Abstract
Transthyretin (TTR), a homotetrameric protein that transports thyroxine and retinol both in plasma and in cerebrospinal (CSF) fluid provides a natural protective response against Alzheimer’s disease (AD), modulates amyloid-β (Aβ) deposition by direct interaction and co-localizes with Aβ in plaques. TTR levels are lower in the CSF of AD patients. Zn2+, Mn2+ and Fe2+ transform TTR into a protease able to cleave Aβ. To explain these activities, monomer dissociation or conformational changes have been suggested. Here, we report that when TTR crystals are exposed to copper or iron salts, the tetramer undergoes a significant conformational change that alters the dimer-dimer interface and rearranges residues implicated in TTR’s ability to neutralize Aβ. We also describe the conformational changes in TTR upon the binding of the various metal ions. Furthermore, using bio-layer interferometry (BLI) with immobilized Aβ(1–28), we observe the binding of TTR only in the presence of copper. Such Cu2+-dependent binding suggests a recognition mechanism whereby Cu2+ modulates both the TTR conformation, induces a complementary Aβ structure and may participate in the interaction. Cu2+-soaked TTR crystals show a conformation different from that induced by Fe2+, and intriguingly, TTR crystals grown in presence of Aβ(1–28) show different positions for the copper sites from those grown its absence.
Introduction
Human transthyretin (TTR), a homotetrameric 127-residue protein, is the main carrier of thyroxine (T4) in celebrospinal fluid (CSF) and the second main carrier in blood1. TTR also interacts with other proteins, such as retinol-binding protein (RBP) and thus contributes to retinol transport2. In aged patients, TTR can be responsible for certain amyloidotic diseases, when sporadically the tetramer dissociates into conformationally unstable TTR monomers prone to aggregate into TTR-amyloid fibrils3,4. Interestingly, TTR also interacts with amyloid-β (Aβ) and plays a protective role in Alzheimer’s disease (AD) by sequestering Aβ and reducing proteopathic stress. Aβ is generated upon sequential cleavage of the amyloid precursor protein (APP), and an imbalance between Aβ production and brain clearance has been postulated as a possible cause of Aβ-amyloid deposition in AD5. TTR binds soluble, oligomeric and Aβ fibrils6,7 playing a role in Aβ clearance8. The precise mechanism by which TTR binds to Aβ remains unknown and is particularly difficult to pinpoint because of the plethora of aggregated forms of Aβ.
Metals ions such as Zn2+, Mn2+, Cu1+,2+, Fe2+,3+ affect Aβ fibril formation and toxicity inducing a profusion of different conformations9–13. The same cations also interact with TTR. High concentrations of Zn2+ and Cu2+ (but not the iron cations) can promote the formation of a TTR-amyloid complex in vitro, while chelators (EDTA or EGTA) can disrupt fibrils composed of aggregated TTR14. Zinc ions boost L55P-TTR fibril formation15 and promote TTR metallopeptidase activity16. Such activity is also induced by other ions, such as Co2+, Mn2+ and Fe2+, but not Cu2+ or Ni2+ 17.
Currently, AD drugs are mainly supportive or palliative. All potentially curative drugs have failed during or before phase III trials18. Recently, trials of two anti-Aβ monoclonal antibodies, bapineuzumab and solanezumab, for AD patients with mild-to-moderate symptoms, were stopped due to a lack of beneficial clinical outcomes19. Clearly, the strategy of inhibiting Aβ is too simplistic. It would be beneficial to comprehend how Aβ is cleared from the brain naturally, and what mechanisms are employed by TTR to scavenge Aβ. The structure of human TTR is well known20–22, and hypothetical models for its interaction with Aβ have been proposed. A recent NMR study places Aβ(12–28) within an external pocket spanning across the epigallocatechin-3-gallate (EGCG) binding site23. Previous studies positioned Aβ in the TTR interior pocket extending towards the short α-helix7, involving L110 and L82, since their mutation (L → A) destroys TTR’s ability to bind Aβ24. L110 is located in the central hydrophobic channel, and for Aβ to bind in proximity, a rearrangement of the TTR monomers would be required, as the available volume is insufficiently small in the channel of the standard TTR tetramer25.
Senile amyloid plaques contain high levels of Cu, Fe and Zn26 that can promote polymorphic Aβ aggregation13,27. We question whether metal-aggregated forms might interact better with TTR, and whether the TTR-Aβ interaction might be modulated by metal ions. Although the zinc binding sites of TTR have already been elucidated in the cryptic peptidase form of TTR28,29, the positioning of Aβ within the TTR proteolytic site remains to be determined. Apart from zinc, the binding of other metal ions to TTR has not yet been characterized crystallographically.
Here we report the interaction between TTR and Aβ in the presence of Cu2+ obtained using bio-layer interferometry (BLI) with a biotinylated peptide comprising the 1–28 region of Aβ (Fig. 1). We also present the crystal structures of Fe2+, Mn2+ and of Cu2+ bound to TTR (Table 1) crystallized in the presence and absence of Aβ(1–28). Soaking TTR crystals with CuCl2 or FeCl2 at acidic pH induces a change in conformation comparable to that observed for the TTR-rhenium complex30 (Fig. 2). We hypothesize that the conformational change induced by Cu and Fe is related to TTR’s ability to bind Aβ.
Figure 1.
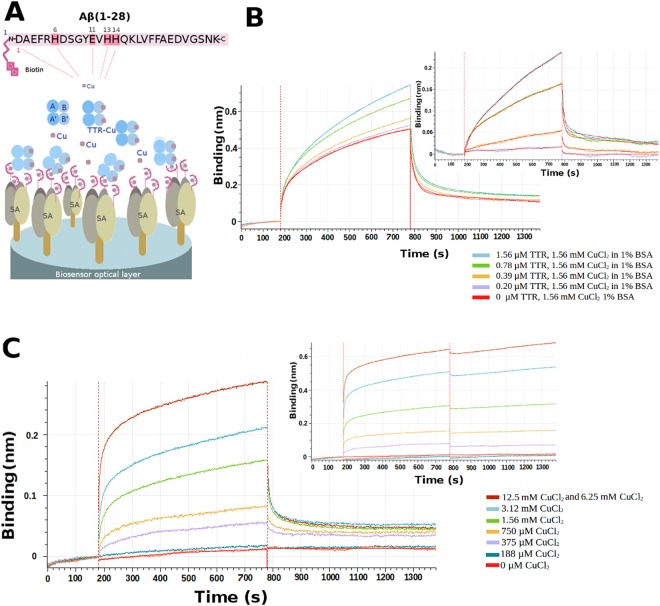
Copper-mediated TTR-Aβ interactions. (A) Overall scenario for bio-layer interferometry (BLI) using a streptavidin (SA) coated optical sensor. Biotinylated Aβ(1–28) was loaded on the BLI bio-sensor tip surface (100 nM). A single biotin is present at the N-terminal position of the Aβ peptide. The tip was then plunged in solutions containing TTR and copper at different concentrations to analyze the interactions. (B) Sensorgram curves colored according to TTR concentration (0 μM–1.56 μM). Inset on the right, represents the specific time dependent binding of TTR on immobilized Aβ(1–28) in the presence of 1% of BSA at pH 5.5, corresponding to the total binding for each TTR concentration minus the nonspecific binding found without TTR in presence of 1.56 mM Aβ and 1% of BSA. (C) Time dependent binding of TTR on immobilized Aβ peptide at a concentration of 1.56 μM of TTR in presence of various CuCl2 concentrations at pH 5.5 with 1% of BSA without CuCl2 during the dissociation. The sensorgram curves are colored according to CuCl2 concentration (0 mM–12.5 mM). Inset on the right, shows the dissociation step using buffer added by various CuCl2 concentrations (0 to 12.5 mM).
Table 1.
