Abstract
N-glycosylation is a post-translational modification that occurs in all three domains. In Archaea, however, N-linked glycans present a degree of compositional diversity not observed in either Eukarya or Bacteria. As such, it is surprising that nonulosonic acids (NulOs), nine-carbon sugars that include sialic acids, pseudaminic acids, and legionaminic acids, are routinely detected as components of protein-linked glycans in Eukarya and Bacteria but not in Archaea. In the following, we report that the N-linked glycan attached to the S-layer glycoprotein of the haloarchaea Halorubrum sp. PV6 includes an N-formylated legionaminic acid. Analysis of the Halorubrum sp. PV6 genome led to the identification of sequences predicted to comprise the legionaminic acid biosynthesis pathway. The transcription of pathway genes was confirmed, as was the co-transcription of several of these genes. In addition, the activities of LegI, which catalyzes the condensation of 2,4-di-N-acetyl-6-deoxymannose and phosphoenolpyruvate to generate legionaminic acid, and LegF, which catalyzes the addition of cytidine monophosphate (CMP) to legionaminic acid, both heterologously expressed in Haloferax volcanii, were demonstrated. Further genome analysis predicts that the genes encoding enzymes of the legionaminic acid biosynthetic pathway are clustered together with sequences seemingly encoding components of the N-glycosylation pathway in this organism. In defining the first example of a legionaminic acid biosynthesis pathway in Archaea, the findings reported here expand our insight into archaeal N-glycosylation, an almost universal post-translational modification in this domain of life.
Keywords: archaea, halophile, Halorubrum, legionaminic acid, N-formylation, N-glycosylation, S-layer glycoprotein
Introduction
Post-translational modifications represent a major source of proteomic expansion. Of the numerous processing events that can modify a protein, N-glycosylation, or the covalent attachment of a glycan to selected Asn residues of a target protein, is the most complex. Long held to be specific to Eukarya, it is now clear that Archaea and Bacteria also perform this post-translational modification. However, while N-glycosylation in Bacteria is believed to solely occur in the delta/epsilon proteobacteria (Nothaft and Szymanski, 2010), such protein processing appears to be an almost universal trait in Archaea (Kaminski et al., 2013a). As insight into N-glycosylation across evolution accumulates, unique aspects of this post-translational modification in each domain are being defined (Jones et al., 2009; Maita et al., 2010; Larkin and Imperiali, 2011; Koomey and Eichler, 2017; Eichler and Imperiali, 2018). For instance, domain-related diversity in the sugar composition of N-linked glycans serves to distinguish eukaryal, bacterial, and archaeal N-glycosylation (Schwarz and Aebi, 2011; Eichler, 2013).
In Eukarya, N-glycosylation begins in the endoplasmic reticulum (ER) and is completed in the Golgi. Whereas glycoproteins leaving the ER bear a common N-linked glycan core (Aebi, 2013), considerable diversity is introduced at the level of the Golgi, largely through the addition of any number of a variety of sialic acids (Angata and Varki, 2002; Chen and Varki, 2010; Cohen and Varki, 2010; Deng et al., 2013). Sialic acids correspond to a large group (>50 members) of structurally distinct molecules that are part of a larger group of nine-carbon sugars termed nonulosonic acid (NulO) sugars (Varki et al., 2017) and which are important for a variety of eukaryal process, including development, recognition, and immune responses (Varki and Gagneux, 2012; Schnaar et al., 2014; Stencel-Baerenwald et al., 2014; Pearce and Läubli, 2016). Although less common, some Bacteria also present sialic acids, either synthesized de novo or scavenged from other cells, as components of cell-surface structures (Vimr et al., 2004; Ferrero and Aparicio, 2010; Vimr, 2013). Indeed, genomic analysis predicts sialic acid biosynthetic pathways in a variety of bacterial groups (Lewis et al., 2009). Bacteria can also synthesize saccharides assigned to two other NulO sugar families, pseudaminic acids, and legionaminic acids, for use in protein glycosylation (Knirel et al., 2003; Vimr et al., 2004; Schoenhofen et al., 2006; McNally et al., 2007; Glaze et al., 2008; Lewis et al., 2009; Morrison and Imperiali, 2014). First identified in the O-polysaccharide of lipopolysaccharide in Legionella pneumophila, the cause of Legionnaires’ disease (Knirel et al., 1994), legionaminic acid (5,7-diacetamido-3,5,7,9-tetradeoxy-D-glycero-D-galacto-NulO) is thought to serve as a molecular mimic of sialic acid, allowing the pathogenic bacteria that synthesize this sugar to evade detection by the immune system of infected eukaryal hosts (Schoenhofen et al., 2009).
Given how the N-linked glycans decorating archaeal glycoproteins present a degree of diversity in sugar composition not found elsewhere (Schwarz and Aebi, 2011; Eichler, 2013; Jarrell et al., 2014), it is surprising that so few Archaea encode enzymes predicted to participate in NulO sugar biogenesis pathways. In surveying 122 archaeal genomes, putative components of sialic acid, pseudaminic acid, or legionaminic acid biosynthesis pathways were only predicted in less than 20% of the cases considered (Kandiba and Eichler, 2013). Indeed, genes putatively encoding complete pathways were only detected in six species. Even less evidence has been offered for any NulO as being associated with an archaeal glycoprotein. Pseudaminic acid was detected in an extract of Methanobrevibacter smithii (Lewis et al., 2009), a methanogen found in the human gut microbiome (Miller et al., 1982), correcting an earlier report of sialic acid being present in this organism (Samuel et al., 2007). This study did not, however, determine whether the detected pseudaminic acid originated from an archaeal protein-linked glycan or if it was somehow linked to protein glycosylation. Later (Kandiba et al., 2012), 5-N-formyl-legionaminic acid, a legionaminic acid derivative, was shown to be the final sugar of a pentasaccharide N-linked to VP4, a glycoprotein component of Halorubrum pleomorphic virus 1 (HRPV-1), a haloarchaeal virus that infects Halorubrum sp. PV6 (Pietilä et al., 2009, 2010). As HRPV-1 does not encode components of a pathway for legionaminic acid biosynthesis, it was predicted that the sugar was in fact synthesized by a pathway found in the archaeal host strain and added to the VP4 protein during viral particle assembly (Kandiba et al., 2012).
In the present report, a set of bioinformatics, mass spectrometry (MS), and genetic and biochemical experiments provide evidence for a legionaminic acid being a component of an N-linked glycan derived from the Halorubrum sp. PV6 S-layer glycoprotein and that Halorubrum sp. PV6 encodes a legionaminic acid biosynthesis pathway. Moreover, the genes encoding the components of this pathway are seemingly part of a larger N-glycosylation gene cluster in this organism.
Materials and Methods
Strains and Growth Conditions
Halorubrum sp. PV6 cells were grown in MGM-broth containing 23% artificial salt water (3.1 M NaCl, 113 mM MgCl2⋅6H2O, 109 mM MgSO4.7H2O, 72 mM KCl, 3.8 mM CaCl2, 61 mM Tris–HCl, pH 7.2), 0.5% (w/v), oxoid peptone, and 0.1% (w/v) yeast extract at 37°C. Haloferax volcanii WR536 (H53) strain cells were grown in complete medium containing 3.4 M NaCl, 0.15 M MgSO4.7H2O, 1 mM MnCl2, 4 mM KCl, 3 mM CaCl2, 0.3% (w/v) yeast extract, 0.5% (w/v) tryptone, 50 mM Tris–HCl, and pH 7.2 (Mevarech and Werczberger, 1985) at 42°C.
Bioinformatics
The Halorubrum sp. PV6 genome sequence was determined (accession number CP030064, Roine, Bamford, and Holm, manuscript in preparation), open reading frames (ORFs) were predicted using Prodigal (Hyatt et al., 2010) and RAST (Aziz et al., 2008; Overbeek et al., 2014) and automatically annotated using RAST and PANNZER 2 (Koskinen et al., 2015). For further annotation, HHpred (Söding et al., 2005) and Phyre2 (Kelley et al., 2015) were employed. The genomic region spanning the N-glycosylation gene cluster has been submitted to the GenBank nucleotide database (accession number MH673034).
Various other bioinformatics tools were used to describe the Halorubrum sp. PV6 S-layer glycoprotein. The signal peptide of the protein was predicted using SignalP 4.1 set for Gram-positive bacteria1. Alignment of the Halorubrum sp. PV6 and Hfx. volcanii S-layer glycoproteins was performed using ClustalW2, with shading being added using the Boxshade tool at the same site. The extent of homology between two proteins was determined using the BL2SEQ tool also found at this site.
Cell Wall Preparation
To enrich the S-layer glycoprotein, stationary phase Halorubrum sp. PV6 cells were collected, washed, and resuspended in 3.4 M NaCl and 27 mM KCl and incubated with shaking at room temperature for 15 min. After centrifugation (Sorvall SA-300, 9,000 rpm, 15 min, 4°C), the supernatant was collected, MgSO4 was added to a final concentration of 160 mM, and incubated at 4°C overnight. S-layer glycoprotein was harvested by pelleting (Sorvall T-1270, 41,000 rpm, 20 min, 4°C). The pellet was resuspended in protein sample buffer, subjected to 12% SDS-PAGE and stained with Coomassie R-250 (Fluka).
