Abstract
Ubiquitinated proteins carried by the extracellular vesicles (EV) released by myeloid-derived suppressor cells (MDSC) have been investigated using proteomic strategies to examine the effect of tumor-associated inflammation. EV were collected from MDSC directly following isolation from tumor-bearing mice with low and high inflammation. Among the 1092 proteins (high inflammation) and 925 proteins (low inflammation) identified, more than 50% were observed as ubiquitinated proteoforms. More than three ubiquitin-attachment sites were characterized per ubiquitinated protein, on average. Multiple ubiquitination sites were identified in the pro-inflammatory proteins S100 A8 and S100 A9, characteristic of MDSC and in histones and transcription regulators among other proteins. Spectral counting and pathway analysis suggest that ubiquitination occurs independently of inflammation. Some ubiquitinated proteins were shown to cause the migration of MDSC, which has been previously connected with immune suppression and tumor progression. Finally, MDSC EV are found collectively to carry all the enzymes required to catalyze ubiquitination, and the hypothesis is presented that a portion of the ubiquitinated proteins are produced in situ.
Keywords: extracellular vesicles, ubiquitome, inflammation, MDSC, LC−MS/MS, tumor-induced immune suppression, top-down proteomics, spectral counting
INTRODUCTION
Myeloid-derived suppressor cells (MDSC) congregate in the tumor microenvironment of individuals and mice with cancer, where they promote tumor progression and inhibit antitumor immunity through mechanisms that include the inhibition of Tcell activation and the polarization of macrophages toward a tumor-promoting phenotype.1 Inflammation, which often accompanies cancer, increases MDSC accumulation and suppressive activity.2–4 We have demonstrated previously that MDSCs release extracellular vesicles (EV) that carry bioactive proteins mirroring those present in the parental MDSC3,5–7 Generally, the incorporation of proteins into EV has been shown to be a nonrandom process whose inventory reflects the functions of parental cells.8,9 Proteins in high abundance in the EV shed by MDSC include the proinflammatory mediators S100 A8 and S100 A9, which have been shown to exert chemotactic activity toward other MDSC and catalyze their migration into the tumor environment.3,10 More than 90% of this autocrine activity is carried in the EV rather than parent cells themselves.3
The studies reported here continue an ongoing investigation of the changes that inflammation causes in MDSC and their EV, seeking to understand how inflammation enhances immune suppression. We have determined that inflammation is associated with an increased number of MDSC in tumorbearing mice which in turn increases suppression of the immune response and facilitates tumor progression.2,10,11 Inflammatory (INF) and conventional (CON) MDSC have been shown to shed EV at similar rates and bottom-up proteomic analyses indicate that both INF and CON EVs carry similar amounts of proinflammatory S100 A8 and S100 A9, which regulate the accumulation of MDSC.10 In contrast, topdown proteomic analysis and spectral counting detect inflammation-related differences in abundances of oxidized proteoforms of S100A8 and A9.3,4 Bioactive roles have been proposed by others for modified proteoforms in tumor EV, most notably phosphorylation.8,12 Since ubiquitination is thought to play a role in the selection of exosomal protein cargo, we have examined and report here the effect of inflammation on ubiquitin-conjugated proteins in EV produced by MDSC.
Ubiquitination is involved in invagination of the plasma membrane, the initial step in the formation of the endosomes that eventually progress to become exosomes, and ubiquitinated plasma membrane and cytoskeletal proteins have been identified in exosome cargo.5 Furthermore, the endosomal sorting complex required for transport (ESCRT), which mediates transformation of endosomes into multivesicular bodies (MVBs), sorts proteins into intraluminal vesicles (ILVs) within the MVBs, based on ubiquitination.9,13,14 Investigators have proposed that ubiquitin is removed from proteins that are sequestered into ILVs destined to be released as exosomes.15–17 However, recent identifications of significant numbers of ubiquitinated proteins in the lysates of exosomes from a variety of cell types challenge this mechanism,5,18,19 and sorting mechanisms in addition to ESCRT have been proposed.20 Previous researchers have reported that free and anchored polyubiquitins in exosomes have significant activities, including inhibiting platelet activation21 and the modulation of immune responses.22
(Poly)ubiquitinated proteins are challenging to study. The modification is very large, and the ubiquitinated proteoforms are usually present at substoichiometric levels compared to their unmodified proteins.23–25 However, tryptic digestion of ubiquitinated proteins also cleaves attached ubiquitins, notably at the ubiquitin amino acid, Arg74, leaving two glycine residues attached to the substrate lysine by an isopeptide bond. The socalled GG-tag on Lys residues adds 114.04 Da to the modified tryptic peptide and marks the site of ubiquitin attachment on the substrate protein.24,26–28
The present study uses state-of-the-art mass spectrometry optimized for bottom-up proteomic analysis to examine the proteomes recovered from EV shed by MDSC isolated from tumor-bearing mice with high and low inflammation and to characterize their respective ubiquitomes. In this study, GGtagged peptides are recovered without antibody or other affinity enrichment.29 Spectral counting is used to estimate relative amounts of ubiquitinated proteins in EV samples from high and low inflammatory mice, and pathway analysis used to provide a functional comparison between CON and INF MDSC EV relative to ubiquitination. A full complement of ubiquitin conjugating enzymes and hydrolases, thirty-three murine proteasome subunits and the five glycolytic enzymes that synthesize ATP were found in similar amounts in samples from animals with high and low inflammation. Previous investigators have demonstrated that these enzymes are active in exosomes. Consequently, we propose that the ubiquitinated protein cargo in MDSC EV is dynamic and modulated in situ.
EXPERIMENTAL SECTION
Myeloid-Derived Suppressor Cells
BALB/c mice were injected in the mammary fat pad with either 7000 wild-type 4T1 mammary carcinoma cells or 4T1 cells stably transfected to express interlukin-1 β (IL-1β). When tumors were greater than approximately 8 mm in diameter (3− 4 weeks after initial inoculation), MDSC were harvested from the blood and analyzed by flow cytometry for expression of MDSC markers (Gr1 and CD 11b), as previously described.3
All MDSC used in these experiments were greater than 90% Gr1+CD11b+. All procedures with animals and animal-derived materials were approved by the UMBC and UMCP Institutional Animal Care and Use Committees.
Isolation and Characterization of Extracellular Vesicles
A total of three biological replicates, each consisting of an independent preparation of EV from MDSC induced by 4T1/ IL-1 β cells, were prepared as high inflammation samples. For low-inflammation samples, three biological replicates were obtained from MDSC induced by non-IL-1 β -transfected 4T1 cells. Each replicate was prepared from approximately 1 × 108 MDSC pooled from 2 or 3 tumor-bearing mice. These primary cells were incubated in serum-free HL-1 medium (BioWhittaker, Walkersville, MD) and maintained at 37 °C with 5% CO2. After 18 h, the cultures were centrifuged at 805g for 5 min (Eppendorf 5210 rotor, Eppendorf, Hamburg, Germany), the pellets were discarded, and the supernatants were centrifuged at 12800g (12 000 rpm) for 30 min (Sorvall RC5C, SS34 rotor, DuPont, Wilmington, DE). The supernatants were then ultracentrifuged at 100000g for 20 h at 10 °C (Beckman L8, SW40Ti rotor, Beckman, Pasadena, CA). The supernatants were discarded, and the pellets containing the EV were resuspended in PBS and stored at −80 °C until used.
