Abstract
Most differentiated cells convert glucose to pyruvate in the cytosol through glycolysis, followed by pyruvate oxidation in the mitochondria. These processes are linked by the Mitochondrial Pyruvate Carrier (MPC), which is required for efficient mitochondrial pyruvate uptake. In contrast, proliferative cells, including many cancer and stem cells, perform glycolysis robustly but limit fractional mitochondrial pyruvate oxidation. We sought to understand the role this transition from glycolysis to pyruvate oxidation plays in stem cell maintenance and differentiation. Loss of the MPC in Lgr5-EGFP positive stem cells, or treatment of intestinal organoids with an MPC inhibitor, increases proliferation and expands the stem cell compartment. Similarly, genetic deletion of the MPC in Drosophila intestinal stem cells also increases proliferation, whereas MPC overexpression suppresses stem cell proliferation. These data demonstrate that limiting mitochondrial pyruvate metabolism is necessary and sufficient to maintain the proliferation of intestinal stem cells.
Introduction
It was first observed almost 100 years ago that, unlike differentiated cells, cancer cells tend to avidly consume glucose, but not fully oxidize the pyruvate that is generated from glycolysis 1. This was originally proposed to be due to dysfunctional or absent mitochondria, but it has become increasingly clear that mitochondria remain functional and critical. Mitochondria are particularly important in proliferating cells because essential steps in the biosynthesis of amino acids, nucleotide and lipid occur therein 2–5. Most proliferating stem cell populations also exhibit a similar glycolytic metabolic program 6–9, which transitions to a program of mitochondrial carbohydrate oxidation during differentiation 10,11. The first distinct step in carbohydrate oxidation is import of pyruvate into the mitochondrial matrix, where it gains access to the pyruvate dehydrogenase complex (PDH) and enters the tricarboxylic acid (TCA) cycle as acetyl-CoA. We, and others, recently discovered the two proteins that assemble to form the Mitochondrial Pyruvate Carrier (MPC) 12,13. This complex is necessary and sufficient for mitochondrial pyruvate import in yeast, flies and mammals, and thereby serves as the junction between cytoplasmic glycolysis and mitochondrial oxidative phosphorylation. We previously showed that decreased expression and activity of the MPC underlies the glycolytic program in colon cancer cells in vitro and that forced re-expression of the MPC subunits increased carbohydrate oxidation and impaired the ability of these cells to form colonies in vitro and tumors in vivo 14. This impairment of tumorigenicity was coincident with a loss of key markers and phenotypes associated with stem cells. This prompted us to examine whether glycolytic non-transformed stem cells might also exhibit low MPC expression and mitochondrial pyruvate oxidation, which must increase during differentiation.
Results
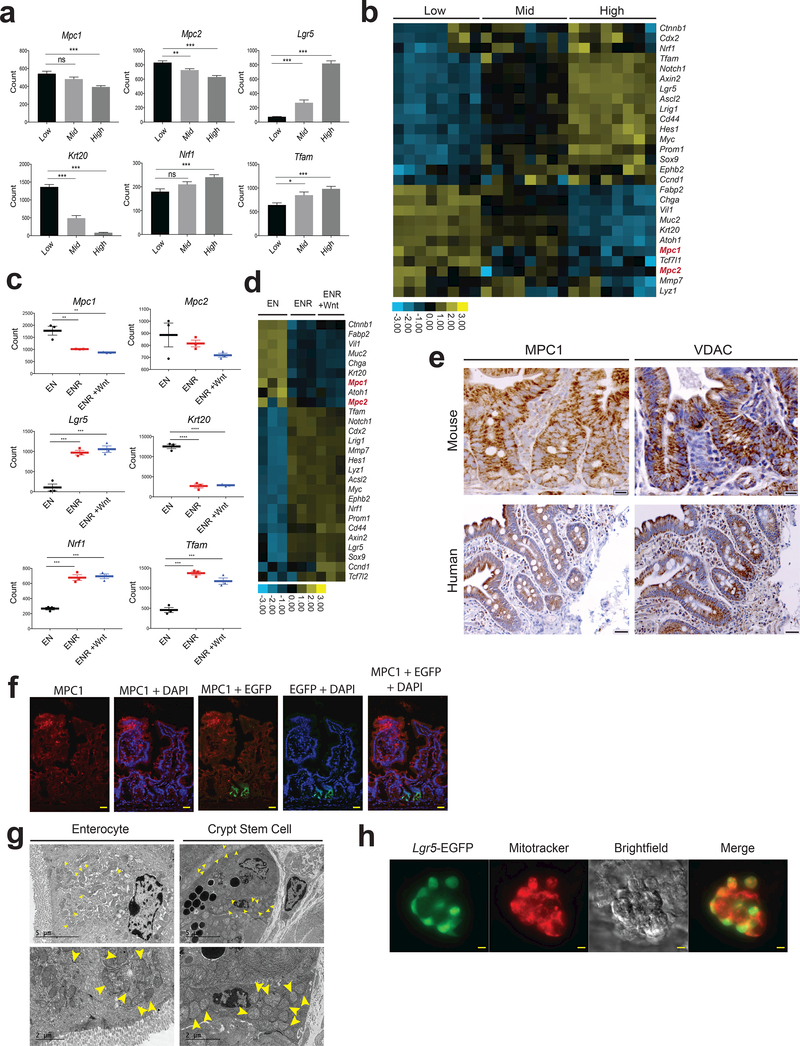
MPC expression is low in intestinal stem cells and increases upon differentiation.
As a model system, we have studied LGR5+ intestinal stem cells (ISCs), which reside near the base of the intestinal crypt and continuously proliferate. Their progeny differentiate and populate the intestinal epithelium, which is completely renewed approximately every 5 days 15,16. Using EGFP fluorescence from an Lgr5-EGFP-IRES-CreERT2 allele, we single-cell sorted intestinal crypt stem cells into low, medium and high LGR5-expressing populations (Supplemental Fig. 1a). As expected Lgr5 mRNA, as well as that of other markers of stem cells, correlated with Lgr5-EGFP expression, while Krt20 and other markers of differentiation anti-correlated with EGFP (Fig. 1a,b; Supplemental Table 1). The pattern of Mpc1 and Mpc2 expression resembled that of differentiation genes, exhibiting lower expression in the more stem-like cells that increased with differentiation. In vitro organoids maintained in stem cell or differentiation-promoting conditions displayed a similar pattern. When grown in basal medium containing EGF and Noggin, organoids exhibit a largely differentiated gene expression pattern, which is progressively more stem-like when R-spondin 1 and Wnt3a are added to the medium (Fig. 1c,d; Supplemental Table 2). Expression of Mpc1 and, to a lesser extent, Mpc2 again correlate with the expression of differentiation genes. Both in vivo and in vitro, the expression of indicators of mitochondrial biogenesis like Tfam and Nrf1 was higher in more stem-like cell populations (Fig. 1a-d) suggesting that the decreased MPC expression is not due to a global suppression of mitochondrial gene expression. Similarly, immunohistochemical analysis of the proximal small intestine (jejunum) revealed that MPC1 was nearly absent from the base of the crypt, the site of LGR5+ ISCs, but strongly expressed through the upper crypt and villus, whereas VDAC, a marker of total mitochondrial mass, was more abundant at the base of the crypt relative to the remainder of the intestinal epithelium in both mouse and human (Fig. 1e). Similar anti-correlation of MPC1 and LGR5 expression was observed by immunofluorescence staining of small intestine (Fig. 1f). This pattern of MPC1 and VDAC expression was consistent throughout the murine small intestine (jejunum and ileum) and NRF1, TFAM, and PDK1 were also more abundant in the crypt cells in human intestine while the differentiation mark CK20 was less abundant17,18 (Supplemental Fig. 1b, c). Electron microscopy also showed high mitochondrial content in crypt stem cells, and isolated Lgr5-EGFP stem cells robustly stain with a dye dependent on mitochondrial membrane potential (Fig. 1g,h). These data are consistent with the hypothesis that crypt stem cells contain functional mitochondrial, but that they are geared toward biosynthetic activities or oxidation of fatty acids or other non-carbohydrate fuels.
Figure 1: MPC expression is low in intestinal stem cells and increases upon differentiation.
a, The indicated transcripts were quantified in Lgr5-EGFP cells sorted for low, mid, and high EGFP intensity (n = 13, low and mid, n = 12 high). b, Heat map of mRNA content from the Lgr5-EGFP populations in (a). c, The indicated transcripts were quantified from organoids transferred to differentiation conditions (EN: EGF and Noggin) compared to stem-maintaining conditions containing RSPO1 (ENR) or RSPO1 and Wnt3A (ENR + Wnt) (n = 3 per treatment). d, Heat map of mRNA content from organoids in (c). e, Antibody stain of MPC1 and VDAC on crypts of proximal small intestine in mouse (top) and human (bottom). f, Immunofluorescence images of mouse proximal small intestine staining for MPC1 (red) and EGFP for Lgr5 intestinal stem cells (green). g, Electron micrographs of enterocytes (left) and crypt stem cells and surrounding paneth cells (right) at low (top) and high (bottom) magnification. Yellow arrows indicate mitochondria. h, Isolated live crypts imaged for Lgr5-EGFP and co-stained with MitoTracker Red CMXRos. Data are mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001. Scale bars, 50 μm (e, human, h) and 20 μm (e (mouse), f). P values for 1a and 1c were calculated by One-way ANOVA with correction for multiple comparisons.
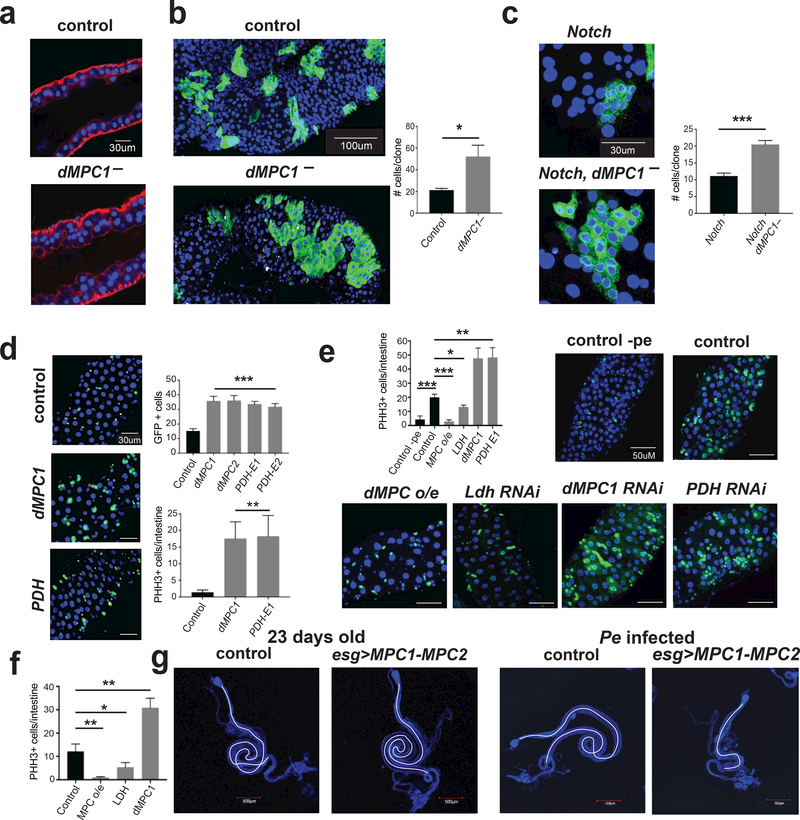
Drosophila MPC regulates intestinal stem cell proliferation.