Crystallization conditions and data collection statistics.
| Structure | TTR-Fe | TTR-Mn | TTR-Cu | TTR-Cu-Aβ |
|---|---|---|---|---|
| PDB code | 5N5Q | 5N62 | 5N7C | Not deposited |
| Metal-Ligand | Fe | Mn | Cu | Cu-Aβ |
| Crystallization | 27% polyethylene glycol 4,000 (PEG4K), 0.2 M imidazole malate, pH 6.0 | 26% PEG4K, 0.2 M imidazole malate, pH 6.0 | 21% PEG4K, 0.14 M imidazole malate, pH 6.0, 3.6% MPEG 5 K, 0.03 M sodium acetate, pH 5.5 | Co-crystallization with Aβ 1–28 and CuCl2 in 21% PEG4K, 0.14 M imidazole malate, pH 6.0, 3.6% MPEG, 5 K, 0.1 M sodium acetate, pH 5.5 |
| Cryoprotectant | 40% SM3*, 25% MPEG 5 K, 0.1 M CHC (90% acid/10% basic), 30 mM FeCl2, 2 h soak. | 40% CM7*, 25% MPEG 5 K, 5 mM MnCl2, 10 min soak. | 40% SM3*, 25% MPEG 5 K, 0.1 M CHC(90% acid/10% basic), 30 mM CuCl2 30 min soak. | 40% CM1*, 25% MPEG 5 K, 0.1 M CHC (90% acid/10% basic), 30 mM CuCl2 30 min soak. |
| Data Collection | anomalous | anomalous | anomalous | non anomalous |
| Source | Soleil Proxima-2 | Soleil Proxima-2 | Soleil Proxima-2 | Soleil Proxima-2 |
| Wavelength (Å) | 1.739153 | 1.891993 | 1.175919 | 0.980035 |
| Space group | P21212 | P21212 | P21212 | P21212 |
| Unit-cell (Å) | 42.82 83.10 65.34 | 43.27 85.83 63.90 | 42.99 82.54 67.77 | 43.10 82.75 67.14 |
| Molec./asym. | 2 | 2 | 2 | 2 |
| Resolution (Å) | 50–2.53/2.68–2.53 | 50–1.8/1.85–1.80 | 50–2.45/2.59–2.45 | 50–2.14/2.27–2.14 |
| CC1/2 (%) | 99.6/30.0 | 99.8/39.0 | 99.8/89.0 | 99.9/52.5 |
| 〈I/σ(I)〉 | 8.28/0.83 | 11.53/1.52 | 14.75/3.29 | 13.0/1.10 |
| R-merge (%) | 20.0/198 | 17.6/217 | 10.7/67.2 | 12.9/215 |
| R-factor (%) | 18.5/182 | 16.3/202 | 9.9/62.1 | 12.4/222 |
| Completeness (%) | 99.2/95.3 | 100/100 | 99.5/96.7 | 99.8/98.9 |
| Multiplicity | 13.21/6.87 (anomalous) | 7.37 (anomalous) | 13.7/7.0 (anomalous) | 13.1 |
| Refinement | REFMAC5 | Phenix | REFMAC5 | REFMAC5 |
| Resolution (Å) | 40.91–2.53/2.60–2.53 | 38.63–1.80/1.89–1.80 | 41.06–2.45/2.51–2.45 | 38.23–2.43/2.49–2.43 |
| No. of reflections | 8113/548 (non-anomalous) | 22723/2658 (anomalous) | 8868/467 (non-anomalous) | 9042/650 (non-anomalous) |
| R-work | 19.2/41.0 | 19.5/28.4 | 16.9/20.3 | 20.9/31.0 |
| R-free | 25.3/40.4 | 24.4/33.2 | 25.4/43.1 | 29.7/44.4 |
| RMSD Bond lengths (Å) | 0.013 | 0.009 | 0.009 | 0.015 |
| RMSD Bond angles (°) | 1.6 | 1.025 | 1.714 | 1.773 |
| Ramachandran favored | 96.0% | 98.0% | 94.0% | 96.9% |
| Ramachandran outliers | 0 | 0 | 0 | 3 |
Cryoprotection: SM1: 12.5% diethylene glycol + 12.5% glycerol + 12.5% 1,2-propanediol + 25% DMSO + 25% 1,4-dioxane; SM3: 25% diethylene glycol + 25% ethylene glycol + 25% glycerol + 25% 1,4-dioxane; CM1: 12.5% diethylene glycol + 37.5% 1,2-propanediol + 12.5% DMSO; CM7: 12.5% di-ethylene glycol + 12.5% ethylene glycol + 12.5% glycerol + 25% 1,2-propanediol + 12.5% DMSO68. Cryoprotectant solution is formulated with 40% v/v mixed compounds (CM), 50% v/v precipitant and 10% v/v buffer69. CHC: Linear mixed buffer composed of citric acid, HEPES and CHES; acid mix at pH 4.0, basic at pH 10.070. CC1/2: Data quality correlation coefficient71. Data collection statistics are from XDS60. Refinement statistics are from REFMAC563 or phenix.refine (Phenix)65.
Figure 2.
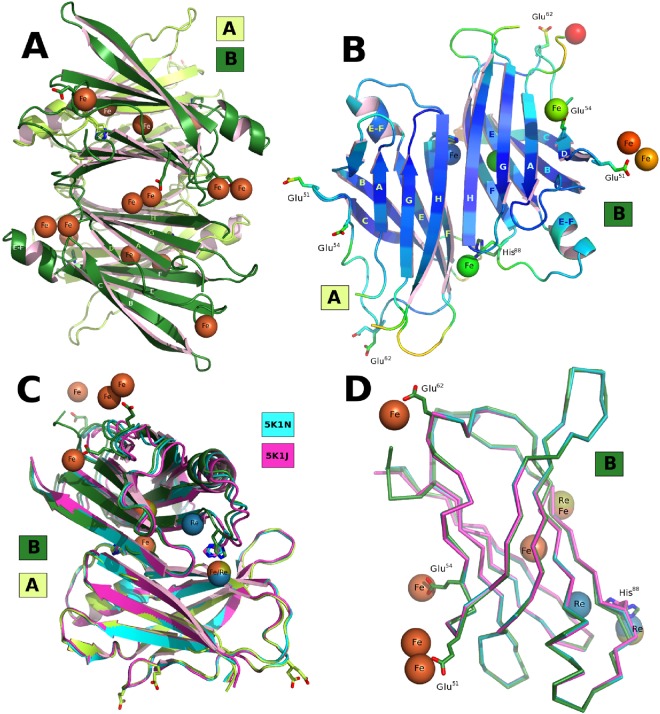
Conformation of the TTR-Fe complex and comparison with the TTR-Re complex. (A) Overall view of the tetrameter highlighting the iron positions with monomer A and B in light and dark green, respectively. (B) Overall view of the hetero-conformational A-B dimer colored according to the crystallographic B-value. (C) Superposition of the dimers of the Fe vs. Re TTR complexes showing that additional changes occur in the B monomer increasing its difference from that of the A monomer. (D) Superimposition of monomers B from the two rhenium complexes against the TTR-Re complex30.
Results
Binding affinity of Transthyretin with wt-Aβ peptide 1–28
Biotin-tagged peptide of the wild type-Aβ fragment 1–28 (wt-Aβ) was immobilized on a streptavidin-coated BLI tip (Octet Red 96 platform, Pall FortèBio) (Fig. 1A). Several solutions of 1.56 mM of CuCl2 in 50 mM acetate buffer, pH 5.5, 1% BSA with variable amounts (0, 0.20, 0.39, 0.78, and 1.56 μM) of TTR were tested and the binding saturation was observed at 1.56 μM. The apparent KD of wt-Aβ, calculated in presence of 1.56 mM of CuCl2 was estimated to be 54.70 ± 1.19 nM (Fig. 1B). In order to evaluate the influence of Cu2+ in the kinetic parameters of this interaction, various concentrations of CuCl2 were tested during the association step (Fig. 1C). In addition, a second experiment has been done to investigate the effect of Cu+2 in the dissociation step (Fig. 1C, insert). Different solutions of 1.56 μM TTR in 50 mM acetate buffer, pH 5.5, 1% BSA with 0, 188, 375, 750 μM and 1.56, 3.12, 6.25 and 12.5 mM of CuCl2 were tested. No significant binding was observed with CuCl2 below 188 μM. Starting at 375 μM, the signal progressively increased reaching a maximum at 6.25 mM and 12.5 mM which represents the saturation signal (Fig. 1C). When the Cu2+ is added in the dissociation step, the trimeric complex remains stable and no dissociation was observed (Fig. 1C).