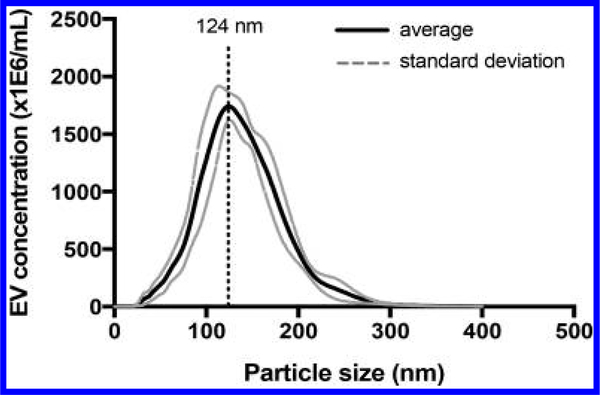
Our MDSC EV were previously characterized using sucrose density gradient measurements and transmission electron microscopy (TEM), which revealed homogeneous circular vesicles about 30 nm in diameter.3 In the present work, nanoparticle tracking analysis using a NanoSight LM-10 apparatus (Nanosight Limited, Amesbury, UK)) provided evidence for the presence of larger vesicles (Figure 1). This nanoparticle tracking analysis (NTA) analysis was made mindful of the conclusion of a study by van der Pol et al.30 that “[t]he minimum detectable vesicle sizes were 70–90 nm for NTA”. The term “extracellular vesicles”, or EV, is used in this report.
Figure 1.
Nanosight analysis of extracellular vesicles.
EVs were diluted to achieve 40–100 particles per frame in a NanoSight LM-10 (Nanosight Limited, Amesbury, UK). Approximately, 500 μL of sample was manually injected into the sample chamber at ambient temperature. Sample was measured in triplicate at camera setting 14 with an acquisition time of 60 s and detection threshold setting of 5. At least 1200 completed tracks were analyzed per video. NTA analytical software version 2.3 was used for capturing and processing the data.
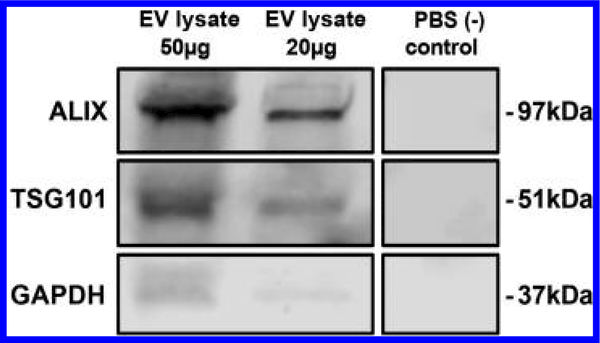
Immunoblotting (Figure 2) revealed the presence of TSG101 and ALIX, biomarkers for exosomes, using antibodies against ALIX (EPR15314; Abcam; ab186429) at 1:1000, TSG101 (C-2; Santa Cruz, sc-7964) at 1:200 and GAPDH (D16H11; cell signaling, 5174) at 1:2000. Goat anti-rabbit IRDye 800CW (925–32210; LICOR) and goat anti-mouse IRDye 680RD (925–68070; LICOR) secondary antibodies were used at a dilution of 1:10 000. Bands were detected with a LICOR Odyssey CLX Imager.
Figure 2.
Immunoblots of EV lysate with antibodies characteristic of exosomes.
Protein Preparation
Extracellular vesicles were lysed by suspension in 8 M urea in 50 mM ammonium bicarbonate with 50 μM deubiquitinase inhibitor PR-619 (LifeSensors, Malvern, PA) and 1% by volume protease inhibitor cocktail (Sigma-Aldrich, Saint Louis, MO). The solution was centrifuged at 13000g for 30 min through a 3 kDa molecular weight cut-off filter (Millipore, Darmstadt, Germany), and the filtrate was discarded. This process was performed three times before the buffer was diluted to 0.8 M urea in 50 mM ammonium bicarbonate. Protein content was measured using the Pierce BCA Assay Kit (Thermo Scientific, San Jose, CA).
A solution containing 25 μg of protein lysate was digested with trypsin. This normalized the amount of total protein between the two samples. Samples were reduced with 20 mM dithiothreitol (Sigma-Aldrich) for 30 min at 56 °C and alkylated with 10 mM methylmethanethiosulfonate (SigmaAldrich) for 45 min. Trypsin (Promega, Madison, WI) was added for a final 1:50 enzyme/protein concentration, and digestion was performed overnight at 37 °C before the addition of 0.1% formic acid. A total of three technical replicates of 1 μg of total protein were prepared and analyzed by liquid chromatography–tandem mass spectrometry (LC–MS/MS) for each of three biological replicates.
LC–MS/MS
A bottom up strategy was used to provide an inclusive and extensive inventory. LC–MS/MS analyses were performed on an Ultimate 3000 nano-HPLC system (Dionex, Sunnyvale, CA) in-line with an orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific, San Jose, CA). A 2 μL aliquot of tryptic peptides was injected onto a C18 precolumn (Dionex, Sunnyvale, CA) and desalted with 10% solvent A (97.5% H2O, 2.5% ACN, and 0.1% formic acid) for 10 min. Peptides were fractionated on a C18 column (Dionex, Sunnyvale, CA) with a 2 h linear gradient at a flow rate of 300 nL/min increasing from 5 to 55% solvent B (97.5% ACN, 2.5% H2O, and 0.1% formic acid) in 90 min, followed by an increase from 55 to 90% solvent B in 5 min, and held at 90% solvent B for 5 min. Precursor scans were acquired in the orbitrap with a resolution of 120,000 at m/z 200. The most-abundant ions, as many as possible, during each 3 s duty cycle were selected for fragmentation by collisional induced dissociation (35% collision energy) in the ion trap, and product ion scans were acquired in the ion trap. A dynamic exclusion of 1 repeat count over 30 s was used.