These observations led us to hypothesize that the MPC plays an active and direct role in regulating ISC proliferation. To begin to test this hypothesis, we examined the role of the MPC in the ISCs of the fruit fly Drosophila, which share key aspects of their biology with mammalian ISCs 19,20. Both MPC1 and MPC2 are expressed in all four cell types of the intestine, with the lowest level of expression in the ISCs and the highest expression in the differentiated enteroendocrine cells 21,22. Confocal imaging of intestines dissected from dMPC1 mutants revealed that the epithelium exhibits multi-layering unlike the normal single cell layer seen in controls (Fig. 2a). This is a classic overgrowth phenotype that is associated with oncogene mutations in Drosophila 20. Accordingly, we used MARCM clonal analysis to determine if a specific loss of the MPC in ISCs leads to an increase in their proliferation 23. On average, newly-divided GFP-marked dMPC1 mutant clones are more than two-fold larger than control clones, which were generated in parallel using a wild-type chromosome, indicating that the MPC is required in the ISC lineage to suppress proliferation (Fig. 2b; Supplemental Table 7). Because GFP-marked clones could include cells that differentiate into mature enterocytes or enteroendocrine cells, we also conducted clonal analysis in the absence of Notch, thereby blocking ISC differentiation 24,25. Under these conditions, we again observed an approximately two-fold increase in the size of dMPC1 mutant ISC clones (Fig. 2c; Supplemental Table 7). To confirm and extend these results, we disrupted MPC function specifically in the ISCs by using the Dl-GAL4 driver in combination with UAS-GFP, which facilitates stem cell identification. Once again, approximately two-fold more GFP-marked stem cells were observed relative to controls when either dMPC1 or dMPC2 expression was disrupted by RNAi along with increased ISC proliferation as detected by staining for phosphorylated histone H3 (pHH3) (Fig. 2d). Similar results were obtained when RNAi was targeted to the E1 or E2 subunits of PDH to specifically disrupt the next step in mitochondrial pyruvate oxidation (Fig. 2d; Supplemental Fig. 2a; Supplemental Table 7). Importantly, an opposite phenotype was seen when Ldh was reduced by RNAi in the ISCs or progenitor cells (Fig. 2e,f; Supplemental Fig. 2a; Supplemental Table 7). Ldh suppression is known to result in a significant increase in pyruvate levels, which can promote pyruvate oxidation 26. Taken together with our results with Pdh RNAi, these observations support the model that the MPC limits stem cell proliferation through promoting oxidative pyruvate metabolism in the mitochondria. It also appears to be sufficient as specific overexpression of MPC1 and MPC2 in ISCs or progenitors caused a reduction in stem cell proliferation, the opposite of the loss-of-function phenotype. This can be seen in either Pseudomonas-infected intestines (Fig. 2e; Supplemental Fig. 2a), which undergo rapid stem cell proliferation 27, or under basal conditions in aged animals (Fig. 2f). Consistent with this, MPC overexpression under basal conditions had no effects on intestinal morphology, while the intestines from infected flies displayed a fully penetrant size reduction, likely due to the inability of ISCs to maintain tissue homeostasis (Fig. 2g; Supplemental Fig. 2b, 7c). Taken together, our results demonstrate that mitochondrial pyruvate uptake and metabolism is both necessary and sufficient in a stem cell autonomous manner to regulate ISC proliferation and maintain intestinal homeostasis in Drosophila.
Figure 2: Drosophila MPC regulates intestinal stem cell proliferation.
a, Intestines from controls and dMPC1 mutants stained with phalliodin (red) and DAPI (blue), (n = 10 control, n = 7 knockout). b, MARCM dMPC1 mutant clones marked by GFP along with quantification of clone size, (n = 13 clones from control, n = 12 clones from knockout, 15 animals assessed for each group. c, MARCM dMPC1 mutant clones with Notch RNAi marked by GFP along with quantification, (n = 31 from control, n = 38 from knockout). d, Dl-GAL4 used to target RNAi as indicated, (Control n = 8, dMPC RNAi n = 8, dMPC2 RNAi n = 8, PDH E1 n = 7, PDH E2 n = 7 experimental replicates). Increased proliferation as detected by staining for phosphorylated histone H3 (pHH3), (PHH3 + cells counted from control n= 8, dMPC1 RNAi n= 8, PDH E1 n= 7). e, Dl-GAL4 used to target RNAi as indicated or MPC overexpression (o/e) under infected conditions, (Control n = 5 (no infection) Control with infection n = 10, MPC O/E n = 10, LDH RNAi n = 10 MPC RNAi n = 10 PDH RNAi n = 10). f, esg-GAL4 was used to target RNAi as indicated or MPC overexpression (o/e) under uninfected basal conditions, 23 days old at 29°C, (Control n = 10, MPC O/E n = 10, LDH RNAi n = 10 MPC RNAi n = 10). g, Intestine size is unaffected in animals overexpressing the MPC under basal conditions in aged flies (left two panels) and reduced in infected animals (right two panels) (n = 10 for Control and MPC O/E). Data are mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001. All p values were calculated using Student’s t-test.
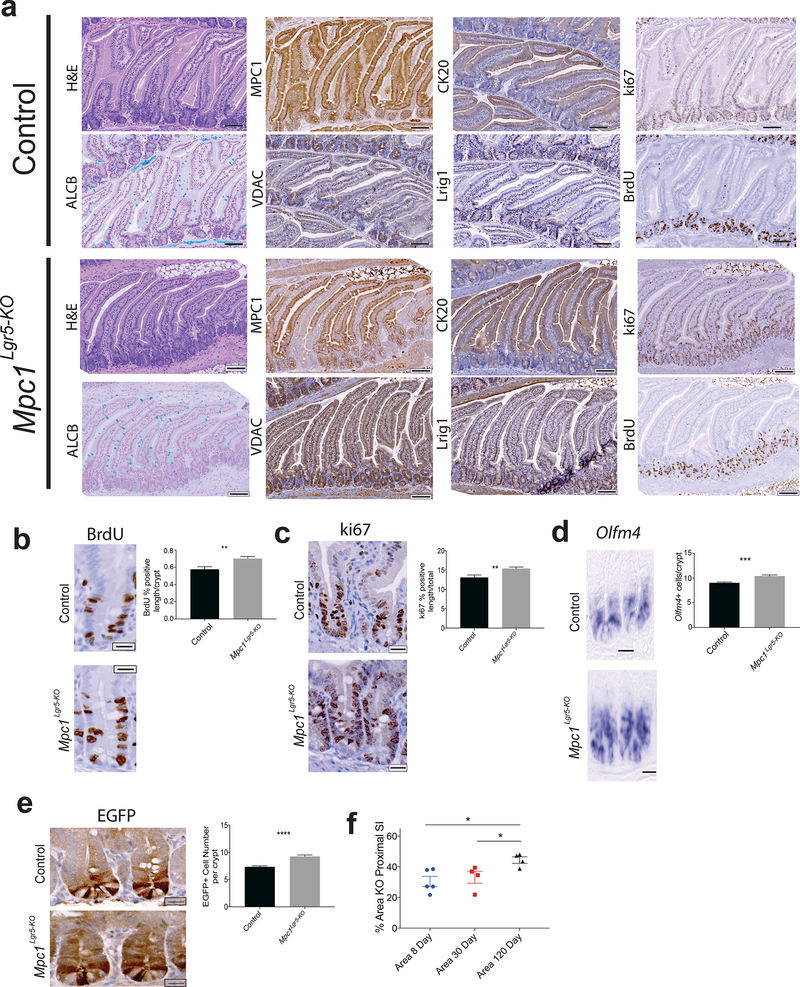
MPC deletion in the Lgr5-EGFP cells expands the intestinal stem cells compartment and increases proliferation.
To test whether loss of the mammalian MPC might have similar effects on ISC homeostasis, we generated mice carrying the Lgr5-EGFP-IRES-CreERT2 allele together with homozygous floxed alleles of Mpc1 (Supplemental Fig. 3a). Tamoxifen treatment resulted in a significant but incomplete (~30%) loss of MPC1 in the proximal small intestine at 30 days (Fig. 3a,f). Loss of MPC1 had no effect on body weight and caused no change in the morphology or height of intestinal crypts or villi (Fig. 3a; Supplemental Fig. 3b,c,d). We also observed no change in Alcian blue staining, which marks goblet cells or perturbation in the mature enterocyte marker CK20 (Fig. 3a; Supplemental Fig. 3e). These animals exhibited, however, a significant increase in the number and zone size of actively proliferating cells in the crypt as measured both by BrdU incorporation and Ki67 staining (Fig. 3b,c; Supplemental Fig. 3f). In addition, Mpc1Lgr5-KO mice had an increased number of crypt stem cells as assessed by Olfm4, Lgr5, and LRIG1 expression as well as by expression of GFP from the Lgr5-EGFP allele (Fig. 3d,e; Supplemental Fig. 3g,h,i). Consistent with these data, we found that the zone of MPC1-deficiency increased over time post-Mpc1 deletion, suggesting positive selection for Mpc1 deleted cells (Fig. 3f). These data support our results in Drosophila and demonstrate that Mpc1Lgr5-KO crypts display increased cell cycling and ISC expansion.
Figure 3: MPC deletion in the Lgr5-EGFP compartment increases intestinal stem cells maintenance and proliferation.
a, Images of H&E, MPC1, CK20, ki67, Alcian Blue, VDAC, Lrig1, and BrdU staining in control and Mpc1Lgr5-KO proximal small intestine. b, Representative images and quantification of BrdU stained percent of crypt nuclei in control and Mpc1Lgr5-KO animals (n = 9 control, n=10 knockout) c, Representative images and quantification of ki67 positive nuclei percent of total in crypts of control and Mpc1Lgr5-KO animals (n = 9 control, n=10 knockout) d, Representative images of Olfm4 in situ hybridization of control and Mpc1Lgr5-KO small intestine with quantification of positive cells per crypt (right) (n = 9 control and knockout) e, Images of GFP stained cells in control and Mpc1Lgr5-KO animals with quantification of GFP positive cells per crypt (n = 9 control and knockout) f, Percent area of proximal small intestine without MPC1 by IHC in animals 8, 30, and 160 days post-tamoxifen treatment (n = 5 for 8 and 30 day, n = 4 for 160 day). Data are mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001. Scale bars, 100 μm (a) and 20 μm (b, c, d, and e). All p values were calculated using Student’s t-test.