Liquid Chromatography–Electrospray Ionization Mass Spectrometry (LC–ESI MS)
For LC-ESI MS analysis of the Halorubrum sp., PV6 S-layer glycoprotein was conducted essentially as reported previously (Calo et al., 2011). In-gel digestion of the S-layer glycoprotein was first conducted. The S-layer glycoprotein-containing band was excised from the gel using a clean scalpel, destained in 400 μl of 50% (vol/vol) acetonitrile (Sigma) in 40 mM NH4HCO3, pH 8.4, dehydrated with 100% acetonitrile, and dried using a SpeedVac drying apparatus. The S-layer glycoprotein was reduced with 10 mM dithiothreitol (Sigma) in 40 mM NH4HCO3 at 56°C for 60 min and then alkylated for 45 min at room temperature with 55 mM iodoacetamide in 40 mM NH4HCO3. The gel pieces were washed with 40 mM NH4HCO3 for 15 min, dehydrated with 100% acetonitrile, and SpeedVac-dried. The gel slices were rehydrated with 12.5 ng/μl of mass spectrometry (MS) grade Trypsin Gold (Promega) in 40 mM NH4HCO3 and incubated overnight at 37°C. The protease-generated peptides were extracted with 0.1% (v/v) formic acid in 20 mM NH4HCO3, followed by sonication for 20 min at room temperature, dehydration with 50% (v/v) acetonitrile, and additional sonication. After three rounds of extraction, the gel pieces were dehydrated with 100% acetonitrile and dried completely with a SpeedVac. Next, 12.5 ng/μl Glu-C (V8) protease (Promega, sequencing-grade) in 40 mM NH4HCO3 were added. After an overnight incubation at 37°C, the sample was dried completely with a SpeedVac, resuspended in 5% (v/v) acetonitrile containing 1% formic acid (v/v) and infused into the mass spectrometer using static nanospray Econotips (New Objective, Woburn, MA, United States). The protein digests were separated online by nano-flow reverse-phase liquid chromatography (LC) by loading onto a 150-mm by 75-μm (internal diameter) by 365-μm (external diameter) Jupifer pre-packed fused silica 5-μm C18 300Å reverse-phase column (Thermo Fisher Scientific, Bremen, Germany). The sample was eluted into the LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific) using a 60-min linear gradient of 0.1% formic acid (v/v) in acetonitrile/0.1% formic acid (1:19, by volume) to 0.1% formic acid in acetonitrile/0.1% formic acid (4:1, by volume) at a flow rate of 300 nl/min.
Reverse Transcriptase Polymer Chain Reaction (RT-PCR)
To assess the transcription and co-transcription of ORFs contributing to a putative legionaminic acid biosynthesis pathway, RT-PCR was performed. Specific forward and reverse oligonucleotide primers were designed to amplify each Halorubrum sp. PV6 ORF under consideration, as well as stretches beginning in a given sequence and ending in the downstream sequence (for primer sequences, see Supplementary Tables S1 and S2). RNA isolation was carried out using an RNeasy mini-kit (Qiagen) according to the manufacturer’s instructions. Contaminating DNA in the RNA samples was eliminated with a RNase-Free DNase Set (Qiagen) during RNA extraction. RNA concentration was determined spectrophotometrically. Single-stranded cDNA was prepared from the extracted RNA using an Oligo(dT) primer in a Superscript IV 1st Strand System (Invitrogen). Together with appropriate forward and reverse primer pairs, the cDNA generated was used for PCR amplification. cDNA amplification was monitored by electrophoresis in 1% agarose gels. To exclude possible contributions from contaminating DNA, control experiments were conducted in which PCR amplification was performed on total RNA prior to cDNA preparation.
Expression and Purification of Halorubrum sp. PV6 LegI and LegF
To generate plasmids pWL-CBD-LegI and pWL-CBD-LegF encoding Clostridium thermocellum cellulose-binding domain (CBD; Morag et al., 1995)-tagged Halorubrum sp. PV6 legionaminic acid synthase (LegI; Schoenhofen et al., 2009) and CMP-legionaminic acid synthase (LegF; Schoenhofen et al., 2009), respectively, HrrPV6_1053 (encoding LegI) and HrrPV6_1048 (encoding LegF) were PCR-amplified from Halorubrum sp. PV6 genomic DNA using primers designed to introduce NdeI and KpnI restriction sites at the 5′ and 3′ ends of the LegI- and LegF-encoding sequences, respectively, as well as 25 bp-long overlaps with plasmid pWL-CBD (Irihimovitch and Eichler, 2003) at each end of the amplified fragments (legI forward primer: CGGTGGCAGTGTAGTAGGAGGTCATATGGAGATTGATGGTACCCG; legI reverse primer: GGCCAGCGGGAGATCCCCGGGTACCTTAATCCTCACAACCAGTACC; legF forward primer: CGGTGGCAGTGTAGTAGGAGGTCATATGTCCACCCGCACATTAGC; legF reverse primer: GGCCAGCGGGAGATCCCCGGGTACCTTAATCCTCACAACCAGTACC). The PCR products were ligated into plasmid pWL-CBD, previously digested with NdeI and KpnI, using the Gibson Assembly Kit (New England BioLabs), according to the manufacturer’s instructions. The assembled plasmids were introduced into Escherichia coli Cloni cells (Lucigen) and then into Hfx. volcanii cells.
Cellulose-binding domain-tagged LegI and LegF were purified as previously described (Irihimovitch and Eichler, 2003). Briefly, 500 ml of Hfx. volcanii cells transformed to express the CBD-tagged LegI or LegF were grown to mid-logarithmic phase, harvested, and resuspended in 60 ml solubilization buffer (1% Triton X-100, 2 M NaCl, 50 mM Tris–HCl, pH 7.2) containing 400 U benzonase nuclease (Novagen) and 0.5 mM PMSF. The solubilized mixture was nutated for 20 min at room temperature and subjected to sonication (2 s on and 1 s off for 90 s, 70% output, Misonix XL2020 ultrasonicator) on ice and centrifugation (10,500 g, 30 min, 4°C). Cellulose (400 μl of a 10% (w/v) solution; Sigma) was added to 10 ml aliquots of the supernatant. After a 1-h nutation at 4°C, the suspensions were centrifuged (2,650 g, 2 min, 4°C), the supernatants were discarded and the cellulose pellets were washed four times with wash buffer containing 2 M NaCl, 50 mM Tris–HCl, pH 7.2. After the final wash, the cellulose beads were centrifuged (2,650 g, 2 min, 4°C), the supernatants were removed and the pellets, containing cellulose beads linked to the CBD-tagged proteins, were resuspended in SDS-PAGE sample buffer, boiled for 5 min and centrifuged (2,000 rpm, 2 min). The solubilized proteins were separated by 12% SDS-PAGE and visualized by InstantBlue (Expedeon) staining. Alternatively, 2 ml of the washed cellulose beads containing the bound CBD-tagged proteins were used for in vitro assaying of LegI and LegF function.
Assaying Halorubrum sp. PV6 LegI Activity
Halorubrum sp. PV6 LegI activity was assessed by confirming the ability of the enzyme to catalyze the same reaction as does NeuB, its homolog in sialic acid biosynthesis (Vann et al., 1997; Glaze et al., 2008; Schoenhofen et al., 2009), as revealed by the thiobarbituric acid assay (Warren, 1959), given that the LegI substrate 2,4-diNac-6-deoxy mannose, is not commercially available. Briefly, reactions (50 μl) containing 12.5 mM N-acetylmannosamine (Sigma), 12.5 mM phosphoenolpyruvate (Sigma), 10 mM MnCl2, 2 M NaCl, 100 mM Tris–HCl, pH 7.2, and cellulose-bound enzyme were incubated at 37°C for 0–4 h, with aliquots being removed once an hour. The reactions were terminated by adding sodium periodate (137 μL of a 2.5 mg/ml stock solution prepared in 57 mM H2SO4; Sigma) and incubating at 37°C for 15 min. Sodium arsenite (50 μL of a 25 mg/ml stock solution prepared in 0.5 M HCl; Sigma) was added and the reactions were shaken until the brown color that developed disappeared. Next, 2-thiobarbituric acid (100 μL of a 71 mg/mL stock solution, pH 9.0 with NaOH; Alfa Aesor) was added and the mixtures were incubated at 100°C for 10 min, transferred to ice for 5 min and then held at room temperature for an additional 5 min. One milliliter of acidic butanol (n-butanol with 5% HCl) was added to each reaction. After shaking for 10 min, the reactions were centrifuged at 13,000 rpm for 10 min at room temperature to separate the organic and inorganic phases. The fluorescence of 100-μl aliqouts of the organic phase (containing the N-acetylneuraminic acid produced) was measured on a Tecan Infinite M200 plate reader (λem/ex = 585/555 nm). Fluorescence measurements were converted into N-acetylneuraminic acid concentrations using a calibration curve prepared with known amounts of this nonulosonic acid (Abnova).
Assaying Halorubrum sp. PV6 LegF Activity
Halorubrum sp. PV6 LegF activity was assessed by confirming the ability of the enzyme to catalyze the same reaction as does NeuA, its homolog in sialic acid biosynthesis (Vann et al., 1987; Haft and Wessels, 1994; Glaze et al., 2008; Schoenhofen et al., 2009), given that the LegF substrate legionaminic acid, is not commercially available. The assay was performed as described (Liu et al., 2004) with several modifications. Briefly, reactions (100 μl) containing 2.8 mM N-acetylneuraminic acid (Acros), 5.5 mM cytidine triphosphate (CTP; Alfa Aesor), 20 mM MgCl2, 1.7 M NaCl, 0.2 mM dithiothreitol (Sigma), 200 mM Tris–HCl, pH 9, and cellulose-bound enzyme were incubated at 37°C for 0–2 h, with aliquots being removed every 30 min. In each aliquot, unreacted N-acetylneuraminic acid was reduced with 20 μl of 1.6 M sodium borohydride (Sigma) at room temperature for 15 min. The reactions were placed on ice and 20 μl of 20 N phosphoric acid were added to decompose the sodium borohydride. After standing at 0°C for 5 min, the reactions were incubated at 37°C for 10 min to cleave the phosphoester bond of the CMP-N-acetylneuraminic acid formed. Free N-acetylneuraminic acid was oxidized with 50 μl of 0.2 M sodium periodate (Sigma) at room temperature for 15 min, at which point 160 μl of 4% sodium arsenite (Sigma) in 0.5 N hydrochloric acid were added. Thiobarbituric acid (400 μl of a 0.6% solution; Alfa Aesor) in 0.5 M sodium sulfate (Acros) were added and the samples were heated in boiling water for 15 min. After the solution had cooled, 400 μl of cyclohexanone were added, and the mixture was shaken and centrifuged at 13,000 rpm for 10 min at room temperature to separate the organic and inorganic phases. The fluorescence of 100-μl aliqouts of the organic phase (containing the N-acetylneuraminic acid produced) was measured on a Tecan Infinite M200 plate reader (λem/ex = 585/555 nm). Fluorescence measurements were converted into N-acetylneuraminic acid concentrations using a calibration curve prepared with known amounts of this nonulosonic acid (Abnova).