A top-down analysis was also performed on the EV lysates. Proteins were analyzed using an Ultimate 3000 RSLC nano ultrahigh-performance liquid chromatograph (Dionex, Sunnyvale, CA). The LC was used in line with the orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific, Waltham, MA). EV lysates were desalted and concentrated in a PepSwift RP-4H monolith trap (100 μm × 5 mm) and then separated using a ProSwift RP-4H monolith column (200 μm × 25 cm). Proteins were desalted and concentrated at 20% solution B and separated using a linear gradient from 20% to 55% B for 145 min. The mass spectrometer was set to intact protein analysis in which the ion routing multipole pressure was maintained at 3 mTorr. Precursor and product ions were analyzed in the Orbitrap with a mass resolution of 120,000. The “top-speed” data-dependent mode was selected for precursor ions fragmentation in which a maximum number of abundant precursors are isolated by the quadrupole in a fixed duty cycle time of 10 s. Precursor ions with a charge state of +8 or higher were selected for fragmentation, and precursors with an undetermined charge state were included as well. Dynamic exclusion was set to 60 s for this analysis. The fragmentation technique used was EThcD with a reaction time of 6 ms for electron transfer dissociation (ETD) and 10% supplemental higher collision energy (HCD). A total of three microscans were averaged for each spectrum generated. Automatic gain control targets were set to 1 × 106 for precursor ions and 5 × 105 for product ions with a 200 ms maximum injection time.
Bioinformatics
Peptide identifications were determined using the PepArML31,32 peptide identification meta-search engine by matching spectra against semispecific peptides from the UniProt mouse database (release 01_2015). The methylthio modification of cysteine was applied as a fixed modification, with Met oxidation, Lys glycinylglycine, and peptide N-terminal cyclic pyrolidone derivative variable modifications. Reversed protein sequence decoys were used to estimate false discovery rates and the peptide-spectral matches filtered at 1% spectral FDR. Global protein parsimony with at least two unshared peptides per protein was used to identify proteins at 3.3% protein FDR.
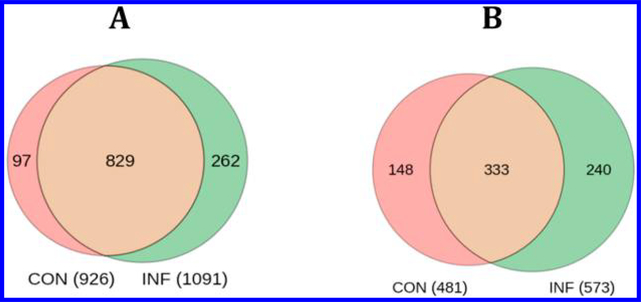
Following rigorous identification of exosomal proteins, all peptide identifications aligned with the identified proteins and spectral FDR at most 10% were recruited for subsequent analysis. Identified proteins with at least one GG-tag peptide were considered ubiquitinated with the ubiquitin protein false discovery rate estimated at 3.8%. We note, with reference to Figure 3, that some proteins observed in both CON and INF samples do not have a GG-tagged peptide in either sample.
Figure 3.
Venn diagrams of the overlap between proteins in (A) overall proteins identified in CON and INF extracellular vesicles and (B) proteins with ubiquitinated proteoforms in CON and INF extracellular vesicles.
Data acquired from the top down analysis was processed using the ProSight node v. 1.1 on Proteome Discoverer v.2.1. The raw data file was searched against the Mus musculus reference proteome downloaded from UniProt (June 2017). The searches were performed with the following parameters: precursor mass tolerance of 40 000 Da, fragment mass tolerance of 10 ppm, precursor m/z tolerance of 0.2, max retention time difference of 0.5 min, and a precursor and fragment signal-to-noise threshold of 3. The target decoy protein spectrum match (PSM) validator was used with a maximum delta correlation (Cn) (the normalized difference between the best score assigned and the second best score for a given spectrum) of 0.05. ProSight Lite33 was used to view the MS/MS ions identified for the proteins, with a fragment tolerance of 10 ppm. Xcalibur v.2.1. was used to view the raw data and generate extracted ion chromatograms.
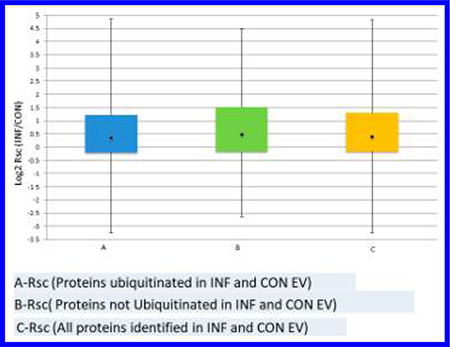
The relative abundances of proteins in CON and INF samples were estimated by spectral counting using in house software. Ratios of spectral count (Rsc) were generated by the method described in Old et al.34 The Fisher exact test was used to generate p-values for differential spectral counts and Benjamini–Hochberg FDR estimates used to correct for multiple testing.35 Spectra of ubiquitinated and nonubiquitinated peptides were counted together to compute relative protein abundance. Rsc values are presented as log2 values.
Pathway enrichment analysis for differentially abundant proteins was carried out with respect to the canonical pathways of the REACTOME database.36 MDSC exosome proteins with significantly increased or decreased spectral counts (|Rsc| ≥ 1) and Fisher’s exact test FDR ≤ 5% (Benjamini–Hochberg multiple-test correction) with respect to CON or INF samples were selected as candidate proteins. EV proteins ubiqutinated in both CON and INF samples satisfying the same criteria (|Rsc| ≥ 1 and FDR ≤ 5%) were selected for ubiquitome-focused pathway-enrichment analysis. In each case, the set of all identified EV proteins, specifically EV proteins ubiquitinated in both CON and INF, were used as the pathway enrichment background. Fisher’s exact test and the Benjamini–Hochberg FDR estimation was used to assess the statistical significance of the number of proteins in common between each canonical pathway and the candidate proteins versus the number expected based on the background protein set. No REACTOME pathways were found to be significantly enriched in increased or decreased EV or ubiquitinated EV proteins (data not shown).
Differential pathway analysis by sample-based peptide counting37 was also applied to canonical pathways from the REACTOME database. The proportion of INF-specific (respectively CON-specific) peptides for each pathway was compared to the overall proportion of INF-specific (respectively CON-specific) peptides, and the statistical significance was computed using Fisher exact test and Benjamini– Hochberg multiple-test correction. The analysis was applied to peptides from the MDSC EV protein set, and the set of MDSC EV proteins was ubiquitinated, in both CON and INF. REACTOME pathways with peptides from at least five proteins, and the FDR was, at most, 10−4 (a significant change) for the sample with an increased (decreased) number of peptides over that expected; an FDR of 1 (no significant change) for the other sample’s peptide increase (and decrease) is shown.
Bioassay for MDSC Chemotaxis
This in vitro bioassay is widely used as a model for migration of MDSC in vivo from the bloodstream to the tumor microenvironment. As previously reported3 MDSC (1 × 106) in 100 μL of migration medium (IMDM supplemented with 3% fetal calf serum) were placed in the upper chamber of a 8.0 μ transwell in 24-well plates. Either 500 uL of fresh medium, 500 uL of medium from cultured tumor cells (conditioned medium; positive control), EV, EV plus 3 μL of polyclonal rabbit antiubiquitin antibody (ThermoFisher PA1–10023), or 3 μL of an irrelevant polyclonal rabbit antibody inactivated by heating at 56 °C for 30 min (Cederlane, Burlington NC C13051) were in the lower chamber. Assembled transwells were incubated for 3 h at 37 °C in air supplemented with 5% CO2. The number of MDSC migrating to the bottom chamber was quantified by hemocytometer. Values for each sample are the average results of triplicate samples and two independent hemocytometer counts per sample. The amount of ubiquitin antibody included in the assay was determined by titering the antibody on MDSC using flow cytometry. Ubiquitin staining was saturating when the polyclonal antibody was used at a 1:200–1:600 dilution. Neither the antiubiquitin antibody nor the control antibody are toxic to MDSC.