The same experiment carried out with ZnCl2 instead of CuCl2 showed no binding. When the experiment is performed at physiological pH (pH 7.4) in presence of CuCl2 the KD is close to the value found at pH 5.5, while with ZnCl2 no association was observed. For iron, it was not possible perform the same experiments due to the iron-induced denaturation of TTR and its subsequent precipitation (see Supplementary Fig. S1).
Complementary experiments were carried out with two control peptides, one with a scrambled sequence (sm-Aβ) and another with the three histidines and a glutamate31 mutated to alanine (mut-Aβ). In presence of 1.56 mM of CuCl2 with 1.56 μM of TTR, the mutated peptide was found to bind with a KD = 685 ± 68 nM that is ten times less than the wt-peptide. No binding was observed for the scrambled peptide.
Identifying specific metal binding sites and key inter-residue distances
Human TTR crystals were soaked directly in metal-containing cryoprotectant solutions. The duration, pH and metal salt concentration were optimized to allow any conformational changes to occur with minimal deterioration to the X-ray diffraction quality. In order to unambiguously identify metal sites, X-ray diffraction data were collected at two X-ray wavelengths just prior and just beyond the X-ray absorption K-edge of the corresponding metal (see Supplementary Information). The phased anomalous difference Fourier maps were used as a guide to identify and position the specific metal ions (see Supplementary Fig. S2).
Three key inter-residue distances can be used to describe the conformational changes induced by metal binding to TTR. Differences in the A-A′ and B-B′ Leu-110–Leu-110 separation (see Supplementary Table S3) measures the asymmetric rotation of the dimers relative to each other. The Asp-38–Asp-38 separation between the A and B monomers is a monitor of the change in shape of the dimer, while the Thr-123A–Gly-83B distance changes as residues 72–92 on monomer B shifts relative to A. This distance varies in response to the conformational change that displaces His-88.
Iron binding to TTR
Human TTR crystals are particularly sensitive to FeCl2 soaking experiments. Once optimized, the crystal packing was found to be affected by the TTR conformational changes, and the unit cell parameters alter from a = 43.3 Å, b = 85.8 Å, c = 63.9 Å, as observed for the TTR-Mn complex, and typical for most human TTR-ligand complexes in the PDB32, to a = 42.8 Å, b = 83.1 Å, c = 65.3 Å, for the TTR-Fe complex (Table 1, PDB code: 5N5Q).
The TTR-Fe structure with the strongest anomalous signal in the phased anomalous difference Fourier map, out of 35 successful soak experiments, is reported here (see Supplementary Table S1). The observed conformational changes surpass those induced by rhenium30. The magnitude of these changes, combined with an increased total absorbed dose at the X-ray absorption K-edge of Fe33 and the higher multiplicity to improve the anomalous signal34, results in a structure with a crystallographic resolution limit lower than that for TTR-Re.
The largest anomalous features for the TTR-Fe complex are located in proximity of Glu-51. On A monomers, the electron density can be fitted by a three-atom Fe-cluster (see Supplementary Fig. S2B) and on B monomers by two Fe atoms (Fig. 2C). An additional Fe site at the entrance of the T4 binding site involves Glu-54 from symmetry related B and B′ monomers belonging to the same tetramer.
Some uninterpreted electron density extends beyond the phased anomalous difference density for the Fe, suggesting the presence of a chelating residue, probably Glu-7 from the disordered N-terminus. An additional unidentified peak, too strong be explained by a water molecule, but without any anomalous signal and thus can not be an Fe atom, is located close to His-88 (see Supplementary Fig. S2D). The TTR tetramer loses the symmetry of the T4-TTR complex (PDB code: 1ICT22) matching the asymmetric conformation of the TTR-Re-complex30. In this conformation, the spatial relationship between monomers A and B is altered giving the dimer a different shape. The conformational change involves only the B monomer and affects the β-strands E and F and the short E-F α-helix. In total, sixteen Fe atoms (including the clusters in interaction with Glu-51) are bound to each tetramer chelated by just three residues from each monomer (see Supplementary Tables S1 and S2).
The conformation of the TTR-Fe complex, with an Asp-38–Asp-38 separation of 25.7 Å is intermediate compared to the two TTR-Re complexes30. The large shift in the stretch of residues 72–92 of B relative to A with a Thr-123A–Gly-83B distance of 27.0 Å and a large Leu-110–Leu110 asymmetry attests to the largest conformational changes observed in TTR structures to date.
Manganese binding to TTR
Although manganese is of minor interest for either AD35 or TTR amyloidogenesis because the Mn concentrations are 1000-fold lower than either Cu or Fe in the CSF36, its position in the periodic table relative to Fe and Re warranted the determination of its complex. When TTR crystals grown at pH 6.0 were soaked in 5 mM MnCl2 under conditions similar to those carried out with Fe, Mn was found bound to Glu-66 and Asn-98 in monomer B and to Asp-99 and Glu-66 in monomer A, with minor local changes. The residues involved in Mn binding are not involved in Fe or Re binding (see Supplementary Fig. S3, PDB code: 5N62). More globally, Mn may influence an A-A′ and B-B′ asymmetry. The separation between the central Leu-110 residues (B-B′ Leu-110–Leu110 separation of only 6.73 Å) suggests a rotation of the TTR dimers in a direction opposite to that observed for Fe and Re-TTR structures (see Supplementary Table S3). The conformation of the TTR-Mn complex with a distance Asp38–Asp38 of 19.2 Å is within the range for distances observed for wt-TTR with ligands (e.g. 5EZP37) and even a wide selection of mutant TTR structures including those subjected to heating (PDB code: 2QEL38) or to a pH as low as 4.6 (PDB code: 2G4G39).
Copper binding to TTR
When TTR crystals grown or soaked in 30 mM CuCl2 the conformation of the TTR-Cu complex yields a distance Asp-38–Asp-38 of 25.0 Å, a Thr-123–Gly-83 distance of 25.7 Å and a Leu-110–Leu-110 asymmetry, which are comparable in magnitude but opposite to that of the TTR-Mn complex. This suggests a more nuanced response to metal binding (see Supplementary Table S2, PDB code: 5N7C).
The overall structure of the complex is illustrated in Fig. 3. The main difference that distinguishes this complex and others is in the positioning of the copper ion chelated between His-88, His-90 and Asp-72 on monomer B (Fig. 3D) and the binding between His-90 and Asp-72 on monomer A (Fig. 3D) which endows the stretch of residues 72–92 with a conformation different from those of the other TTR-metal complexes.
Figure 3.