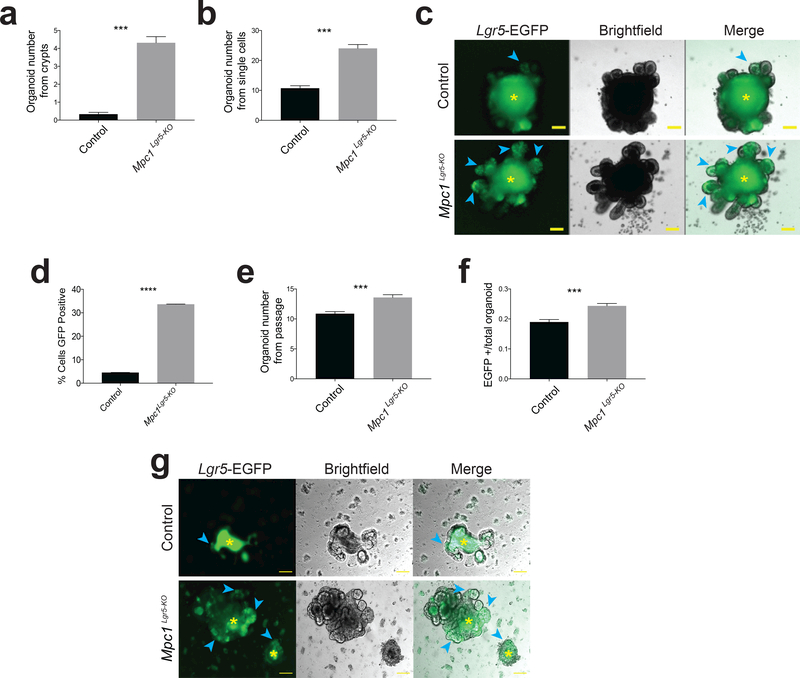
In vitro loss of the MPC increases stem cell function and organoid formation.
A dramatic increase in organoid forming potential was observed after plating crypts isolated from Mpc1Lgr5-KO mice, supporting the conclusion that these animals have an enlarged population of proliferating ISCs (Fig. 4a; Supplemental Table 7). Similar results were seen when organoids were established from single Lgr5+ stem cells treated with tamoxifen in vitro to eliminate MPC1 (Fig. 4b; Supplemental Table 7), exhibiting more Lgr5-EGFP-positive branches (Fig. 4c). When cells from these organoids were dissociated and sorted, Mpc1Lgr5-KO organoids maintained a higher fraction of GFP-positive cells (Fig. 4d; Supplemental Fig. 4a; Supplemental Table 7). When cells from these wild-type and Mpc1Lgr5-KO organoids were dissociated, but not sorted for EGFP, those from Mpc1Lgr5-KO organoids maintained their increased organoid forming ability (Fig. 4e) and were more likely to be Lgr5-EGFP positive (Fig. 4f). This increase in organoid formation of single Mpc1Lgr5-KO cells was maintained over extended passaging over two months (Supplemental Fig. 4b). Moreover, Mpc1Lgr5-KO organoids transferred to basal medium lacking the drugs that maintain stemness rapidly lost EGFP fluorescence, while Mpc1Lgr5-KO organoids retained significant EGFP expression surrounding the auto-fluorescent lumen (Fig. 4g).
Figure 4: In vitro loss of the MPC increases stem cell function and organoid formation.
a, Quantitation of organoid formation from intestinal crypts isolated from control and Mpc1 KO mice treated with tamoxifen (n = 3 control, n=4 knockout animals with 10 separate organoid cultures from each animal). b, Quantitation of organoid formation from single Lgr5-EGFP+ cells from Mpc1fl/fl intestines after in vitro vehicle (control) or tamoxifen (KO) treatment (n = 20 experimental replicates). c, Images of mature organoids grown from cells in (b). Blue arrows denote Lgr5-EGFP+ crypt domains and yellow asterisks indicate autofluorescent lumens. d, Flow cytometry for cells in (b) showing percent GFP positive (n = 5 control, = 6 knockout). e, Secondary organoid formation from single cells passaged from (b) as single live cells (n = 20 experimental replicates). f, Fraction of Lgr5-EGFP expressing secondary organoids in control and Mpc1 KO cultures (n = 20 experimental replicates). g, Images of control and Mpc1 KO organoids upon withdrawal of stem cell maintaining drugs CHIR99021 and valproic acid (representative of 2 experiments). Blue arrows denote Lgr5-EGFP+ cells and yellow asterisks indicate autofluorescent lumens. Data are mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001. Scale bars, 100 μm (c, g). All p values were calculated using Student’s t-test.
In vitro loss of the MPC alters protein expression and metabolism.
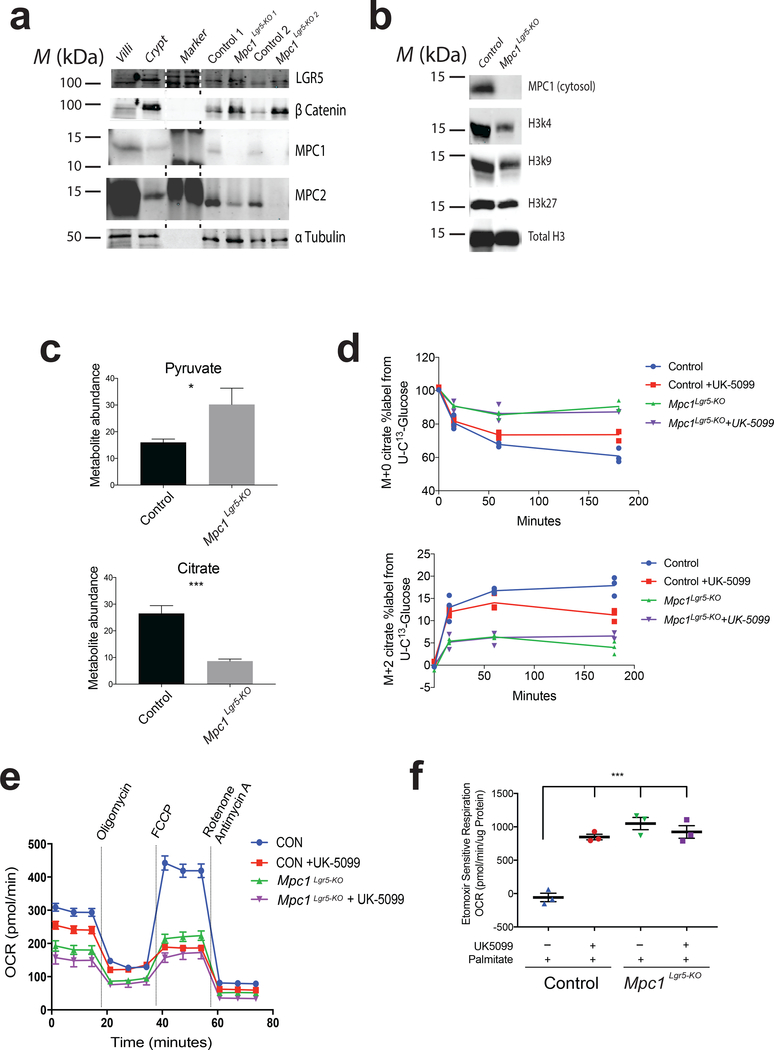
To further define the molecular effects of MPC loss, we isolated wild-type and Mpc1Lgr5-KO organoid populations and subjected them to protein analysis. As expected, Mpc1-deleted organoids had an essentially complete loss of MPC1 protein, and a significant reduction in MPC2, which is dependent on MPC1 for its stability (Fig. 5a; Supplemental Fig. 4c, 7a,d) 14,28–30. Both the LGR5 and beta-catenin stem cell markers were elevated in Mpc1Lgr5-KO organoids (Fig. 5a; Supplemental Fig. 7a), consistent with effects on the mRNAs encoding stem and differentiation marks (Supplemental Fig. 4d, Supplemental Table 3). We also observed that histone acetylation, and specifically H3K27 and H3K4 acetylation marks, were decreased in Mpc1-deleted organoids (Fig. 5b; Supplemental Fig. 4e, 7b). As expected, Mpc1Lgr5-KO organoids exhibited a steady-state increase in pyruvate and a decrease in citrate, malate and alpha-ketoglutarate, consistent with a failure of cytoplasmic pyruvate to enter the mitochondrial TCA cycle (Fig. 5c; Supplemental Fig. 4f). Mpc1Lgr5-KO organoids also showed a significant impairment in flux of 13C-glucose into the TCA cycle as measured by M+0 loss and M+2 accrual in the citrate pool (Fig. 5d; Supplemental Table 7). Oxygen consumption was also markedly impaired under both basal and FCCP-stimulated (maximal) conditions in both Mpc1Lgr5-KO organoids and wild-type organoids treated with UK-5099, which is a specific and efficacious MPC inhibitor 12,13,31 (Fig. 5e). No further decrease in oxygen consumption was observed when Mpc1Lgr5-KO organoids were treated with the inhibitor. This led us to explore whether fatty acid oxidation might be increased to compensate for loss of Mpc1-dependent carbohydrate oxidation. In fact, we found that etomoxir-sensitive respiration (i.e. fatty acid oxidation) was profoundly increased by either inhibition or genetic deletion of MPC function (Fig. 5f).
Figure 5: In vitro loss of the MPC alters protein expression and metabolism.
a, Immunoblot of replicate control and Mpc1Lgr5-KO organoid cultures with villi and crypt extract as negative and positive controls respectively. b, Immunoblot on isolated nuclei from control and Mpc1Lgr5-KO organoids for acetyl Histone 3 marks. c,. Steady state abundance of citrate and pyruvate in control and Mpc1Lgr5-KO organoids (n = 6 experimental replicates). d, Flux metabolic labeling of citrate (M+0 on top, M+2 on bottom) at times following U-13C-glucose addition in control and Mpc1Lgr5-KO organoids with and without UK-5099 treatment (n = 1 for 0 hour per condition and n= 3 for each additional time point). e, Oxygen consumption of control and Mpc1 KO organoids with and without UK-5099 utilizing glucose for respiration (n = 8 experimental replicates). f, Etomoxir sensitive oxygen consumption of control and Mpc1Lgr5-KO organoids with and without UK-5099 treatment utilizing palmitate for respiration (n = 3 experimental replicates). Data are mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001. All p values were calculated using Student’s t-test. Unprocessed western blots are provided in Supplementary Figure 7.