RESULTS AND DISCUSSION
Ubiquitinated peptides and Proteoforms in MDSC Extracellular Vesicles
In extracellular vesicles from mice with either high or low inflammation, we found that more than 50% of the proteins identified contributed ubiquitinated proteoforms. This is the highest percentage reported in EV thus far. A total of 1092 proteins were identified in the inflammatory MDSC EV lysate (Supplementary Table 1), of which 573 were shown to be ubiquitinated (Supplementary Table 2). Among 926 proteins identified in the conventional sample (Supplementary Table 1), 481 were shown to contribute ubiquitinated proteoforms (Supplementary Table 2). There is considerable overlap between the pairs of samples (see Figure 3), and the overall total is composed of 753 ubiquitinated proteins with 2322 sites of attachment. The GG-modified peptide sequences are reported in Supplementary Table 2. In many cases, the same peptide is identified with and without a GG-tag. Hyperubiquitination, defined here as at least three conjugation sites per ubiquitinated protein, on average, is evident with 3.15 sites per protein observed. The identification of 747 ubiquitinated peptides from 593 proteins has been reported in urinary tract exosomes by Huebner et al.,19 and it appears that hyperubiquitination characterizes some types of exosomes. Topdown studies are underway to determine whether ubiquitination occurs concurrently at multiple locations.
The number of ubiquitinated proteins reported here is much larger than we reported in an earlier study of exosomes from the same kind of mice,5 and we point out that the recent samples were processed in the presence of deubiquitinase inhibitors, the lysate was analyzed without affinity fractionation, and a new model mass spectrometer was used. Nonetheless, the earlier work adds another 39, excluding duplications, bringing the overall total to 776. The actual number is likely to be still higher because many GG-tagged peptides may be present but not identified in bottom-up studies.
As can be seen in Supplementary Table 2, about half of the GG-tagged tryptic peptides carry the modification on a terminal lysine. Such cleavage products have been observed previously,5,19,27,38 and a mechanism has been proposed that allows the enzyme to carry out the two cleavages without releasing the substrate.39 The presence of polyubiquitin side chains or unanchored polymers in both EV samples was confirmed by identification of peptides from ubiquitin that carry GG tags on K6, K11, K33, K48, or K63. Proteoforms of the proinflammatory S100 A8 and S100 A9 proteins characteristic of MDSC were characterized as ubiquitin conjugates (Table 1). Ubiquitination is a large modification, and the question arises of whether the biologically active A8/A9 heterodimer11 can still be formed and whether it is functional when one or both proteins are modified with ubiquitin. Other proteins of interest with multiple conjugation sites include many nucleic acid binding proteins, e.g., histones and transcription regulators (Supplementary Table 2). In both INF and CON exosomes, transcription regulators contribute 14% of the inventoried proteins, and histones comprise ∼1%.
Table 1.
GG-Tagged Peptides from Proteins Characteristic of MDSC
| protein | peptide | spectral counts INF/CON |
|---|---|---|
| protein S100-A8 (P27005) | 1MPSELEK(gg)ALSNLIDVYHNYSNIQGNHHALYK | 1/– |
| 8ALSNLIDVYHNYSNIQGNHHALYK(gg) | 2/2 | |
| 36K(gg)MVTTECPQFVQNINIENLFR | 4/4 | |
| 57ELDINSDNAINFEEFLAMVIK(gg)VGVASHK | 3/– | |
| protein S100-A9 (P31725) | 3NK(gg)APSQMER | 1/– |
| 37EFRQMVEAQLATFMK(gg) | 1/2 | |
| 54RNEALINDIMEDLDTNQDNQLSFEECMMLMAK(gg)L | 23/14 | |
| 88IFACHEK(gg)LHENNPR | 1/– |
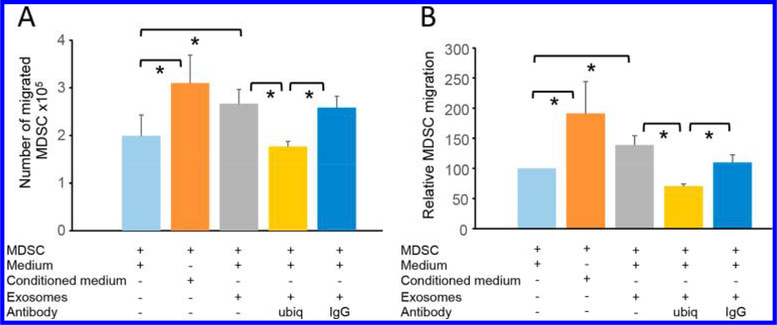
To determine if ubiquitinated proteins present in MDSC EV are biologically functional, we used a migration assay based on the knowledge that extracellular vesicles shed by MDSC facilitate the migration of parental MDSC.1,3,40 This in vitro assay models the migration in vivo of MDSC from blood to the tumor microenvironment where the density of MDSC correlates well with immune suppression and tumor growth.2,10,11,40 Migration was assessed by quantifying the movement of parental MDSC through a semipermeable membrane toward a suspension of MDSC-derived EV. The impact of ubiquitin on molecules that signal the migration process was evaluated by blocking with a ubiquitin-specific antibody. Figure 4A shows a representative experiment; Figure 4B shows the pooled results of six independent experiments. Inclusion of conditioned medium containing EV and soluble proteins (positive control) or purified MDSC-derived EV promoted MDSC migration. However, when antiubiquitin antibody was included, migration was inhibited with statistical significance. An irrelevant control antibody had no effect. Because the antiubiquitin antibody and EV are in the lower chamber and the assay time is relatively short (3 h), the antibody is most likely reacting with the EV. However, the antibody could potentially diffuse to the upper chamber and bind to the MDSC. Therefore, these results suggest that at least some of the ubiquitinated molecules carried by MDSC-derived EV or the MDSC themselves are biologically active and drive MDSC chemotaxis.
Figure 4.
Ubiquitinated proteins of MDSC and EV shed by MDSC and chemotactic activity. MDSC were placed in the upper compartment of transwells, and either tumor-conditioned medium, nonconditioned medium, or MDSC-shed EV with or without antibodies to ubiquitin were placed in the lower compartment. The number of MDSC migrating to the lower compartment was determined by hemocytometer count after 3 h of incubation. Asterisks indicate pair-wise significance at the 0.05 level (after Bonferroni multiple test correction). (A) Representative individual experiment consisting of three replicates and two hemocytometer counts per sample. (B) Pooled data of six independent experiments. (A) Standard deviations and (b) standard errors are shown.