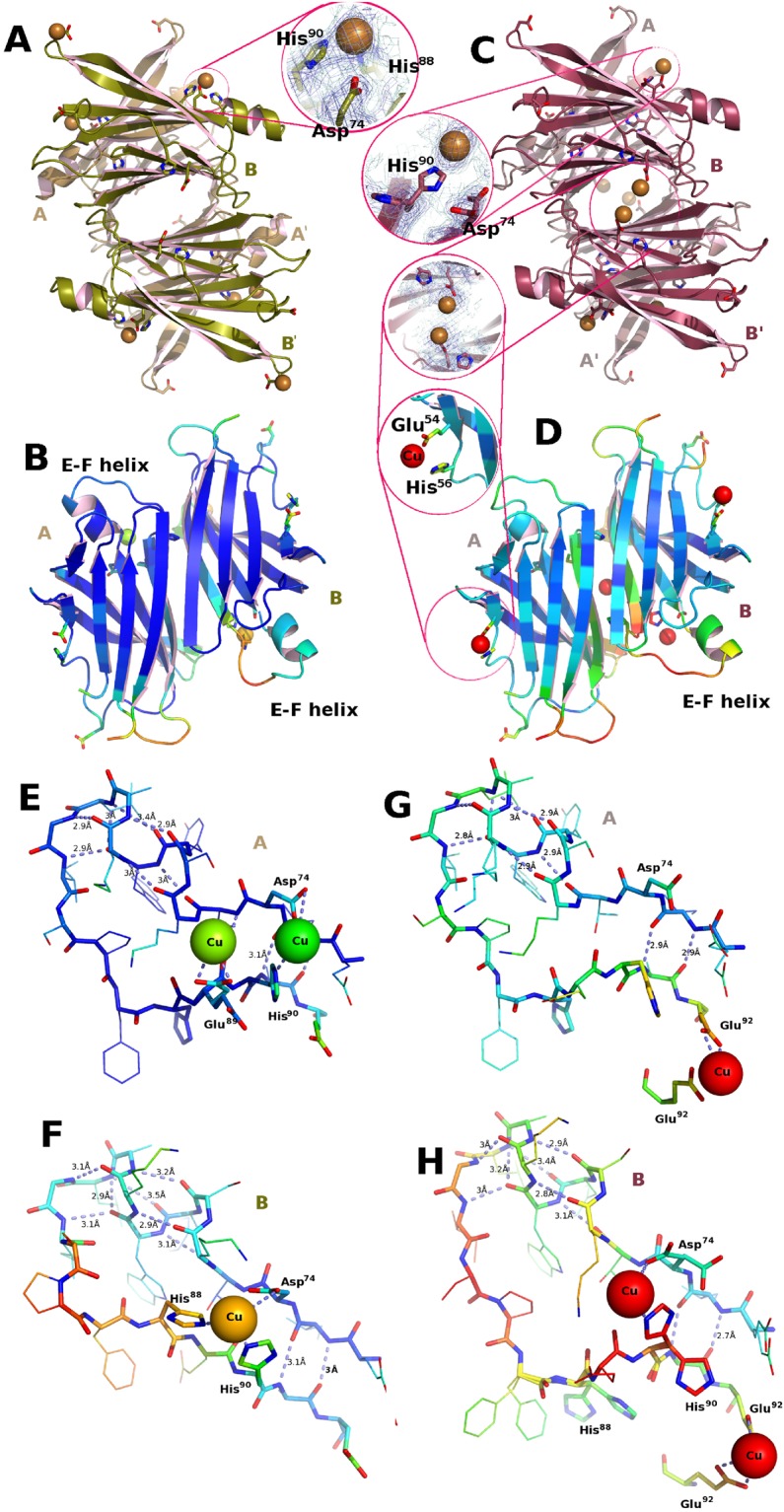
Conformation induced by copper binding to TTR. (A,B) Structure obtained in the absence of Aβ. (C,D) Structure obtained using crystals grown in the presence of Aβ(1–28). Copper soaking into TTR crystals stimulates in both cases a change in conformation similar to that observed for iron and for rhenium. The amplitude of the changes are greater in TTR crystals grown in the absence of Aβ. (A) Overall view of the hetero-conformational A-B/A′-B′ tetramer rebuilt using the 2-fold crystallographic symmetry operation. (B) Crystallographic A-B dimer colored according to B-value to highlight the increased mobility that occurs in the 74–92 stretch that includes the E-F helix. (C) In the crystals obtained in the presence of Aβ(1–28) the positioning of the copper is different from those grown in the absence of the amyloid peptide. The binding is probably weakened from copper chelation by disordered Aβ(1–28) in the solvent channels of the crystal. Residual copper binding is observed in proximity of Asp-74 with minor cooperation of His-90 and at the entrance of the ligand binding channel mediated by Glu-92 with His-56 in proximity. (D) The overall mobility of the protein structure for the TTR crystals grown in the presence of Aβ(1–28) is greater than for those grown without the peptide. (E) In the crystals obtained in the presence of Aβ(1–28) the positioning of the copper is different from those grown in the absence of the amyloid peptide. Residual copper binding is observed in proximity of Asp-74 with minor cooperation of His-90 and at the entrance of the ligand binding channel mediated by Glu-92 with His-56 in proximity. (F) The overall mobility of the structure for the crystals grown in the presence of Aβ(1–28) is greater than for those grown without the peptide. (G) Copper binds only at the entrance of the ligand binding channel on monomer A. (H) The disorder of stretch of residues 72–92 of monomer B is probably due to an incomplete transition towards the conformation observed in (D).
Copper binding to TTR crystals grown in presence of Aβ
Contrary to other metal ions, Cu does not transform TTR into a protease that can cleave Aβ17, and this permits their co-crystallization without the possibility of Aβ proteolytic degradation. Crystallographic analysis of the crystals obtained from solutions of TTR and Aβ do not show any convincing electron density corresponding to the Aβ peptide, although some uninterpretable density is apparent nearby. Similarly, crystals grown in the presence of Aβ and CuCl2 also failed to incorporate the peptide in an ordered manner. However, it is noteworthy that when soaked with CuCl2 (conditions as for the TTR-Cu complex PDB code: 5N7C; this work), the crystals grown in the presence of Aβ show a reduced binding of copper and an incomplete transformation, with B-values higher than those for other complexes (PDB code: not deposited, see Fig. 3F).
Based on the magnitude of the three key distances, the extent of the conformational changes is intermediate between the TTR-Mn and the TTR-Cu complexes (see Supplementary Table S3). The electron density for the 72–92 zone in proximity of His-90 suggests the coexistence of modified and unmodified B monomers. The conformation of the unmodified monomers matches that of the A monomers, while the B monomers adopt a conformation different from that found in the TTR-Cu complex (Fig. 3 compare zoom A vs. zoom E and D vs. H). Note that His-88 does not participate in metal chelation and a Cu2+ ion is chelated by Glu-92 (Fig. 3G,H) and Glu-54 (Fig. 3E,F) at the A–B the B–B′ interfaces, respectively. The latter TTR-metal interaction is common to the TTR-Fe complex (see Supplementary Tables S1 and 2; Fig. 2).
Discussion
TTR functions as a transporter in plasma and in the brain of several molecules, such as T4 and retinol. In the CSF, TTR is also recognized as the main Aβ binder, and, it is able to transport Aβ across the brain-blood barrier in the brain-to-blood direction8. The details of this clearance mechanism, however, remain to be elucidated.
The concentration of TTR is different in the blood and in the CSF: 3–4 μM and 0.1–0.4 μM, respectively24. Under physiological conditions, the soluble Aβ peptide is secreted into the synaptic cleft of normal subjects and AD patients, and its concentration is in the nanomolar range40. In the brain, the concentration of Aβ is about six-times higher than in plasma. Perturbations in the efflux of Aβ out of the brain could affect the levels of soluble Aβ in the CSF12,41. Moreover, when metals ions are released into the synaptic space during neurotransmission, they can modify the stability of the Aβ peptide. In the AD affected brain, the total concentration of Cu and other metals contained in amyloid plaque deposits has been established at 400 μM26.
In this work, we have investigated the interaction between TTR and Aβ by using a segment (residues 1–28) of the full length Aβ peptide as it is more soluble and stable in buffer solutions. More importantly, however, Aβ(1–28) contains the hydrophobic core VFF (residues 18–20), which is recognized by TTR7,23. Furthermore, Aβ(1–28) holds the residues that chelate metal ions31 and that can mediate the interaction between TTR and Aβ. It is not known if TTR binds the entire sequence of Aβ or only the fragments of Aβ cleaved by TTR6,42, but the segment 1–28 of Aβ is exposed in amyloid fibrils of Aβ(1–42)43.
Our BLI study has revealed that the affinity of TTR for Aβ(1–28) is modulated by copper. The binding of TTR to immobilized Aβ(1–28) increases with CuCl2 concentration from 0 to 12.5 mM (Fig. 1). When CuCl2 is included in the BLI dissociation buffer, no dissociation is observed Fig. 1C, consistent with the hypothesis that Cu2+ plays a key role in the stability of the TTR-Cu-Aβ complex.