Results
The Halorubrum sp. PV6 S-Layer Glycoprotein Is N-Glycosylated by a Legionaminic Acid-Containing Pentasaccharide
In earlier work, we reported that VP4, the major structural protein of the haloarchaeal pleomorphic virus HRPV-1, is N-glycosylated by a pentasaccharide comprising glucose, glucuronic acid, mannose, sulphated glucuronic acid, and a terminal 5-N-formyl-legionaminic acid residue, when directly isolated from virions produced by the host strain, Halorubrum sp. PV6 (Kandiba et al., 2012). The present study began with efforts to determine whether native glycoproteins of Halorubrum sp. PV6 are also N-glycosylated by the same glycan.
The S-layer glycoprotein has served as a useful reporter of archaeal N-glycosylation (Jarrell et al., 2010). Accordingly, N-glycosylation of the predicted Halorubrum sp. PV6 S-layer glycoprotein was considered here. In Halorubrum sp. PV6, the predicted S-layer glycoprotein (HrrPV6_1002) corresponds to an 805 residue-long protein, preceded by an apparent 31 residue-long signal peptide. Alignment of the amino acid sequence of the predicted Halorubrum sp. PV6 S-layer glycoprotein with its Hfx. volcanii counterpart (HVO_2072) revealed that the two share 40% identity and 59% similarity (E value = 4e-169; score = 579 bits over the entire sequence length; Supplementary Figure 1). Both proteins present seven putative N-glycosylation sites, namely Asn-94, Asn-138, Asn-163, Asn-267, Asn-273, Asn-315 and Asn-357 in the Halorubrum sp. PV6 protein and Asn-13, Asn-83, Asn-274, Asn-279, Asn-370, Asn-498, and Asn-732 in the Hfx. volcanii protein. In the latter, several of these sites have been experimentally confirmed as being modified (Mengele and Sumper, 1992; Abu-Qarn et al., 2007; Kaminski et al., 2013b; Parente et al., 2014; Kandiba et al., 2016). The alignment further revealed that several of these putative or known N-glycosylation sites coincide or are found in close proximity to each other in the two proteins. Both S-layer glycoproteins also present a string of O-glycosylation targets near their C-terminus. In the Hfx. volcanii protein, an unspecified number of these Thr residues are decorated with a disaccharide comprising glucose and galactose (Sumper et al., 1990). Finally, both proteins share a C-terminal region that includes a transmembrane domain and a motif putatively recognized by ArtA, the archaeal archaeosortase involved in replacing this domain with a lipid anchor (Abdul Halim et al., 2013; Kandiba et al., 2013).
To determine whether the predicted Halorubrum sp. PV6 S-layer glycoprotein is modified by the same N-linked glycan as is the virus HRPV-1 VP4 glycoprotein when synthesized in Halorubrum sp. PV6 (Kandiba et al., 2012), proteolytic fragments of the predicted S-layer glycoprotein were examined by LC-ESI MS. Such analysis revealed a peak of m/z 1168.58 (Figure 1A), corresponding to the [M+2H]2+ ion of the predicted S-layer glycoprotein-derived peptide 78TGSYAIGGPDAADGAFNVTVVTPR101 (calculated mass, m/z 1168.58), containing the putative N-glycosylation site Asn-94. Peaks of m/z 1249.61, 1337.62, 1418.65, 1546.65, and 1697.70 were also detected, consistent with calculated masses of the same Asn-94-containing peptide modified by a hexose (calculated mass m/z 1249.58; Figure 1B); a hexose and a hexuronic acid (calculated mass m/z 1337.58; Figure 1C); a hexose, a hexuronic acid, and a hexose (m/z 1418.58; Figure 1D); a hexose, a hexuronic acid, a hexose, and a sulphated/phosphorylated hexuronic acid (m/z 1546.58; Figure 1E); and a hexose, a hexuronic acid, a hexose, a sulphated/phosphorylated hexuronic acid, and 5-N-formyl-legionaminic acid (m/z 1697.58; Figure 1F). Glycosylation of Asn-94 was verified by MS/MS analysis of the [M+2H]2+ base peak of the same predicted S-layer glycoprotein-derived peptide modified by a hexose observed at m/z 1249.61. The product ion spectrum contains a series of y-ion fragments that confirmed the presence of a hexose attached at the Asn-94 position (Figure 2).
FIGURE 1.
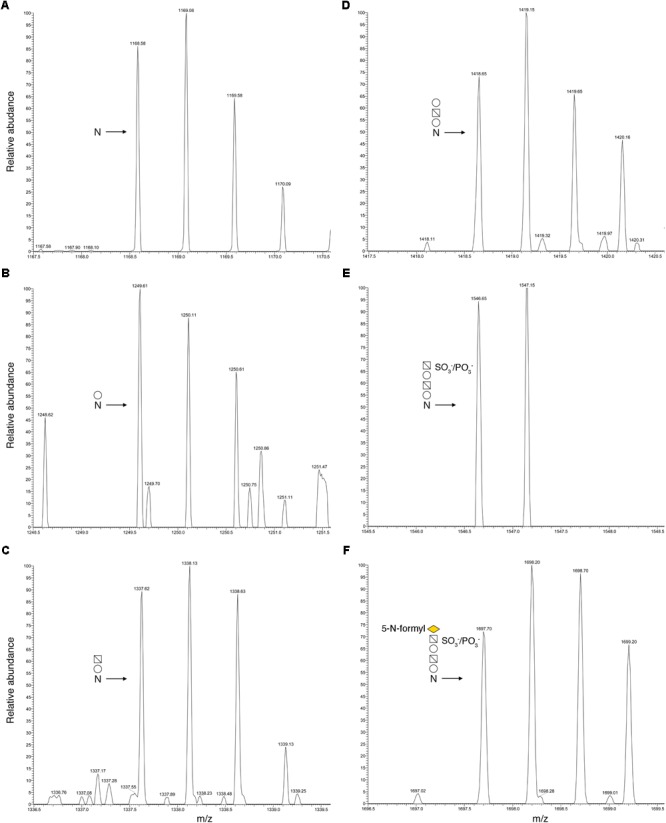
N-glycosylation profile of Halorubrum sp. PV6 S-layer glycoprotein Asn-94. LC-ESI/MS analysis of the Asn-94-containing peptide derived from the Halorubrum sp. PV6 S-layer glycoprotein following digestion with trypsin and GluC protease was performed. Shown are doubly charged [M+2H]2+ ion peaks corresponding to (A) the 78TGSYAIGGPDAADGAFNVTVVTPR101 peptide (m/z 1168.58), and the same peptide successively modified by (B) a hexose (m/z 1249.61), (C) a hexuronic acid (m/z 1337.62), (D) a hexose (m/z 1418.65), (E) a sulfated/phosphorylated hexuronic acid (m/z 1546.65), and (F) a 5-N-formyl-legionaminic acid (m/z 1697.70). In each panel, the N-glycosylation status of the peptide is schematically depicted, where “N” corresponds to Asn-94. Employing symbol nomenclature for glycans guidelines (Varki et al., 2015), open circles correspond to hexoses, open squares containing a diagonal correspond to hexuronic acids, and the yellow diamond corresponds to 5-N-formyl-legionaminic acid.
FIGURE 2.
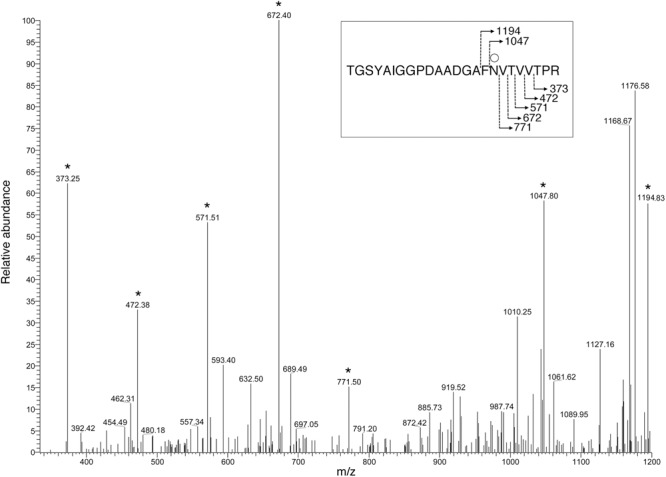
MS/MS verification of Halorubrum sp. PV6 S-layer glycoprotein Asn-94 glycosylation. To verify that glycosylation of the hexose-modified S-layer glycoprotein-derived peptide, 78TGSYAIGGPDAADGAFNVTVVTPR101, occurs on Asn-94, the MS spectrum of the [M+2H]2+ base peak of this peptide, observed at m/z 1249.61, was obtained via mass-dependent acquisition. The y-ion series includes fragments that contain the hexose-modified Asn-94 residue. The inset shows the fragmentation scheme, with the Asn94-bound hexose being represented by the open circle. Peaks corresponding to y-ion series fragments are indicated by stars.