In vitro inhibition of MPC activity increases organoid formation.
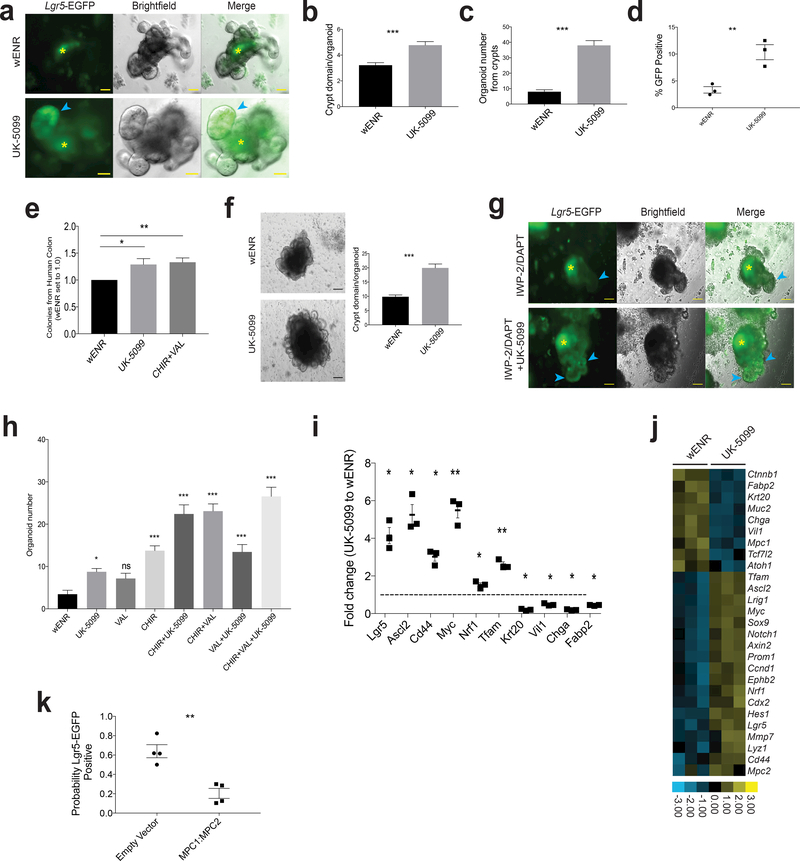
In addition to genetically MPC-deficient organoids, we next utilized the MPC inhibitor UK-5099 to examine the effects of acute MPC loss. Similar to Mpc1Lgr5-KO organoids, we found that treatment of crypt-derived organoids with UK-5099 led to enhanced maintenance of EGFP expression in the epithelial lining (Fig. 6a) and a larger number of crypt-like branches per organoid, indicative of increased stem cell activity (Fig. 6b; Supplemental Table 7). We also observed that UK-5099 treatment caused an ~4-fold increase in the efficiency of organoid formation from proximal small intestine (Fig. 6c; Supplemental Table 7). When treated organoids were dissociated and assessed by flow, UK-5099 treatment significantly increased the percentage of GFP positive cells (Fig. 6d; Supplemental Table 7). MPC inhibition enhanced organoid formation from all regions of the small intestine and from the colon to a degree similar to that of the canonical organoid maintaining condition containing valproic acid and CHIR99021 (Supplemental Fig. 5a). We also observed that human colon organoid colony formation and growth was promoted by UK-5099 treatment (Fig. 6e; Supplemental Table 7).
Figure 6: In vitro inhibition of MPC activity increases organoid formation.
a, Images of organoids from crypts with and without UK-5099. Blue arrow points to Lgr5-EGFP+ crypt domain and purple asterisks indicate autofluorescent lumens. b, Quantification of crypt domains per organoid with and without UK-5099 (n = 6 experimental replicates). c, Quantification of organoid formation from crypts from proximal small intestine with and without UK-5099 (n = 16 wENR, n= 14 UK-5099 experimental replicates). d, Flow cytometry for cells in (c) showing percent GFP positive (n = 3 replicates per condition). e, Human ascending colon crypts treated with wENR basal media (set to 1) or media containing UK-5099 or CHIR+VAL (n = 5 patients with 6–8 individual cultures per patient sample). f, Representative images and quantification of crypt domains in organoids maintained with and without UK-5099 for 12 months (wENR n = 46 organoids from 2 separate wells, UK-5099 n = 26 organoids from 2 separate wells). g, Images of organoids treated with IWP-2 and DAPT to promote differentiation with and without UK-5099. Blue arrows point to crypt domains and yellow asterisks indicate autofluorescent lumens. h, Organoid formation from crypts treated with the indicated single, double, and triple drug combinations (CHIR = CHIR99021 and VAL = valproic acid) (n = 10 experimental replicates, p values are for each treatment compared to wENR). i, Transcript abundance in organoids showing the ratio of UK5099 treated to control (n = 3 experimental replicates, p values calculated for each transcript compared to wENR). j, Heat map for additional transcripts from (i). k, Probability of cells infected with virus containing empty vector or both MPC1-iRFP and MPC2-mCherry also being GFP positive (n = 4 independent organoids with at least 15 infected cells counted). Data are mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001. Scale bars, 50 μm (a,) and 100 μm (f, g). P values for 6h were calculated by One-way ANOVA with correction for multiple comparisons. All other p values were calculated using Student’s t-test.
Interestingly, organoids could be maintained for at least one year of continuous UK-5099 treatment and they continued to display an increase in crypt domains per organoid (Fig. 6f). The addition of UK-5099 also improved passaging efficiency of early crypts (Supplemental Fig. 5b). When organoids were transferred to differentiation conditions, treatment with UK-5099 resulted in maintenance of Lgr5-EGFP expression in the crypt domains (Fig. 6g). Similarly, when removing stem cell maintaining factors and allowing passive differentiation, UK-5099 delayed the emergence of highly autofluorescent organoid lumens suggesting fewer or lower activity of differentiated secretory cells (Supplemental Fig. 5c). We also examined the effect of MPC inhibition in comparison and combination with the HDAC inhibitor valproic acid and/or the GSK-3beta inhibitor CHIR99021, both of which promote stem cell maintenance independent of the Paneth cell niche 32. UK-5099 and valproic acid caused a similar increase in organoid formation from crypts relative to controls, and resulted in a similar additive effect when combined with CHIR99021 (Fig. 6h; Supplemental Table 7) with each drug being used at its optimal concentration (Supplemental Fig. 5d). The GSK-3beta inhibitor had stronger effects than either of the other two drugs alone and was also additive with all other combinations. We conclude that MPC inhibition has profound effects on stem cell maintenance that are at least partially mechanistically distinct from those caused by HDAC or GSK-3beta inhibition. Given the previous suggestion that valproic acid might inhibit the MPC 33,34, we examined 13C-glucose flux with and without valproic acid and found that valproic acid had no effect on the kinetics of citrate labeling demonstrating that at the concentrations used, this drug does not inhibit MPC activity (Supplemental Fig. 6a).
These observations of enhanced stem cell function and previously described metabolic effects (Fig. 5d,e,f) of UK-5099 treatment were accompanied by changes in the expression of molecular markers of stemness. UK-5099 treatment in mixed organoid populations led to a robust increase in several stem cell markers, including Lgr5, Ascl2, Cd44 and Myc, along with a substantial decrease in the abundance of the Krt20, Villin1, Chga and Fabp2 differentiation markers (Fig. 6i, j, Supplemental Table 4). These data demonstrate that loss of mitochondrial pyruvate import leads to enhanced maintenance of molecular and functional markers of ISCs (and decreased markers of differentiation) to a degree similar to well-described stem cell maintaining agents.
Finally, we performed the converse of prior experiments and tested the hypothesis that forced expression of Mpc1 and Mpc2 in LGR5+ ISCs causes a loss of stemness. Indeed, using a previously validated system for overexpression 14, we found that those cells expressing exogenous Mpc1 and Mpc2, which are both required for MPC function, were much less likely to maintain Lgr5 expression, as indicated by EGFP fluorescence (Fig. 6k; Supplemental Fig. 6b; Supplemental Table 7). This is consistent with work suggesting that stimulating glucose oxidation with the small molecule dichloroacetate (DCA) or by substitution of glucose with galactose promotes differentiation 10.
Discussion
Our studies in Drosophila, intestinal organoids, and mice provide strong evidence that the MPC is necessary and sufficient in a cell autonomous manner to suppress stem cell proliferation. Consistent with this, we have demonstrated that ISCs maintain low expression of the subunits that comprise the MPC, which enforces a mode of carbohydrate metabolism wherein glucose is metabolized in the cytosol to pyruvate and other biosynthetic intermediates. This glycolytic metabolic program appears to be sufficient to drive robust and continuous stem cell proliferation 35. We also observed high mitochondrial content in ISCs, which must be geared primarily toward biosynthetic functions and/or oxidation of other substrates like fatty acids. Increased fatty acids, the metabolism of which is enhanced in MPC-deficient and MPC-inhibited organoids, have been shown to promote ISC expansion and proliferation via enhanced beta-Catenin signaling and increasing tumor initiating capacity 36. MPC expression increases upon differentiation, consistent with the shift in demand from macromolecule biosynthesis to ATP production in support of post-mitotic differentiated cell function. A similar switch in MPC expression can be seen upon differentiation of embryonic stem cells, hematopoietic stem cells, and trophoblast stem cells 37–41. Conversely, MPC expression is reduced upon reprogramming fibroblasts to iPSCs 42–45. This suggests that the effects of altering pyruvate flux that we observe herein might not be restricted to ISCs, but instead be representative of similar effects on multiple stem cell populations (see also accompanying manuscript on hair follicle stem cells). Interestingly, Myc is known to drive a metabolic program that is similar to that observed upon MPC loss, characterized by increased glycolysis and reliance on glutamine and fatty acid oxidation with reduced glucose oxidation 46–48. This suggests that Myc may play a role in repressing the MPC in stem cells, possibly acting downstream of Wnt/beta-Catenin signaling 49. Consistent with this, Myc and its repressive co-factors localize to the Mpc1 promoter and Myc expression is strongly anti-correlated with Mpc1 expression (Supplemental Fig. 6c) 14,50–53.
