. The looping interaction from the rs58618031 annotated enhancer element implicates ASB15. Members of the ASB family feature as protein components of the ubiquitin–proteasome system, intriguingly a therapeutic target in MM27–29.
Second, candidate genes encoding proteins involved in chromatin remodelling were implicated at three of the MM risk loci, supported by promoter variants at 2q31.1, 7q36.1 and 22q13.1. The new locus at 2q31.1 implicates SP3, encoding a TF, which through promoter interaction, has a well-established role in B-cell development influencing the expression of germinal centre genes, including activation-induced cytidine deaminase AID30,31.
Third, the central role IRF4-MYC-mediated apoptosis/autophagy in MM oncogenesis is supported by variation at five loci, including eQTL effects WAC (at 10p12.1) and Hi-C looping interactions (at 8q24.21 and 16q23.1). The 7p15.3 association ascribable to rs4487645 has been documented to influence expression of c-MYC-interacting CDCA7L through differential IRF4 binding32. Similarly, the long-range interaction between CCAT1 (colon cancer-associated transcript 1) and MYC provides an attractive biological basis for the 8q24.21 association, given the notable role of MYC in MM33,34. It is noteworthy that the promising risk locus at 6p25.3 contains IRF4. At the new locus 19p13.11, the missense variant (NP_057354.1:p.Leu104Pro) and the correlated promoter SNP rs11086029 implicates KLF2 in MM biology. Demethylation by KDM3A histone demethylase sustains KLF2 expression and influences IRF4-dependent MM cell survival35. The new 16p11.2 risk locus contains a number of genes including Proline-Rich Protein 14 (PRR14), which is implicated in PI3-kinase/Akt/mTOR signalling, a therapeutic target in myelomatous plasma cells36.
Fourth, loci related to B cell and plasma cell differentiation and function are supported by variation at three loci, including eQTL effects (ELL2 at 5q15) and Hi-C looping interactions (at 6q21). As previously inferred from GM12878 cell line data, the region at 6q21 (rs9372120, ATG5) participates in intra-chromosome looping with the B-cell transcriptional repressor PRDM1 (alias BLIMP1)4. Additionally, SNP rs34562254 at 17p11.2 is responsible for the amino acid substitution (NP_036584.1:p.Pro251Leu) in TNFRSF13B, a key regulator of normal B-cell homoeostasis, which has an established role in MM biology37–42.
Discussion
Our meta-analysis of a new GWAS series in conjunction with previously published MM data sets has identified six novel risk loci. Together, the new and previously reported loci explain an estimated 16% of the SNP heritability for MM in European populations. Ancestral differences in the risk of developing MM are well recognised, with a greater prevalence of MM in African Americans as compared with those with European ancestry43. It is plausible that the effects of MM risk SNPs may differ between Europeans and non-Europeans and hence contribute to differences in prevalence rates. Thus far, there has only been limited evaluation of this possibility with no evidence for significant differences44.
Integration of Hi-C data with ChIP-seq chromatin profiling from MM and lymphoblastoid cell lines and naive B cells and eQTL analysis, using patient expression data, has allowed us to gain preliminary insight into the biological basis of MM susceptibility. This analysis suggests a model of MM susceptibility based on transcriptional dysregulation consistent with altered B-cell differentiation, where dysregulation of autophagy/apoptosis and cell cycle signalling feature as recurrently modulated pathways. Specifically, our findings implicate mTOR-related genes ULK4, ATG5 and WAC, and by virtue of the role of IRF4-MYC related autophagy, CDCA7L, DNMT3A, CBX7 and KLF2 in MM development (Supplementary Data 1). Further investigations are necessary to decipher the functional basis of risk SNPs, nevertheless we highlight mTOR signalling and the ubiquitin–proteasome pathway, targets of approved drugs in MM. As a corollary of this, genes elucidated via the functional annotation of GWAS that discovered MM risk loci may represent promising therapeutic targets for myeloma drug discovery. Finally, our estimation of sample sizes required to identify a larger proportion of the heritable risk of MM attributable to common variation underscore the need for further international collaborative analyses.
Methods
Ethics
Collection of patient samples and associated clinico-pathological information was undertaken with written informed consent and relevant ethical review board approval at respective study centres in accordance with the tenets of the Declaration of Helsinki. Specifically for the Myeloma-IX trial by the Medical Research Council (MRC) Leukaemia Data Monitoring and Ethics committee (MREC 02/8/95, ISRCTN68454111), the Myeloma-XI trial by the Oxfordshire Research Ethics Committee (MREC 17/09/09, ISRCTN49407852), HOVON65/GMMG-HD4 (ISRCTN 644552890; METC 13/01/2015), HOVON87/NMSG18 (EudraCTnr 2007-004007-34, METC 20/11/2008), HOVON95/EMN02 (EudraCTnr 2009-017903-28, METC 04/11/10), University of Heidelberg Ethical Commission (229/2003, S-337/2009, AFmu-119/2010), University of Arkansas for Medical Sciences Institutional Review Board (IRB 202077), Lund University Ethical Review Board (2013/54), the Norwegian REK 2014/97, the Danish Ethical Review Board (no. H-16032570) and Icelandic Data Protection Authority (2,001,010,157 and National Bioethics Committee 01/015).
The diagnosis of MM (ICD-10 C90.0) in all cases was established in accordance with World Health Organization guidelines. All samples from patients for genotyping were obtained before treatment or at presentation.