Under pathological conditions, the pH dramatically decreases in the CNS, in plasma and in cells44–46. It is known that copper binding to histidines enhances Aβ aggregation at the pH typical of physiological acidosis47. The pH dependence of this interaction supports the involvement of the histidine side-chains. Consequently, the majority of our BLI and crystallographic experiments were done at acid pH (pH 5.5). Indeed, acidic pH and elevated CuCl2 concentration in the crystal soaking experiments (Table 1) favors a change in the TTR conformation in the stretch of residues 72–92 (see Supplementary Fig. S3).
The metal ion concentration needed to induce the conformational change in TTR crystals is necessarily elevated compared to the conditions in solution as the system is constrained by lattice packing forces. When TTR crystals of the P21212 crystal form are soaked in FeCl2 at acidic pH, only the B monomer in the asymmetric unit changes its conformation, whereby the E-F helix and the stretch of residues 85–92 undergo a rearrangement that is accompanied by a variation in the dimer-dimer interface. A similar change in conformation was observed when P21212 TTR crystals were soaked with a Re-complex30. Although the binding of various metals provokes similar changes, the changes induced differ in their extent. Three key distances can be used to evaluate the magnitude of the conformational change (see Supplementary Table S3): the separation between the four Ile-110 residues in the tetramer (to monitor the rotation of the dimers relative to each other), the distance between Gly-83B on the E-F loop from Thr-123A (to measure the shift of the E-F helix), and the separation between two Asp-38 residues (to assess the relationship between the monomers that form the dimer). This last measure provides a distinctive signature of metal binding. The Asp-38–Asp-38 separation, typically 21.5 Å in the absence of metals, shortens to 20.3 Å in the TTR-Mn complex and to 17.8 Å for the TTR-Zn complex (PDB code: 3DGD28), while in the TTR-Cu complex it increases to 25.7 Å and 26.3 Å in the TTR-Re complex (PDB code: 5K1N30) up to a maximum of 27.0 Å for the TTR-Fe complex (see Supplementary Table S3).
Metal chelation by proteins often involves the repositioning of certain residues. When considering TTR-Aβ complexes it is important to take into account that both partners in the interaction may alter their conformation in a dynamic and polymorphic manner. Fe promotes Aβ aggregation48 that interferes with the dynamics of amyloid formation49, and its metal binding is pH-dependent and affects oligomerization50. Morphological differences may affect toxicity. Aβ aggregates containing Zn, Fe or Cu are neurotoxic51,52.
Studies in vitro are focused on Aβ interactions with Cu and Zn, even if Mn also binds Aβ peptides31 and plays a role in neurodegeneration. In order to investigate the importance of Mn in the TTR-Aβ interactions, TTR crystals were soaked in 5 mM of MnCl2 at pH 6.0. No significant conformational changes were observed, suggesting that even if Mn binds Aβ31, it does not play a relevant role in the interactions between TTR and Aβ.
Although Cu2+ induces a conformational change in TTR (Fig. 3; X-ray anomalous differences validate the presence Cu sites see Supplementary Table S2) which resembles those revealed by the Fe2+ and Re2+ experiments, the manner in which Cu2+ is chelated differs from those observed for Fe2+ or Re (Fig. 2). Even if both His-88 and His-90 are involved in Cu and Zn chelation, the interaction from the third residue is different, being Asp-74 for Cu2+ (Fig. 3) and Glu-92 in the case of Zn2+ at pH 4.628. The network of residues that intervene in metal binding is quite extensive (see Supplementary Fig. S3), and most residues have been recognized as possible zinc binders. The mutation of His-90 to Ala results in the loss of TTR catalytic activity and its ability to disrupt fibrils42.
Despite the fact that the TTR-Aβ interaction can be demonstrated in the presence of Cu by BLI, TTR crystals grown in the presence of CuCl2 and Aβ did not show any ordered Aβ peptides. This implies that the crystallization forces of TTR molecules disrupt the TTR-Aβ complex. Given the size of the Aβ(1–28) peptide with respect to TTR monomers (127 aa), one would expect the TTR-Aβ complex to crystallize in rather different unit cell constants and/or space group.
However, the TTR crystal structure contains 30Å -wide diamond-shaped solvent channels, which run the length of the crystal, and the likely presence of Aβ in the crystal interstices could be inferred from lower Cu2+ binding due to Aβ chelation. Under identical soak conditions, the TTR conformational change is characterized by a smaller Asp-38–Asp-38 separation (23.9 Å) compared to that seen in the absence of Aβ (25.0 Å). An additional Cu binding site is located in proximity of Glu-54 (Fig. 3E,F). This residue has been identified in regards to Cr3+ binding, and its involvement in Cu2+ binding has been suggested53. Thus, Glu-54 could act as the gatekeeper to the tunnel that leads to the four Leu-110 residues at the center of the tetramer, a residue that when mutated to alanine abolishes TTR’s Aβ scavenging activity. A contact between the Aβ peptide and Leu-110 would require a transit in front of Glu-54.
Our results show that TTR changes its conformation in response to binding Fe2+ 54 (as opposed to Fe3+ as confirmed by XANES experiments, see Supplementary Information Fig. S4) and Cu2+ at acidic pH, and that Cu is essential for the recognition of Aβ(1–28) by TTR. Only certain divalent metals ions (Cu2+, Fe2+) provoke the crystallographic conformational change. When TTR is soaked with trivalent metal ions under acid pH conditions (Al3+, Gd3+, or Fe3+), no conformational changes were observed.
The ability of Cu to promote the formation of the TTR-Aβ complex, and presumably the cerebral clearance of Aβ, is consistent with the report that mice with a defective Cu transporter (which removes Cu) have higher Cu levels, a reduced number of amyloid plaques and diminished plasma Aβ55. The lack of binding Aβ by TTR in the absence of Cu, may appear to be in contradiction with experiments carried out with 125I-labeled Aβ (1–40) and Aβ(1–28) peptides by Schwarzman et al., when it was shown that TTR recognizes Aβ in CSF fluid, inhibits its aggregation and prevents fibril formation5. However, excluding the possibility that the iodinated peptide used might interact via any of the three TTR halogen binding pockets in the TTR central tunnel, the results may also imply that in the absence of a metal, different polymorphic forms of Aβ are recognized by TTR. By stabilizing a Aβ conformation with mild affinity for TTR, Cu2+ ions rescue the interaction. TTR may also bind with higher affinity polymeric Aβ which acquires an alternative and stable conformation through Aβ-Aβ interactions. The immobilization method used here involves Aβ(1–28) mono-biotinylated at the N-terminus (Fig. 1B), and the results are consistent with studies showing that non-aggregating Aβ fragments do not quench TTR tryptophan fluorescence56. It is possible that the same peptide, if allowed to dimerize via the LVFFA stretch, may also adopt a conformation that is recognized by TTR in the absence of Cu. The polymorphic diversity of the longer Aβ (1–40/1–42) peptides, with extensive hydrophobic stretches, is likely to generate stable conformations with higher affinities for TTR. Modifying the BLI to function with more complex aggregates may be challenging, but will be necessary to guide the crystallographic structure determination of the TTR-Aβ complex with and without metals.
To conclude, the TTR-Cu conformation provides an alternative starting point for the design of molecules that aim to stabilize a form of TTR with an enhanced Aβ-scavenging activity to counter the reduction of TTR expression in AD patients57. As such, these results emphasize Cu as a “ forgotten factor” upon which the TTR-Aβ interaction may depend. Furthermore, metal chelation therapies, which are useful to reduce oxidative stress, should also take into account the essential levels of Cu required for Aβ clearance.