Based on these MS results, combined with those of earlier analyses (Kandiba et al., 2012), it can be concluded that the predicted Halorubrum sp. PV6 S-layer glycoprotein is indeed modified by the same glycan as is N-linked to the viral VP4 glycoprotein expressed in this strain. As such, this represents the first example of a NulO sugar, and in particular, a legionaminic acid, as a component of an N-linked glycan decorating a native archaeal glycoprotein.
Halorubrum sp. PV6 Encodes Components of a Legionaminic Acid Biosynthesis Pathway
As the predicted Halorubrum sp. PV6 S-layer glycoprotein is seemingly modified by an N-linked glycan that includes 5-N-formyl-legionaminic acid, the genomic sequence of this organism (GenBank accession number CP030064; Roine, Bamford, and Holm, manuscript in preparation) was scanned for ORFs corresponding to genes encoding components of a legionaminic acid biosynthetic pathway (Figure 3). Accordingly, the components of the two known pathways of legionaminic acid biosynthesis in Bacteria, namely, that of L. pneumophila, where UDP-conjugated intermediates are generated (Glaze et al., 2008), and that of Campylobacter jejuni, where GDP-conjugated intermediates are generated (Schoenhofen et al., 2009), served as queries in BLAST searches of the Halorubrum sp. PV6 genome. Such analysis identified sequences (see Supplementary Material) putatively encoding components of a Halorubrum sp. PV6 legionaminic acid biosynthetic pathway comprising NDP-N-acetylglucosamine (GlcNAc)-4,6-dehydratase (LegB; HrrPV6_1047), NDP-GlcNAc aminotransferase (LegC; HrrPV6_1046), NDP-GlcNAc N-acetyltransferase (LegH; HrrPV6_1014), NDP-GlcNAc-hydrolase/2-epimerase [LegG (a NeuC homolog); HrrPV6_1049], legionaminic acid synthase [LegI (a NeuB homolog); HrrPV6_1053], and CMP-legionaminic acid synthase [LegF (a NeuA homolog); HrrPV6_1048]. A schematic depiction of the region of the Halorubrum sp. PV6 containing these genes is presented in Figure 4, while information on the ORFs predicted to catalyze steps of the putative Halorubrum sp. PV6 legionaminic acid biosynthesis pathway is provided in Table 1.
FIGURE 3.
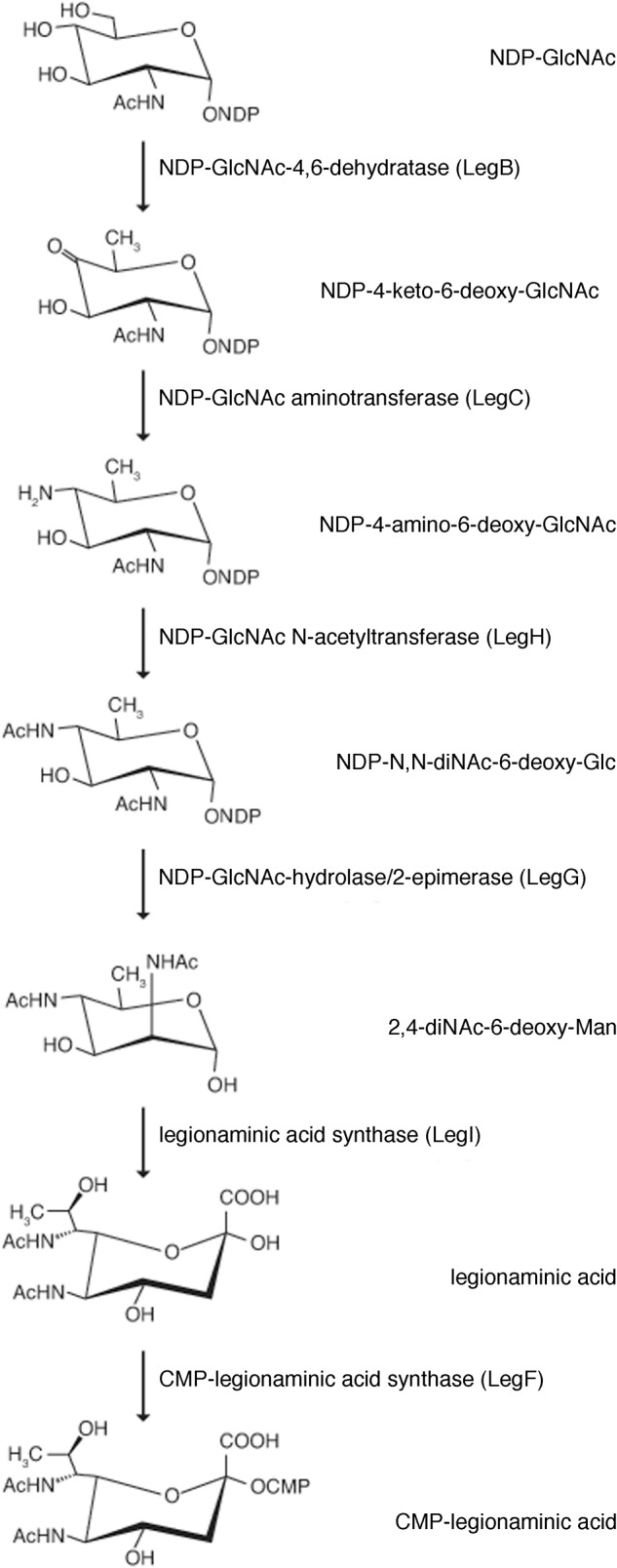
The predicted pathway of Halorubrum sp. PV6 legionaminic acid biosynthesis. The pathway (Glaze et al., 2008; Schoenhofen et al., 2009) starts with the dehydratase LegB that generates NDP-4-keto-6-deoxy-N-acetylglucosamine (GlcNAc) from NDP-GlcNAc. The aminotransferase LegC next produces the amino sugar NDP-4-amino-6-deoxy-GlcNAc. The acetyltransferase LegH subsequently generates NDP-N,N-diacetamido-6-deoxy-glucose (NDP-diacetamido-basillosamine), which is then converted into 2,4-diacetamido-6-deoxy-mannose by the hydrolyzing 2-epimerase LegG. The legionaminic synthase LegI now condenses this sugar with pyruvate to yield legionaminic acid. Finally, the sugar is activated by the actions of LegF, the CMP-legionaminic acid synthase. In the diagram, the enzymes that catalyze each pathway step, as well as the products formed, are listed.
FIGURE 4.
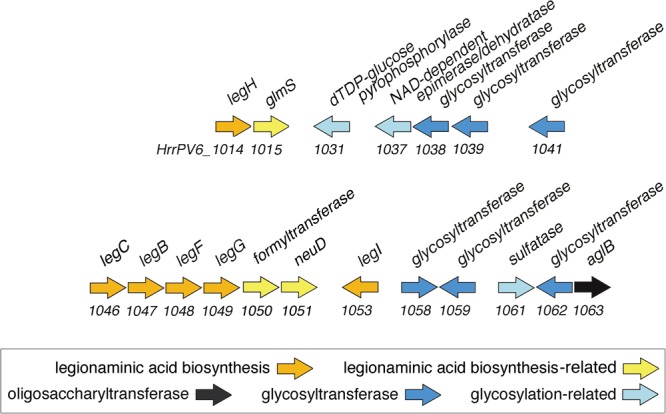
Schematic depiction of the region of the Halorubrum sp. PV6 genome containing ORFs encoding putative components involved in 5-N-formyl-legionaminic acid biosynthesis and N-glycosylation. The scheme presents the orientation but not the actual length of each ORF. Sequence information can be found at GenBank accession number MH673034.
Table 1.
Sequences putatively comprising the Halorubrum sp. PV6 legionaminic acid biosynthesis pathway.
| Sequence | Current annotation | Revised annotation |
|---|---|---|
| HrrPV6_1014 | Nucleotidyltransferase | LegH (NDP-GlcNAc N-acetyltransferase) |
| HrrPV6_1046 | PLP-dependent aminotransferase | LegC (NDP-GlcNAc aminotransferase) |
| HrrPV6_1047 | Nucleotide sugar epimerase/dehydratase | LegB (NDP-GlcNAc-4,6-dehydratase) |
| HrrPV6_1048 | N-acetylneuraminate cytidylyltransferase (NeuA) | LegF (CMP-legionaminic acid synthase) |
| HrrPV6_1049 | UDP-N-GlcNAc- 2-epimerase (NeuC) | LegG (NDP-GlcNAc-hydrolase/2-epimerase) |
| HrrPV6_1053 | N-acetylneuraminate synthase (NeuB) | LegI (legionaminic acid synthase) |
This region of the genome also includes additional sequences encoding enzymes possibly related to 5-N-formyl legionaminic acid biosynthesis, such as HrrPV6_1051, encoding a homolog of the O-acetyltransferase NeuD involved in sialic acid biogenesis in E. coli (Daines et al., 2000). HrrPV6_1050 possibly encodes the N-formyltransferase involved in modifying legionaminic acid. Finally, HrrPV6_1015 is predicted to encode the glucosamine-6-phosphate synthase GlmS.