Effects of Inflammation on Ubiquitination
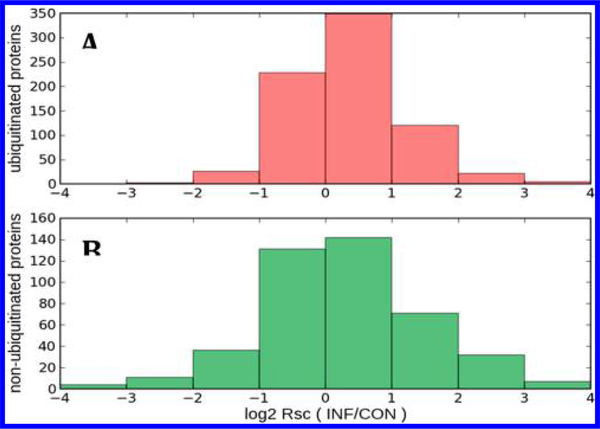
Spectral counting was used to study the effect of inflammation on the relative abundance profiles of ubiquitinated and nonubiquitinated proteins. The ratio of CON versus INF spectral counts (shown as log2 Rsc) are summarized in Figure 5, in which the distribution of CON versus INF relative abundance, or relative abundance profiles, for ubiquitinated and nonubiquitinated proteins are shown as aligned histograms. Surprisingly, despite the CON versus INF treatment difference, the relative abundance profiles of ubiquitinated versus nonubiquitinated proteins are quite similar, suggesting that inflammation and ubiquitination act independently on EV, even though the ubiquitinated proteins have a reduced range of relative abundances and a small bias toward positive Rsc (increased abundance in inflammation).
Figure 5.
Relative abundance profiles (INF vs CON as log2 Rsc) for (A) 753 ubiquitinated proteins and (B) 435 nonubiquitinated proteins. Positive log2 Rsc indicates increased abundance in INF samples.
Finally, the effect of inflammation on metabolic pathways was evaluated by analyzing the increase, or decrease, of distinct peptides in a sample versus overall distinct peptides for peptide sets associated, via their UniProt proteins, with Reactome pathways.31 Table 2 shows pathways whose proteins have an increased or decreased number of distinct peptides (FDR ≤ 10−4 and peptides from at least 5 proteins) in the ubiquitome of extracellular vesicles from mice with high and low inflammation. Table 3 provides pathways found to be significantly changed between high and low inflammation for the entire MDSC protein cohort. Of particular interest in Table 2, inflammation increases protein abundances in the neutrophil degranulation pathway in the ubiquitome.
Table 2.
Reactome Pathways with Significant Increase or Decrease of Distinct Peptides from the Ubiquitome in Response to Inflammationa
| pathways | protein | CON peptides | INF peptides | Fisher FDR | |
|---|---|---|---|---|---|
| meiotic synapsis | ↑CON | 14 | 372 (+57) | 392 | 2.88 × 10−6 |
| resolution of sister chromatid cohesion | ↓INF | 5 | 118 | 98 (−31) | 2.53 × 10−5 |
| DNA damage recognition in GG-NER | ↑INF | 5 | 158 | 225 (+32) | 2.01 × 10−6 |
| activated PKN1 stimulation of the transcription of AR-regulated genes KLK2 and KLK3 | ↑INF | 5 | 97 | 131 (+24) | 1.07 × 10−6 |
| neutrophil degranulation | ↑INF | 70 | 1556 | 1976 (+105) | 8.09 × 10−8 |
| translocation of GLUT4 to the plasma membrane | ↑INF | 6 | 270 | 367 (+55) | 3.40 × 10−11 |
The size of the observed increase (decrease) over the expected number of peptides in the sample is shown in parentheses.
Table 3.
Canonical Pathways with Significant Increase or Decrease of All Distinct Peptides in Response to Inflammationa
| pathways | proteins | CON peptides | INF peptides | Fisher FDR | |
|---|---|---|---|---|---|
| meiotic synapsis | ↑CON | 21 | 455 (+74) | 484 | 2.20 × 10−8 |
| resolution of sister chromatid cohesion | ↓INF | 18 | 220 | 212 (−44) | 1.05 × 10−5 |
| COPI-independent Golgi-to-ER retrograde traffic | ↓INF | 10 | 158 | 137 (−37) | 1.05 × 10−5 |
| pentose phosphate pathway (hexose monophosphate shunt) | ↑INF | 7 | 128 | 195 (+27) | 4.87 × 10−5 |
| HSF1-dependent transactivation | ↑INF | 5 | 90 | 130 (+22) | 2.08 × 10−5 |
| attenuation phase | ↑INF | 6 | 91 | 137 (+23) | 6.36 × 10−6 |
| oxidative-stress-induced senescence | ↑INF | 13 | 143 | 205 (+30) | 2.25 × 10−6 |
| antimicrobial peptides | ↑INF | 8 | 103 | 165 (+27) | 1.54 × 10−6 |
| TP53 regulation of metabolic genes | ↑INF | 14 | 152 | 225 (+34) | 2.73 × 10−7 |
| regulation of TLR by endogenous ligand | ↑INF | 7 | 124 | 205 (+41) | 2.65 × 10−14 |
The size of the observed increase (and decrease) compared to the expected number of peptides for the sample is shown in parentheses.
The EVs studied here derived from animals bearing inflammatory and conventional tumors have been compared based on the abundances of ubiquitinated and nonubiquitinated proteins, and pathways significantly perturbed by inflammation have been cataloged. Pathways “meiotic synapsis” and “resolution of sister chromatid cohesion” are observed with a strengthened significance when all EV proteins are considered, suggesting that for these pathways, inflammatory treatment is acting independently of ubiquitination status. On the other hand, neutrophil degranulation is observed as significant when we focus on the ubiquitinated proteins, suggesting that ubiquitination status and neutrophil degranulation may be somehow connected. The absence of a strong, consistent inflammation effect on ubiquitome-focused pathways suggests that the abundant ubiquitome represent a largely independent sampling of the total proteome (with contributions3 from conjugates formed in invagination and endosomal sorting). Consistent with this pattern, in the following discussion we consider the possibility that ubiquitin conjugation occurs inside the EV.
Ubiquitin-Utilizing Enzymes
Among the many proteins identified in lysates from INF and CON EV, three families of ubiquitin-utilizing enzymes are of special interest. Identifications of multiple enzymes involved in the ubiquitin conjugation pathway are listed in Table 4. Abundances were similar in INF and CON samples. Ubiquitinactivating enzyme E1 is the major ubiquitin activation enzyme in eukaryotic cells. Ubiquitin-conjugating enzyme E2 L3 is also frequently involved in conjugation. Ubiquitin-conjugating enzyme E2 N catalyzes the synthesis of poly-ubiquitin chains that are linked through K63s and are implicated in the regulation of Toll-like, NOD-like, and RIG-I-like inflammatory pathways.41 A total of four E3 type ligases were identified in the INF sample from among several hundred active in various cells. One of these, E3 ubiquitin–protein ligase UBR4 (A2AN08) is reported to form meshwork structures with clathrin (also identified in MDSC exosomes) and may be involved in invagination.42 The conjugation ongoing in situ by this suite of enzymes could offer one explanation for the extensive and hyper-ubiquitination observed.