Methods
Synthesis of biotinylated Aβ peptides
Fmoc-amino acids, Fmoc-pseudoproline dipeptide, and 2-(6-chloro-1H-benzotriazole-1-yl)-1,1,3,3,-tetra-methylaminium hexafluorophosphate (HCTU) were obtained from Novabiochem (Darmstadt, Germany). N-biotine-NH-(PEG)11-COOH and biotinamidohexanoic acid N-hydroxysuccinimide ester were purchased at Merck and Sigma Aldrich respectively. The resin and all the peptide synthesis grade reagents (N-methylpyrrolidone (NMP), N-methylmorpholine (NMM), dichloromethane, piperidine, trifluoroacetic acid (TFA), anisole, thioanisole, and triisopropylsilane) were purchased from Sigma (Saint-Quentin Fallavier, France). The synthesis of peptide Aβ(1–28) wild type (DAEFRHDSGYEVHHQKLVFFAEDVGSNK), scrambled Aβ peptide (1–28) (LNHQEKGDRFSDYEVAFSVGHKVDFHEA) and mutant (DAEFRADSGYAVAAQKLVFFAEDVGSNK) were performed on a Protein Technologies, Inc., Prelude Synthesizer at a 25 μmol scale using a 10-fold excess of Fmoc-amino acid relative to the preloaded Fmoc-Lys(Boc)-wang-LLresin (0.33 μmol/g) or Fmoc-Ala-wang-LLresin (0.33 μmol/g). Fmoc-protected amino acids were used with the following side chain protections: tert-butyl ester (Glu and Asp), tert-butyl ether (Ser and Tyr), trityl (His, Asn, and Gln), tertbutoxycarbonyl (Lys), and 2,2,5,7,8-pentamethyl-chromane-6-sulfonyl (Arg). Amino acids were coupled twice for 5 min using 1:1:2 amino acid/HCTU/NMM in NMP. In scrambled Aβ peptide (1–28) a pseudoproline dipeptide (FS) was used at positions 10–11 and 17–18 and coupled twice for 10 min. After incorporation of each residue, the resin was acetylated for 5 min using a 50-fold excess of a mixture of acetic anhydride and NMM in NMP. Fmoc deprotection was performed twice for 3 min using 20% piperidine in NMP, and 30 sec NMP top washes were performed between deprotection and coupling and after acetylation steps. Biotinylation of Aβ peptides (1–28) was done with 50 mg resin after Fmoc deprotection of the N-terminal residue, using a 10-fold excess of either N-biotin-NH-(PEG)11-COOH (Aβ peptide 1–28 wild type and mutant) and HCTU and NMM as coupling reagents (see above) or biotinamidohexanoic acid N-hydroxysuccinimide ester in NMP (scrambled Aβ peptide). After completion, the peptidyl-resins were treated with a mixture of TFA/thioanisole/anisole/TPS/water (82:5:5:2.5:5) for 2 h. The crude peptides were obtained after precipitation and washes in cold ethyl ether followed by dissolution in 10% acetic acid and lyophilization. The different peptides were purified by reverse phase HPLC using an X-Bridge BHE C18-300-5 semi-preparative column (Waters, USA) (250 × 4.6 mm; 4 mL × min−1; solvent A: H2O/TFA 0.1%; solvent B: acetonitrile/TFA 0.1% using a gradient of 0–60% solvent B into A in 60 min). The purity of each peptide was checked by mass spectrometry using ESI-MS (Bruker,Germany). Biotinylated Aβ peptide (1–28) wild type calculated m/z: 4088.5, found: 4088.0; biotinylated mutant Aβ peptide (1–28) calculated m/z: 3832.2, found m/z: 3832.1 and biotinylated scrambled Aβ peptide (1–28) calculated m/z: 3601.5, found m/z: 3601.8.
Binding affinity measurements by bio-layer interferometry
Binding between TTR and Aβ peptides (wild type, scrambled and mutant) in absence or presence of copper was performed in real time by bio-layer interferometry using an Octet Red 96 platform instrument (Pall FortéBio Corp., Menlo Park, CA). Streptavidin bio-sensors were hydrated in 50 mM acetate, pH 5.5, 1% BSA (buffer A) for over 20 minutes before the start of each run and then loaded with 100 nM of biotinylated Aβ(1–28). A single run was subdivided into five distinct steps as follows58: (i) Baseline determination, in which the streptavidin bio-sensor tip was immersed in buffer A for 180 s to set the baseline; (ii) Loading step, during which 100 nM Aβ was immobilized on to the streptavidin-coated bio-sensor tip for 80 s; (iii) Back-soak step, during which the bio-sensor tip was again immersed in buffer A for 180 s to remove excess unbound Aβ; (iv) Association step, during which the Aβ loaded bio-sensor tip was immersed in TTR solutions for 600 s; (v) Dissociation step, during which the tip with TTR-Aβ bound was immersed in the same buffer for 600 s to dissociate the TTR. The data from these two last steps were used to estimate differences in interference caused by binding of TTR to the Aβ loaded tip. A blank run was carried out with buffer A for calibration purposes. A series of 5 runs was performed with CuCl2 at constant 1.56 mM, the first without TTR followed by 4 runs with increasing amounts of TTR (0, 0.20, 0.39, 0.78, 1.56 μM. Another series of 8 runs was performed with TTR at constant 1.56 μM, the first without CuCl2 followed by 7 runs with increasing amounts of CuCl2 (0, 188, 375, 750, 1.56 μM and 1.56, 3.12, 6.25, 12.5 mM. Affinity (KD) was calculated using the FortéBio Data Analysis HT v10.0.1.7 software.
Crystal preparation and structure determination
TTR crystals were grown by sitting drop vapor diffusion as previously reported59 using lyophilized human TTR (Calbiochem, Merck Millipore, Darmstadt, Germany) dissolved, 1 mg in 100 mL of 0.02% (w/v) NaN3 and dialyzed overnight against 0.1 M NaCl, 50 mM sodium acetate, pH 5.5. The reservoir solution consisted of 21% polyethylene glycol 4,000 (PEG4K), 0.14 M imidazole malate, pH 6.0 and the second one from 21% polyethylene glycol 4,000 (PEG4K), 0.14 M imidazole malate, pH 6.0, 3.6% polyethylene glycol monomethylether (MPEG5K), 30 mM sodium acetate, pH 5.5. Streak seeding was used to induce nucleation. The cryoprotectant solution was composed of 40% of SM2 (12.5% ethylene glycol, 12.5% glycerol, 12.5% 1,2-propanediol, 25% DMSO and 37.5% 1,4-dioxane) 25% PEG 8K and 30 mM of CuCl2, MnCl2 or FeCl2, to which 2.5% H2O2 or 0.8 mM Methylene Blue (MB) was added in some experiments. The combination of H2O2 and MB resulted in the loss of the blue color of MB. Crystals were flash cooled by in liquid nitrogen and data were collected on beamline Proxima-2A at the Soleil storage ring in Saint Aubin, France, on a Dectris Eiger 9M detector. Data processing was carried out at the synchrotron facility using XDS60 with the xdsme script61. The structure was solved by molecular replacement using Phaser62 followed by refinement using REFMAC563 starting with a TTR model without inhibitor. The electron density maps were viewed and fitted in COOT64. The structures were subjected to over ten cycles of rebuilding and refinement with REFMAC563 and PHENIX65. The phased anomalous difference Fourier maps were calculated with the program ANODE66. Data processing and refinement statistics are given in Table 1. The coordinates for the TTR wild-type and various mutants, see Supplementary Table S3, were retrieved from the Protein Data Bank32. The figures were made with PyMOL67. Structures deposited into the PDB Data Bank: 5N5Q, 5N62 and 5N7C.
Electronic supplementary material
Acknowledgements
This work was partially supported by grant of the Italian Ministero dell’Istruzione, dell’Universitá e della Ricerca: PRIN 20109MXHMR_007.
Author Contributions
L.C., W.S. and E.A.S. wrote the manuscript and designed the experiment, L.C. and G.M. synthesized the peptides, L.C. and C.F.-G. performed Blitz experiment, L.C. prepared crystals, L.C., M.S., W.S. and E.A.S. carried out data collections at SOLEIL, L.C., W.S. and E.A.S. analyzed data and solved the structures, S.N., E.O. and D.S. contributed to the discussion of the results. All the authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-31808-5.