To determine whether the Leg pathway is widely distributed in the genus Halorubrum, the different Leg protein sequences identified here were used as queries in a series of BLAST searches of completed genome sequences of other Halorubrum species. This revealed the presence of the complete set of Leg proteins in only two species, namely, Halorubrum ezzemoulense and Halorubrum sodomense. This suggests that legionaminic acid is not widely produced in this genus, in agreement with earlier efforts showing the limited distribution of NulO biosynthesis pathway genes in Archaea (Kandiba and Eichler, 2013). Future efforts will, however, be needed to confirm that Hrr. ezzemoulense and Hrr. sodomense indeed synthesize and employ legionaminic acids in N-glycosylation.
Several Genes Involved in Legionaminic Acid Biosynthesis Are Co-transcribed
Because transcription of an ORF is a good indicator of that sequence being a true protein-encoding gene, the generation of RNA from the sequences corresponding to legH (HrrPV6_1014), legC (HrrPV6_1046), legB (HrrPV6_1047), legF (HrrPV6_1048), legG (HrrPV6_1049), a predicted formyltransferase-encoding gene (HrrPV6_1050), neuD (HrrPV6_1051), and legI (HrrPV6_1053) was considered by RT-PCR. Accordingly, cDNA prepared from RNA isolated from exponentially growing Halorubrum sp. PV6 cells using reverse transcriptase served as template in a series of PCR amplifications involving primers designed to generate PCR products corresponding to each ORF. In each case, products of the expected lengths (HrrPV6_1014: 1,368 bp; HrrPV6_1046: 1,139 bp; HrrPV6_1047: 1,025 bp; HrrPV6_1048: 722 bp; HrrPV6_1049: 1,163 bp; HrrPV6_1050: 953 bp; HrrPV6_1051: 629 bp; and HrrPV6_1053: 1,053 bp) were obtained (Figure 5A, upper panel). As negative controls, the same PCR amplifications were repeated, this time using DNase-treated RNA as template (i.e., without reverse transcriptase treatment). Now, no PCR products were obtained (Figure 5A, lower panel).
FIGURE 5.
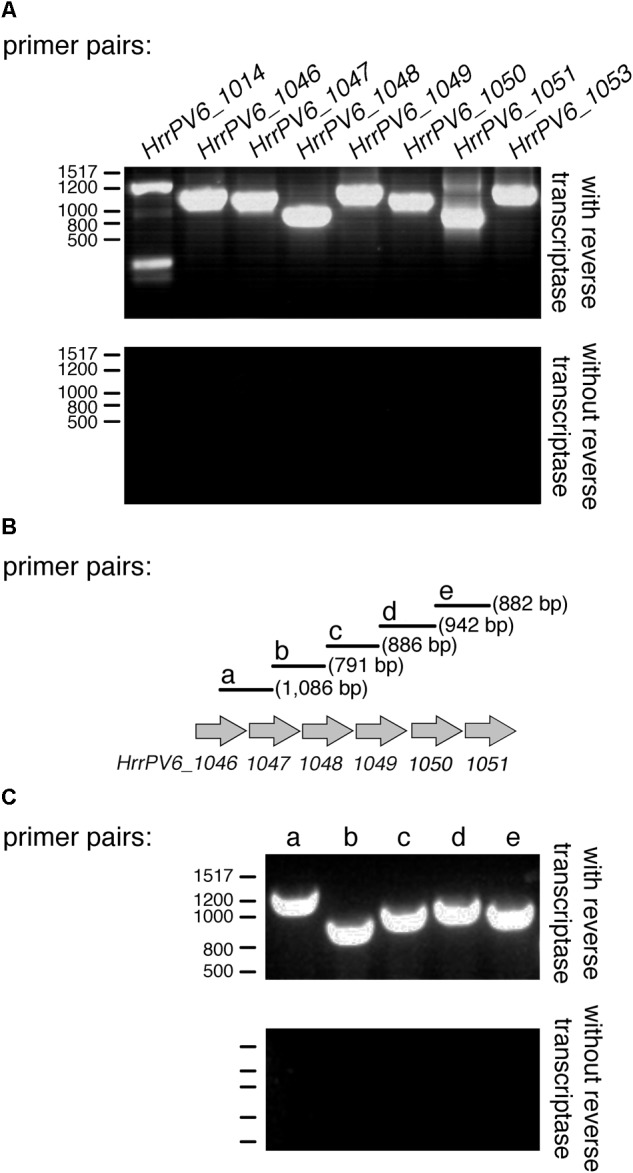
Genes putatively involved in legionaminic acid biosynthesis are co-transcribed. (A) RT-PCR products obtained using primer pairs designed to amplify the indicated ORF and cDNA (with reverse transcriptase; upper panel) or RNA pre-treated with DNase but not with reverse transcriptase (without reverse transcriptase; lower panel) as template. (B) Schematic depiction of the Halorubrum sp. PV6 genome containing genes encoding putative components involved in legionaminic acid biosynthesis suspected of being co-transcribed. Primer pairs beginning within the upstream sequence and ending within the downstream sequence are schematically depicted. Primer pairs a, b, c, d, and e are designed to amplify products of the listed sizes. (C) RT-PCR products obtained using each primer pair and cDNA (with reverse transcriptase; upper panel) or RNA pre-treated with DNase but not with reverse transcriptase (without reverse transcriptase; lower panel) as template. In (A,C), the positions of bp markers are provided on the left of each panel and values are listed on the left of the upper profile.
Examination of the region of the Halorubrum sp. PV6 genome that spans HrrPV6_1046 to HrrPV6_1051 revealed these genes to be not only adjacent but also to be similarly oriented. As such, the possibility that several or all of these genes are co-transcribed was next considered by RT-PCR in a series of amplifications involving primers designed to generate PCR products spanning regions within and between adjacent ORFs (Figure 5B). When PCR was performed with these primers pairs and the cDNA template, products of the expected sizes were obtained in each case (Figure 5C, upper panel). As negative controls, the same PCR amplifications were repeated, this time using extracted RNA pre-treated with DNase but not with reverse transcriptase as template. Now, no PCR products were obtained (Figure 5C, lower panel).
It can thus be concluded that HrrPV6_1046, HrrPV6_1047, HrrPV6_1048, HrrPV6_1049, HrrPV6_1050, HrrPV6_1051, and HrrPV6_1053 correspond to protein-encoding genes and, moreover, that HrrPV6_1046, HrrPV6_1047, HrrPV6_1048, HrrPV6_1049, HrrPV6_1050, and HrrPV6_1051 are co-transcribed.
Biochemical Characterization of Halorubrum sp. PV6 LegI and LegF
To verify the actions of proteins assigned to the predicted Halorubrum sp. PV6 legionaminic acid biosynthesis pathway, biochemical studies were undertaken. Hfx. volcanii cells were transformed to express the predicted legionaminic acid synthase (LegI) bearing an N-terminal C. thermocellum CBD tag. Hfx. volcanii was used as a heterologous expression platform in these experiments since no genetic tools presently exist for the manipulation of Halorubrum sp. PV6. The presence of this CBD tag allows for cellulose-based purification in the presence of molar concentrations of salt, as required for the proper folding and activity of haloarchaeal enzymes (Irihimovitch and Eichler, 2003). Cellulose-based purification of CBD-tagged LegI (59.5 kDa) and LegF (45.3 kDa) from a total cell lysate of Hfx. volcanii cells transformed to express each protein is presented in Figures 6A,B, respectively.
FIGURE 6.
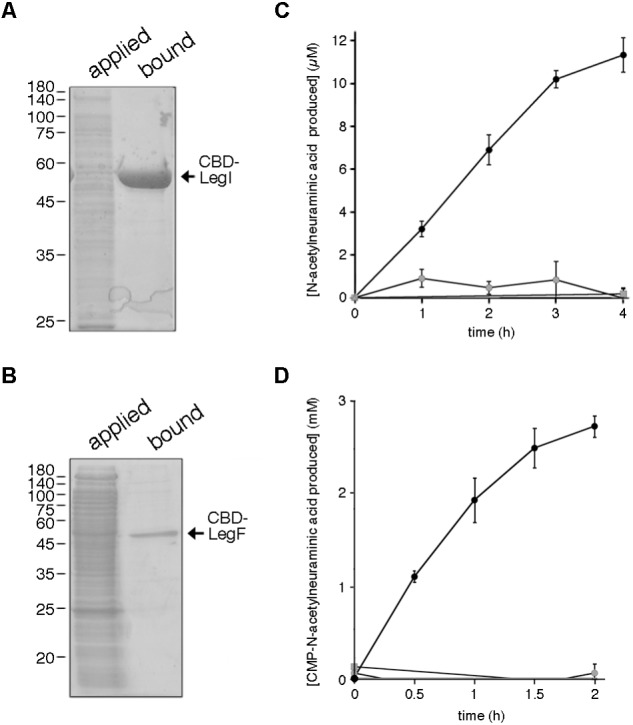
Halorubrum sp. PV6 LegI and LegF contribute to legionaminic acid biosynthesis. (A) Halorubrum sp. PV6 LegI and (B) LegF were purified from total lysates of Hfx. volcanii expressing each CBD-tagged protein (applied) on cellulose (bound). Aliquots of each pool were separated by 12% SDS-PAGE and Coomassie stained. In each panel, the positions of molecular weight markers are indicated on the left, while the position of the purified protein is indicated on the right. (C) LegI activity was confirmed in reactions containing cellulose-bound CBD-LegI, N-acetylmannosamine, and phosphoenolpyruvate (black circles) or in which the N-acetylmannosamine (gray circles) or phosphoenolpyruvate (gray squares) were omitted, or where cellulose incubated with the lysate of Hfx. volcanii not expressing the CBD-tagged protein was added instead of CBD-LegI-bearing beads (gray triangles). Each point is the average of three repeats ± SEM. (D) LegF activity was confirmed in reactions containing cellulose-bound CBD-LegF, N-acetylneuraminic acid and CTP (black circles) or in which the N-acetylneuraminic acid (gray circles) or CTP (gray squares) were omitted, or where cellulose incubated with the lysate of Hfx. volcanii not expressing the CBD-tagged protein was added instead of CBD-LegF-bearing beads (gray triangles). The amount of N-acetylneuraminic acid produced in each reaction was determined every 30 min over a 2 h interval, except in the latter two controls, when measurements were only taken at the start and the end of the experiment. Each point is the average of three repeats ± SEM.