Primary GWAS
We analysed constitutional DNA (EDTA-venous blood derived) from 931 cases ascertained through the UK Myeloma XI trial; detailed in Supplementary Table 1. Cases were genotyped using the Illumina OncoArray (Illumina Inc. San Diego, CA 92122, USA). Controls were also genotyped using the OncoArray and comprised: (1) 2976 cancer-free men recruited by the PRACTICAL Consortium—the UK Genetic Prostate Cancer Study (UKGPCS) (age <65 years), a study conducted through the Royal Marsden NHS Foundation Trust and SEARCH (Study of Epidemiology & Risk Factors in Cancer), recruited via GP practices in East Anglia (2003–2009) and (2) 4446 cancer-free women across the UK, recruited via the Breast Cancer Association Consortium (BCAC).
Standard quality-control measures were applied to the GWAS45. Specifically, individuals with low SNP call rate (<95%) as well as individuals evaluated to be of non-European ancestry (using the HapMap version 2 CEU, JPT/CHB and YRI populations as a reference) were excluded (Supplementary Fig. 8). For apparent first-degree relative pairs, we excluded the control from a case–control pair; otherwise, we excluded the individual with the lower call rate. SNPs with a call rate <95% were excluded as were those with a MAF <0.01 or displaying significant deviation from Hardy–Weinberg equilibrium (P < 10−5). GWAS data were imputed to >10 million SNPs using IMPUTE2 v2.346 software in conjunction with a merged reference panel consisting of data from 1000 Genomes Project47 (phase 1 integrated release 3 March 2012) and UK10K48. Genotypes were aligned to the positive strand in both imputation and genotyping. We imposed predefined thresholds for imputation quality to retain potential risk variants with MAF >0.01 for validation. Poorly imputed SNPs with an information measure <0.80 were excluded. Tests of association between imputed SNPs and MM was performed under an additive model in SNPTESTv2.549. The adequacy of the case–control matching and possibility of differential genotyping of cases and controls was evaluated using a Q–Q plot of test statistics (Supplementary Fig. 1). The inflation λ was based on the 90% least-significant SNPs50 and assessment of λ1000. Details of SNP QC are provided in in Supplementary Table 2.
Published GWAS
The data from six previously reported GWAS2–5 are summarised in Supplementary Table 1. All these studies were based on individuals with European ancestry and comprised: UK-GWAS (2282 cases, 5197 controls), Swedish-GWAS (1714 cases, 10,391 controls), German-GWAS (1508 cases, 2107 controls), Netherlands-GWAS (555 cases, 2669 controls), US-GWAS (780 cases, 1857 controls) and Iceland (480 cases, 212,164 controls).
Replication studies and technical validation
To validate promising associations, we analysed three case–control series from Germany, Sweden and Denmark, summarised in Supplementary Table 3. The German replication series comprised 911 cases collected by the German Myeloma Study Group (Deutsche Studiengruppe Multiples Myeloma (DSMM)), GMMG, University Clinic, Heidelberg and University Clinic, Ulm. Controls comprised 1477 healthy German blood donors recruited between 2004 and 2007 by the Institute of Transfusion Medicine and Immunology, University of Mannheim, Germany. The Swedish replication series comprised 534 MM cases from the Swedish National Myeloma Biobank and the Danish replication series comprised 332 MM cases from the University Hospital of Copenhagen. As controls, we analysed 2382 Swedish blood donors and 2229 individuals from Denmark and Skåne County, Sweden (the southernmost part of Sweden adjacent to Denmark). Replication genotyping of German and Scandinavian samples was performed using competitive allele-specific PCR KASPar chemistry (LGC, Hertfordshire, UK). Call rates for SNP genotypes were >95% in each of the replication series. To ensure the quality of genotyping in all assays, at least two negative controls and duplicate samples (showing a concordance of >99%) were genotyped at each centre. The fidelity of imputation was assessed by directly sequencing a set of 147 randomly selected samples from the UK OncoArray case series. Imputation was found to be robust; concordance was >90% (Supplementary Table 13). Genotyping and sequencing primers are detailed in Supplementary Table 14 and 15, respectively.
Meta-analysis
Meta-analyses were performed using the fixed-effects inverse-variance method using META v1.651. Cochran’s Q-statistic to test for heterogeneity and the I2 statistic to quantify the proportion of the total variation due to heterogeneity was calculated. Using the meta-analysis summary statistics and LD correlations from a reference panel of the 1000 Genomes Project combined with UK10K, we implemented Genome-wide Complex Trait Analysis52 to perform conditional association analysis. Association statistics were calculated for all SNPs conditioning on the top SNP in each loci showing genome-wide significance. This was carried out step-wise.
For borderline associations, the BFDP10 was calculated based on a plausible OR of 1.2 and a prior probability of association of 0.0001. For both promising associations, the BFDP was <10%.
Fluorescence in situ hybridisation
Fluorescence in situ hybridisation (FISH) and ploidy classification of UK and German samples were performed as previously described53,54. Logistic regression in case-only analyses was used to assess the relationship between SNP genotype and IgH translocations or tumour ploidy.
eQTL analysis
eQTL analyses were performed using CD138-purified plasma cells from 183 UK MyIX trial patients and 658 German GMMG patients32. Briefly, German and UK data were pre-processed separately, followed by analysis using a Bayesian approach to probabilistic estimation of expression residuals to infer broad variance components, accounting for hidden determinants influencing global expression. The association between genotype of SNPs and expression of genes within 1 Mb either side of each MM risk locus was evaluated based on the significance of linear regression coefficients. We pooled data from each study under a fixed-effects model.
The relationship between SNP genotype and gene expression was carried out using SMR analysis as per Zhu et al.2 Briefly, if bxy is the effect size of x (gene expression) on y (slope of y regressed on the genetic value of x), bzx is the effect of z on x and bzy be the effect of z on y, bxy (bzy/bzx) is the effect of x on y. To distinguish pleiotropy from linkage where the top associated cis-eQTL is in LD with two causal variants, one affecting gene expression and the other affecting a trait, we tested for heterogeneity in dependent instruments (HEIDI), using multiple SNPs in each cis-eQTL region. Under the hypothesis of pleiotropy, bxy values for SNPs in LD with the causal variant should be identical. For each probe that passed significance threshold for the SMR test, we tested the heterogeneity in the bxy values estimated for multiple SNPs in the cis-eQTL region using HEIDI.
