Abstract
Predictable seasonal change in photoperiod triggers a sequential change in the daily activity-rest pattern, adaptive for migration in several bird species. The night-migratory black-headed bunting (Emberiza melanocephala) is day active under short photoperiods (8 h light:16 h dark, short day sensitive). Under long photoperiods (16 h light:8 h dark), the buntings are initially day active (long day premigratory) but subsequently become intensely night active (long day migratory) and after few weeks again return to a day active pattern (long day refractory). However, it is unclear how the daily expression of circadian genes changes during photoperiod-induced seasonal life-history states (LHSs). We measured period 2 (Per2), cryptochrome 1 (Cry1), brain and muscle arnt-like protein 1 (Bmal1), and circadian locomotor output cycles kaput (Clock) mRNA expressions in various neural and peripheral tissues of buntings in different LHSs and discovered differences of ∼2 to 6 h in the phase and 2- to 4-fold in amplitude of circadian oscillations of Per2, Cry1, and Bmal1 between photoperiod-induced LHSs. Phase relationship in mRNA oscillations was altered between oscillator components in the circadian pacemaker system (retina, pineal, hypothalamus) as well as in the peripheral (liver, muscle) tissues. These results show for the first time altered waveforms of clock gene expressions in all tissues in parallel with behavioral shifts and suggest the involvement of circadian system in photoperiod induction of seasonal LHSs in a migratory species.—Singh, D., Trivedi, A. K., Rani, S., Panda, S., Kumar, V. Circadian timing in central and peripheral tissues in a migratory songbird: dependence on annual life-history states.
Keywords: black-headed bunting, Bmal1, circadian genes, Clock, cryptochrome
In long-lived animals, seasonal recurrence of biologic activities (phenology) like migration, hibernation, reproduction, and molt is achieved by adjusting the period of endogenous clocks to the annual change in the photoperiod (1, 2). Responding to the environmental photoperiod cycle demands behavioral and physiologic plasticity, as shown by the development of seasonal phenotypes in many vertebrates (3). This is particularly evident in latitudinal migratory species, which respond to photoperiod change with a precise succession of change in the behavior for accurate seasonal timing of their migration and reproduction (1). In captivity, migratory birds exhibit winter (nonmigratory, nonreproductive) and spring/summer (migratory/reproductive) phenotypes under short and long days, respectively (4, 5). When continuously exposed to long days, short day photosensitive (SD-S) migratory birds show successive seasonal life-history states (LHSs), characterized by premigratory (initiation of lipogenesis and gametogenesis; LD-pM), migratory (fat deposition and increased body mass, recrudesced gonads, and nocturnal migratory restlessness, known as Zugunruhe; LD-M) (6), and postmigratory [fat stores depleted and lean body mass, regressed gonads, return to day activity, and development of photorefractoriness; long day refractory (LD-R)] phenotypes (7–10).
Although several studies have shown the involvement of circadian rhythms in photoperiod-induced seasonal phenotypes to be linked with migration and reproduction in photoperiodic species (1, 8, 9), the change in molecular circadian oscillator with seasonal behavior in nonmodel organisms is less well understood. Circadian pacemaker in the suprachiasmatic nuclei shows phase plasticity with winter and summer photoperiod in seasonal mammals (11). In vertebrates, the circadian molecular oscillator is based on transcriptional-translational feedback loops, in which periods (Per1, Per2) and cryptochromes (Cry1, Cry2) are negative regulators of their own transcription driven by brain and muscle arnt-like protein 1 (Bmal1) and circadian locomotor output cycles kaput (Clock) transcriptional activators. Nuclear hormone receptors Rors and Rev-erbs function as positive and negative elements driving oscillations in Bmal1 and adjust the phase of oscillations of other clock genes. There is emerging evidence for the involvement of this molecular oscillator in the regulation of seasonality in birds. For example, Clock polymorphism has been found to be associated with breeding in the blue tit (Cyanistes caeruleu) (12) and migratory barn swallows (Hirundo rustica) (13). Similarly, midday and midnight Ror-α and Rev-erb-α mRNA expressions have been found to vary with photoperiod-induced phenotypes in the night-migratory black-headed bunting (Emberiza melanocephala) (10).
However, it remains largely unknown how the circadian clock connects to photoperiod-induced seasonal change in physiology and behavior in vertebrates. We hypothesized phase and amplitude plasticity in the oscillations of Per2, Cry1, Bmal1, and Clock genes with photoperiod-induced phenotypes in a seasonal species. A migratory species like the black-headed bunting is an ideal model system to test this because photoperiod-induced LHSs in buntings can be easily and reproducibly induced within a few weeks in controlled laboratory conditions. We predicted that differences in the phase and/or amplitude of circadian gene oscillations between photoperiod-induced LHSs would accompany the diurnal behavioral shifts in buntings. Corresponding with LHSs, a change was also expected in the relationship between circadian molecular oscillators constituting the central circadian pacemaker system (hypothalamus, retina, pineal) (1, 14) as well as between the central clock and peripheral (liver, muscle) tissues in buntings.
MATERIALS AND METHODS
Animals and experiment
The experiment was carried out at the Department of Zoology, University of Lucknow (Lucknow, India), in accordance with the guidelines of the Institutional Animal Ethics Committee. Adult male black-headed buntings (body mass, 23–27 g) procured from an overwintering flock and maintained under short days (8 h light:16 h darkness, photosensitive; n = 72) or long days (16 h light:8 h dark, photorefractory; n = 24) and constant temperature (22 ± 2°C) were used. To examine photoperiod induced changes, birds were singly housed in activity cages (size, 60 × 45 × 35 cm) and placed individually in photoperiodic chambers maintained under 8 h light or 16 h light photoperiods. After 1 wk, the birds on short days were continued for another week on short days (group 1; SD-S). Birds on long days were continued (16 h light:8 h dark) under that condition for 1 wk, during which birds initiated fat deposition and testis recrudescence but were still day active (group 2, premigratory; LD-pM), or until the time (11–18 d) they exhibited 7 nights of Zugunruhe, characterized by intense night activity and wing whirring (6), increased body weight, and matured testes (group 3, migratory; LD-M). A separate group of birds were maintained under long days for up to 40 wk, during which they went through premigratory and migratory or night active phases and reverted back to day active (photorefractory birds, group 4; LD-R). Although these long photoperiod-induced phenotypes also overlapped with breeding status [nonbreeding, prebreeding, breeding, and postbreeding (4)], we refer these LHSs with respect to their more distinct migration behavior. The experimental protocol is shown in Fig. 1A.