In legionaminic acid biosynthesis, LegI catalyzes the condensation of 2,4-di-N-acetyl-6-deoxymannose and phosphoenolpyruvate to generate legionaminic acid (Glaze et al., 2008; Schoenhofen et al., 2009). As this is essentially the same reaction as catalyzed by the LegI homolog NeuB (sialic acid synthase) during sialic acid biosynthesis (Vann et al., 1997), the ability of cellulose-bound CBD-tagged Halorubrum sp. PV6 LegI to condense N-acetylmannosamine and phosphoenolpyruvate, yielding N-acetylneuraminic acid, was determined. As reflected in Figure 6C, incubation of cellulose-bound LegI with the two substrates led to a linear increase in sialic acid levels over the first 3 h, activity which began to taper off during the fourth hour of incubation. By contrast, no N-acetylneuraminic acid was generated when either N-acetylmannosamine or phosphoenolpyruvate were omitted from the reaction, or when cellulose beads pre-incubated with a lysate prepared from Hfx. volcanii cells not transformed to express CBD-tagged LegI were tested.
The conversion of legionaminic acid to CMP-legionaminic acid by Halorubrum sp. PV6 LegF (Glaze et al., 2008; Schoenhofen et al., 2009) was next considered by addressing the ability of the enzyme to catalyze the parallel NeuA-mediated reaction that occurs in sialic acid biogenesis (Vann et al., 1987). Accordingly, the ability of cellulose-bound CBD-tagged LegF to generate CMP-N-acetylneuraminic acid from N-acetylneuraminic acid and CTP was determined. Incubation of cellulose-bound LegF with the two substrates led to a constant increase in CMP-N-acetylneuraminic acid levels that appeared to approach saturation over the course of the experiment (Figure 6D). By contrast, no N-acetylneuraminic acid was generated when either N-acetylneuraminic acid or CTP were omitted from the reaction, or when cellulose beads pre-incubated with a lysate prepared from Hfx. volcanii cells not transformed to express CBD-tagged LegF were tested.
Putative Halorubrum sp. PV6 N-Glycosylation Pathway Components
In seeking ORFs putatively encoding components of a legionaminic acid biosynthesis pathway in Halorubrum sp. PV6, it was noted that the predicted product of HrrPV6_1063 is annotated as an oligosaccharyltransferase. Indeed, HrrPV6_1063 shares 46% identity and 59% similarity (87% coverage; score 757 bits) with Hfx. volcanii AglB, the confirmed oligosaccharyltransferase in this organism (Abu-Qarn et al., 2007). As such, additional putative N-glycosylation pathway component-encoding sequences were sought in the neighboring ORFs, given how in halophiles, as in certain other Archaea, genes encoding N-glycosylation pathway components tend to cluster around aglB (Magidovich and Eichler, 2009; Kaminski et al., 2013a; Kandiba and Eichler, 2015). In this manner, Halorubrum sp. PV6 ORFs predicted to encode glycosyltransferases and other enzymes putatively involved in N-linked glycan biosynthesis and assembly were identified in the immediate and near vicinity of the putative AglB encoding sequence (Table 2). It is reasonable to assume that these sequences, together with those sequences encoding the legionaminic acid biosynthesis pathway, comprise the bulk of the Halorubrum sp. PV6 N-glycosylation pathway (Figure 4). That portion of the Halorubrum sp. PV6 genome that contains genes encoding N-glycosylation pathway components, as well as genes encoding the legionaminic acid biosynthesis pathway, have been submitted to GenBank and assigned accession number MH673034.
Table 2.
Sequences putatively encoding components of a Halorubrum sp. PV6 N-glycosylation pathway.
| Sequence | Current annotation |
|---|---|
| HrrPV6_1031 | dTDP-glucose pyrophosphorylase |
| HrrPV6_1037 | NAD-dependent epimerase/hydratase |
| HrrPV6_1038 | Glycosyltransferase |
| HrrPV6_1039 | Glycosyltransferase |
| HrrPV6_1041 | Glycosyltransferase |
| HrrPV6_1058 | Glycosyltransferase |
| HrrPV6_1059 | Glycosyltransferase |
| HrrPV6_1061 | Sulfatase |
| HrrPV6_1062 | Glycosyltransferase |
| HrrPV6_1063 | Oligosaccharyltransferase (AglB) |
Discussion
Although currently limited in numeric terms, those N-linked glycans decorating archaeal glycoproteins for which compositional and/or structural data is available reveal a degree of diversity not seen in their eukaryotic or bacterial counterparts (Schwarz and Aebi, 2011; Eichler, 2013). The list of sugars found as part of N-linked glycans in Archaea has been expanded to include 5-N-formyl-legionaminic acid, first detected in the glycan of a major structural protein of a virus that infects Halorubrum sp. PV6 (Kandiba et al., 2012) and now inferred to be the terminal sugar of the glycan N-linked the S-layer glycoprotein in this haloarchaea. The present report, moreover, delineated a pathway for legionaminic acid biosynthesis in Halorubrum sp. PV6, demonstrated the transcription of pathway genes (as well as the co-transcription of several of these genes) and confirmed the enzymatic activities of two pathway components, LegI and LegF. Finally, the delineation of a pathway for legionaminic acid biosynthesis in Halorubrum sp. PV6 implies that Archaea are capable of generating NDP-di-N-acetylbacillosamine (2,4-diacetamido-2,4,6-trideoxy-D-glucose) through the actions of LegH. UDP-di-N-acetylbacillosamine is important in bacterial protein glycosylation, providing the first sugar of both the O-linked glycan decorating glycoproteins in Neisseria gonorrhoeae (Aas et al., 2007; Hartley et al., 2011) and the N-linked glycan in C. jejuni (Young et al., 2002; Glover et al., 2006).
In Halorubrum sp. PV6, the components of a predicted legionaminic acid biosynthesis pathway share a common region spanning some 67 kB of the genome with ORFs predicted to encode enzymes that participate in the biogenesis of the remaining sugars of the N-linked pentasaccharide decorating the S-layer glycoprotein. These could include the legionaminic acid transferase that has yet to be identified in any organism employing this sugar in protein glycosylation. However, because no genetic tools yet exist for the manipulation of Halorubrum sp. PV6, it is not yet possible to rely on gene deletion to directly test the involvement of any these components in either legionaminic acid biosynthesis or N-glycosylation. Instead, sequences of interest can be expressed in other halophilic archaea for which appropriate molecular techniques, such as transformation, exist, including Hfx. volcanii. Indeed, Halorubrum sp. PV6 LegF and LegI characterized here were expressed in Hfx. volcanii. For those ORFs encoding proteins that are assigned various roles in Halorubrum sp. PV6 N-glycosylation, studies in which the abilities of these sequences to complement Hfx. volcanii strains deleted of the homologous sequence, thereby restoring the missing activity, are ongoing. This approach has been previously used to confirm the actions of oligosaccharyltransferase-encoding aglB sequences from Haloarcula marismortui, Halobacterium salinarum, and Haloferax mediterranei (Cohen-Rosenzweig et al., 2014) and to identify other components of a Hbt. salinarum N-glycosylation pathway (Kandiba and Eichler, 2015).
While various functions have been assigned to the NulO sugars found as components of N-linked glycans in Eukarya and Bacteria (Kelm and Schauer, 1997; Schauer, 1982; Chen and Varki, 2010; Varki and Gagneux, 2012; Morrison and Imperiali, 2014; Schnaar et al., 2014; Varki et al., 2017), the roles served by these sugars in Archaea remains an open question, given the overall lack of information on NulO sugars as components of the N-linked glycans that decorate archaeal glycoproteins. At the same time, previous reports provided evidence for 5-N-formyl-legionaminic acid as being involved in the interaction between Halorubrum sp. PV6 and HRPV-1 that infects these cells (Kandiba et al., 2012). The HRPV-1 VP4 spike protein that protrudes from the viral membrane surface and which is thought to mediate host recognition and the initial stages of infection is modified by the same N-linked glycan that is apparently bound to the Halorubrum sp. PV6 S-layer glycoprotein (Pietilä et al., 2009; Kandiba et al., 2012). Viral adsorption, as reflected in the level of virus production in infected cells, was reduced in the presence of the sialic acid N-acetylneuraminic acid. The added sialic acid likely competed with the virus for a binding site on the Halorubrum sp. PV6 host that recognizes the 5-N-formyl-legionaminic acid that caps the N-linked glycan decorating both the S-layer glycoprotein and VP4. This raises questions as to the intended role played by this binding site in Halorubrum sp. PV6. Recently, glycosylation of the S-layer glycoprotein was shown to be important for cell-cell recognition and mating in H. volcanii (Shalev et al., 2017). It is tempting to speculate that Halorubrum sp. PV6 relies on the 5-N-formyl-legionaminic acid unit of the S-layer glycoprotein N-linked pentasaccharide for a similar purpose.
In summary, the findings presented here further expand the growing list of sugars employed in the archaeal version of N-glycosylation and provides new information on how one such sugar, 5-N-formyl-legionaminic acid, is generated. Such knowledge could help provide novel insight into the importance of this sugar, and indeed, roles played by N-glycosylation in archaea.
Author Contributions
ER and JE conceived the study. MZ performed the experiments. MZ, ER, and JE analyzed the data. JE wrote the manuscript. All authors read and commented on the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Lina Kandiba for early contributions to this study.