References
- 1.Hamilton JA, Benson MD. Transthyretin: a review from a structural perspective. Cellular and molecular life sciences. 2001;58:1491–521. doi: 10.1007/PL00000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Naylor HM, Newcomer ME. The structure of human retinol-binding protein (RBP) with its carrier protein transthyretin reveals an interaction with the carboxy terminus of RBP. Biochemistry. 1999;38:2647–53. doi: 10.1021/bi982291i. [DOI] [PubMed] [Google Scholar]
- 3.Damas AM, Saraiva MJ. Review: TTR amyloidosis-structural features leading to protein aggregation and their implications on therapeutic strategies. Journal of structural biology. 2000;130:290–9. doi: 10.1006/jsbi.2000.4273. [DOI] [PubMed] [Google Scholar]
- 4.Tantau, A., Laszlo, M. & Laszlo, I. Transthyretin amyloidosis: an over review. Cardiovascular Regenerative Medicine, 10.14800/crm.952, 1–9 (2015).
- 5.Schwarzman AL, et al. Transthyretin sequesters amyloid beta protein and prevents amyloid formation. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:8368–72. doi: 10.1073/pnas.91.18.8368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ribeiro CA, Saraiva MJ, Cardoso I. Stability of the transthyretin molecule as a key factor in the interaction with a-beta peptide-relevance in alzheimer’s disease. PloS one. 2012;7:e45368. doi: 10.1371/journal.pone.0045368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X, et al. Mechanisms of transthyretin inhibition of β-amyloid aggregation in vitro. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2013;33:19423–33. doi: 10.1523/JNEUROSCI.2561-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alemi M, et al. Transthyretin participates in beta-amyloid transport from the brain to the liver- involvement of the low-density lipoprotein receptor-related protein 1? Scientific reports. 2016;6:20164. doi: 10.1038/srep20164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brzyska M, Trzesniewska K, Wieckowska A, Szczepankiewicz A, Elbaum D. Electrochemical and conformational consequences of copper (cui and cuii) binding to β-amyloid (1–40) Chem Bio Chem. 2009;10:1045–1055. doi: 10.1002/cbic.200800732. [DOI] [PubMed] [Google Scholar]
- 10.Pietropaolo A, Satriano C, Strano G, La Mendola D, Rizzarelli E. Different zinc (ii) complex species and binding modes at aβ n-terminus drive distinct long range cross-talks in the aβ monomers. Journal of inorganic biochemistry. 2015;153:367–376. doi: 10.1016/j.jinorgbio.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 11.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta crystallographica. Section D, Biological crystallography. 2011;67:235–42. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leal SS, Botelho HM, Gomes CM. Metal ions as modulators of protein conformation and misfolding in neurodegeneration. Coordination Chemistry Reviews. 2012;256:2253–2270. doi: 10.1016/j.ccr.2012.04.004. [DOI] [Google Scholar]
- 13.Chen W-T, Liao Y-H, Yu H-M, Cheng IH, Chen Y-R. Distinct effects of Zn2+, Cu2+, Fe3+, and Al3+ on amyloid-beta stability, oligomerization, and aggregation: amyloid-beta destabilization promotes annular protofibril formation. The Journal of biological chemistry. 2011;286:9646–56. doi: 10.1074/jbc.M110.177246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilkinson-White LE, Easterbrook-Smith SB. Characterization of the binding of Cu(II) and Zn(II) to transthyretin: effects on amyloid formation. Biochemistry. 2007;46:9123–32. doi: 10.1021/bi700607z. [DOI] [PubMed] [Google Scholar]
- 15.Castro-Rodrigues AF, Gales L, Saraiva MJ, Damas AM. Structural insights into a zinc-dependent pathway leading to Leu55Pro transthyretin amyloid fibrils. Acta Crystallographica Section D: Biological Crystallography. 2011;67:1035–1044. doi: 10.1107/S090744491104491X. [DOI] [PubMed] [Google Scholar]
- 16.Gouvea IE, et al. Studies on the peptidase activity of transthyretin (TTR) Biochimie. 2013;95:215–23. doi: 10.1016/j.biochi.2012.09.014. [DOI] [PubMed] [Google Scholar]
- 17.Liz MA, et al. Transthyretin is a metallopeptidase with an inducible active site. Biochemical Journal. 2012;443:769–778. doi: 10.1042/BJ20111690. [DOI] [PubMed] [Google Scholar]
- 18.Amanatkar HR, Papagiannopoulos B, Grossberg GT. Analysis of recent failures of disease modifying therapies in alzheimer’s disease suggesting a new methodology for future studies. Expert review of neurotherapeutics. 2017;17:7–16. doi: 10.1080/14737175.2016.1194203. [DOI] [PubMed] [Google Scholar]
- 19.Salloway S, et al. Two phase 3 trials of bapineuzumab in mild-to-moderate alzheimer’s disease. New England Journal of Medicine. 2014;370:322–333. doi: 10.1056/NEJMoa1304839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blake C, et al. An X-ray study of the subunit structure of prealbumin. Journal of Molecular Biology. 1971;61:217–224. doi: 10.1016/0022-2836(71)90218-X. [DOI] [PubMed] [Google Scholar]
- 21.Blake CC, Geisow MJ, Oatley SJ, Rérat B, Rérat C. Structure of prealbumin: secondary, tertiary and quaternary interactions determined by Fourier refinement at 1.8 A. Journal of molecular biology. 1978;121:339–56. doi: 10.1016/0022-2836(78)90368-6. [DOI] [PubMed] [Google Scholar]
- 22.Wojtczak A, Cody V, Luft JR, Pangborn W. Structure of rat transthyretin (rTTR) complex with thyroxine at 2.5 Å resolution: first non-biased insight into thyroxine binding reveals different hormone orientation in two binding sites. Acta Crystallographica Section D Biological Crystallography. 2001;57:1061–1070. doi: 10.1107/S0907444901007235. [DOI] [PubMed] [Google Scholar]
- 23.Gimeno A, et al. Insights on the interaction between transthyretin and aβ in solution. a saturation transfer difference (std) nmr analysis of the role of iododiflunisal. Journal of Medicinal Chemistry. 2017;60:5749–5758. doi: 10.1021/acs.jmedchem.7b00428. [DOI] [PubMed] [Google Scholar]
- 24.Du J, Cho PY, Yang DT, Murphy RM. Identification of beta-amyloid-binding sites on transthyretin. Protein engineering, design & selection: PEDS. 2012;25:337–45. doi: 10.1093/protein/gzs026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mangrolia, P., Yang, D. T. & Murphy, R. M. Transthyretin variants with improved inhibition of β-amyloid aggregation. Protein engineering, design & selection: PEDS, 10.1093/protein/gzw008, 1–10 (2016). [DOI] [PMC free article] [PubMed]
- 26.Lovell MA, Robertson JD, Teesdale WJ, Campbell JL, Markesbery WR. Copper, iron and zinc in Alzheimer’s disease senile plaques. Journal of the neurological sciences. 1998;158:47–52. doi: 10.1016/S0022-510X(98)00092-6. [DOI] [PubMed] [Google Scholar]
- 27.Miller Y, Ma B, Nussinov R. Zinc ions promote Alzheimer Aβ aggregation via population shift of polymorphic states. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9490–5. doi: 10.1073/pnas.0913114107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palmieri LDC, et al. Novel Zn2+-binding sites in human transthyretin: implications for amyloidogenesis and retinol-binding protein recognition. The Journal of biological chemistry. 2010;285:31731–41. doi: 10.1074/jbc.M110.157206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Costa, R., Ferreira-da Silva, F., Saraiva, M. J. & Cardoso, I. Transthyretin protects against A-beta peptide toxicity by proteolytic cleavage of the peptide: A mechanism sensitive to the kunitz protease inhibitor. Plos One3, 10.1371/journal.pone.0002899 (2008). [DOI] [PMC free article] [PubMed]
- 30.Ciccone L, Policar C, Stura EA, Shepard W. Human TTR conformation altered by rhenium tris-carbonyl derivatives. Journal of structural biology. 2016;195:353–364. doi: 10.1016/j.jsb.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 31.Wallin C, et al. Characterization of mn (ii) ion binding to the amyloid-β peptide in alzheimer’s disease. Journal of Trace Elements in Medicine and Biology. 2016;38:183–193. doi: 10.1016/j.jtemb.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 32.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic acids research. 2007;35:D301–3. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Owen RL, Rudiño-Piñera E, Garman EF. Experimental determination of the radiation dose limit for cryocooled protein crystals. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4912–7. doi: 10.1073/pnas.0600973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dauter Z. Estimation of anomalous signal in diffraction data. Acta crystallographica. Section D, Biological crystallography. 2006;62:867–76. doi: 10.1107/S0907444906023481. [DOI] [PubMed] [Google Scholar]
- 35. Wallin, C., Luo, J., Jarvet, J., Wärmländer, S. K. T. S. & Gräslund, A. The Amyloid-β Peptide in Amyloid Formation Processes: Interactions with Blood Proteins and Naturally Occurring Metal Ions. Israel Journal of Chemistry (2017).