Figure 1.
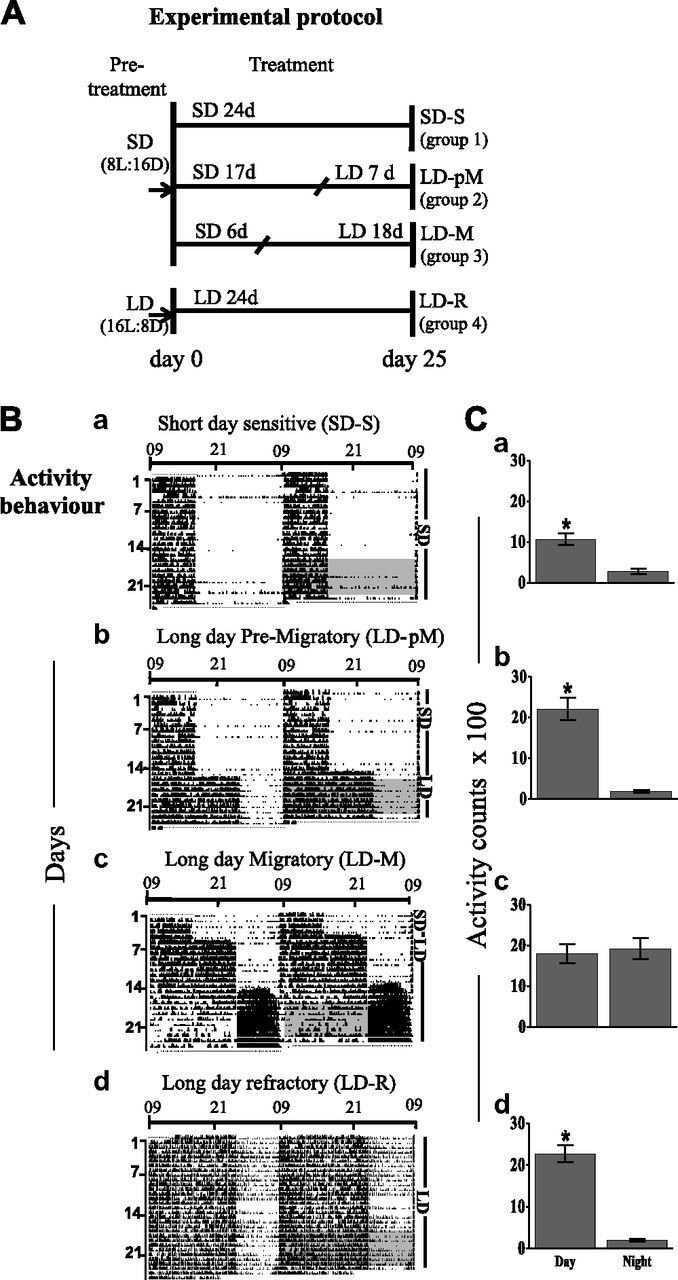
A) Experimental protocol. Photosensitive black-headed buntings maintained on short days (SDs; 8 h light:16 h darkness, pretreatment) were divided in 3 groups and singly housed in activity recording cages. Group 1 was retained on SD throughout the 24 d treatment period (SD-S), while groups 2 and 3 were exposed first to SDs and then to long days (LDs; 16 h light:8 h dark) for varying durations [group 2 (LD-pM) – SD 17 d + LD 7 d; group 3 (LD-M) – SD 6 d + LD 18 d]. A group of photorefractory buntings previously maintained on LDs were simultaneously housed individually in activity recording cages and maintained on the same long photoperiod for the next 24 d (LD-R). On d 25, all birds were humanely killed and tissues collected for measurement of gene expressions. B, C) Change in activity behavior with photoperiod-induced seasonal phenotypes. Representative double-plotted activity record (B, actogram) and percentage daily activity distribution between light day and dark period (C) in black-headed buntings with photoperiodically induced seasonal states under SDs (Ba, Ca, SD-S) and LDs (Bb, Cb, LD-pM; Bc, Cc, LD-M; Bd, Cd, LD-R). Gray boxed portion of actogram (B) indicates 7 d segment for which total activity during day and night was calculated. Asterisk on bar (C) indicates significant difference in distribution of activity between light and dark periods of 24 h LD cycle (P < 0.05, Bonferroni correction).
The activity behavior of each bird was longitudinally monitored over the entire duration of the experiment. Each activity cage was provided with 2 perches and mounted with an infrared sensor that continuously detected the movement of the bird within its cage. Activity data were collected in 5-min bins in a computerized data-logging system. The Chronobiology Kit software program (Stanford Software Systems, Santa Cruz, CA, USA) was used to collect, plot, and analyze daily activity data. Daily activity records over the period of treatment (actogram) were double plotted, with each successive day repeated sideways, to illustrate a better visual representation of activity behavior of individual birds. We also averaged hourly activity over 7 d, and from this, we calculated total activity during the day and night for each condition. The measurement on gene expression was done in each LHS at 6 times of day, with reference to lights on [zeitgeber time (ZT)—ZT1, ZT5, ZT9, ZT13, ZT17, and ZT21; ZT0 = lights on].
Measurement of gene expression level by real-time quantitative PCR
The retina, pineal, hypothalamus, liver, and muscle were quickly dissected and immediately stored in RNAlater (AM7020; Ambion, Austin, TX, USA), first overnight at 4°C and then at −80° until RNA extraction. Total RNA was extracted using Tri reagent (AM9738; Ambion), as per the manufacturer’s protocol, and a 1 µg RNA aliquot treated with RQ1 DNase (M6101; Promega, Madison, WI, USA) was used for cDNA synthesis using the first strand cDNA synthesis kit (K1622; Thermo Fisher Scientific, Waltham, MA, USA). Gene-specific primers were designed from the cDNA sequences using the Eurofins MWG Operon primer design (http://www.operon.com/tools/oligo-analysis-tool.aspx) online program. Transcript levels were measured in 10 ng/µl (hypothalamus, retina, liver, muscle, testes) or 5 ng/µl (pineal, fat tissue) by real-time PCR [7200 SDS Thermal cycler; Applied Biosystems (ABI), Foster City, CA, USA] using Power SYBR Green PCR Master Mix (ABI 4387669), with β-actin as a reference gene. A fold change in expression of each sample (in duplicate) was calculated as 2−(ΔΔCt) (15, 16).