GWAS summary statistics files were generated from the meta-analysis. We set a threshold for the SMR test of PSMR < 1 × 10−3 corresponding to a Bonferroni correction for 45 tests, i.e. 45 probes which demonstrated an association in the SMR test. For all genes passing this threshold, we generated plots of the eQTL and GWAS associations at the locus, as well as plots of GWAS and eQTL effect sizes (i.e. input for the HEIDI heterogeneity test). HEIDI test P values < 0.05 were considered as reflective of heterogeneity. This threshold is, however, conservative for gene discovery because it retains fewer genes than when correcting for multiple testing. SMR plots for significant eQTLs are shown in Supplementary Fig. 9 and 10, and a summary of results are shown in Supplementary Table 16.
Promoter CHi-C
To map risk SNPs to interactions involving promoter contacts and identify genes involved in MM susceptibility, we analysed publicly accessible promoter CHi-C data on the naive B cells downloaded from Blueprint Epigenome Project. Additionally, we also analysed promoter CHi-C data that we have previously generated for the MM cell line KMS1112. Interactions were called using the CHiCAGO pipeline to obtain a unique list of reproducible contacts55 and those with a −log(weighted P) ≥5 were considered significant.
Chromatin state annotation
Variant sets (i.e. sentinel risk SNP and correlated SNPs, r2>0.8) were annotated for putative functional effect based upon histone mark ChIP-seq data for H3K27ac, H3K4Me1, H3K27Me3, H3K9Me3, H3K36Me3 and H3K27Me3 from KMS11 cell lines, generated in-house and naive B cells from Blueprint Epigenome Project56. We used ChromHMM to infer chromatin states by integrating information on these histone modifications, training the model on three MM cell lines; KMS11, MM1S and JJN3. Genome-wide signal tracks were binarized (including input controls for ChIP-seq data), and a set of learned models were generated using ChromHMM software57. A 12-state model was suitable for interpretation, and biological meaning was assigned to the states based on chromatin marks that use putative rules as previously described (Supplementary Fig. 11).
TF and histone mark enrichment analysis
To examine enrichment in specific TF binding across risk loci, we adapted the method of Cowper-Sal lari et al.58. Briefly, for each risk locus, a region of strong LD (defined as r2>0.8 and D′>0.8) was determined, and these SNPs were considered the associated variant set (AVS). Publically available data on TF ChIP-seq uniform peak data were obtained from ENCODE for the GM12878 cell line, including data for 82 TF and 11 histone marks59. In addition, ChIP-seq peak data for six histone marks from KMS11 cell line were generated in-house, and naive B-cell ChIP-seq data were downloaded from Blueprint Epigenome Project56. For each mark, the overlap of the SNPs in the AVS and the binding sites was assessed to generate a mapping tally. A null distribution was produced by randomly selecting SNPs with the same characteristics as the risk-associated SNPs, and the null mapping tally calculated. This process was repeated 10,000 times, and P values were calculated as the proportion of permutations, where null mapping tally was greater or equal to the AVS mapping tally. An enrichment score was calculated by normalising the tallies to the median of the null distribution. Thus, the enrichment score is the number of standard deviations of the AVS mapping tally from the median of the null distribution tallies. Enrichment plots are shown in Supplementary Fig. 6 and 7.
Functional annotation
For the integrated functional annotation of risk loci, variant sets (i.e. all SNPs in LD r2 > 0.8 with the sentinel SNP) were annotated with: (i) presence of a Hi-C contact linking to a gene promoter, (ii) presence of an association from SMR analysis, (iii) presence of a regulatory ChromHMM state, (iv) evidence of transcription factor binding and (v) presence of a nonsynonymous coding change. Candidate causal genes were then assigned to MM risk loci using the target genes implicated in annotation tracks (i), (ii), (iiii) and (iv). If the data supported multiple gene candidates, the gene with the highest number of individual functional data points was considered as the candidate. Where multiple genes have the same number of data points, all genes are listed. Direct non-synonymous coding variants were allocated additional weighting. Competing mechanisms for the same gene (e.g. both coding and promoter variants) were permitted.
Heritability analysis
We used LDAK to estimate the polygenic variance (i.e. heritability) ascribable to all genotyped and imputed GWAS SNPs from summary statistic data. SNP-specific expected heritability, adjusted for LD, MAF and genotype certainty was calculated from the UK10K and 1000 Genomes data. Samples were excluded with a call rate <0.99 or if individuals were closely related or of divergent ancestry from CEU. Individual SNPs were excluded if they showed deviation from HWE with P < 1 × 10−5, an individual SNP genotype yield <95%, MAF <1%, SNP imputation score <0.99 and the absence of the SNP in the GWAS summary statistic data. This resulted in a total 1,254,459 SNPs which were used to estimate the heritability of MM.
To estimate the sample size required for a given proportion of the GWAS heritability, we implemented a likelihood-based approach to model the effect-size distribution15 using association statistics, from the MM meta-analysis, and LD information, obtained from individuals of European ancestry in the 1000 Genomes Project Phase 3. LD values were based on an r2 threshold of 0.1 and a window size of 1 MB. The goodness of fit of the observed distribution of P values against the expected from a two-component model (single normal distribution) and a three-component model (mixture of two normal distributions) were assessed15, and a better fit was observed for the latter model (Supplementary Figure 4). The percentage of GWAS heritability explained for a projected sample size was determined using this model and is based on power calculations for the discovery of genome-wide significant SNPs. The genetic variance explained was calculated as the proportion of total GWAS heritability explained by SNPs reaching genome-wide significance at a given sample size. The 95% confidence intervals were determined using 10,000 simulations.