Table 4.
Enzymes Involved in the Ubiquitin Conjugation Pathwaya
| accession no. | protein | spectral counts, INF(-GG)a | spectral counts, CON(-GG)a |
|---|---|---|---|
| Q02053 | ubiquitin-activating enzyme E1 | 490 (6) | 356 (3) |
| P68037 | ubiquitin-conjugating enzyme E2 L3 | 38(−) | 12 (−) |
| P61089 | ubiquitin-conjugating enzyme E2 N | 8 (−) | 6 (−) |
| P22682 | E3 ubiquitin-protein ligase CBL | 6 (1) | 2 (−) |
| A2AN08 | E3 ubiquitin-protein ligase UBR4 | 19 (6) | 29 (5) |
| Q6PAV2 | probable E3 ubiquitin-protein ligase HERC4 | 5 (1) | 3 (−) |
| O08759 | ubiquitin-protein ligase E3A | 6 (1) | 2 (−) |
| Q9D906 | ubiquitin-like modifier-activating enzyme 1 | 38 (1) | 39 (2) |
| Q8C7R4 | ubiquitin-like modifier-activating enzyme 6 | 3 (−) | 4 (−) |
Entries in parentheses indicate the number of GG-tagged peptides identified.
A total of eight deubiquitinase enzymes were also identified in comparable abundances in the lysate of INF EV and CON EV, as listed in Table 5. One of these, ubiquitin carboxyl terminal hydrolase 15 is reported to act as a negative regulator of T-cell activation.22 The presence of deubiquitinases highlights the importance of deubiquitinase inhibitors during the preparation of EV samples for ubiquitome analyses. The relative abundances of ubiquitin ligases and hydrolases are similar in INF and CON EV.
Table 5.
Enzymes Involved in Hydrolyses of Ubiquitina
| accession no. | protein | spectral counts INF(-GG)a | spectral counts CON(-GG)a |
|---|---|---|---|
| Q9JMA1 | ubiquitin carboxyl-terminal hydrolase 14 | 39 (2) | 13 (2) |
| P70398 | probable ubiquitin carboxyl-terminal hydrolase FAF-X | 39 (6) | 52 (2) |
| P56399 | ubiquitin carboxyl-terminal hydrolase 5 | 43 (2) | 41 (3) |
| Q7TQI3 | ubiquitin thioesterase OTUB1 | 22 (−) | 22 (−) |
| Q6A4J8 | ubiquitin carboxyl-terminal hydrolase 7 | 72 (−) | 45 (1) |
| Q8R5H1 | ubiquitin carboxyl-terminal hydrolase 15 | 6 (3) | 5 (1) |
| B1AY13 | ubiquitin carboxyl-terminal hydrolase 24 | 7 (2) | 11 (3) |
| Q9WUP7 | ubiquitin carboxyl-terminal hydrolase isozyme L5 | 3 (−) | 9 (−) |
Entries in parentheses indicate the number of GG-tagged peptides identified.
The 26S proteasome is another protein family that hydrolyses ubiquitin conjugates and is readily identified in MDSC-derived and other EV. A total of 33 subunits were identified here (Supplementary Table 3). Spectral count ratios are included; however, the FDR values are poor and support differential abundances in only one case. High numbers of subunits of the 26S proteasome have also been identified in at least six other large-scale proteomic studies of exosomes shed by mouse and human cells,18,19,43–46 and we have suggested3 that secretion in exosomes is part of the turnover mechanism for proteasome subunits. Proteolytic activity of the 26S proteasome has been demonstrated in exosomes from mesenchymal stem cells44 and in lysate from exosomes released by tumor-associated macrophages.18 A top-down analysis in the present investigation has identified many truncated proteoforms from various proteins. A total of 192 truncated proteoforms were assigned to 29 proteins (see Supplementary Table 4.). This finding is consistent with the possibility of truncation by proteasome activity and also consistent with previous observations8,19 of extensively truncated proteins in exosome lysates. Relevant to the proinflammatory effects of MDSC EV on potential receiver cells, the proteasome has been shown to help regulate cytokine induction and to activate NFKB, which is critical for inflammation.47,48 Inflammation requires active proteasomes.
Ubiquitin conjugation, ubiquitin hydrolysis, and proteasome activity all require ATP. Extracellular vesicles do not contain mitochondria to synthesize ATP by oxidative phosphorylation; however, the five enzymes that synthesize ATP in the glycolysis pathway were all reliably identified in both INF and CON MDSC EV (Table 6). Abundances are similar. This enzyme family has previously been identified in exosomes derived from mesenchymal stem cells, where enzyme activity was confirmed for three of the proteins43 and in exosomes released by normal and cancerous prostate cells, in which ATP synthesis has been demonstrated.49 Ronquist et al. have also demonstrated that ATP is also required for internalization of exosomes (prostasomes) into receiver cells.49
Table 6.
Proteins Identified from the ATP-Generating Glycolysis Pathwaya
| accession no. | protein name | spectral counts INF(-GG)a | spectral counts CON(-gg)a |
|---|---|---|---|
| P16858 | glyceraldehyde 3-phosphate dehydrogenase | 1072 (1) | 1627 (2) |
| P09411 | phosphoglycerate kinase | 251 (3) | 215 (4) |
| Q9DBJ1 | phosphoglycerate mutase | 160 (1) | 68 (2) |
| P17182 | Enolase I AKA α enolase | 906 (1) | 1245 (2) |
| P52480 | pyruvate kinase | 941 (1) | 1945 (2) |
Entries in parentheses indicate the number of GG-tagged peptides identified.
CONCLUSIONS
This bottom-up study finds that more than 50% of the proteins carried by MDSC-derived EV contribute ubiquitinated proteo forms to the cargo and that, on average, three GG-tagged peptides represent each of these ubiquitinated proteins. It is not yet known if more than one site is ubiquitinated concurrently in a given proteoform or if K63-linked polyubiquitin conjugates dominate, as suggested previously19 for urinary tract exosomes. Our experiments indicate that at least some of the ubiquitinated proteoforms in the MDSC EV contribute to MDSC chemotaxis. We observe no correlation between the extent of ubiquitination in EV shed by MDSC and the high and low levels of inflammation in the tumor-bearing animal hosts.
Enhanced metabolic pathways defined by Reactome software found that neutrophil degranulation pathway proteins are strongly enhanced by inflammation in the ubiquitinated cohort from MDSC-derived EV. Polymorphonuclear MDSC, the dominant population induced by the 4T1 tumor, shares many characteristics with neutrophils,50 and MDSC accumulation and suppressive potency are driven by inflammation. Thus, it is not unexpected that the MDSC EV generated in the heightened inflammatory setting have enhanced levels of neutrophil degranulation pathway molecules. What remains unknown is which molecules contribute to the increased suppressive potency.