Statistical analysis
One-way ANOVA followed by Newman-Keuls post hoc test assessed significance in daily variation in gene expression levels in a photoperiod-induced phenotype. Two-way ANOVA tested the effect of LHSs on activity behavior and clock gene expression levels (factor 1 = LHS; time of day = factor 2). Post hoc comparison used Bonferroni correction for the significance level. To determine circadian oscillations, the data were fitted to a sine wave equation, y = baseline + amplitude × sin (frequency x + phase shift), and gene expression frequency was fixed to 24 h (Cosinor analysis) (17). The significance of regression analysis determined at P < 0.05; the value was calculated using the number of samples, R2 value, and numbers of predictors (16). The amplitude and peak expression times, as determined by Cosinor analysis, were graphed in polar (SigmaStat, version 12 software; SigmaStat, San Jose, CA, USA) to show phase relationships in the mRNA expressions between photoperiod-induced phenotypes and between central clock and peripheral tissues. Further, the extra sum of squares F test using P < 0.05 as a threshold level of significance determined significant differences in the phase and amplitude of gene oscillations between LHSs (17). Unless specified otherwise, the statistics were performed by GraphPad Prism software, version 5.0 (GraphPad Software, La Jolla, CA, USA).
RESULTS
Daily activity pattern changes with photoperiod-induced states
All the individuals showed the same activity pattern. Buntings were day active under short days, as expected, with significantly higher activity during the day than at night (P < 0.05, Bonferroni correction; Fig. 1Ba, Ca). Upon exposure to long days, they maintained their daily activity pattern initially for 10 to 14 d, with significantly higher activity during the day compared with night (Fig. 1Bb, Cb; P < 0.05, Bonferroni correction). Thereafter, there was a behavioral shift, with birds becoming predominantly night active; they exhibited intense activity at night, resembling Zugunruhe (Fig. 1Bc). At this stage, there was no day-night difference in the activity distribution (Fig. 1Cc). After a few weeks under long day conditions, the birds became photorefractory. Photorefractory birds had daily activity pattern behavior similar to those under short days, with activity significantly higher during the day than at night (Fig. 1Bd) (P < 0.05, Bonferroni correction; Fig. 1Cd). Thus, there was a reversal of the activity pattern between the nonmigratory (SD-S, LD-pM, and LD-R) and migratory (LD-M) states (cf. Fig. 1B, C). Two-way ANOVA revealed a significant effect of LHSs (F3,104 = 14.38, P < 0.0001) and time (F1,104 = 86.84, P < 0.0001), as well as LHSs by time interaction (F3,104 = 17.06, P < 0.0001) on activity behavior (Fig. 1B, C).
Daily mRNA oscillations in central clock and peripheral clock tissues
We measured the mRNA expression level of 4 different clock components in 5 different tissues collected at 6 different time points from birds at 4 different life-history states. Such comprehensive and parallel analyses offered, for the first time, how circadian clock components’ expression changed in both neural and metabolic tissues of a migratory species. There were significant variations in daily mRNA expression levels of both the negative (Per2 and Cry1) and positive (Bmal1 but not Clock) elements of the transcriptional-translational feedback loops, albeit with tissue-specific and LHS-dependent expression patterns (P < 0.05, 1-way ANOVA; Fig. 2).
Figure 2.
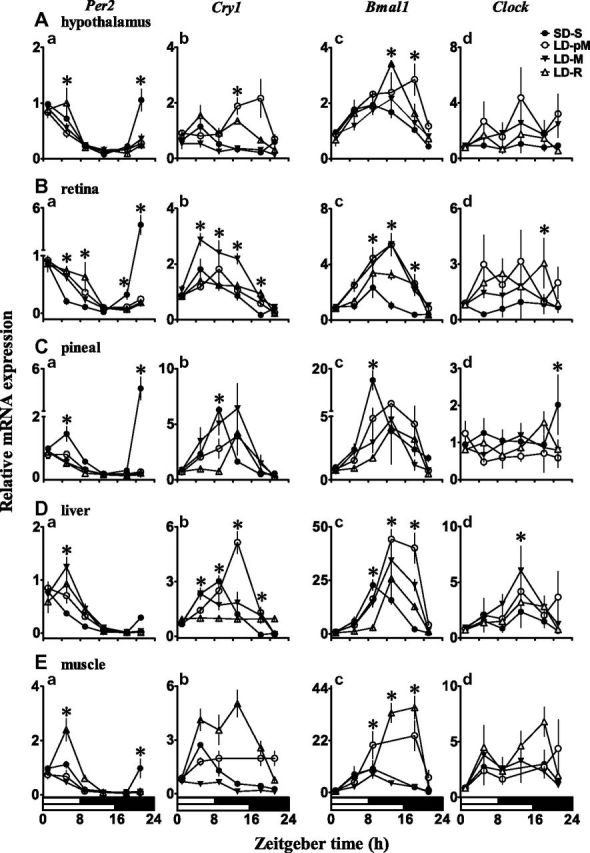
Daily expression profile of circadian clock genes in central clock and peripheral tissues. Relative mRNA expression of Per2 (Aa, Ba, Ca, Da, Ea), Cry1 (Ab, Bb, Cb, Db, Eb), Bmal1 (Ac, Bc, Cc, Dc, Ec), and Clock (Ad, Bd, Cd, Dd, Ed) genes in central [hypothalamus (A); retina (B); pineal (C)] and peripheral [liver (D); muscle (E)] clock tissues in photoperiod induced seasonal (behavioral) states in black-headed buntings under short days (SD-S) and long days (LD-pM, LD-M, and LD-R). mRNA expression levels were measured with reference to light-dark ZT at 6 times of day: ZT1, ZT5, ZT9, ZT13, ZT17, and ZT21 (ZT0 = lights on, indicated on x axis underneath bottom LD bars). Each data point represents mean (±se, n = 4). Asterisk on symbol indicates significant difference between groups at P < 0.05 level, as indicated by Bonferroni correction after 2-way ANOVA (factor 1, photoperiod-induced state; factor 2, time of day). Note differences in y axis scales between graphs.