PRS for familial MM (n = 38) from 25 families were compared with sporadic MM (n = 1530) and population-based controls (n = 10,171); first as a simple sum of risk alleles and secondly as sum of risk alleles weighted by their log-transformed ORs. Family member scores were averaged. A one-sided Student’s t-test was used to assess difference between groups. The genetic data have been previously described5,60 with the familial MM cases having been identified by linkages of Swedish registry information.
Data availability
SNP genotyping data that support the findings of this study have been deposited in Gene Expression Omnibus with accession codes GSE21349, GSE19784, GSE24080, GSE2658 and GSE15695; in the European Genome-phenome Archive (EGA) with accession code EGAS00000000001; in the European Bioinformatics Institute (Part of the European Molecular Biology Laboratory) (EMBL-EBI) with accession code E-MTAB-362 and E-TABM-1138; and in the database of Genotypes and Phenotypes (dbGaP) with accession code phs000207.v1.p1. Expression data that support the findings of this study have been deposited in GEO with accession codes GSE21349, GSE2658, GSE31161 and EMBL-EBI with accession code E-MTAB-2299. The remaining data are contained within the paper and Supplementary Files or are available from the author upon request. KMS11 Hi-C data used in this manuscript are deposited in EGA under accession number EGAS00001002614. The accession number for the KMS11 ChIP-seq data reported in this paper is EGA: S00001002414. Naive B-cell HiC data used in this work is publically available from Blueprint Blueprint Epigenome Project [https://osf.io/u8tzp/]. ChIP-seq data for H3K27ac, H3K4Me1, H3K27Me3, H3K9Me3, H3K36Me3 and H3K27Me3 from naive B cells are publically available and were obtained from Blueprint Epigenome Project [http://www.blueprint-epigenome.eu/].
Electronic supplementary material
Description of Additional Supplementary Files
Acknowledgements
In the United Kingdom, Myeloma UK and Bloodwise provided principal funding. Additional funding was provided by Cancer Research UK (C1298/A8362 supported by the Bobby Moore Fund) and The Rosetrees Trust. M.W. is supported by funding from Mr Ralph Stockwell. A.S. is supported by a clinical fellowship from Cancer Research UK and charitable funds from the Royal Marsden Hospital. N.W. was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number P20GM125503. This study made use of genotyping data on the 1958 Birth Cohort generated by the Wellcome Trust Sanger Institute (http://www.wtccc.org.uk). We thank the High-Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics (funded by Wellcome Trust grant reference 090532/Z/09/Z) for the generation of UK myeloma OncoArray data. The BCAC study would not have been possible without the contributions of the following: Manjeet K. Bolla, Qin Wang, Kyriaki Michailidou and Joe Dennis. BCAC is funded by Cancer Research UK (C1287/A10118, C1287/A16563). For the BBCS study, we thank Eileen Williams, Elaine Ryder-Mills and Kara Sargus. The BBCS is funded by Cancer Research UK and Breast Cancer Now and acknowledges NHS funding to the National Institute of Health Research (NIHR) Biomedical Research Centre (BRC), and the National Cancer Research Network (NCRN). We thank the participants and the investigators of EPIC (European Prospective Investigation into Cancer and Nutrition). The coordination of EPIC is financially supported by the European Commission (DG-SANCO) and the International Agency for Research on Cancer. The national cohorts are supported by: Ligue Contre le Cancer, Institut Gustave Roussy, Mutuelle Générale de l’Education Nationale, Institut National de la Santé et de la Recherche Médicale (INSERM) (France); German Cancer Aid, German Cancer Research Center (DKFZ), Federal Ministry of Education and Research (BMBF) (Germany); the Hellenic Health Foundation, the Stavros Niarchos Foundation (Greece); Associazione Italiana per la Ricerca sul Cancro-AIRC-Italy and National Research Council (Italy); Dutch Ministry of Public Health, Welfare and Sports (VWS), Netherlands Cancer Registry (NKR), LK Research Funds, Dutch Prevention Funds, Dutch ZON (Zorg Onderzoek Nederland), World Cancer Research Fund (WCRF), Statistics Netherlands (The Netherlands); Health Research Fund (FIS), PI13/00061 to Granada, PI13/01162 to EPIC-Murcia, Regional Governments of Andalucía, Asturias, Basque Country, Murcia and Navarra, ISCIII RETIC (RD06/0020) (Spain); Cancer Research UK (14136 to EPIC-Norfolk; C570/A16491 and C8221/A19170 to EPIC-Oxford), Medical Research Council (1000143 to EPIC-Norfolk, MR/M012190/1 to EPIC-Oxford) (UK). We thank the SEARCH and EPIC teams which were funded by a programme grant from Cancer Research UK [A16561] and supported by the UK NIHR BRC at the University of Cambridge. We thank Breast Cancer Now and The Institute of Cancer Research (ICR) for support and funding of the UKBGS, and the study participants, study staff, and the doctors, nurses and other health care providers and health information sources who have contributed to the study. We acknowledge NHS funding to the Royal Marsden/ICR NIHR BRC. UKGPCS would like to thank The Institute of Cancer Research and The Everyman Campaign for funding support. The UKGPCS acknowledges The Prostate Cancer Research Foundation, Prostate Action, The Orchid Cancer Appeal, The National Cancer Research Network UK, The National Cancer Research Institute (NCRI), the NIHR funding to the NIHR Biomedical Research data managers and consultants for their work in the UKGPCS study and urologists and other persons involved in the planning, and data collection of the CAPS study. Genotyping of the OncoArray was funded by the US National Institutes of Health (NIH) [U19 CA 148537 for ELucidating Loci Involved in Prostate cancer SuscEptibility (ELLIPSE) project and X01HG007492 to the Center for Inherited Disease Research (CIDR) under contract number HHSN268201200008I]. Additional analytic support was provided by NIH NCI U01 CA188392 (PI: Schumacher). The PRACTICAL consortium was supported by Cancer Research UK