Taken together, our studies demonstrate that changes in the MPC and mitochondrial pyruvate metabolism are required to properly orchestrate the proliferation and homeostasis of intestinal stem cells. Importantly, this metabolic program—mediated at least partially by the MPC—appears to be instructive for, rather than a downstream consequence of, cell fate. Future work will define the extent to which the results presented herein relate to those showing that diet quality and quantity can modulate ISC behavior. Is ISC metabolism used as a signal for increased or decreased demand for intestinal epithelium? Perhaps of most importance will be to define the mechanisms whereby altered partitioning of pyruvate metabolism affects stem cell proliferation and fate. We speculate that the robust changes in fatty acid oxidation and histone acetylation, likely downstream of altered metabolite utilization for acetyl-CoA production, that we observed play an important role 36,54–56. While the mechanisms are not as yet defined, these studies establish a paradigm wherein mitochondrial metabolism does not merely provide a permissive context for proliferation or differentiation, but rather plays a direct and instructive role in controlling stem cell fate.
Materials and Methods
Mice.
Heterozygous Lgr5-EGFP-IRES-CreERT2 mice were obtained from Jackson Labs 1. MPC1 fl/fl mice were generated as previously described 2. Control animals consisted of Lgr5-EGFP-IRES-CreERT2 × MPC1 wt/wt or MPC1 fl/fl mice negative for Lgr5-EGFP-IRES-CreERT2. For in vivo knockout 6- to 8-week-old mice were injected intraperitoneally with 100ul of 20mg/ml tamoxifen (Sigma Aldrich T5648) dissolved in peanut oil (Sigma Aldrich P2144) for three successive days and sacrificed 8, 30, or 160 days later. BrdU (Invitrogen b23151) was dissolved in PBS to 10mg/ml and 100uL was injected intraperitoneally 4 hours before sacrifice to label dividing cells. Injections were carried out in the morning (between 7am and 11am) and tissue was collected 4 hours later +/− 10 minutes from the time of injection. A 2cm section was used for crypt dissociation and organoid formation as described below. The remainder of the small intestine was divided and fixed in buffered 10% formalin (Fisher Scientific 23–245-685) as previously described 3 for 48 hours, transferred to 70% ethanol and paraffin embedded for sectioning and staining. No formal randomization was performed for animal studies, animals were used as they came available, paired with littermate controls where possible, there was no preference for male or female animals, both genders were included in each analysis. All IACUC guidelines were followed and protocol submitted for animal experiments carried out in this study to the University of Utah Institutional Animal Care and Use Committee.
Drosophila Stocks.
Drosophila stocks were maintained on standard food containing 3% sucrose, 6% glucose, 8% yeast, and 1% agar in a 25°C incubator. For dMPC1 genetic studies, control flies were homozygous for a precise excision of the P(XP)CG14290[d00809] P-element insertion, and mutant flies were transheterozygous for the two deletion alleles, dMPC11 and dMPC12, as described {Bricker, 2012 #9}. Dl-GAL4 (a gift from B. Edgar) was used to target RNAi to the ISCs and esg-GAL4 (a gift from B. Edgar) was used to target RNAi to the ISCs and EBs. RNAi transgenic stocks are as follows: UAS-dMPC1 RNAi and UAS-dMPC2-RNAi {Bricker, 2012 #9}, UAS-PDH-E1 (VDRC101856), UAS-PDH-E2 (VDRC 102893), and UAS-Notch-RNAi (a gift from C. Micchelli). Temperature-sensitive Tub-GAL80ts was utilized to restrict RNAi to adults. For temperature shift experiments, animals were raised at 18°C and 4–5 day adults were transferred to 29°C for days after which intestines were dissected for analysis. Pseudomonas infections were performed as described 4. The Pseudomonas strain was a gift from B. Edgar. Overexpression of the MPC was achieved by inserting the open reading frames for dMPC1 and dMPC2 downstream from the UAS promoter in the pUAST-attB vector, with the coding sequence for the P2A peptide joining the two MPC coding regions 5. This was inserted into the attP40 site on the second chromosome using standard methods, and expression was achieved using GAL4 drivers specific to the ISCs or progenitor cells. All Drosophila studies were done following standard ethical guidelines for working with this organism. As is standard in studies of Drosophila intestinal stem cells, all experiments were performed in females, which have a higher rate of basal stem cell proliferation than males. Experiments were conducted in adults aged four days to three weeks after eclosion.
Mosaic analysis.
The MARCM system was used to generate wild-type and dMPC1 mutant lineages as previously described 6. Four to five day old adult female flies of the following genotypes were subjected to two 38°C heat pulses for 30–40 minutes: y, w, hs-FLP, UAS-GFP / w; +; Tub-GAL4, FRT82B, Tub-Gal80 / FRT82B (control), y, w, hs-FLP, UAS-GFP / w; +; Tub-GAL4, FRT82B, Tub-Gal80 / FRT82B dMPC11 (mutant). Clones were analyzed three weeks after induction. The number of nuclei in GFP-expressing clones were used for quantification. For the dMPC1 MARCM studies in the presence of Notch RNAi, intestines were analyzed five days after clone induction. Intestines were stained with antibodies to detect GFP and Prospero, with DAPI to mark nuclei. Animals of the following genotype were used: y, w, hs-FLP, UAS-GFP / w; UAS-Notch-RNAi; Tub-GAL4, FRT82B, Tub-GAL80 / FRT82B (control, Notch RNAi only), y, w, hs-FLP,UAS-GFP / w; UAS-Notch-RNAi; Tub-GAL4, FRT82B, Tub-GAL80 / FRT82B dMPC11 (Notch RNAi and dMPC1 mutant clones). Only clones that were negative for EEs (Prospero negative) and ECs (larger nuclei) were included in the analysis.
Histology and immunostaining.
Adult flies were dissected in 1×PBS and the gastrointestinal tract was fixed in 4% formaldehyde (Polysciences Inc, EM grade) overnight at 4°C. Tissues were washed four times with 0.1% Triton, 1×PBS (PBST) and incubated with PBST and primary antibodies overnight, washed again, and then incubated for 3–4 hours with secondary antibodies in PBST. Samples were mounted using Vectashield (Vector, USA) with DAPI. Antibody information is found in Supplemental Table 5. Images were acquired using an Olympus FV1000 confocal microscope and assembled into Z stack projections for the figures. For the RNAi experiments, GFP-positive cells were quantified per 40x frame in the mid-R2 region of the intestine. For the number of dividing cells, the average number of PHH3-positive cells for the entire midgut was reported.
Crypt and single-cell isolation.
Small intestinal crypts and single GFP positive cells were isolated as previously described 7. The entire small intestine was harvested, opened longitudinally and washed with PBS. Mucus and villi were scraped and discarded using a thin glass coverslip and tissue was cut into small (approximately 2cm) sections. These were washed again in PBS and incubated in 2.5 mM EDTA diluted in PBS for 1 hour with gentle agitation at 4 degree C. Crypts were liberated and collected by centrifugation (400g for 5 minutes at 4 degree C) followed by washing with PBS to remove EDTA. Crypts in PBS were passed through a 70-um cell strainer and centrifuged as before. This pellet was resuspended in 50% culture media (described below) and 50% Corning Growth Factor Reduced matrigel (Fisher Scientific #356231) for culture or dissociated for single cells. Single cell isolation was carried out as described previously 8 by resuspending crypts in TrypLE Express (Invitrogen #1260413) with DNase I (Gold Biotechnology #D-300–1) for 10 minutes at 37 degree C. Dissociated cells were washed in culture media without growth factors, centrifuged at 700g, resuspended and passed through a 35-um strainer (Fisher Scientific #08–771-23) and analyzed by FACS. Neagtive staining was carried out with DAPI and single live cells were collected, pelleted, and snap frozen for analysis or resuspended for culture.
Culture of crypts and single cells.
Crypts or single cells were maintained as previously described 7 with minimal modification. For regional specific assays of organoid formation the intestine was divided up as follows: the duodenum was classified as the first 4–5 cm, jejunum was classified as the proximal half, and ileum the distal half. The entire colon, excluding the ceacum and rectum were pooled and crypts isolated. Media consisted of advanced DMEM/F12 (Invitrogen #12634028) media prepared with HEPES (Invitrogen #15630–080), Glutamax (Invitrogen #35050061), and Penicillin-Streptomycin (Invitrogen #15140122) with N-2 supplement (Invitrogen #17502048), B27 serum free supplement (Invitrogen #17504044) N-Acetyl-L-cysteine (1 mM) containing all growth factors (Wnt3A to final concentration 2.5ng/ml: Peprotech #315–20, Noggin to final concentration 100ng/ml: Peprotech #250–38, EGF to final concentration 50ng/ml: Gold Biotechnology #1350–04-500, mRSPO1-Fc conditioned media from 293T cells provided by Calvin Kuo at Stanford University as previously described 9) and 50% matrigel (Fisher Scientific #356321). Crypts were resuspended in media and matrigel were spotted in the center of wells in a 48-well plate and allowed to polymerize at 37 degree C prior to overlaying with media. Inhibitors CHIR99021 (3 uM, R & D Systems #4423), Valproic acid (1 mM made fresh each day, R & D Systems #28–151-100), and UK-5099 (10 uM, R & D Systems #4186) used to compare organoid formation from crypts were added to the crypt matrigel mixture as well as to the media overlay and changed every 2–3 days. These drug concentrations were identified as standard from previous studies (CHIR99021: 3 uM, Valproic Acid: 1 mM, and UK-5099 10uM) but were titrated for efficacy at dosaged of 0.5x, 1.0x, 3.0x, and 9.0x. Total organoid number as well as number of crypt domains per organoid were counted after 9 days. For single cells CHIR99021 and Valproic acid were added to maintain Lgr5 stem cells as previously described 10 along with Rho Kinase inhibitor Y-27613 (10 uM Fisher Scientific #125410), Jagged-1 (1 uM AnaSpec, Inc #AS-61298).
Organoid knockout.
Single Lgr5-EGFP positive cells from MPC1 fl/fl × Lgr5-EGFP-IRES-CreERT2 were sorted and plated as above. Control genetically matched sampled received DMSO and genetic recombination was induced using (Z)-4-Hydroxytamoxifen (200nM, Fisher Scientific #34–1210). Organoids grown from these were re-sorted for live EGFP positive cells and plated as above or collected for mRNA analysis. Prior to use of knockout organoid culture in further experiments, loss of MPC1 was verified by western blot where we also looked for a significant destabilization of MPC2. If we could still detect MPC1 protein, cultures were retreated with (Z)-4-Hydroxytamoxifen and purified by flow cytometry. In addition, we routinely assessed MPC1 when blotting for other targets to ensure the knockout was stable and no wild-type escaper cells repopulated the culture.
Secondary organoids.
Organoids were passaged to single cells as above. Cell viability was determined by exclusion of 0.2% Trypan Blue (Sigma Aldrich) and both cell number and viability counted using the Cellometer Auto T4 (Nexcelom). Based on live cell passaged, organoid formation and EGFP positivity was measured after 7–9 days once mature organoids had formed.
Organoid differentiation.