In general, Per2 and Cry1 mRNA expression patterns were significantly varied in all the tissues during all LHSs (P < 0.001, 1-way ANOVA; Fig. 2Aa, b; Ba, b; Ca, b), except in the hypothalamic Cry1 mRNA expression during the LD-pM (P = 0.0695) and LD-M (P = 0.061) states (Fig. 2Ab). Per2 mRNA expression levels peaked in the morning hours, while Cry1 mRNA levels peaked early in the evening in the central clock (Fig. 2Ab, Bb, Cb) and at variable times in the peripheral (Fig. 2Db, Eb) tissues. Similarly, a significant daily variation in Bmal1 mRNA expression with peak levels later in the day (ZT8 to ZT14, depending on LHS) was found in all tissues (P < 0.01, P < 0.001; Fig. 2Ac, Bc, Cc). Clock mRNA expression, however, lacked a significant oscillation during all the LHSs, except LD-R, in which pineal, hypothalamus, liver, and muscle had significant Clock oscillations (P < 0.05, P < 0.01; Fig. 2Ad, Bd, Cd). There were variable Clock mRNA expression peaks distributed from the early morning to evening, particularly in liver and muscle (Fig. 2Dd, Ed).
The Cosinor analysis confirmed Per2, Cry1, and Bmal1 mRNA rhythmicity, with tissue- and LHS-specific peak expression times (Table 1). In general, Per2 peaks were scattered between ZT21 and ZT5, Cry1 peaks between ZT3 and ZT12, and Bmal1 peaks between ZT8 and ZT14 (Table 1). At the same time, Clock peaks were less distinguished.
TABLE 1.
Rhythm parameters (means ± sem; Cosinor analyses) of Per2, Cry1, and Bmal1 oscillations in central clock (hypothalamus, retina, pineal) and peripheral (liver, muscle) tissues in black-headed bunting with photoperiod induced with difference seasonal LHSs
| Tissue | Gene |
SD-S |
LD-pM |
LD-M |
LD-R |
||||
|---|---|---|---|---|---|---|---|---|---|
| Acrophase | Amplitude | Acrophase | Amplitude | Acrophase | Amplitude | Acrophase | Amplitude | ||
| Hypothalamus | Per2 | 0.3 ± 0.4 | 0.5 ± 0.06 | 1.9 ± 0.6 | 0.3 ± 0.05 | 1.9 ± 0.5 | 0.3 ± 0.05 | 3.1 ± 0.6 | 0.47 ± 0.08 |
| Cry1 | 3.8 ± 0.9 | 0.4 ± 0.09 | 12.5 ± 2.5 | 0.7 ± 0.45 | 4.5 ± 2 | 0.12 ± 0.06 | 8.1 ± 1.2 | 0.45 ± 0.13 | |
| Bmal1 | 9.0 ± 0.4 | 0.7 ± 0.08 | 13 ± 1 | 0.9 ± 0.23 | 11.5 ± 0.5 | 0.64 ± 0.08 | 12.0 ± 0.5 | 1.2 ± 0.22 | |
| Retina | Per2 | 21.6 ± 1 | 1.4 ± 0.39 | 3.0 ± 0.4 | 0.4 ± 0.06 | 2.9 ± 0.5 | 0.39 ± 0.05 | 4.2 ± 0.6 | 0.4 ± 0.07 |
| Cry1 | 6.2 ± 0.1 | 0.72 ± 0.1 | 8.3 ± 0.5 | 0.7 ± 0.09 | 8.1 ± 0.4 | 1.2 ± 0.15 | 8.8 ± 0.7 | 0.5 ± 0.08 | |
| Bmal1 | 8.3 ± 1 | 0.8 ± 0.21 | 11.4 ± 0.5 | 2.8 ± 0.30 | 11.2 ± 0.4 | 2.2 ± 0.23 | 11.7 ± 0.8 | 1.6 ± 0.35 | |
| Pineal | Per2 | 23 ± 1 | 1.4 ± 0.40 | 3.2 ± 0.5 | 0.4 ± 0.05 | 2.8 ± 0.7 | 0.3 ± 0.06 | 2.3 ± 0.6 | 0.34 ± 0.05 |
| Cry1 | 8.4 ± 0.9 | 2.3 ± 0.36 | 11.9 ± 1 | 1.6 ± 0.36 | 9.9 ± 0.9 | 2.8 ± 0.65 | 12.4 ± 1.6 | 1.2 ± 0.53 | |
| Bmal1 | 10 ± 1 | 5.3 ± 1.3 | 13.0 ± 0.6 | 3.6 ± 0.56 | 11.0 ± 0.8 | 1.86 ± 0.35 | 13.7 ± 0.98 | 1.74 ± 0.45 | |
| Liver | Per2 | 1.6 ± 0.6 | 0.4 ± 0.05 | 3.7 ± 0.5 | 0.4 ± 0.06 | 4.6 ± 0.5 | 0.57 ± 0.07 | 4.7 ± 0.6 | 0.47 ± 0.07 |
| Cry1 | 7.7 ± 0.3 | 1.5 ± 0.12 | 11.6 ± 0.6 | 2.0 ± 0.32 | 8.9 ± 0.9 | 0.91 ± 0.22 | 6.1 ± 3.3 | 0.02 ± 0.02 | |
| Bmal1 | 10.1 ± 0.5 | 11 ± 1.28 | 14.1 ± 0.4 | 23.5 ± 2.6 | 13.4 ± 0.5 | 16.6 ± 2.3 | 14.0 ± 0.8 | 11.0 ± 2.3 | |
| Muscle | Per2 | 1.6 ± 0.5 | 0.66 ± 0.1 | 2.9 ± 0.7 | 0.4 ± 0.08 | 2.4 ± 0.7 | 0.33 ± 0.06 | 4.7 ± 0.6 | 1.0 ± 0.16 |
| Cry1 | 5.9 ± 0.6 | 1.0 ± 0.16 | 13.4 ± 1.46 | 0.5 ± 0.26 | 4.9 ± 0.6 | 0.3 ± 0.05 | 10.4 ± 0.6 | 2.0 ± 0.36 | |
| Bmal1 | 8.9 ± 0.9 | 5.1 ± 1.2 | 13.4 ± 0.5 | 14.2 ± 2.2 | 10.3 ± 0.7 | 3.0 ± 0.52 | 14.3 ± 0.55 | 18.1 ± 2.6 | |
Effect of photoperiod-induced seasonal phenotypes
The mRNA oscillation of clock genes significantly varied with time of day as well as with development of seasonal phenotypes, as induced by the photoperiod change. In both the central clock (retina, pineal, hypothalamus) and peripheral (liver, muscle) tissues, Per2, Cry1, and Bmal1 mRNA expressions showed a significant effect of the LHS and time of day as well as the interaction between the two (P < 0.05, P < 0.001, 2-way ANOVA; LHS, factor 1; time of day, factor 2; Fig. 2). However, there were tissue-specific effects of these factors on Clock mRNA expression levels (Fig. 2). Clock expression was significantly affected by the LHS, not by the time of day or LHSs by time of day interaction in all the central clock tissues (P < 0.05, P < 0.01), except for the time of day effect in retinal expression (P < 0.05, 2-way ANOVA). There were variable effects of these 2 factors in liver and muscle. The LHS and time of day, not their interaction, had an effect in the liver Clock mRNA expression (P < 0.001), and there was only the effect of the time of day in muscle (P < 0.02, 2-way ANOVA; Fig. 2Dd, Ed).