Grants C5047/A7357, C1287/A10118, C1287/A16563, C5047/A3354, C5047/A10692, C16913/A6135, European Commission’s Seventh Framework Programme grant agreement no. 223175 (HEALTH-F2-2009-223175) and The National Institute of Health (NIH) Cancer Post-Cancer GWAS initiative grant no. 1 U19 CA 148537-01 (the GAME-ON initiative). We would also like to thank the following for funding support: The Institute of Cancer Research and The Everyman Campaign, The Prostate Cancer Research Foundation, Prostate Research Campaign UK (now Prostate Action), The Orchid Cancer Appeal, The National Cancer Research Network UK, The National Cancer Research Institute (NCRI) UK. We are grateful for support of NIHR funding to the NIHR Biomedical Research Centre at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust. The APBC BioResource, which forms part of the PRACTICAL consortium, consists of the following members: Wayne Tilley, Gail Risbridger, Renea Taylor, Judith A Clements, Lisa Horvath, Vanessa Hayes, Lisa Butler, Trina Yeadon, Allison Eckert, Pamela Saunders, Anne-Maree Haynes, Melissa Papargiris. We thank the staff of the CTRU University of Leeds and the NCRI haematology Clinical Studies Group. The US GWAS was supported by a grant from the National Institutes of Health (P01CA055819). The German study was supported by the Dietmar-Hopp-Stiftung, Germany, the German Cancer Aid (110,131), the German Ministry of Education and Science (CLIOMMICS 01ZX1309), The German Research Council (DFG; Project SI 236/81, SI 236/)-1, ER 155/6-1 and the DFG CRI 216), the Harald Huppert Foundation and the Multiple Myeloma Research Foundation. The patients were collected by the GMMG and DSMM studies. The German GWAS made use of genotyping data from the population-based HNR study, which is supported by the Heinz Nixdorf Foundation (Germany). The genotyping of the Illumina HumanOmni-1 Quad BeadChips of the HNR subjects was financed by the German Center for Neurodegenerative Disorders (DZNE), Bonn. We are grateful to all investigators who contributed to the generation of this data set. The German replication controls were collected by Peter Bugert, Institute of Transfusion Medicine and Immunology, Medical Faculty Mannheim, Heidelberg University, German Red Cross Blood Service of Baden-Württemberg-Hessen, Mannheim, Germany. This work was supported by research grants from the Swedish Foundation for Strategic Research (KF10-0009), the Marianne and Marcus Wallenberg Foundation (2010.0112), the Knut and Alice Wallenberg Foundation (2012.0193), the Swedish Research Council (2012–1753), the Royal Swedish Academy of Science, ALF grants to the University and Regional Laboratories (Labmedicin Skåne), the Siv-Inger and Per-Erik Andersson Foundation, the Medical Faculty at Lund University, the Borås foundation for cancer research, and the Swedish Society of Medicine. We thank Jörgen Adolfsson, Tomas Axelsson, Anna Collin, Ildikó Frigyesi, Patrik Magnusson, Bertil Johansson, Jan Westin and Helga Ögmundsdóttir for their assistance. This work was supported by Center for Translational Molecular Medicine (BioCHIP), a clinical research grant from the European Hematology Association, an EMCR Translational Research Grant, a BMBF grant from CLIOMMICS (01ZX1309A) and FP7 grant MSCNET (LSHC-Ct-2006-037602). We thank the staff of the HOVON, as well as patients and physicians at participating sites. In addition, we also thank Jasper Koenders, Michael Vermeulen, André Uitterlinden and Nathalie van der Velde for their assistance. We are indebted to the clinicians who contributed samples to Swedish, Icelandic, Norwegian and Danish biobanks. We are indebted to the patients and other individuals who participated in the project.
Author contributions
M.W. and R.S.H. designed the study. M.W. and R.S.H. drafted the manuscript with contributions from K.H., B.N., A.S. and N.W. M.W. performed principal statistical and bioinformatics analyses. A.S., N.L., P.L., D.C.J., J.S.M. and G.O. performed additional bioinformatics analyses. S.K. performed in situ CHi-C. P.B. coordinated UK laboratory analyses. M.W. and A.H. performed sequencing of UK samples. D.C.J. managed and prepared Myeloma IX and Myeloma XI Case Study DNA samples. M.K., G.J.M., F.E.D., W.A.G. and G.H.J. performed ascertainment and collection of Case Study samples. B.A.W. performed UK expression analyses. F.M.R. performed UK fluorescence in situ hybridisation analyses. The PRACTICAL consortium, D.E., P.P., A.D., J.P., F.C., R.E., Z.K.-J., K.M. and N.P. provided control samples for the UK OncoArray GWAS. H.G., U.B., J.H., J.N. and N.W. coordinated and managed Heidelberg samples. C.L. and H.E. coordinated and managed Ulm/Wurzburg samples. A.F. coordinated German genotyping. C.C. and O.R.B. performed German genotyping. P.H. and M.M.N. performed GWAS of German cases and controls. H.T., B.C. and M.I.d.S.F. carried out statistical analysis. K.H. coordinated the German part of the project. M.M.N. generated genotype data from the Heinz-Nixdorf recall study. K.-H.J. contributed towards the Heinz-Nixdorf control data set. N.N. from Bonn and K.-H.J. provided samples for the German GWAS. M.H. and B.N. coordinated the Swedish/Norwegian part of the project. M.A. and B.-M.H. performed data analysis. E.J., A.-K.W., U.-H.M., H.N., A.V., N.F.A., A.W., I.T. and U.G. performed sample acquisition, sample preparation, clinical data acquisition and additional data analyses of Sweden/Norway samples. In Iceland, G.T. and D.F.G. performed statistical analysis. S.Y.K. provided clinical information. T.R. performed additional analyses. U.T. and K.S. performed project oversight. M.v.D., P.S., A.B. and R.K. coordinated and prepared HOVON65/GMMG-HD4, HOVON87/NMSG18 and HOVON95/EMN02 studies for participating in this study, and coordinated genotyping and pre-processing. At the Myeloma Institute, University of Arkansas for Medical Sciences, N.W. coordinated the US part of the project and performed statistical and eQTL analyses. O.W.S. and N.W. managed Case Study samples. G.J.M. and F.E.D. performed ascertainment and collection of Case Study samples.