Organoids grown for differentiation were passaged to single cells and allowed to establish for 6–8 days in standard media then transferred to media lacking Wnt3A and RSPO1 conditioned media (EN), Wnt3A (ENR), or media with 90ng/ml Wnt3A (ENR +Wnt). Organoids were maintained in this media for 48 hours with fresh media added after 24 hours. To promote robust differentiation IWP-2 (2 uM Fisher Scientific #35–331-0) and DAPT (10 uM Fisher Scientific #26–341-0) were added to organoid media along with vehicle (DMSO) or UK-5099 (20 uM). Images were taken of live organoids to assess the loss of GFP signal. For passive differentiation, crypts were plated identically and allowed to form in wENR with CHIR and Valproic Acid. Media was then changed to basal media (wENR) treated with vehicle (DMSO) or UK-5099 (10 uM) and the emergence of hyperintense autofluorescent organoid lumens were observed and counted.
MPC Overexpression.
Viral infection of Lgr5 stem cells was carried out as previously described with modifications 11. Organoids were dissociated to single cells as described above with TrypLE and DNase followed by resuspension, filtering, and pelleting. This cell pellet was then resuspended in 2x wENR that contained nicotinamide (10 mM), Y-27613 (10 uM), polybrene (10 ug/mL), and CHIR-99021 (3 uM). This cell suspension was added 1:1 with concentrated virus. The cell:virus mixture was incubated for 10 minutes at 37 degree, spun for 30 minutes at 200 × g at room temp, resuspended and plated into 48 well plates and incubated in a tissue culture incubator for 4 hours. After infection cells were collected from wells, centrifuged at 700 × g for 10 minutes, washed and resuspended in fresh cold media. This was mixed with matrigel as described above and spotted for organoid growth. Lentivirus was produced as previously described 12. hMPC1 was subcloned into LeGO-iG2 from Addgene #27341 with GFP substituted for iRFP713 from Addgene #45468. hMPC2 was subcloned into LeGO-iC2 from Addgene #27345 (plasmid without MPC2 also used as empty vector control). Lentiviral packaging and expression vectors were transfected into 293T with Lipofectamine 2000 (Invitrogen #11668019) to begin viral production. Supernatant was collected and pooled every 24 hours from 24–96hr. This was then spun at 1500 × g for 10 minutes to remove cell debris, passed through a 0.45 um filter, and centrifuged in ~30 mL aliquots overnight at 4 degree at 14,000 × g to concentrate viral particles. This was then resuspended in ~ 500 uL HBSS and used for infection or aliquots and frozen at −80 degrees. Organoids from single cell infection were then allowed to form and imaged 6–8 days later for analysis.
Immunohistochemistry.
All tissue processing, staining, and analysis was done in a blinded manner. Paraffin sections (3–4 um for IHC, 7 um for ISH) were cut on positively charged slides (BBC), placed at 60 degrees C for 30 minutes, and placed on automated slide stained (BenchMark, Discovery, Ventana Medical Systems) for processing. Slides were de-paraffinized with EZ prep solution, pre-treated with Cell Conditioner 1, pH 8.5, and stained with primary antibody for 1 hour at 37 degrees C (Supplemental Table 5). Secondary antibody (rabbit 1:100 for VDAC1 and MPC1, rat 1:100 for BrdU) was applied and detection performed using IView DAB detection kit followed by hematoxylin counterstaining. Slides were scanned using an automated slide scanner (Zeiss Axio Scan.Z1). For analysis of Olfm4 and Lgr5 staining, images of swiss rolled proximal small intestine were used. Areas with a high density of crypts in cross section that were continuous with villi were used for counting number of stained cells. Multiple crypts from at least 3 different areas were counted and the average cell number was calculated per mouse. At least 9 control and 9 knockout animals were used and images were unused only if staining did not work or tissue orientation in cross section was inadequate for counting. For histological analysis all small intestinal tissue was processed, stained, and imaged blinded. Animals were excluded from analysis if tissue orientation, staining, or acquisition failed and precluded analysis and for no other reason. For analysis, images of the proximal small intestine were selected that contained areas with crypts in cross-section continuous with villi. 3 separate areas and a minimum of 10 total crypts were analyzed and averaged to give a value for that animal used for statistics. IRB for histology of human intestine: U of UT IRB 91019.
Immunofluorescence staining of mouse small intestine.
Tissue for immunofluorescence was prepared as follows: the jejunum from LGR5 Cre+ mice was extracted and fixed at room temp in 4% PFA × 2.5h. This was followed by washing in PBS and equilibrating in OCT on ice then transferred to a chilled mold that was frozen on dry ice and stored at −80˚C. From this block 10um sections were cut and transferred to slides, which were cured for 30 minutes at room temp in the dark. Slides were then soaked to wash OCT by placing horizontally in a humid box at 37˚C OCT 2× 5 min. with PBS+Ca+Mg. Sections were blocked/permeablized by treating for 1 hour in humid dark box in 1×PBS+Ca+Mg, 0.1% Saponin, 10% Donkey Serum, 3% BSA. For staining: anti-MPC1 sigma cat #HPA045119 in 1×PBS + 10% Nml Donkey Serum,+3% BSA – no Saponin at 1:100, incubated in the dark overnight at 4˚C. The next morning, slides were washed 2 times with PBS for 2 minutes each. Secondary antibody (1:100 (10μg/ml Donkey anti-Rabbit IgG Alexa 594 conjugate) +5μg/ml DAPI in 1 × PBS +3% BSA) was incubated at room temp for 3 hours. Slides were then washed twice with PBS as above and mounted using 100mg/ml PVP mounting medium.(Tris buffer +0.02% Azide,+5% DABCO,+100mg/ml Polyvinylpyrrolidone+ 1/25 volume glycerol) and cured for 2 hours at room temp.
Human colon crypt isolation and growth.
Human colon biopsy samples for organoid cultures were collected under IRB 00051140, Molecular markers of sporadic hyperplastic colon polyps. Patients with a history of inflammatory bowel disease or patients with a personal or family history of familial colon cancer were excluded from the study. Briefly, study patients underwent screening colonoscopy as part of their regular clinical care. Participants scheduled for colonoscopy were not recruited for the study alone. Informed consent was obtained prior to any sedation. The study was approved by the IRB at the University of Utah and VA medical center. All samples were labeled with code numbers to protect confidentiality. All procedures were clinically indicated and recommended by the patients’ physician or consultant. As per US preventative services task force (USPSTF), colon cancer screening is recommended for subjects who are 50 years and older for detection and management of colon polyps and colon cancer.
Removing colon polyps and obtaining colonic tissue is a standard procedure during colonoscopy. The biopsy forceps used to obtain colon biopsies are 2 to 3 mm in diameter. Endoscopic biopsies are commonly taken as part of regular clinical care to diagnose problems and evaluate suspicious tissues. The risks associated with this are minor; patients do not feel the biopsies because the colon does not have pain receptors (just stretch receptors), and bleeding is self-limited. In all situations, minimizing any potential risk to the participants was a priority. If in the clinical judgment of the physician, biopsies should not be taken, they are not. In addition, if taking biopsies will make the endoscopic procedure too long, they will not be taken.
Crypt imaging.
Intestinal crypts were isolated as above and transferred to basal media lacking growth factors. Mitotracker Red CMXRos (Invitrogen #M7512) was prepared according to manufacturers instructions and added to suspended crypts in media to a final concentration of 50 nM and incubated at 37 degree C for 20 minutes.
Organoid imaging.
Live organoids were imaged on the Axio Observer Z1 imaging system (Carl Zeiss) fitted with a heated humidity controlled stage (LiveCell 05–11-0032 Rev B) and analyzed using Zen 2 software (Carl Zeiss). Laser intensity was kept constant when making comparisons of organoids and adjustments for brightness and contrast were made to the entire image equally across groups being compared.
In situ hybridization.
For detection of Lgr5 and Olfm4 mRNA, we designed riboprobes targeting the coding sequence to 3’UTR region of these genes (Supplemental Table 5). Targeted probe region DNA were amplified using standard PCR, with the amplicon length of about 500 bp. These DNA amplicons were used to generate DIG-labeled riboprobes using T7 RNA polymerase. For staining, Formalin fixed paraffin embedded tissue sections were deparaffinized with Citrisolv, washed in 100% ethanol, and air dried. Slides were blocked with Boeringer Blocking Reagent (Roche) and yeast tRNA at 68°C for 1 hour. 0.5 ng/mL probes were hybridized to sections at 68°C overnight. After washing, sections were again blocked with Boeringer Blocking Reagent and 10% sheep serum at room temperature for 1 hour. Alkaline phosphatase labeled sheep anti-DIG antibody (Roche) was incubated with sections at 1:2000 dilution at 4°C overnight. After washing, positive signals were developed using NBT/BCIP substrate (Thermo). Slides were then fixed in 10% neutral buffered formalin and mounted with aqueous mounting medium.
RNA Extraction and Analysis.
Lgr5-GFP sorted cells were directly lysed in buffer RLT (Qiagen # 79216) containing 2-mercaptoethanol (Sigma Aldrich #M6250) and total lysate assayed for transcript count as per manufacturers instructions. Total RNA was purified from organoids using the RNeasy Mini Kit (Qiagen) with 100 ng used for the assay.
mRNA analysis via NanoString.
Transcript abundance was compared using NanoString Elements platform. A custom gene set was created containing intestinal stem cell and differentiation genes including 5 housekeeping genes (ActB, B2m, Cltc, GusB, Rpl19, Tbp, and Ywhaz). nSolver analysis software was used for data analysis with raw counts normalized to positive control probes and housekeeping genes. Only samples passing quality control were used for normalization and analysis. Data was visualized with Java Treeview and graphed using Prism 7 (GraphPad). Source data for the generation of heat maps in the manuscript are provided in Supplemental Tables 1, 2, 3, and 4. Sequences for probe design and source data for the generation of heat maps has been uploaded as Supplemental Table 6.
Immunoblot analysis.
Biological replicate organoids from independent single cell isolations were harvested, washed in cold PBS and snap frozen. Pellets were resuspended in tissue lysis buffer containing protease and phosphatase inhibitors as previously described 13. Samples were disrupted by sonication, cleared by centrifugation at 10000g for 5 minutes, and protein concentration determined by BCA assay (Thermo Scientific) and diluted to allow for loading equal volumes. For histone acetylation assays organoids were collected and nuclei purified using the NE-PER Nuclear and Cytoplasmic Extraction Kit (ThermoFisher Catalog #78835) and following all instructions. Samples were resolved by SDS-PAGE, and analyzed by immunoblotting for organoid experiments and Tris-Tricine Gel for Drosophila experiments. Primary antibodies were obtained from Abcam or Cell Signaling Technology: Supplemental Table 5. Corresponding secondary antibodies (Rabbit:680 and Mouse:800 channel) were visualized using the Odyssey scanner (Li-Cor).
Steady state metabolomics.