Funding. ER was supported by a University of Helsinki 3-year grant (2010–2012). JE was supported by the Israel Science Foundation (grant 109/16).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02133/full#supplementary-material
References
- Aas F. E., Vik A., Vedde J., Koomey M., Egge-Jacobsen W. (2007). Neisseria gonorrhoeae O-linked pilin glycosylation: functional analyses define both the biosynthetic pathway and glycan structure. Mol. Microbiol. 65 607–624. 10.1111/j.1365-2958.2007.05806.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdul Halim M. F., Pfeiffer F., Zou J., Frisch A., Haft D., Wu S., et al. (2013). Haloferax volcanii archaeosortase is required for motility, mating, and C-terminal processing of the S-layer glycoprotein. Mol. Microbiol. 88 1164–1175. 10.1111/mmi.12248 [DOI] [PubMed] [Google Scholar]
- Abu-Qarn M., Yurist-Doutsch S., Giordano A., Trauner A., Morris H. R., Hitchen P., et al. (2007). Haloferax volcanii AglB and AglD are involved in N-glycosylation of the S-layer glycoprotein and proper assembly of the surface layer. J. Mol. Biol. 374 1224–1236. 10.1016/j.jmb.2007.10.042 [DOI] [PubMed] [Google Scholar]
- Aebi M. (2013). N-linked protein glycosylation in the ER. Biochim. Biophys. Acta 1833 2430–2437. 10.1016/j.bbamcr.2013.04.001 [DOI] [PubMed] [Google Scholar]
- Angata T., Varki A. (2002). Chemical diversity in sthe sialic acids and related alpha-keto acids: an evolutionary perspective. Chem. Rev. 102 439–469. 10.1021/cr000407m [DOI] [PubMed] [Google Scholar]
- Aziz R. K., Bartels D., Best A. A., DeJongh M., Disz T., Edwards R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo D., Guan Z., Naparstek S., Eichler J. (2011). Different routes to the same ending: comparing the N-glycosylation processes of Haloferax volcanii and Haloarcula marismortui, two halophilic archaea from the Dead Sea. Mol. Microbiol. 81 1166–1177. 10.1111/j.1365-2958.2011.07781.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Varki A. (2010). Advances in the biology and chemistry of sialic acids. ACS Chem. Biol. 5 163–176. 10.1021/cb900266r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M., Varki A. (2010). The sialome-far more than the sum of its parts. OMICS 14 455–464. 10.1089/omi.2009.0148 [DOI] [PubMed] [Google Scholar]
- Cohen-Rosenzweig C., Guan Z., Shaanan B., Eichler J. (2014). Substrate promiscuity: AglB, the archaeal oligosaccharyltransferase, can process a variety of lipid-linked glycans. Appl. Environ. Microbiol. 80 486–496. 10.1128/AEM.03191-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daines D. A., Wright L. F., Chaffin D. O., Rubens C. E., Silver R. P. (2000). NeuD plays a role in the synthesis of sialic acid in Escherichia coli K1. FEMS Microbiol. Lett. 189 281–284. 10.1111/j.1574-6968.2000.tb09244.x [DOI] [PubMed] [Google Scholar]
- Deng L., Chen X., Varki A. (2013). Exploration of sialic acid diversity and biology using sialoglycan microarrays. Biopolymers 99 650–665. 10.1002/bip.22314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler J. (2013). Extreme sweetness: protein glycosylation in Archaea. Nat. Rev. Microbiol. 11 151–156. 10.1038/nrmicro2957 [DOI] [PubMed] [Google Scholar]
- Eichler J., Imperiali B. (2018). Stereochemical divergence of polyprenol phosphate glycosyltransferases. Trends Biochem. Sci. 43 10–17. 10.1016/j.tibs.2017.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrero M. A., Aparicio L. R. (2010). Biosynthesis and production of polysialic acids in bacteria. Appl. Microbiol. Biotechnol. 86 1621–1635. 10.1007/s00253-010-2531-5 [DOI] [PubMed] [Google Scholar]
- Glaze P. A., Watson D. C., Young N. M., Tanner M. E. (2008). Biosynthesis of CMP-N,N’-diacetyllegionaminic acid from UDP-N,N’-diacetylbacillosamine in Legionella pneumophila. Biochemistry 47 3272–3282. 10.1021/bi702364s [DOI] [PubMed] [Google Scholar]
- Glover K. J., Weerapana E., Chen M. M., Imperiali B. (2006). Direct biochemical evidence for the utilization of UDP-bacillosamine by PglC, an essential glycosyl-1-phosphatetransferase in the Campylobacter jejuni N-linked glycosylation pathway. Biochemistry 45 5343–5350. 10.1021/bi0602056 [DOI] [PubMed] [Google Scholar]
- Haft R. F., Wessels M. R. (1994). Characterization of CMP-N-acetylneuraminic acid synthetase of group B streptococci. J. Bacteriol. 176 7372–7374. 10.1128/jb.176.23.7372-7374.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley M. D., Morrison M. J., Aas F. E., Børud B., Koomey M., Imperiali B. (2011). Biochemical characterization of the O-linked glycosylation pathway in Neisseria gonorrhoeae responsible for biosynthesis of protein glycans containing N,N′-diacetylbacillosamine. Biochemistry 50 4936–4948. 10.1021/bi2003372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D., Chen G.-L., LoCascio P. F., Land M. L., Larimer F. W., Hauser L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irihimovitch V., Eichler J. (2003). Post-translational secretion of fusion proteins in the halophilic archaea Haloferax volcanii. J. Biol. Chem. 278 12881–12887. 10.1074/jbc.M210762200 [DOI] [PubMed] [Google Scholar]
- Jarrell K. F., Ding Y., Meyer B. H., Albers S. V., Kaminski L., Eichler J. (2014). N-linked glycosylation in Archaea: a structural, functional, and genetic analysis. Microbiol. Mol. Biol. Rev. 78 304–341. 10.1128/MMBR.00052-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell K. F., Jones G. M., Kandiba L., Nair D. B., Eichler J. (2010). S-layer glycoproteins and flagellins: reporters of archaeal posttranslational modifications. Archaea 2010:612948. 10.1155/2010/612948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. B., Rosenberg J. N., Betenbaugh M. J., Krag S. S. (2009). Structure and synthesis of polyisoprenoids used in N-glycosylation across the three domains of life. Biochim. Biophys Acta 1790 485–494. 10.1016/j.bbagen.2009.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski L., Lurie-Weinberger M. N., Allers T., Gophna U., Eichler J. (2013a). Phylogenetic- and genome-derived insight into the evolution of N-glycosylation in Archaea. Mol. Phylogenet. Evol. 68 327–339. 10.1016/j.ympev.2013.03.024 [DOI] [PubMed] [Google Scholar]
- Kaminski L., Guan Z., Yurist-Doutsch S., Eichler J. (2013b). Two distinct N-glycosylation pathways process the Haloferax volcanii S-layer glycoprotein upon changes in environmental salinity. mBio 4:e00716-13. 10.1128/mBio.00716-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandiba L., Aitio O., Helin J., Guan Z., Permi P., Bamford D. H., et al. (2012). Diversity in prokaryotic glycosylation: an archaeal-derived N-linked glycan contains legionaminic acid. Mol. Microbiol. 84 578–593. 10.1111/j.1365-2958.2012.08045.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandiba L., Eichler J. (2013). Analysis of genes involved in the biosynthesis of sialic acids and other nonulosonic acid sugars in Archaea reveals a complex evolutionary history. FEMS Microbiol. Lett. 345 110–120. 10.1111/1574-6968 [DOI] [PubMed] [Google Scholar]
- Kandiba L., Eichler J. (2015). Deciphering a pathway of Halobacterium salinarum N-glycosylation. Microbiologyopen 4 28–40. 10.1002/mbo3.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandiba L., Guan Z., Eichler J. (2013). Lipid modification gives rise to two distinct Haloferax volcanii S-layer glycoprotein populations. Biochim. Biophys. Acta 1828 938–943. 10.1016/j.bbamem.2012.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandiba L., Lin C.-W., Aebi M., Eichler J., Guerardel Y. (2016). Structural characterization of the N-linked pentasaccharide decorating glycoproteins of the halophilic archaeon Haloferax volcanii. Glycobiology 26 745–756. 10.1093/glycob/cww014 [DOI] [PubMed] [Google Scholar]
- Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J. E. (2015). The phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10 845–858. 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelm S., Schauer R. (1997). Sialic acids in molecular and cellular interactions. Int. Rev. Cytol. 175 137–240. 10.1016/S0074-7696(08)62127-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knirel Y. A., Rietschel E. T., Marre R., Zähringer U. (1994). The structure of the O-specific chain of Legionella pneumophila serogroup 1 lipopolysaccharide. Eur. J. Biochem. 221 239–245. 10.1111/j.1432-1033.1994.tb18734.x [DOI] [PubMed] [Google Scholar]
- Knirel Y. A., Shashkov A. S., Tsvetkov Y. E., Jansson P. E., Zãhringer U. (2003). 5,7-diamino-3,5,7,9-tetradeoxynon-2-ulosonic acids in bacterial glycopolymers: chemistry and biochemistry. Adv. Carbohydr. Chem. Biochem. 58 371–417. 10.1016/S0065-2318(03)58007-6 [DOI] [PubMed] [Google Scholar]
- Koomey J. M., Eichler J. (2017). Sweet new roles for protein glycosylation in prokaryotes. Trends Microbiol. 25 662–672. 10.1016/j.tim.2017.03.001 [DOI] [PubMed] [Google Scholar]
- Koskinen P., Törönen P., Nokso-Koivisto J., Holm L. (2015). PANNZER - High-throughput functional annotation of uncharacterized proteins in an error-prone environment. Bioinformatics 31 1544–1552. 10.1093/bioinformatics/btu851 [DOI] [PubMed] [Google Scholar]