Competing interests
G.T., D.F.G., T.R., K.S. and U.T. are employed by deCode Genetics/Amgen Inc. The remaining authors declare no competing interests.
Footnotes
Deceased: Brian E. Henderson
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Molly Went, Amit Sud, Asta Försti, Britt-Marie Halvarsson, Niels Weinhold.
These authors jointly supervised the work: Hartmut Goldschmidt, Kari Stefansson, Gareth J. Morgan, Björn Nilsson, Kari Hemminki, Richard S. Houlston.
Change history
1/10/2019
The original version of this Article contained an error in the spelling of a member of the PRACTICAL Consortium, Manuela Gago-Dominguez, which was incorrectly given as Manuela Gago Dominguez. This has now been corrected in both the PDF and HTML versions of the Article. Furthermore, in the original HTML version of this Article, the order of authors within the author list was incorrect. The PRACTICAL consortium was incorrectly listed after Richard S. Houlston and should have been listed after Nora Pashayan. This error has been corrected in the HTML version of the Article; the PDF version was correct at the time of publication.
Contributor Information
Kari Hemminki, Email: K.Hemminki@dkfz-heidelberg.de.
Björn Nilsson, Email: bjorn.nilsson@med.lu.se.
Richard S. Houlston, Email: richard.houlston@icr.ac.uk
The PRACTICAL consortium:
Brian E. Henderson, Christopher A. Haiman, Sara Benlloch, Fredrick R. Schumacher, Ali Amin Al Olama, Sonja I. Berndt, David V. Conti, Fredrik Wiklund, Stephen Chanock, Victoria L. Stevens, Catherine M. Tangen, Jyotsna Batra, Judith Clements, Henrik Gronberg, Johanna Schleutker, Demetrius Albanes, Stephanie Weinstein, Alicja Wolk, Catharine West, Lorelei Mucci, Géraldine Cancel-Tassin, Stella Koutros, Karina Dalsgaard Sorensen, Eli Marie Grindedal, David E. Neal, Freddie C. Hamdy, Jenny L. Donovan, Ruth C. Travis, Robert J. Hamilton, Sue Ann Ingles, Barry Rosenstein, Yong-Jie Lu, Graham G. Giles, Adam S. Kibel, Ana Vega, Manolis Kogevinas, Kathryn L. Penney, Jong Y. Park, Janet L. Stanford, Cezary Cybulski, Børge G. Nordestgaard, Hermann Brenner, Christiane Maier, Jeri Kim, Esther M. John, Manuel R. Teixeira, Susan L. Neuhausen, Kim De Ruyck, Azad Razack, Lisa F. Newcomb, Davor Lessel, Radka Kaneva, Nawaid Usmani, Frank Claessens, Paul A. Townsend, Manuela Gago-Dominguez, Monique J. Roobol, Florence Menegaux, Kay-Tee Khaw, Lisa Cannon-Albright, Hardev Pandha, and Stephen N. Thibodeau
Electronic supplementary material
Supplementary Information accompanies this paper at 10.1038/s41467-018-04989-w.