For steady state metabolite accumulation control and knockout organoids were passaged to single cells and replated in basal media without drug (wENR). These organoids were grown for 8 days with media changes every 2–3 days and approximately 12 hours prior to harvest media was refreshed. For harvest organoids in matrigel were mechanically disrupted and dissolved using ice cold 0.9% sodium chloride, spun to 5000g for 30 seconds, supernatant aspirated and samples snap froze with liquid nitrogen. Metabolites were extracted as previously described 14. Briefly 450 uL of cold 90% methanol solution with internal standards (1 μg of d4-succinic acid) was added to each sample, vortexed briefly and sonicated for 3 minutes. This was kept at −20 degrees C for 1 hour, centrifuged at 20,000g for 5 minutes at 4 degrees C, and the supernatant transferred to a new tube, and sample was dried in a speed-vac. All GC-MS analysis was performed with a Waters GCT Premier mass spectrometer fitted with an Agilent 6890 gas chromatograph and a Gerstel MPS2 autosampler. Dried samples were suspended in 40 μL of a 40 mg/mL O-methoxylamine hydrochloride (MOX) in pyridine and incubated for one hour at 30°C. Metabolites were identified and their peak area was recorded using QuanLynx. This data was transferred to an Excel spread sheet (Microsoft, Redmond WA). Metabolite identity was established using a combination of an in house metabolite library developed using pure purchased standards and the commercially available NIST library.
Flux metabolic labeling and Cell Lines.
Organoids were passaged and grown as above. Multiple (~30) 5 uL organoid matrigel spots were plated into wells of a 6 well plate, overlaid with media, and allowed to establish for approximately 1 week. 16 hours before beginning flux labeling, media was changed to minimal media (wENR as above) with and without UK-5099 (10 uM). At time 0 sample was taken for reference and media was removed, organoid plates washed gently with warm PBS and changed to flux media consisting of SILAC Advanced DMEM/F-12 (ThermoFisher #A2494301) with 10 mM U-13C6-Glucose (CLM-1396–10), growth factors and supplements as above but lacking glutamine, unlabeled glucose, or amino acids. Flux labeling of cell lines was carried out as above in DMEM lacking any carbon source (Invitrogen 11966025) supplemented with 10 mM U-13C6-Glucose (CLM-1396–10), Glutamax (Invitrogen #35050061), and 10% Fetal Calf Serum. For drug treatment 10 uM UK-5099 and 1 mM Valproic acid were added to above media. C2C12 cells were purchased from ATCC (CRL-1772) and tMEFs were a gift from Dean Tantin. No cell lines used in this study were found in the database of commonly misidentified cell lines that is maintained by ICLAC and NCBI Biosample. The cell lines were not authenticated and were not tested for mycoplasma contamination.
Electron microscopy.
Tissue was saved for electron microscopy by cutting approximately 1mm donuts and fixed overnight at 4 degree C in fresh 2.5% glutaraldehyde (Electron Microscopy Sciences #16320) diluted in PBS. The next day tissue was washed 3 times in Cacodylate buffer. Secondary fixation was carried out in 2% osmium tetroxide at room temperature for 1 hour followed by 2 washes with cacodylate buffer followed by 1 wash with water. Staining used saturated uranyl acetate for 1 hour at room temperature, washed 3 times with water, and dehydrated with a graded ethanol series (30%, 50%, 70%, 95% × 2, 100% × 3, and finally absolute acetone). Infiltration with Epon epoxy resin (30% for 5 hours, 70% overnight, 100% for 8 hours with 3 changes, and embedded with 100% fresh resin. Polymerization in fresh resin was carried out at 60 degrees C for 48 hours followed by ultracutting (Leica UC 6ultratome) at 70 nm. Images were collected using a JEM-1400 Plus Transmission Electron Microscope (JEOL).
Seahorse.
Control and MPC1 knockout organoids for oxygen consumption studies were passaged by mechanical disruption rather than complete dissociation to single cells. Organoid fragments were spotted into a 6 well dish with multiple <10uL spots per well and overlaid with 2 mL of organoid growth media. Two days following plating at the first media change vehicle of UK5099 was added. For seahorse analysis organoids were extracted from Matrigel with gentle pipetting to minimize disaggregation. An XF96 well plate was coated with a 1:10 dilution of matrigel in PBS and allowed to sit at RT for 1 hour. Organoids were evenly seeded in each well with the respective media (wENR CV +/− UK5099). The following day organoids were run on a XF96e Analyzer for a Mito Stress Test in standard assay media (DMEM, 25mM Glucose, 2mM pyruvate, 2mM Glutamine, pH 7.4) using manufacturers protocol and standard drug concentrations (Oligomycin 2uM, FCCP 2uM, Rotenone 0.5uM, and Antimycin A 0.5uM).
For Fatty Acid Oxidation:
Substrate limited media was prepared as adapted from the stem cell recipe (all concentrations were kept the same except B27 supplement was 66.6uL, D-Glucose 0.5mM, 1mM Glutamine and Carnitine 0.5mM per 5mL of 4X media). The following drugs were added to substrate-limited media: 3uM CHIR99021, 1mM Valproic Acid, +/− 10uM UK5099. Media was changed and the organoids were maintained for 6 hrs in substrate-limited conditions. FAO Assay running buffer was standard 1X KHB (111mM NaCl, 4.7mM KCl, 1.25mM CaCl2, 2.0mM MgSO4, 1.2mM Na2HPO4) + 2.5mM D-Glucose, 0.5mM L-Carnitine, and 5mM HEPES Buffer (pH 7.4). The following drugs were added to 1X KHB: 3uM CHIR99021, 1mM Valproic Acid, +/− 10uM UK5099. Following media change into 1X KHB, the plate was maintained at 37°C in a non- CO2 incubator and experimental conditions and controls were added 30min (40uM final Etomoxir, R&D Systems cat #: 4539) and 45min (BSA or Palmitate, Agilent cat #: 102720–100) following the media change added. Following the addition of the BSA or Palmitate substrates, the assay was run in the XF96e Analyzer for the Mito Stress Test. Assay protocol was adapted as follows (3 measurements per phase, acute injection followed by 3 min mixing, 3 min waiting, 3 min measuring) with the final concentrations of drugs as follow (Oligomycin 2uM, FCCP 2uM, Rotenone 0.5uM, and Antimycin A 0.5uM). Normalization to cellular protein was quantified by Pierce BCA assay (Fisher Scientific cat #: 23227) and read on a photometric plate reader (Varioskan Flash). Results were analyzed in WAVE software and processed through the XF Mito Stress Test Report and Glycolysis Stress Test Generators.
ENCODE ChIP-Seq for MPC1 Promoter.
This track shows ChIP-seq data from the Myers Lab at the Hudson Alpha Institute for Biotechnology and by the labs of Michael Snyder, Mark Gerstein, Sherman Weissman at Yale University, Peggy Farnham at the University of Southern California, Kevin Struhl at Harvard, Kevin White at the University of Chicago, and Vishy Iyer at the University of Texas, Austin. These data were processed into uniform peak calls by the ENCODE Analysis Working Group pipeline developed by Anshul Kundaje. The clustering of the uniform peaks was performed by UCSC (https://genome.ucsc.edu/cgi-bin/hgGateway). The Factorbook motif identifications and localizations (and valuable assistance with interpretation) were provided by Jie Wang, Bong Hyun Kim and Jiali Zhuang of the Zlab (Weng Lab) at UMass Medical School.
Statistics and Reproducibility.
No pre-specified effect size was calculated, no statistical method was used to predetermine sample size. For comparisons of multiple groups an ordinary one-way ANOVA with Dunnett or Holm-Sidak correction for multiple comparisons (Figure 1a groups compared to low, 1c groups compared to EN, 4f groups compared to wENR). Statistical tests were appropriate for comparisons being made, for all t-tests and ANOVA data meet assumptions of the tests, assessment of variation was carried out but not included. For mouse studies, an n of 10 was chosen for control (equal Lgr5-EGFP heterozygous × Mpc1 wt/wt and Lgr5-EGFP wild-type × Mpc1 fl/fl) and knockout (Lgr5-EGFP heterozygous × Mpc1 fl/fl) comprised of both males and females. No statistical method was used to predetermine sample size. Experiments were not randomized. Investigators were not blinded to allocation during experiments and outcome assessment. Reproducibility: Fig. 2a: representative of two experiments; Fig. 2b: representative of two experiments; Fig. 2c representative of three experiments (pooled); Fig. 2d representative of three experiments; Fig. 2e representative of two experiments; Fig. 2f representative of two experiments; Fig. 2g representative of three experiments; Fig. 4a: representative of two experiments; Fig. 4b: representative of two experiments; Fig. 4c: representative of two experiments; Fig. 4d: representative of two experiments; Fig. 4g: representative of two experiments; Fig. 5d: representative of two experiments; Fig. 6a: representative of two experiments; Fig. 6b: representative of two experiments; Fig. 6c: representative of two experiments; Fig. 6d: representative of two experiments; Fig. 6e: four individual patients, n = 6 replicates, averaged and pooled; Fig. 6g: representative of two experiments; Fig. 6h: representative of two experiments; Fig. 6k: two independent infections, data pooled; Fig. S2a: representative of three experiments; Fig. S4b: representative of three experiments; Fig. S6a: representative of two experiments; Fig. S6b: two independent infections, data pooled (as in Fig. 6k).
Supplementary Material
Acknowledgements
We thank B. Edgar for stocks and reagents, C. Micchelli for providing the Notch RNAi line, K. Beebe for helpful advice and comments on the Drosophila intestinal studies, and G. Lam for establishing the Drosophila MPC overexpression strain, Omer Yilmaz and David Sabatini for assistance and insight into intestinal stem cell metabolism, Dean Tantin for critiques and comments, members of the Rutter lab for assistance and advice, John O’Shea, Ryan Orbus, and Christopher DeHeer for assistance with NanoString, Wojciech Swiatek for mouse assistance, ARUP Institute for Clinical and Experimental Pathology, Sheryl R. Tripp and Erica Hammond for histology, Linda Nikolova at the University of Utah Electron Microscopy Core Laboratory performed electron microscopy, Mass spectrometry analysis was performed at the Mass Spectrometry and Proteomics Core Facility at the University of Utah. Mass spectrometry equipment was obtained through NCRR Shared Instrumentation Grant # 1 S10 RR020883–01, 1 S10 RR025532–01A1, NIH 1 S10OD021505–01 (Cox) and the Diabetes and Metabolism Center at the University of Utah. This study was conducted with support from the Biorepository and Molecular Pathology Shared Resource supported by the Cancer Center Support Grant awarded to the Huntsman Cancer Institute by the National Cancer Institute of the National Institutes of Health. Nanostring transcript analysis utilized the Molecular Diagnostics Section of the Biorepository and Molecular Pathology Shared Resource and was supported by the National Cancer Institute of the National Institutes of Health under Award Number P30CA042014 (the content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH), James Marvin at the University of Utah Flow Cytometry Facility carried our flow sorting (National Cancer Institute through Award Number 5P30CA042014–24, National Center for Research Resources of the National Institutes of Health under Award Number 1S10RR026802–01), HHMI (JR), Treadwell (JR) and RO1GM094232 (to JR and CT). J.C.S. was supported by an NIH Developmental Biology Training Grant (5T32 HD07491) and a University of Utah Graduate Research Fellowship. D.R.W. was supported by a University of Utah Graduate Research Fellowship.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
Code Availability.