- 36.Molina JA, et al. Cerebrospinal fluid levels of transition metals in patients with Alzheimer’s disease. Journal of neural transmission (Vienna, Austria: 1996) 1998;105:479–488. doi: 10.1007/s007020050071. [DOI] [PubMed] [Google Scholar]
- 37.Polsinelli I, et al. A new crystal form of human transthyretin obtained with a curcumin derived ligand. Journal of structural biology. 2016;194:8–17. doi: 10.1016/j.jsb.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 38.Karlsson A, Sauer-Eriksson AE. Heating of proteins as a means of improving crystallization: A successful case study on a highly amyloidogenic triple mutant of human transthyretin. Acta Crystallographica Section F: Structural Biology and Crystallization Communications. 2007;63:695–700. doi: 10.1107/S1744309107033957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pasquato N, et al. Acidic ph-induced conformational changes in amyloidogenic mutant transthyretin. Journal of Molecular Biology. 2007;366:711–719. doi: 10.1016/j.jmb.2006.11.076. [DOI] [PubMed] [Google Scholar]
- 40.Hu X, et al. Amyloid seeds formed by cellular uptake, concentration, and aggregation of the amyloid-beta peptide. Proceedings of the National Academy of Sciences. 2009;106:20324–20329. doi: 10.1073/pnas.0911281106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bates K, et al. Clearance mechanisms of alzheimer’s amyloid-β peptide: implications for therapeutic design and diagnostic tests. Molecular psychiatry. 2009;14:469. doi: 10.1038/mp.2008.96. [DOI] [PubMed] [Google Scholar]
- 42.Silva CS, et al. Transthyretin neuroprotection in alzheimer’s disease is dependent on proteolysis. Neurobiology of aging. 2017;59:10–14. doi: 10.1016/j.neurobiolaging.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 43.Gremer L, et al. Fibril structure of amyloid-β (1–42) by cryo–electron microscopy. Science. 2017;358:116–119. doi: 10.1126/science.aao2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tyrtyshnaia AA, et al. Acute neuroinflammation provokes intracellular acidification in mouse hippocampus. Journal of neuroinflammation. 2016;13:283. doi: 10.1186/s12974-016-0747-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Menkin, V., Springfield, I. & Thomas, C. The role of hydrogen ion concentration and the cytology of an exudate. Biochemical Mechanisms in Inflammation. Springfield, IL: Charles C Thomas 66–103 (1956).
- 46.Lardner A. The effects of extracellular ph on immune function. Journal of leukocyte biology. 2001;69:522–530. [PubMed] [Google Scholar]
- 47.Atwood CS, et al. Dramatic aggregation of Alzheimer abeta by Cu(II) is induced by conditions representing physiological acidosis. The Journal of biological chemistry. 1998;273:12817–26. doi: 10.1074/jbc.273.21.12817. [DOI] [PubMed] [Google Scholar]
- 48.Mantyh PW, et al. Aluminum, iron, and zinc ions promote aggregation of physiological concentrations of beta-amyloid peptide. Journal of neurochemistry. 1993;61:1171–1174. doi: 10.1111/j.1471-4159.1993.tb03639.x. [DOI] [PubMed] [Google Scholar]
- 49.Liu B, et al. Iron promotes the toxicity of amyloid beta peptide by impeding its ordered aggregation. The Journal of biological chemistry. 2011;286:4248–56. doi: 10.1074/jbc.M110.158980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghalebani L, Wahlström A, Danielsson J, Wärmländer SKTS, Gräslund A. pH-dependence of the specific binding of Cu(II) and Zn(II) ions to the amyloid-β peptide. Biochemical and biophysical research communications. 2012;421:554–60. doi: 10.1016/j.bbrc.2012.04.043. [DOI] [PubMed] [Google Scholar]
- 51.Bishop GM, Robinson SR. The amyloid paradox: amyloid-beta-metal complexes can be neurotoxic and neuroprotective. Brain pathology (Zurich, Switzerland) 2004;14:448–52. doi: 10.1111/j.1750-3639.2004.tb00089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sarell CJ, Wilkinson SR, Viles JH. Substoichiometric levels of cu(2+) ions accelerate the kinetics of fiber formation and promote cell toxicity of amyloid-β from alzheimer disease. Journal of biological chemistry. 2010;285:41533–41540. doi: 10.1074/jbc.M110.171355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sato T, et al. Chromium (iii) ion and thyroxine cooperate to stabilize the transthyretin tetramer and suppress in vitro amyloid fibril formation. FEBS Letters. 2006;580:491–496. doi: 10.1016/j.febslet.2005.12.047. [DOI] [PubMed] [Google Scholar]
- 54.Sigfridsson KG, et al. Rapid x-ray photoreduction of dimetal-oxygen cofactors in ribonucleotide reductase. Journal of Biological Chemistry. 2013;288:9648–9661. doi: 10.1074/jbc.M112.438796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Phinney AL, et al. In vivo reduction of amyloid-beta by a mutant copper transporter. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:14193–8. doi: 10.1073/pnas.2332851100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu L, Murphy RM. Kinetics of Inhibition of β-Amyloid Aggregation by Transthyretin. Biochemistry. 2006;45:15702–15709. doi: 10.1021/bi0618520. [DOI] [PubMed] [Google Scholar]
- 57.Sousa JC, Cardoso I, Marques F, Saraiva MJ, Palha JA. Transthyretin and alzheimer’s disease: where in the brain? Neurobiology of aging. 2007;28:713–718. doi: 10.1016/j.neurobiolaging.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 58.Dhandhukia J, Weitzhandler I, Wang W, MacKay JA. Switchable elastin-like polypeptides that respond to chemical inducers of dimerization. Biomacromolecules. 2013;14:976–985. doi: 10.1021/bm301558q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ciccone L, Tepshi L, Nencetti S, Stura EA. Transthyretin complexes with curcumin and bromo-estradiol: evaluation of solubilizing multicomponent mixtures. New biotechnology. 2015;32:54–64. doi: 10.1016/j.nbt.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 60.Kabsch W. XDS. Acta crystallographica. Section D, Biological crystallography. 2010;66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Legrand, P. Xdsme: Xds made easier. https://github.com/legrandp/xdsme (2010).
- 62.McCoy AJ, et al. Phaser crystallographic software. Journal of applied crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta crystallographica. Section D, Biological crystallography. 2011;67:355–67. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica. Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biological crystallography. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thorn A, Sheldrick GM. Anode: anomalous and heavy-atom density calculation. Journal of applied crystallography. 2011;44:1285–1287. doi: 10.1107/S0021889811041768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.DeLano, W. L. The PyMOL Molecular Graphics System (Schrödinger, L., New York Editor, 2010).
- 68.Ciccone L, et al. Multicomponent mixtures for cryoprotection and ligand solubilization. Biotechnology Reports. 2015;7:120–127. doi: 10.1016/j.btre.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vera L, Stura EA. Strategies for Protein Cryocrystallography. Crystal Growth & Design. 2014;14:427–435. doi: 10.1021/cg301531f. [DOI] [Google Scholar]
- 70.Newman J. Novel buffer systems for macromolecular crystallization. Acta crystallographica. Section D, Biological crystallography. 2004;60:610–2. doi: 10.1107/S0907444903029640. [DOI] [PubMed] [Google Scholar]
- 71.Diederichs K, Karplus PA. Better models by discarding data? Acta crystallographica. Section D, Biological crystallography. 2013;69:1215–22. doi: 10.1107/S0907444913001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.