Change in phase and amplitude of gene oscillations
Because of the lack of significant daily variations in Clock expression in central clock tissues, we calculated and plotted the phase and amplitude in Per2, Cry1, and Bmal1 mRNA oscillations in relation to the photoperiod-induced LHS in different tissues. Hypothalamic Cry1 mRNA expression did not have a significant oscillation in the LD-pM and LD-M states, and they were thus discounted from the phase and amplitude analysis and plots.
The peak expression time was considered to be the phase reference point of the gene oscillation. There was a significant change in the phase of Per2, Cry1, and Bmal1 oscillations by ∼2 to 6 h between the SD-S and other 3 long day states (LD-pM, LD-M, LD-R), although with tissue-specific differences (Fig. 3). For example, compared with the time in SD-S, Per2 peaks in all 3 long day states were delayed by ∼2 to 5 h in the central and by ∼2 h in peripheral clock tissues (P < 0.05; F test; Fig. 3Aa, Ba, Ca, Da, Ea), except in muscle, in which Per2 peak was significantly delayed in the LD-R by ∼3 h (Fig. 3Ea). Only in the retina were the phases of expression of Per2, Cry1, and Bmal1 under all 3 long day conditions coherent (Fig. 3), while in other tissues the phases under 3 long day conditions differed. A parsimonious explanation is that the phase of the circadian oscillator in the retina is primarily dictated by the photoperiod, while in other tissues the metabolic state and photoperiod interact in setting the phase. Phase change in the Cry1 oscillation was relatively less consistent. For example, compared with SD-S, there was a delay of ∼2 to 3.5 h in the central tissues and ∼4 to 6.5 h in the peripheral tissues. In particular, there was significant delay of ∼2 to 2.5 h in the Cry1 peak in LD-pM, LD-M, and LD-R in retina (P = 0.007, P = 0.007, P = 0.009) and ∼3 to 7 h in LD-R in hypothalamus (P = 0.008), LD-pM and LD-R in pineal (P = 0.017, P = 0.008) and muscle (P = 0.038, P = 0.02), and LD-pM in liver (P < 0.001, F test; Fig. 3Ab, Bb, Cb, Db, Eb). However, phase change in the Bmal1 oscillations between photoperiod-induced phenotypes was similar to the Per2 mRNA. Compared with the peak expression time in the SD-S, there was a significant delay of ∼3 to 4 h in Bmal1 mRNA peaks in all 3 long day states LD-pM, LD-M, and LD-R in the hypothalamus (P = 0.002, P = 0.0004, P = 0.005), retina (P = 0.046, P = 0.027, P = 0.05), and liver (P < 0.0001, P = 0.0002, P = 0.0002), but only in LD-pM in pineal (P = 0.04) and LD-pM and LD-R in the muscle (P = 0.007, P = 0.027, F test; Fig. 3Ac, Bc, Cc, Dc, Ec).
Figure 3.