References
- 1.Altieri A, Chen B, Bermejo JL, Castro F, Hemminki K. Familial risks and temporal incidence trends of multiple myeloma. Eur. J. Cancer. 2006;42:1661–1670. doi: 10.1016/j.ejca.2005.11.033. [DOI] [PubMed] [Google Scholar]
- 2.Broderick P, et al. Common variation at 3p22.1 and 7p15.3 influences multiple myeloma risk. Nat. Genet. 2011;44:58–61. doi: 10.1038/ng.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chubb D, et al. Common variation at 3q26.2, 6p21.33, 17p11.2 and 22q13.1 influences multiple myeloma risk. Nat. Genet. 2013;45:1221–1225. doi: 10.1038/ng.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchell JS, et al. Genome-wide association study identifies multiple susceptibility loci for multiple myeloma. Nat. Commun. 2016;7:12050. doi: 10.1038/ncomms12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swaminathan B, et al. Variants in ELL2 influencing immunoglobulin levels associate with multiple myeloma. Nat. Commun. 2015;6:7213. doi: 10.1038/ncomms8213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinhold N, et al. The CCND1 c.870G>A polymorphism is a risk factor for t(11;14)(q13;q32) multiple myeloma. Nat. Genet. 2013;45:522–525. doi: 10.1038/ng.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell JS, et al. Implementation of genome-wide complex trait analysis to quantify the heritability in multiple myeloma. Sci. Rep. 2015;5:12473. doi: 10.1038/srep12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amos CI, et al. The OncoArray Consortium: a network for understanding the genetic architecture of common cancers. Cancer Epidemiol. Biomark. Prev. 2017;26:126–135. doi: 10.1158/1055-9965.EPI-16-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 10.Wakefield J. A Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am. J. Hum. Genet. 2007;81:208–227. doi: 10.1086/519024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erickson SW, et al. Genome-wide scan identifies variant in 2q12.3 associated with risk for multiple myeloma. Blood. 2014;124:2001–2003. doi: 10.1182/blood-2014-07-586701. [DOI] [PubMed] [Google Scholar]
- 12.Li N, et al. Genetic predisposition to multiple myeloma at 5q15 is mediated by an ELL2 enhancer polymorphism. Cell Rep. 2017;20:2556–2564. doi: 10.1016/j.celrep.2017.08.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Speed D, et al. Reevaluation of SNP heritability in complex human traits. Nat. Genet. 2017;49:986–992. doi: 10.1038/ng.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frampton MJ, et al. Implications of polygenic risk for personalised colorectal cancer screening. Ann. Oncol. 2016;27:429–434. doi: 10.1093/annonc/mdv540. [DOI] [PubMed] [Google Scholar]
- 15.Chatterjee, N. Estimation of complex effect-size distributions using summary-level statistics from genome-wide association studies across 32 complex traits and implications for the future. Preprint at https://www.biorxiv.org/content/early/2017/08/11/175406 (2017). [DOI] [PubMed]
- 16.Chatterjee N, et al. Projecting the performance of risk prediction based on polygenic analyses of genome-wide association studies. Nat. Genet. 2013;45:400–405. doi: 10.1038/ng.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freedman ML, et al. Principles for the post-GWAS functional characterization of cancer risk loci. Nat. Genet. 2011;43:513–518. doi: 10.1038/ng.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Javierre BM, et al. Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell. 2016;167:1369–1384.e19. doi: 10.1016/j.cell.2016.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Risca VI, Greenleaf WJ. Unraveling the 3D genome: genomics tools for multiscale exploration. Trends Genet. 2015;31:357–372. doi: 10.1016/j.tig.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu Z, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 2016;48:481–487. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 21.Comartin D, et al. CEP120 and SPICE1 cooperate with CPAP in centriole elongation. Curr. Biol. 2013;23:1360–1366. doi: 10.1016/j.cub.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 22.Rice C, et al. Structural and functional analysis of the human POT1-TPP1 telomeric complex. Nat. Commun. 2017;8:14928. doi: 10.1038/ncomms14928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinzaru AM, et al. Telomere replication stress induced by POT1 inactivation accelerates tumorigenesis. Cell Rep. 2016;15:2170–2184. doi: 10.1016/j.celrep.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang S. Cancer chromosomes going to POT1. Nat. Genet. 2013;45:473–475. doi: 10.1038/ng.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Speedy HE, et al. A genome-wide association study identifies multiple susceptibility loci for chronic lymphocytic leukemia. Nat. Genet. 2014;46:56–60. doi: 10.1038/ng.2843. [DOI] [PubMed] [Google Scholar]
- 26.Speedy HE, et al. Germ line mutations in shelterin complex genes are associated with familial chronic lymphocytic leukemia. Blood. 2016;128:2319–2326. doi: 10.1182/blood-2016-01-695692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDaneld TG, Hannon K, Moody DE. Ankyrin repeat and SOCS box protein 15 regulates protein synthesis in skeletal muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2006;290:R1672–R1682. doi: 10.1152/ajpregu.00239.2005. [DOI] [PubMed] [Google Scholar]
- 28.McDaneld TG, Spurlock DM. Ankyrin repeat and suppressor of cytokine signaling (SOCS) box-containing protein (ASB) 15 alters differentiation of mouse C2C12 myoblasts and phosphorylation of mitogen-activated protein kinase and Akt. J. Anim. Sci. 2008;86:2897–2902. doi: 10.2527/jas.2008-1076. [DOI] [PubMed] [Google Scholar]
- 29.Kile BT, et al. The SOCS box: a tale of destruction and degradation. Trends Biochem. Sci. 2002;27:235–241. doi: 10.1016/S0968-0004(02)02085-6. [DOI] [PubMed] [Google Scholar]
- 30.Park SR, et al. HoxC4 binds to the promoter of the cytidine deaminase AID gene to induce AID expression, class-switch DNA recombination and somatic hypermutation. Nat. Immunol. 2009;10:540–550. doi: 10.1038/ni.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steinke JW, et al. Identification of an Sp factor-dependent promoter in GCET, a gene expressed at high levels in germinal center B cells. Mol. Immunol. 2004;41:1145–1153. doi: 10.1016/j.molimm.2004.06.031. [DOI] [PubMed] [Google Scholar]
- 32.Li N, et al. Multiple myeloma risk variant at 7p15.3 creates an IRF4-binding site and interferes with CDCA7L expression. Nat. Commun. 2016;7:13656. doi: 10.1038/ncomms13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holien T, Vatsveen TK, Hella H, Waage A, Sundan A. Addiction to c-MYC in multiple myeloma. Blood. 2012;120:2450–2453. doi: 10.1182/blood-2011-08-371567. [DOI] [PubMed] [Google Scholar]