In addition, we have identified enzymes from the E1, E2, and E3 families required for ubiquitination and nine different ubiquitin hydrolases in both INF and CON exosomes. The identification of ubiquitin ligases, deubiquitin hydrolases, glycolytic enzymes for ATP synthesis, and the 26S proteasome repertoire in MDSC EV, in concert with peer reviewed reports of intraexosomal activity for the latter two families, support the hypothesis that ubiquitination and proteolysis occur within MDSC EV. We propose that MDSC EV carry an internally synthesized, possibly dynamic cohort of ubiquitinated proteins, some of which contribute to the immune suppressive mechanisms by which MDSC promote tumor progression.
Supplementary Material
ACKNOWLEDGMENTS
The research was supported by grants from the National Institutes of Health (grant nos. GM021248, HL112905, and OD019938) and a fellowship from the American Heart Association 16PRE30770016. We thank Mr. Arsh Chauhan for assistance with some of the figures and Dr. Meghan Burke for helpful discussions.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.7b00585.
Tables showing proteins identified in the MDSC-derived extracellular vesicle lysate, ubiquitinated proteins identified in the MDSC extracellular vesicles, proteasome subunits identified and ratios of spectral counts calculated, and truncated proteoforms identified by topdown mass spectrometry. (PDF)
REFERENCES
- (1).Gabrilovich DI; Ostrand-Rosenberg S; Bronte V Coordinated regulation of myeloid cells by tumours. Nat. Rev. Immunol 2012, 12 (4), 253–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Bunt SK; Yang L; Sinha P; Clements VK; Leips J; Ostrand-Rosenberg S Reduced inflammation in the tumor microenvironment delays the accumulation of myeloid-derived suppressor cells and limits tumor progression. Cancer Res. 2007, 67 (20), 10019–10026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Burke M; Choksawangkarn W; Edwards N; OstrandRosenberg S; Fenselau C Exosomes from Myeloid-Derived Suppressor Cells Carry Biologically Active Proteins. J. Proteome Res 2014, 13 (2), 836–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Geis-Asteggiante L; Ostrand-Rosenberg S; Fenselau C; Edwards NJ Evaluation of spectral counting for relative quantitation of proteoforms in top-down proteomics. Anal. Chem 2016, 88 (22), 10900–10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Burke MC; Oei MS; Edwards NJ; Ostrand-Rosenberg S; Fenselau C Ubiquitinated proteins in exosomes secreted by myeloidderived suppressor cells. J. Proteome Res 2014, 13 (12), 5965–5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Geis-asteggiante L; Dhabaria A; Edwards N; Ostrandrosenberg S; Fenselau C International Journal of Mass Spectrometry Top – down analysis of low mass proteins in exosomes shed by murine myeloid-derived suppressor cells. Int. J. Mass Spectrom 2015, 378, 264–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Chauhan S; Danielson S; Clements V; Edwards N; OstrandRosenberg S; Fenselau C Surface Glycoproteins of Exosomes Shed by Myeloid-Derived Suppressor Cells Contribute to Function. J. Proteome Res 2017, 16 (1), 238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Moreno-Gonzalo O; Villarroya-Beltri C; Sanchez-Madrid F´ Post-translational modifications of exosomal proteins. Front. Immunol 2014, 5, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Trajkovic K; Hsu C; Chiantia S; Rajendran L; Wenzel D; Wieland F; Schwille P; Brugger B; Simons M Ceramide Triggers Budding of Exosome Vesicles into Multivesicular Endosomes. Science 2008, 319 (5867), 1244–1247. [DOI] [PubMed] [Google Scholar]
- (10).Sinha P; Okoro C; Foell D; Freeze HH; OstrandRosenberg S; Srikrishna G Proinflammatory S100 Proteins Regulate the Accumulation of Myeloid-Derived Suppressor Cells. J. Immunol 2008, 181 (7), 4666–4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Bunt SK; Sinha P; Clements VK; Leips J; OstrandRosenberg S Inflammation induces myeloid-derived suppressor cells that facilitate tumor progression. J. Immunol 2006, 176, 284–290. [DOI] [PubMed] [Google Scholar]
- (12).Chen I; Xue L; Hsu C; Paez JSP; Pan L; Andaluz H; Wendt MK; Iliuk AB; Zhu J-K; Tao WA Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proc. Natl. Acad. Sci. U. S. A 2017, 114 (12), 3175–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Piper RC; Luzio JP Ubiquitin-dependent sorting of integral membrane proteins for degradation in lysosomes. Curr. Opin. Cell Biol 2007, 19 (4), 459–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Smith VL; Jackson L; Schorey JS Ubiquitination as a Mechanism To Transport Soluble Mycobacterial and Eukaryotic Proteins to Exosomes. J. Immunol 2015, 195 (6), 2722–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Hurley JH; Odorizzi G Get on the exosome bus with ALIX. Nat. Cell Biol 2012, 14 (7), 654–655. [DOI] [PubMed] [Google Scholar]
- (16).Piper RC; Lehner PJ Endosomal transport via ubiquitination. Trends Cell Biol. 2011, 21 (11), 647–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Amerik AY; Nowak J; Swaminathan S; Hochstrasser M Doa4 Deubiquitinating Enzyme Is Functionally Linked to the Vacuolar Protein-sorting and Endocytic Pathways 2000, 11, 3365–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Zhu Y; Chen X; Pan Q; Wang Y; Su S; Jiang C; Li Y; Xu N; Wu L; Lou X; et al. A Comprehensive Proteomics Analysis Reveals a Secretory Path- and Status-Dependent Signature of Exosomes Released from Tumor-Associated Macrophages. J. Proteome Res 2015, 14 (10), 4319–4331. [DOI] [PubMed] [Google Scholar]
- (19).Huebner AR; Cheng L; Somparn P; Knepper MA; Fenton RA; Pisitkun T Deubiquitylation of Protein Cargo Is Not an Essential Step in Exosome Formation. Mol. Cell. Proteomics 2016, 15 (5), 1556–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Raposo G; Stoorvogel W Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol 2013, 200 (4), 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Srikanthan S; Li W; Silverstein RL; Mcintyre TM Exosome poly-ubiquitin inhibits platelet activation, downregulates CD36 and inhibits pro-atherothombotic cellular functions. J. Thromb. Haemostasis 2014, 12 (11), 1906–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Sujashvili R Advantages of Extracellular Ubiquitin in Modulation of Immune Responses. Mediators Inflammation 2016, 2016, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Kirkpatrick DS; Denison C; Gygi SP Weighing in on ubiquitin: the expanding role of mass-spectrometry-based proteomics. Nat. Cell Biol 2005, 7 (8), 750–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Peng J; Schwartz D; Elias JE; Thoreen CC; Cheng D; Marsischky G; Roelofs J; Finley D; Gygi SP A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol 2003, 21 (8), 921–926. [DOI] [PubMed] [Google Scholar]