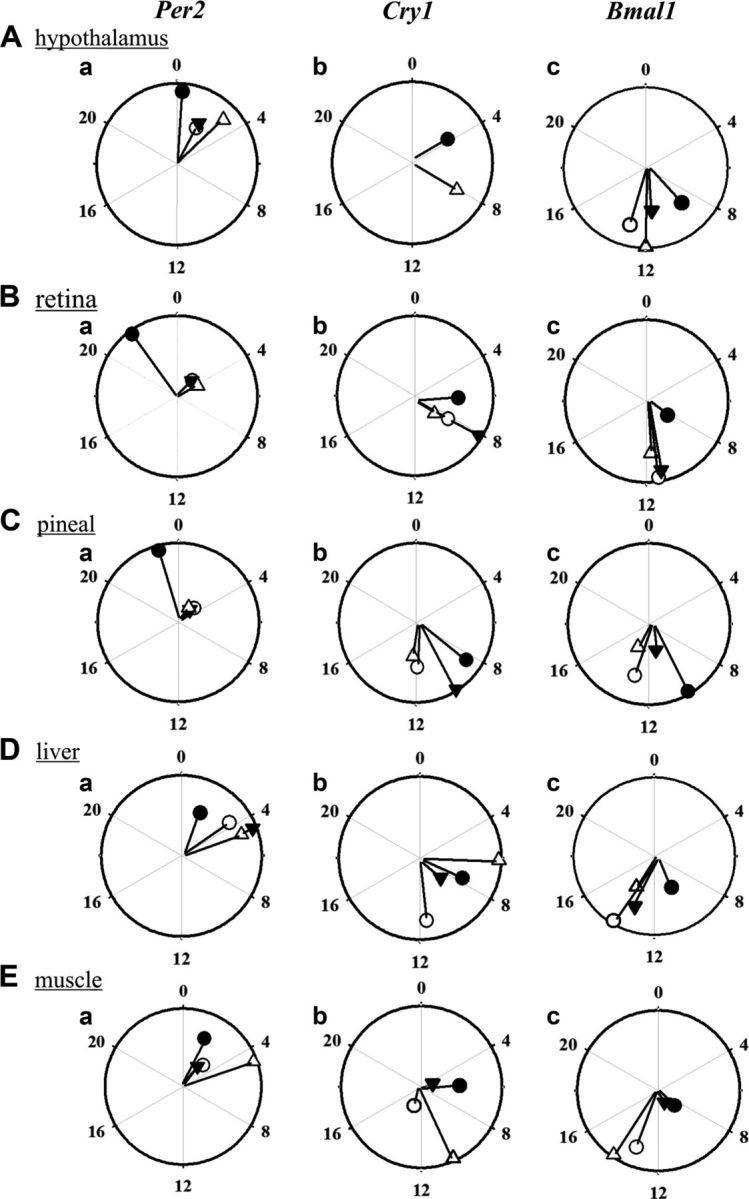
Relative presentation of phase and amplitude of clock gene oscillations in photoperiod-induced seasonal phenotypes. Central clock [hypothalamus (A), retina (B), pineal (C)] and peripheral [liver (D), muscle (E)] tissues in different photoperiod-induced behavioral states in black-headed buntings (n = 4) under short days (SD-S, solid circle) and long days (LD-pM, open circle; LD-M, solid triangle and LD-R, open triangle), as determined by Cosinor analysis. Amplitude (line) and peak time (bullets) of Per2 (Aa, Ba, Ca, Da, Ea), Cry1 (Ab, Bb, Cb, Db, Eb), and Bmal1 (Ac, Bc, Cc, Dc, Ec) mRNA expressions are shown. Long line indicates higher amplitude of mRNA expression.
The amplitude of Per2, Cry1, and Bmal1 oscillations significantly varied by ∼2- to 4-fold between the photoperiod-induced phenotypes, but with tissue-specific differences. In all 3 central clock tissues, Per2 oscillations were 2- to 4-fold higher in amplitude in SD-S than in LD-pM, LD-M, and LD-R (except in hypothalamus in LD-R) states (P < 0.05, F test; Fig. 3Aa, Ba, Ca). In contrast, the amplitude of Per2 oscillation was reduced by half in SD-S than LD-M and LD-pM in liver (P = 0.019) and muscle (P = 0.046, P = 0.008, respectively) (Fig. 3Da, Ea). However, muscle Cry1 oscillations remained significantly larger by 2- to 3-fold in SD-S and LD-M (P < 0.0001, P = 0.01, F test; Fig. 3Eb). Like phase, the amplitude variation in Cry1 oscillations was also less consistent (Fig. 3Ab, Eb). There was no difference in the hypothalamic Cry1 oscillation amplitudes between the SD-S and LD-R states. Retinal Cry1 oscillation amplitudes were ∼2-fold higher in LD-M than in SD-S (P = 0.007), LD-R (P < 0.001), and LD-pM (P = 0.002) states; LD-R oscillations were lowest in amplitude, SD-S > LD-R (P < 0.05, F test; Fig. 3Ab, Bb). There was no difference in Cry1 oscillation amplitudes in the pineal (Fig. 3Cb). In liver and muscle, Cry1 oscillations were ∼2- to 6-fold higher in amplitude in LD-R than the SD-S, LD-pM, and LD-M; lowest-amplitude Cry1 oscillations were found in the LD-M (P < 0.05, F test; Fig. 3Db, Eb). Similarly, Bmal1 oscillation amplitude showed variations. Hypothalamic Bmal1 oscillations did not vary with the photoperiod-induced states, except a 2-fold-higher amplitude occurred in LD-R rather than LD-M (P = 0.02, F test; Fig. 3Ac). Retinal Bmal1 oscillations were, however, ∼2- to 3-fold higher in amplitude in all 3 long day–induced states (LD-pM, LD-M, and LD-R) compared with SD-S (P < 0.001, P < 0.001, P < 0.05, F test; Fig. 3Bc). In the pineal, the amplitude of Bmal1 oscillations was higher by ∼2- to 3-fold in SD-S and LD-pM than the LD-M and LD-R (P < 0.01, F test; Fig. 3Cc). Similar differences were found in the peripheral clock tissues. Liver Bmal1 oscillations were higher in amplitude in LD-pM than the SD-S, LD-M, and LD-R (P < 0.0001, P < 0.05, P < 0.001), and in LD-M than the SD-S (P = 0.037, F test; Fig. 3Dc). In the muscle, however, Bmal1 oscillations were higher by ∼3- to 6-fold in the LD-pM and LD-R than the SD-S and LD-M states (P < 0.001, F test; Fig. 3Ec).
Tissue-specific phase relationships in circadian genes
The times of peak mRNA expressions was also used to show phase-relationships in Per2, Cry1, and Bmal1 between photoperiod-induced states in the central clock as well as the peripheral tissues. In all LHSs, Per2 mRNA expression peaked early in the day, whereas Bmal1 and Cry1 peaked later in the day (Fig. 3). The times of transcriptional peaks were relatively advanced in SD-S than the other 3 states in all the tissues (cf. Fig. 3). Also in SD-S, Per2 peaked earlier in the pineal and retina than in the hypothalamus (cf. Fig. 3). Further, Per2, Cry1 (except in hypothalamus), and Bmal1 mRNA peaks were delayed on transfer from short (SD-S) to long (LD-pM) days (cf. Fig. 3). Between the SD-S and LD-R, both nonstimulated states, mRNA peaks were delayed in the latter (Fig. 3). However, mRNA expression peak times did not differ between the photostimulated phases LD-pM and LD-M (Fig. 3). Interestingly, the phase relationship between Per2 and Cry1 genes, with increased interval between their peak expression times, was altered with the induction of the long day phenotype—that is, when buntings were transferred from short (SD-S) to long (LD-pM) days (Fig. 3).