The software tools used for ENCODE are freely available at: https://genome.ucsc.edu/ENCODE/encodeTools.html.
Data Availability.
Previously published ENCODE ChIP-Seq data for Myc that were re-analysed here are available under UCSC accession code wgEncodeEH000545, wgEncodeEH001867, wgEncodeEH000670, wgEncodeEH002800, wgEncodeEH000536, wgEncodeEH001133.
Data corresponding to mRNA transcript abundance have been deposited at GEO under the accession numbers GSE100830 and GSE100831.
Metabolomics data have been deposited in Figshare and can be accessed under doi.org/10.6084/m9.figshare.5117269.v1.
Source data for the figures have been provided as Supplementary Table 7.
All other data supporting the findings of this study are available from the corresponding author on reasonable request.
References
- 1.WARBURG O On respiratory impairment in cancer cells. Science 124, 269–270 (1956). [PubMed] [Google Scholar]
- 2.Birsoy K et al. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell 162, 540–551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sullivan LB et al. Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell 162, 552–563 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flavell RB Mitochondrion as a multifunctional organelle. Nature 230, 504–506 (1971). [DOI] [PubMed] [Google Scholar]
- 5.Martínez-Reyes I et al. TCA Cycle and Mitochondrial Membrane Potential Are Necessary for Diverse Biological Functions. Molecular Cell 61, 199–209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simsek T et al. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell 7, 380–390 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito K & Suda T Metabolic requirements for the maintenance of self-renewing stem cells. Nat Rev Mol Cell Biol 15, 243–256 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stringari C et al. Metabolic trajectory of cellular differentiation in small intestine by Phasor Fluorescence Lifetime Microscopy of NADH. Sci. Rep 2, 568 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan Y-Y et al. A bioassay to measure energy metabolism in mouse colonic crypts, organoids, and sorted stem cells. Am. J. Physiol. Gastrointest. Liver Physiol 309, G1–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodríguez-Colman MJ et al. Interplay between metabolic identities in the intestinal crypt supports stem cell function. Nature 543, 424–427 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Berger E et al. Mitochondrial function controls intestinal epithelial stemness and proliferation. Nature Communications 7, 13171 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bricker DK et al. A Mitochondrial Pyruvate Carrier Required for Pyruvate Uptake in Yeast, Drosophila, and Humans. Science (2012). doi: 10.1126/science.1218099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herzig S et al. Identification and functional expression of the mitochondrial pyruvate carrier. Science 337, 93–96 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Schell JC et al. A role for the mitochondrial pyruvate carrier as a repressor of the Warburg effect and colon cancer cell growth. Molecular Cell 56, 400–413 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato T et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Sato T & Clevers H Primary mouse small intestinal epithelial cell cultures. Methods Mol. Biol. 945, 319–328 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Uhlen M et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol 28, 1248–1250 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Uhlen M, Pontén F & Lindskog C Charting the human proteome: Understanding disease using a tissue-based atlas. Science 347, 1274–1274 (2015). [Google Scholar]
- 19.Jiang H & Edgar BA Intestinal stem cell function in Drosophila and mice. Curr. Opin. Genet. Dev 22, 354–360 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H & Jasper H Gastrointestinal stem cells in health and disease: from flies to humans. Dis Model Mech 9, 487–499 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dutta D et al. Regional Cell-Specific Transcriptome Mapping Reveals Regulatory Complexity in the Adult Drosophila Midgut. Cell Rep 12, 346–358 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Buchon N et al. Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell Rep 3, 1725–1738 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Wu JS & Luo L A protocol for mosaic analysis with a repressible cell marker (MARCM) in Drosophila. Nature Protocols 1, 2583–2589 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Micchelli CA & Perrimon N Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 439, 475–479 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Ohlstein B & Spradling A The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 439, 470–474 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Li H et al. Drosophila larvae synthesize the putative oncometabolite L-2-hydroxyglutarate during normal developmental growth. Proc. Natl. Acad. Sci. U.S.A 114, 1353–1358 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Micchelli CA Whole-mount immunostaining of the adult Drosophila gastrointestinal tract. Methods 68, 273–279 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vigueira PA et al. Mitochondrial pyruvate carrier 2 hypomorphism in mice leads to defects in glucose-stimulated insulin secretion. Cell Rep 7, 2042–2053 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vacanti NM et al. Regulation of substrate utilization by the mitochondrial pyruvate carrier. Molecular Cell 56, 425–435 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bender T, Pena G & Martinou J-C Regulation of mitochondrial pyruvate uptake by alternative pyruvate carrier complexes. The EMBO Journal 34, 911–924 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang C et al. Glutamine oxidation maintains the TCA cycle and cell survival during impaired mitochondrial pyruvate transport. Molecular Cell 56, 414–424 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin X et al. Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nature Methods 11, 106–112 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aires CCP et al. Pyruvate uptake is inhibited by valproic acid and metabolites in mitochondrial membranes. FEBS LETTERS 582, 3359–3366 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Benavides J, Martin A, Ugarte M & Valdivieso F Inhibition by valproic acid of pyruvate uptake by brain mitochondria. Biochem. Pharmacol 31, 1633–1636 (1982). [DOI] [PubMed] [Google Scholar]
- 35.Dailey MJ Nutrient-induced intestinal adaption and its effect in obesity. Physiol. Behav 136, 74–78 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beyaz S et al. High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature 531, 53–58 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao F et al. Transcriptional and functional profiling of human embryonic stem cell-derived cardiomyocytes. PLoS ONE 3, e3474 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen B-Z et al. Identification of microRNAs expressed highly in pancreatic islet-like cell clusters differentiated from human embryonic stem cells. Cell Biol. Int 35, 29–37 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Ralston A et al. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development 137, 395–403 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Muntean AG et al. The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer Cell 17, 609–621 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keller MA et al. Transcriptional regulatory network analysis of developing human erythroid progenitors reveals patterns of coregulation and potential transcriptional regulators. Physiological Genomics 28, 114–128 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Tateno H et al. Glycome diagnosis of human induced pluripotent stem cells using lectin microarray. J. Biol. Chem. 286, 20345–20353 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishino K et al. DNA methylation dynamics in human induced pluripotent stem cells over time. PLoS Genet 7, e1002085 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saito S et al. Possible linkages between the inner and outer cellular states of human induced pluripotent stem cells. BMC Syst Biol 5 Suppl 1, S17 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X-M et al. The gene expression profiles of induced pluripotent stem cells from individuals with childhood cerebral adrenoleukodystrophy are consistent with proposed mechanisms of pathogenesis. Stem Cell Res Ther 3, 39 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Camarda R et al. Inhibition of fatty acid oxidation as a therapy for MYC-overexpressing triple-negative breast cancer. Nature Medicine 22, 427–432 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boroughs LK & DeBerardinis RJ Metabolic pathways promoting cancer cell survival and growth. Nature Cell Biology 17, 351–359 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edmunds LR et al. c-Myc programs fatty acid metabolism and dictates acetyl-CoA abundance and fate. J. Biol. Chem 289, 25382–25392 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pate KT et al. Wnt signaling directs a metabolic program of glycolysis and angiogenesis in colon cancer. The EMBO Journal (2014). doi: 10.15252/embj.201488598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gerstein MB et al. Architecture of the human regulatory network derived from ENCODE data. Nature 489, 91–100 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J et al. Factorbook.org: a Wiki-based database for transcription factor-binding data generated by the ENCODE consortium. Nucleic Acids Research 41, D171–6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang J et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 22, 1798–1812 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karolchik D et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Research 42, D764–70 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCool KW, Xu X, Singer DB, Murdoch FE & Fritsch MK The role of histone acetylation in regulating early gene expression patterns during early embryonic stem cell differentiation. J. Biol. Chem 282, 6696–6706 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Roostaee A, Benoit YD, Boudjadi S & Beaulieu J-F Epigenetics in Intestinal Epithelial Cell Renewal. J. Cell. Physiol 231, 2361–2367 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Golob JL, Paige SL, Muskheli V, Pabon L & Murry CE Chromatin remodeling during mouse and human embryonic stem cell differentiation. Developmental Dynamics 237, 1389–1398 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barker N et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Gray LR et al. Hepatic Mitochondrial Pyruvate Carrier 1 Is Required for Efficient Regulation of Gluconeogenesis and Whole-Body Glucose Homeostasis. Cell Metab. 22, 669–681 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moolenbeek C & Ruitenberg EJ The ‘Swiss roll’: a simple technique for histological studies of the rodent intestine. Lab. Anim 15, 57–59 (1981). [DOI] [PubMed] [Google Scholar]
- 60.Micchelli CA Whole-mount immunostaining of the adult Drosophila gastrointestinal tract. Methods 68, 273–279 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Daniels RW, Rossano AJ, Macleod GT & Ganetzky B Expression of multiple transgenes from a single construct using viral 2A peptides in Drosophila. PLoS ONE 9, e100637 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu JS & Luo L A protocol for mosaic analysis with a repressible cell marker (MARCM) in Drosophila. Nature Protocols 1, 2583–2589 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Sato T et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009). [DOI] [PubMed] [Google Scholar]
- 64.Sato T et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469, 415–418 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ootani A et al. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nature Medicine 15, 701–706 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yin X et al. Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nature Methods 11, 106–112 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simmini S et al. Transformation of intestinal stem cells into gastric stem cells on loss of transcription factor Cdx2. Nature Communications 5, 5728 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weber K, Mock U, Petrowitz B, Bartsch U & Fehse B Lentiviral gene ontology (LeGO) vectors equipped with novel drug-selectable fluorescent proteins: new building blocks for cell marking and multi-gene analysis. Gene Ther 17, 511–520 (2010). [DOI] [PubMed] [Google Scholar]
- 69.Schell JC et al. A role for the mitochondrial pyruvate carrier as a repressor of the Warburg effect and colon cancer cell growth. Molecular Cell 56, 400–413 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bricker DK et al. A Mitochondrial Pyruvate Carrier Required for Pyruvate Uptake in Yeast, Drosophila, and Humans. Science (2012). doi: 10.1126/science.1218099 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.