- (25).Danielsen JMR; Sylvestersen KB; Bekker-Jensen S; Szklarczyk D; Poulsen JW; Horn H; Jensen LJ; Mailand N; Nielsen ML Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Mol. Cell. Proteomics 2011, 10 (3), M110003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Van Nocker S; Vierstra RD Cloning and characterization of a 20-kDa ubiquitin carrier protein from wheat that catalyzes multiubiquitin chain formation in vitro. Proc. Natl. Acad. Sci. U. S. A 1991, 88 (22), 10297–10301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Denis NJ; Vasilescu J; Lambert JP; Smith JC; Figeys D Tryptic digestion of ubiquitin standards reveals an improved strategy for identifying ubiquitinated proteins by mass spectrometry. Proteomics 2007, 7 (6), 868–874. [DOI] [PubMed] [Google Scholar]
- (28).Valkevich E; Sanchez N; Ge Y; Strieter E Middle-Down Mass Spectrometry Enables Characterization of Branched Ubiquitin Chains. Biochemistry 2014, 53 (30), 4979–4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Adams K; Wang Y; Ostrand-Rosenberg S; Fenselau C Ubiquitinated proteins in MDSC exosomes analyzed by highresolution tandem mass spectrometry. Proceedings of the Annual Conference of the American Society for Mass Spectrometry; American Society for Mass Spectrometry: Santa Fe, NM, 2016. [Google Scholar]
- (30).vander Pol E; Coumans FA; Grootemaat AE; Gardiner C; Sargent IL; Harrison P; sturk A; van Leeuwen TG; Nieuwland R Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis, and resistive pule sensing. J. Thromb. Haemostasis 2014, 12, 1182–1192. [DOI] [PubMed] [Google Scholar]
- (31).Edwards N; Wu X; Tseng CW An unsupervised, modelfree, machine-learning combiner for peptide identifications from tandem mass spectra. Clin. Proteomics 2009, 5 (1), 23–36. [Google Scholar]
- (32).Edwards N PepArML: A meta-search peptide identification platform. Current Protocols in Bioinformatics 2013, 44, 13231–13232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Fellers RT; Greer JB; Early BP; Yu X; Leduc RD; Kelleher NL; Thomas PM ProSight Lite: Graphical software to analyze top-down mass spectrometry data. Proteomics 2015, 15 (7), 1235–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Old WM; Meyer-Arendt K; Aveline-Wolf L; Pierce KG; Mendoza A; Sevinsky JR; Resing K a; Ahn, N. G. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics 2005, 4 (10), 1487–1502. [DOI] [PubMed] [Google Scholar]
- (35).Benjamini Y; Hochberg Y Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B 1995, 57 (1), 289–300. [Google Scholar]
- (36).Croft D; O’Kelly G; Wu G; Haw R; Gillespie M; Matthews L; Caudy M; Garapati P; Gopinath G; Jassal B; et al. Reactome: A database of reactions, pathways and biological processes. Nucleic Acids Res. 2011, 39 (1), 691–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Choksawangkarn W; Graham LM; Burke M; Lee SB; Ostrand-Rosenberg S; Fenselau C; Edwards NJ Peptide-based systems analysis of inflammation induced myeloid-derived suppressor cells reveals diverse signaling pathways. Proteomics 2016, 16 (13), 1881–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Xu G; Paige JS; Jaffrey SR Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat. Biotechnol 2010, 28, 868–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Burke MC; Wang Y; Lee AE; Dixon EK; Castaneda CA; Fushman D; Fenselau C Unexpected Trypsin Cleavage at Ubiquitinated Lysines. Anal. Chem 2015, 87 (16), 8144–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Ostrand-Rosenberg S; Sinha P Myeloid-Derived Suppressor Cells: Linking Inflammation and Cancer. J. Immunol 2009, 182 (8), 4499–4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Corn JE; Vucic D Ubiquitin, 28 inflammation: the right linkage makes all the difference. Nat. Struct. Mol. Biol 2014, 21 (4), 297–300. [DOI] [PubMed] [Google Scholar]
- (42).Nakatani Y; Konishi H; Vassilev A; Kurooka H; Ishiguro K; Sawada J; Ikura T; Korsmeyer SJ; Qin J; Herlitz AM P600, a Unique Protein Required for Membrane Morphogenesis and Cell Survival. Proc. Natl. Acad. Sci. U. S. A 2005, 102 (42), 15093–15098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Buschow SI; van Balkom BW; Aalberts M; Heck AJ; Wauben M; Stoorvogel W MHC class II-associated proteins in Bcell exosomes and potential functional implications for exosome biogenesis. Immunol. Cell Biol 2010, 88 (8), 851–856. [DOI] [PubMed] [Google Scholar]
- (44).Lai RC; Tan SS; Teh BJ; Sze SK; Arslan F; de Kleijn DP; Choo A; Lim SK Proteolytic Potential of the MSC Exosome Proteome: Implications for an Exosome-Mediated Delivery of Therapeutic Proteasome. Int. J. Proteomics 2012, 2012 (45), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).de Jong OG; Verhaar MC; Chen Y; Vader P; Gremmels H; Posthuma G; Schiffelers RM; Gucek M; van Balkom BWM Cellular stress conditions are reflected in the protein and RNA content of endothelial cell-derived exosomes. J. Extracell. Vesicles 2012, 1 (1), 18396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Tan SS; Chen TS; Tan KH; Lim SK An overview of the proteomic and miRNA cargo of MSC-derived exosomes. Mesenchymal Stem Cell Derived Exosomes 2015, 21–36. [Google Scholar]
- (47).Ichikawa HT; Conley T; Muchamuel T; Jiang J; Lee S; Owen T; Barnard J; Nevarez S; Goldman BI; Kirk CJ; et al. Beneficial effect of novel proteasome inhibitors in murine lupus via dual inhibition of type i interferon and autoantibody-secreting cells. Arthritis Rheum. 2012, 64 (2), 493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Qureshi N; Morrison DC; Reis J Proteasome protease mediated regulation of cytokine induction and inflammation. Biochim. Biophys. Acta, Mol. Cell Res 2012, 1823 (11), 2087–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Ronquist KG; Sanchez C; Dubois L; Chioureas D; Fonseca P; Larsson A; Ullen A; Yachnin J; Ronquist G; Panaretakis T. Energy-requiring uptake of prostasomes and PC3 cell-derived exosomes into non-malignant and malignant cells. J. Extracell. Vesicles 2016, 5 (1–11), 29877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Bronte V; Brandau S; Chen SH; Colombo MP; Frey AB; Greten TF; Mandruzzato S; Murray PJ; Ochoa A; OstrandRosenberg S; Rodriguez PC; Sica A; Umansky V; Vonderheide RH; Gabrilovich ID Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat. Commun. 2016, 7, 12150–12154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.