- Larkin A., Imperiali B. (2011). The expanding horizons of asparagine-linked glycosylation. Biochemistry 50 4411–4426. 10.1038/nchembio.1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A. L., Desa N., Hansen E. E., Knirel Y. A., Gordon J. I., Gagneux P., et al. (2009). Innovations in host and microbial sialic acid biosynthesis revealed by phylogenomic prediction of nonulosonic acid structure. Proc. Natl. Acad. Sci. U.S.A. 106 13552–13557. 10.1073/pnas.0902431106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Jin C., Jin C. (2004). CMP-N-acetylneuraminic acid synthetase from Escherichia coli K1 is a bifunctional enzyme: identification of minimal catalytic domain for synthetase activity and novel functional domain for platelet-activating factor acetylhydrolase activity. J. Biol. Chem. 279 17738–17749. 10.1074/jbc.M400143200 [DOI] [PubMed] [Google Scholar]
- Magidovich H., Eichler J. (2009). Glycosyltransferases and oligosaccharyltransferases in Archaea: putative components of the N-glycosylation pathway in the third domain of life. FEMS Microbiol. Lett. 300 122–130. 10.1111/j.1574-6968.2009.01775.x [DOI] [PubMed] [Google Scholar]
- Maita N., Nyirenda J., Igura M., Kamishikiryo J., Kohda D. (2010). Comparative structural biology of eubacterial and archaeal oligosaccharyltransferases. J. Biol. Chem. 285 4941–4950. 10.1074/jbc.M109.081752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally D. J., Aubry A. J., Hui J. P., Khieu N. H., Whitfield D., Ewing C. P., et al. (2007). Targeted metabolomics analysis of Campylobacter coli VC167 reveals legionaminic acid derivatives as novel flagellar glycans. J. Biol. Chem. 282 14463–14475. 10.1074/jbc.M611027200 [DOI] [PubMed] [Google Scholar]
- Mengele R., Sumper M. (1992). Drastic differences in glycosylation of related S-layer glycoproteins from moderate and extreme halophiles. J. Biol. Chem. 267 8182–8185. [PubMed] [Google Scholar]
- Mevarech M., Werczberger R. (1985). Genetic transfer in Halobacterium volcanii. J. Bacteriol. 162 461–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T. L., Wolin M. J., Conway de Macario E., Macario A. J. (1982). Isolation of Methanobrevibacter smithii from human feces. Appl. Environ. Microbiol. 43 227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morag E., Lapidot A., Govorko D., Lamed R., Wilchek M., Bayer E. A., et al. (1995). Expression, purification, and characterization of the cellulose-binding domain of the scaffoldin subunit from the cellulosome of Clostridium thermocellum. Appl. Environ. Microbiol. 61 1980–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison M. J., Imperiali B. (2014). The renaissance of bacillosamine and its derivatives: pathway characterization and implications in pathogenicity. Biochemistry 53 624–638. 10.1021/bi401546r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothaft H., Szymanski C. M. (2010). Protein glycosylation in bacteria: sweeter than ever. Nat. Rev. Microbiol. 8 765–778. 10.1038/nrmicro2383 [DOI] [PubMed] [Google Scholar]
- Overbeek R., Olson R., Pusch G. D., Olsen G. J., Davis J. J., Disz T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42 D206–D214. 10.1093/nar/gkt1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parente J., Casabuono A., Ferrari M. C., Paggi R. A., De Castro R. E., Couto A. S., et al. (2014). A rhomboid protease gene deletion affects a novel oligosaccharide N-linked to the S-layer glycoprotein of Haloferax volcanii. J. Biol. Chem. 289 11304–11317. 10.1074/jbc.M113.546531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce O. M., Läubli H. (2016). Sialic acids in cancer biology and immunity. Glycobiology 26 111–128. 10.1093/glycob/cwv097 [DOI] [PubMed] [Google Scholar]
- Pietilä M. K., Laurinavicius S., Sund J., Roine E., Bamford D. H. (2010). The single-stranded DNA genome of novel archaeal virus halorubrum pleomorphic virus 1 is enclosed in the envelope decorated with glycoprotein spikes. J. Virol. 84 788–798. 10.1128/JVI.01347-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietilä M. K., Roine E., Paulin L., Kalkkinen N., Bamford D. H. (2009). An ssDNA virus infecting archaea: a new lineage of viruses with a membrane envelope. Mol. Microbiol. 72 307–319. 10.1111/j.1365-2958.2009.06642.x [DOI] [PubMed] [Google Scholar]
- Samuel B. S., Hansen E. E., Manchester J. K., Coutinho P. M., Henrissat B., Fulton R., et al. (2007). Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proc. Natl. Acad. Sci. U.S.A. 104 10643–10648. 10.1073/pnas.0704189104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer R. (1982). Chemistry, metabolism, and biological functions of sialic acids. Adv. Carbohydr. Chem. Biochem. 40 131–234. 10.1016/S0065-2318(08)60109-2 [DOI] [PubMed] [Google Scholar]
- Schnaar R. L., Gerardy-Schahn R., Hildebrandt H. (2014). Sialic acids in the brain: gangliosides and polysialic acid in nervous system development, stability, disease, and regeneration. Physiol. Rev. 94 461–518. 10.1152/physrev.00033.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenhofen I. C., McNally D. J., Brisson J. R., Logan S. M. (2006). Elucidation of the CMP-pseudaminic acid pathway in Helicobacter pylori: synthesis from UDP-N-acetylglucosamine by a single enzymatic reaction. Glycobiology 16 8C–14C. 10.1093/glycob/cwl010 [DOI] [PubMed] [Google Scholar]
- Schoenhofen I. C., Vinogradov E., Whitfield D. M., Brisson J. R., Logan S. M. (2009). The CMP-legionaminic acid pathway in campylobacter: biosynthesis involving novel GDP-linked precursors. Glycobiology 19 715–725. 10.1093/glycob/cwp039 [DOI] [PubMed] [Google Scholar]
- Schwarz F., Aebi M. (2011). Mechanisms and principles of N-linked protein glycosylation. Curr. Opin. Struct. Biol. 21 576–582. 10.1016/j.sbi.2011.08.005 [DOI] [PubMed] [Google Scholar]
- Shalev Y., Turgeman-Grott I., Tamir A., Eichler J., Gophna U. (2017). Cell surface glycosylation is required for efficient mating of Haloferax volcanii. Front. Microbiol. 8:1253. 10.3389/fmicb.2017.01253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söding J., Biegert A., Lupas A. N. (2005). The HHpred interactive server for protein homology detection and structure prediction. Nucleic. Acids Res. 33 W244–W248. 10.1093/nar/gki408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stencel-Baerenwald J. E., Reiss K., Reiter D. M., Stehle T., Dermody T. S. (2014). The sweet spot: defining virus-sialic acid interactions. Nat. Rev. Microbiol. 12 739–749. 10.1038/nrmicro3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumper M., Berg E., Mengele R., Strobel I. (1990). Primary structure and glycosylation of the S-layer protein of Haloferax volcanii. J. Bacteriol. 172 7111–7118. 10.1128/JB.172.12.7111-7118.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vann W. F., Silver R. P., Abeijon C., Chang K., Aaronson W., Sutton A., et al. (1987). Purification, properties, and genetic location of Escherichia coli cytidine 5′-monophosphate N-acetylneuraminic acid synthetase. J. Biol. Chem. 262 17556–17562. [PubMed] [Google Scholar]
- Vann W. F., Tavarez J. J., Crowley J., Vimr E., Silver R. P. (1997). Purification andcharacterization of the Escherichia coli K1 neuB gene product N-acetylneuraminic acid synthetase. Glycobiology 7 697–701. 10.1093/glycob/7.5.697 [DOI] [PubMed] [Google Scholar]
- Varki A., Cummings R. D., Aebi M., Packer N. H., Seeberger P. H., Esko J. D., et al. (2015). Symbol nomenclature for graphical representations of glycans. Glycobiology 25 1323–1324. 10.1093/glycob/cwv091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A., Gagneux P. (2012). Multifarious roles of sialic acids in immunity. Ann. N. Y. Acad. Sci. 1253 16–36. 10.1111/j.1749-6632.2012.06517.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A., Schnaar R. L., Schauer R. (2017). “Sialic acids and other nonulosonic acids,” in Essentials of Glycobiology [Internet] 3rd Edn eds Varki A., Cummings R. D., Esko J. D., Stanley P., Hart G. W., Aebi M.et al. (Cold) Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ) 2015–2017. [PubMed] [Google Scholar]
- Vimr E. R. (2013). Unified theory of bacterial sialometabolism: how and why bacteria metabolize host sialic acids. ISRN Microbiol. 2013:816713. 10.1155/2013/816713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimr E. R., Kalivoda K. A., Deszo E. L., Steenbergen S. M. (2004). Diversity of microbial sialic acid metabolism. Microbiol. Mol. Biol. Rev. 68 132–153. 10.1128/MMBR.68.1.132-153.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L. (1959). The thiobarbituric acid assay of sialic acids. J. Biol. Chem. 234 1971–1975. [PubMed] [Google Scholar]
- Young N. M., Brisson J. R., Kelly J., Watson D. C., Tessier L., Lanthier P. H., et al. (2002). Structure of the N-linked glycan present on multiple glycoproteins in the gram-negative bacterium, Campylobacter jejuni. J. Biol. Chem. 2277 42530–42539. 10.1074/jbc.M206114200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