DISCUSSION
We for the first time show the persistence of circadian molecular oscillations in the central clock as well as the peripheral tissues during all photoperiod-induced LHSs in a migratory species (Fig. 2). Present results on mRNA oscillations support an interactive central circadian clock system in buntings (14) and indicate a functional hierarchy between oscillators in the retina, pineal, and hypothalamus as well as between these oscillators and those in the peripheral tissues. There were advanced circadian transcription peaks in the retina and pineal relative to those in the hypothalamus (Fig. 3). Similarly, a delayed Per2 peak in peripheral tissues may account for the time lag between central pacemaker output and entrained oscillations in the peripheral tissues (18). This suggests a close interaction between the central and peripheral clocks, with the former influencing the latter (cf. Fig. 3). However, in general, bunting’s clock gene oscillations were of higher amplitude in peripheral rather than central clock tissues (Figs. 2 and 3). We speculate that this was related to the difference in the homogeneity of cellular oscillations in central clock and peripheral oscillators. Heterogeneity in cellular oscillations, a known feature of the suprachiasmatic nuclei clock in mammals (19), may account for larger variations in the phase and period of cellular oscillations and in turn the low amplitude oscillations in bunting’s retinal, pineal, and hypothalamic clocks (20). At the same time, the peripheral tissues with relatively homogenous cell population and low variations in the phase and period may have generated the high-amplitude molecular oscillations.
Phased core clock gene mRNA oscillations with long day cycle, irrespective of the LHS (Fig. 3), are suggestive of the interaction of light with a specific light-responsive element in the promoter region, a clock gene (21). Circadian transcription is regulated mainly by 3 clock-controlled DNA promoter region elements: E/E0 box in the morning (22, 23), D box during the day (23, 24), and the Rev-Erb/ROR-binding element at night (23, 25, 26). A Per2 peak early in the day may thus be the result of an interaction of light with the morning light–responsive element, and Cry1 and Bmal1 mRNA peaks later during the day are perhaps the results of an interactive effect of light with all 3 promoter elements (23). Further, phase relationships between the circadian gene oscillations may determine long day–induced physiologic response in black-headed buntings, as suggested in the other photoperiodic species (27, 28). Increased phase intervals between the Per2 and Cry1 peaks in buntings when they were transferred from short to long days are evidence of this (Fig. 3).
This study asked whether circadian transcriptions were dependent on photoperiod-regulated annual LHSs in migratory black-headed buntings. By and large, the answer is yes, mainly on the basis of observed differences in the phase and amplitude of Per2, Cry1, and Bmal1 mRNA oscillations between the short and long photoperiods and between different LHSs (Fig. 3). In particular, the advanced phase and the relatively larger amplitude of Per2 oscillation in SD-S compared to those in the LD-pM, LD-M or LD-R states suggest photoperiod effects on circadian molecular oscillations in buntings. This is also consistent with the suggested photoperiod-dependent seasonal plasticity in the suprachiasmatic nuclei clock gene expression waveforms in mammals, with narrow- and high-amplitude oscillations under short days, and broad- and low-amplitude oscillations under long days (11).
Among all clock components examined, Bmal1 expression was largely rhythmic in different tissues, and its expression level, phase, and amplitude showed tissue- and photoperiod-specific changes. The change in molecular oscillator can thus be exemplified with the Bmal1 expression. In general, we found the amplitude of Bmal1 oscillation in all tissues (except pineal) increased and the phase of oscillation delayed under long days relative to the short day. However, the phase of Bmal1 oscillation was similar in all central clock and peripheral tissues, irrespective of the LHS. However, Bmal1 mRNA oscillations in all 5 tissues examined were distinct from each other in all 3 long day–induced LHSs: LD-pM, LD-M, and LD-R. This observation is novel in that it suggests that both photoperiod and internal factors interact to specify the daily expression pattern of clock components. Further, the overt activity rest pattern is perhaps a poor predictor of tissue-level daily oscillation of clock genes, particularly Bmal1. Although the birds were mostly equally active between day and night (akin to arrhythmicity), Bmal1 mRNA expression clearly showed daily oscillations in all tissues in birds during the migratory state (LD-M). It will be interesting to test in a future study whether this oscillation is driven by the photoperiod or any sustained rhythm in feeding. Furthermore, despite the overt similarity in the activity patterns under pre- and postmigratory long day conditions, the distinct pattern of Bmal1 expression in all tissues suggests that the metabolic states of the birds at these 2 LHSs might contribute to differences in Bmal1 expression.
In summary, this is the first comprehensive study at the transcriptional level showing the involvement of the circadian system in photoperiod-induced changes in seasonal LHSs in a photoperiodic species. There were altered waveforms of clock gene expression in parallel with photoperiod-induced change in the activity behavior. We propose that this is a potential mechanism underlying photoperiodism in songbirds in particular and vertebrates in general.
Acknowledgments
The authors are grateful to M. Gorman (University of California, San Diego, CA, USA) who helped to analyze the present results. Generous funding from the Department of Biotechnology [New Delhi, India (BT/PR4984/MED/30/752/2012)], Science and Engineering Research Board [New Delhi, India (IR/SO/LU-02/2005, SR/SO/AS-50/2010)], Indo–U.S. Joint Center on Biological Timing, and University of Delhi–Department of Science and Technology Promotion of University Research and Scientific Excellence (DU-DST PURSE) funding is acknowledged.