- 34.Kuehl WM, Bergsagel PL. MYC addiction: a potential therapeutic target in MM. Blood. 2012;120:2351–2352. doi: 10.1182/blood-2012-08-445262. [DOI] [PubMed] [Google Scholar]
- 35.Ohguchi H, et al. The KDM3A-KLF2-IRF4 axis maintains myeloma cell survival. Nat. Commun. 2016;7:10258. doi: 10.1038/ncomms10258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang M, Lewinska M, Fan X, Zhu J, Yuan ZM. PRR14 is a novel activator of the PI3K pathway promoting lung carcinogenesis. Oncogene. 2016;35:5527–5538. doi: 10.1038/onc.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moreaux J, et al. APRIL and TACI interact with syndecan-1 on the surface of multiple myeloma cells to form an essential survival loop. Eur. J. Haematol. 2009;83:119–129. doi: 10.1111/j.1600-0609.2009.01262.x. [DOI] [PubMed] [Google Scholar]
- 38.Moreaux J, et al. The level of TACI gene expression in myeloma cells is associated with a signature of microenvironment dependence versus a plasmablastic signature. Blood. 2005;106:1021–1030. doi: 10.1182/blood-2004-11-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ju S, et al. Correlation of expression levels of BLyS and its receptors with multiple myeloma. Clin. Biochem. 2009;42:387–399. doi: 10.1016/j.clinbiochem.2008.10.024. [DOI] [PubMed] [Google Scholar]
- 40.Novak AJ, et al. Expression of BCMA, TACI, and BAFF-R in multiple myeloma: a mechanism for growth and survival. Blood. 2004;103:689–694. doi: 10.1182/blood-2003-06-2043. [DOI] [PubMed] [Google Scholar]
- 41.Mackay F, Schneider P. TACI, an enigmatic BAFF/APRIL receptor, with new unappreciated biochemical and biological properties. Cytokine Growth Factor Rev. 2008;19:263–276. doi: 10.1016/j.cytogfr.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 42.Moreaux J, et al. TACI expression is associated with a mature bone marrow plasma cell signature and C-MAF overexpression in human myeloma cell lines. Haematologica. 2007;92:803–811. doi: 10.3324/haematol.10574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Landgren O, Weiss BM. Patterns of monoclonal gammopathy of undetermined significance and multiple myeloma in various ethnic/racial groups: support for genetic factors in pathogenesis. Leukemia. 2009;23:1691–1697. doi: 10.1038/leu.2009.134. [DOI] [PubMed] [Google Scholar]
- 44.Rand KA, et al. A meta-analysis of multiple myeloma risk regions in African and European ancestry populations identifies putatively functional loci. Cancer Epidemiol. Biomark. Prev. 2016;25:1609–1618. doi: 10.1158/1055-9965.EPI-15-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anderson, C.A. et al. Data quality control in genetic case-control association studies. Nat. Protoc.5, 1564-1573 (2010). [DOI] [PMC free article] [PubMed]
- 46.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS. Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abecasis GR, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang J, et al. Improved imputation of low-frequency and rare variants using the UK10K haplotype reference panel. Nat. Commun. 2015;6:8111. doi: 10.1038/ncomms9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 50.Clayton DG, et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat. Genet. 2005;37:1243–1246. doi: 10.1038/ng1653. [DOI] [PubMed] [Google Scholar]
- 51.Liu JZ, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat. Genet. 2010;42:436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neben K, et al. Combining information regarding chromosomal aberrations t(4;14) and del(17p13) with the International Staging System classification allows stratification of myeloma patients undergoing autologous stem cell transplantation. Haematologica. 2010;95:1150–1157. doi: 10.3324/haematol.2009.016436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walker BA, et al. A compendium of myeloma-associated chromosomal copy number abnormalities and their prognostic value. Blood. 2010;116:e56–e65. doi: 10.1182/blood-2010-04-279596. [DOI] [PubMed] [Google Scholar]
- 55.Cairns J, et al. CHiCAGO: robust detection of DNA looping interactions in Capture Hi-C data. Genome Biol. 2016;17:127. doi: 10.1186/s13059-016-0992-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fernandez JM, et al. The BLUEPRINT data analysis portal. Cell Syst. 2016;3:491–495.e5. doi: 10.1016/j.cels.2016.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cowper-Sal lari R, et al. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat. Genet. 2012;44:1191–1198. doi: 10.1038/ng.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de Souza N. The ENCODE project. Nat. Methods. 2012;9:1046. doi: 10.1038/nmeth.2238. [DOI] [PubMed] [Google Scholar]
- 60.Halvarsson BM, et al. Direct evidence for a polygenic etiology in familial multiple myeloma. Blood Adv. 2017;1:619–623. doi: 10.1182/bloodadvances.2016003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Via M, Gignoux C, Burchard EG. The 1000 genomes project: new opportunities for research and social challenges. Genome Med. 2010;2:3. doi: 10.1186/gm124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
SNP genotyping data that support the findings of this study have been deposited in Gene Expression Omnibus with accession codes GSE21349, GSE19784, GSE24080, GSE2658 and GSE15695; in the European Genome-phenome Archive (EGA) with accession code EGAS00000000001; in the European Bioinformatics Institute (Part of the European Molecular Biology Laboratory) (EMBL-EBI) with accession code E-MTAB-362 and E-TABM-1138; and in the database of Genotypes and Phenotypes (dbGaP) with accession code phs000207.v1.p1. Expression data that support the findings of this study have been deposited in GEO with accession codes GSE21349, GSE2658, GSE31161 and EMBL-EBI with accession code E-MTAB-2299. The remaining data are contained within the paper and Supplementary Files or are available from the author upon request. KMS11 Hi-C data used in this manuscript are deposited in EGA under accession number EGAS00001002614. The accession number for the KMS11 ChIP-seq data reported in this paper is EGA: S00001002414. Naive B-cell HiC data used in this work is publically available from Blueprint Blueprint Epigenome Project [https://osf.io/u8tzp/]. ChIP-seq data for H3K27ac, H3K4Me1, H3K27Me3, H3K9Me3, H3K36Me3 and H3K27Me3 from naive B cells are publically available and were obtained from Blueprint Epigenome Project [http://www.blueprint-epigenome.eu/].