Glossary
- Clock
circadian locomotor output cycles kaput
- Cry
cryptochrome
- LD-M
long day migratory
- LD-pM
long day premigratory
- LD-R
long day refractory
- LHS
life-history state
- Per
period
- SD-S
short day sensitive
- ZT
zeitgeber time
REFERENCES
- 1.Kumar V., Wingfield J. C., Dawson A., Ramenofsky M., Rani S., Bartell P. (2010) Biological clocks and regulation of seasonal reproduction and migration in birds. Physiol. Biochem. Zool. , 827–835 [DOI] [PubMed] [Google Scholar]
- 2.Helm B., Ben-Shlomo R., Sheriff M. J., Hut R. A., Foster R., Barnes B. M., Dominoni D. (2013) Annual rhythms that underlie phenology: biological time-keeping meets environmental change. Proc. Biol. Sci. , 20130016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stevenson T. J., Ball G. F. (2011) Information theory and the neuropeptidergic regulation of seasonal reproduction in mammals and birds. Proc. Biol. Sci. , 2477–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Misra M., Rani S., Singh S., Kumar V. (2004) Regulation of seasonality in the migratory male blackheaded bunting (Emberiza melanocephala). Reprod. Nutr. Dev. , 341–352 [DOI] [PubMed] [Google Scholar]
- 5.Ramenofsky M., németh Z. (2014) Regulatory mechanisms for the development of the migratory phenotype: roles for photoperiod and the gonad. Horm. Behav. , 148–158 [DOI] [PubMed] [Google Scholar]
- 6.Gwinner E., Czeslik D. (1978) On the significance of spring migratory restlessness in caged birds. Oikos , 364–372 [Google Scholar]
- 7.Gwinner E. (1986) Circannual Rhythms, Springer-Verlag, Heidelberg, Berlin [Google Scholar]
- 8.Bartell P. A., Gwinner E. (2005) A separate circadian oscillator controls nocturnal migratory restlessness in the songbird Sylvia borin. J. Biol. Rhythms , 538–549 [DOI] [PubMed] [Google Scholar]
- 9.Rani S., Malik S., Trivedi A. K., Singh S., Kumar V. (2006) A circadian clock regulates migratory restlessness in the blackheaded bunting, Emberiza melanocephala. Curr. Sci. , 1093–1096 [Google Scholar]
- 10.Trivedi A. K., Kumar J., Rani S., Kumar V. (2014) Annual life history–dependent gene expression in the hypothalamus and liver of a migratory songbird: insights into the molecular regulation of seasonal metabolism. J. Biol. Rhythms , 332–345 [DOI] [PubMed] [Google Scholar]
- 11.Lincoln G., Messager S., Andersson H., Hazlerigg D. (2002) Temporal expression of seven clock genes in the suprachiasmatic nucleus and the pars tuberalis of the sheep: evidence for an internal coincidence timer. Proc. Natl. Acad. Sci. USA , 13890–13895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liedvogel M., Szulkin M., Knowles S. C. L., Wood M. J., Sheldon B. C. (2009) Phenotypic correlates of Clock gene variation in a wild blue tit population: evidence for a role in seasonal timing of reproduction. Mol. Ecol. , 2444–2456 [DOI] [PubMed] [Google Scholar]
- 13.Caprioli M., Ambrosini R., Boncoraglio G., Gatti E., Romano A., Romano M., Rubolini D., Gianfranceschi L., Saino N. (2012) Clock gene variation is associated with breeding phenology and maybe under directional selection in the migratory barn swallow. PLoS One , e35140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar V., Singh B. P., Rani S. (2004) The bird clock: a complex, multi-oscillatory and highly diversified system. Biol. Rhythm Res. , 121–144 [Google Scholar]
- 15. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta c(t)) method. Methods , 402–408 [DOI] [PubMed] [Google Scholar]
- 16.Singh D., Rani S., Kumar V. (2013) Daily expression of six clock genes in central and peripheral tissues of a night-migratory songbird: evidence for tissue-specific circadian timing. Chronobiol. Int. , 1208–1217 [DOI] [PubMed] [Google Scholar]
- 17.Kiessling S., Eichele G., Oster H. (2010) Adrenal glucocorticoids have a key role in circadian resynchronization in a mouse model of jet lag. J. Clin. Invest. , 2600–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cermakian N., Sassone-Corsi P. (2002) Environmental stimulus perception and control of circadian clocks. Curr. Opin. Neurobiol. , 359–365 [DOI] [PubMed] [Google Scholar]
- 19.Welsh D. K., Logothetis D. E., Meister M., Reppert S. M. (1995) Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron , 697–706 [DOI] [PubMed] [Google Scholar]
- 20.Peirson S. N., Butler J. N., Duffield G. E., Takher S., Sharma P., Foster R. G. (2006) Comparison of Clock gene expression in SCN, retina, heart, and liver of mice. Biochem. Biophys. Res. Commun. , 800–807 [DOI] [PubMed] [Google Scholar]
- 21.Chen R., Schirmer A., Lee Y., Lee H., Kumar V., Yoo S. H., Takahashi J. S., Lee C. (2009) Rhythmic per abundance defines a critical nodal point for negative feedback within the circadian clock mechanism. Mol. Cell , 417–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gekakis N., Staknis D., Nguyen H. B., Davis F. C., Wilsbacher L. D., King D. P., Takahashi J. S., Weitz C. J. (1998) Role of the Clock protein in the mammalian circadian mechanism. Science , 1564–1569 [DOI] [PubMed] [Google Scholar]
- 23.Ukai-Tadenuma M., Yamada R. G., Xu H., Ripperger J. A., Liu A. C., Ueda H. R. (2011) Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell , 268–281 [DOI] [PubMed] [Google Scholar]
- 24.Falvey E., Marcacci L., Schibler U. (1996) DNA-binding specificity of PAR and C/EBP leucine zipper proteins: a single amino acid substitution in the C/EBP DNA-binding domain confers PAR-like specificity to C/EBP. Biol. Chem. , 797–809 [PubMed] [Google Scholar]
- 25.Harding H. P., Lazar M. A. (1993) The orphan receptor Rev-ErbA alpha activates transcription via a novel response element. Mol. Cell. Biol. , 3113–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ueda H. R., Chen W., Adachi A., Wakamatsu H., Hayashi S., Takasugi T., Nagano M., Nakahama K., Suzuki Y., Sugano S., Iino M., Shigeyoshi Y., Hashimoto S. (2002) A transcription factor response element for gene expression during circadian night. Nature , 534–539 [DOI] [PubMed] [Google Scholar]
- 27.Lincoln G. A., Andersson H., Loudon A. (2003) Clock genes in calendar cells as the basis of annual timekeeping in mammals—a unifying hypothesis. J. Endocrinol. , 1–13 [DOI] [PubMed] [Google Scholar]
- 28.Yasuo S., Watanabe M., Tsukada A., Takagi T., Iigo M., Shimada K., Ebihara S., Yoshimura T. (2004) Photoinducible phase-specific light induction of Cry1 gene in the pars tuberalis of Japanese quail. Endocrinology , 1612–1616 [DOI] [PubMed] [Google Scholar]


