Summary
Neuronal polarity in the developing cortex begins during the early stages of neural progenitor migration toward the cortical plate and culminates with the specification of the axon and dendrites. Here, we demonstrate that the Ran-dependent nucleocytoplasmic transport machinery is essential for the establishment of cortical neuron polarity. We found that Ran-binding protein 1 (RanBP1) regulates axon specification and dendritic arborization in cultured neurons in vitro and radial neural migration in vivo. During axonogenesis, RanBP1 regulates the cytoplasmic levels of the polarity protein LKB1/Par4, and this is dependent on the nuclear export machinery. Our results show that downstream of RanBP1, LKB1 function is mediated by the STK25-GM130 pathway, which promotes axonogenesis through Golgi regulation. Our results indicate that the nucleocytoplasmic transport machinery is a main regulator of neuron polarity, including radial migration, and that the regulated export of LKB1 through RanBP1 is a limiting step of axonogenesis.
Keywords: neuron polarity, Ran, RanBP1, LKB1, STK25, nuclear export, axon, axonogenesis
Graphical Abstract
Highlights
-
•
The nucleocytoplasmic transport machinery regulates neuron polarity
-
•
Ran/RanBP1 regulates LKB1 availability during axonogenesis
-
•
Ran/RanBP1 couples nucleocytoplasmic transport and Golgi regulation
How neurons acquire their polarity is not fully understood. Mencarelli et al. show that the nucleocytoplasmic transport machinery is a key regulator of neuron polarity, including radial migration and axonogenesis. They envisage that regulated nucleocytoplasmic transport of polarity factors may act as a general mechanism of cell polarity in eukaryotes.
Introduction
Neuron polarity underpins neural circuit formation and function. During cortex development, neuron polarity arises as the neuroepithelial progenitors exit the cell cycle and migrate radially from the ventricular zone toward the cortical plate. Radial migration is initiated by the polarized distribution of N-cadherin, which concentrates at one pole of newly differentiating neurons and defines the position of the first of two opposite neurites (Calderon de Anda et al., 2008, Gärtner et al., 2012, Hatanaka and Murakami, 2002, Pollarolo et al., 2011). This early polarity event influences the latter specification of the axon and dendrites (Noctor et al., 2004). Following radial migration, axonogenesis begins as the cells reach the subventricular zone (SVZ), and neurons acquire their mature morphology as they reach the cortical plate (Hatanaka and Murakami, 2002, Noctor et al., 2004, Shoukimas and Hinds, 1978).
Axon specification is a hallmark of neuron polarity, and in cultured neurons it is regulated by a set of conserved polarity proteins, which includes the partitioning-defective protein Par4/LKB1 (Barnes et al., 2007, Shelly et al., 2007). In cultured neurons, this serine/threonine kinase is necessary to promote polarity through the adaptor kinase STK25, the Golgi protein GM130, and the SAD1/2 kinases, which regulate microtubule stability (Barnes et al., 2007, Matsuki et al., 2010, Shelly et al., 2007). In mammalian cells, a fraction of LKB1 is in the nucleus, and its nuclear export is regulated by the Ran pathway via CRM1/exportin 7 and the exportin co-receptor STRADα (Baas et al., 2003, Dorfman and Macara, 2008). However, whether regulated nuclear export of LKB1 is a regulatory step of neuron polarity has not been examined.
In eukaryotes, nucleocytoplasmic transport of cargo macromolecules through nucleopores depends on a concentration gradient of Ran-guanosine triphosphate (GTP), with high levels of RanGTP in the nucleus, where the Ran-guanine nucleotide exchange factor (GEF) RCC1 is localized (Bischoff and Ponstingl, 1991, Nemergut et al., 2001), and low levels in the cytosol, where Ran-guanosine diphosphate (GDP) is the predominant form. In the cytosol, the karyopherin proteins importin-α and importin-β1 form a cargo receptor, which drives cargo import to the nucleus. In the nucleus, RanGTP promotes cargo release by binding to the importins (Bischoff and Görlich, 1997). Conversely, nuclear export of cargoes relies on the assembly of export complexes that include RanGTP, a karyopherin that acts as a cargo receptor, and the cargo. In the cytosol, Ran-binding protein 1 (RanBP1) dissociates the export complex, releasing the cargo and allowing the recycling of RanGTP to RanGDP through Ran-GTPase-activating protein (GAP) (Bischoff and Ponstingl, 1995, Kuhlmann et al., 1997, Lounsbury and Macara, 1997, Seewald et al., 2003). Thus, RanBP1 regulates cargo release during nuclear export and maintains low levels of RanGTP in the cytosol, both of which are essential for nuclear import.
In addition to their function in mediating nucleocytoplasmic transport, Ran and RanBP1 regulate spindle assembly (Kalab et al., 1999, Zhang et al., 2014) and primary cilium initiation (Fan et al., 2011). Furthermore, Ran and RanBP1 regulate nerve repair in peripheral sensory neurons (Yudin et al., 2008). In these neurons, RanBP1 is translated at the site of axonal injury and allows for importin-α and importin-β1 to form a complex that is linked to the microtubule motor dynein. This complex is believed to enable retrograde signaling by transporting signaling cargoes back to the nucleus (Lai et al., 2008, Yudin et al., 2008). Finally, recent work has shown that RanGTP can be detected at the tip of extending neurites, where it regulates acentrosomal microtubule nucleation through TPX2 (Chen et al., 2017). In this context, whether RanBP1-dependent cargo unloading during nucleocytoplasmic transport plays a role in regulating neuron polarity remains unknown.
Results
Components of the Ran Pathway Regulate Axonogenesis in Drosophila
To identify factors regulating axon growth in vivo, we performed a genetic screen in the Drosophila melanogaster larval visual system, which is referred to as the Bolwig’s organ. This bilateral sensory organ consists of approximately 12 photoreceptors that project axons to the optic lobe during the early larval stages (Figure S1A). As the larva grows, the Bolwig’s nerve elongates proportionally in a manner that is similar to that of the sciatic nerve in humans after post-embryonic development. A screen of 4,000 RNAi lines covering all cytoskeletal regulators, kinases and phosphatases, the small GTPases and their regulators, as well as transmembrane proteins predicted in the fly genome, revealed 83 candidate genes that affected Bolwig’s nerve growth (Table S1). We identified among the genes that led to a shortening of the Bolwig’s nerve several regulators of the Ran pathway, including Ran (Figures S1B–S1I), indicating a role for this pathway during axon growth.
Localization of Ran, RCC1, RanGAP, and RanBP1 in Mammalian Cortical Neurons
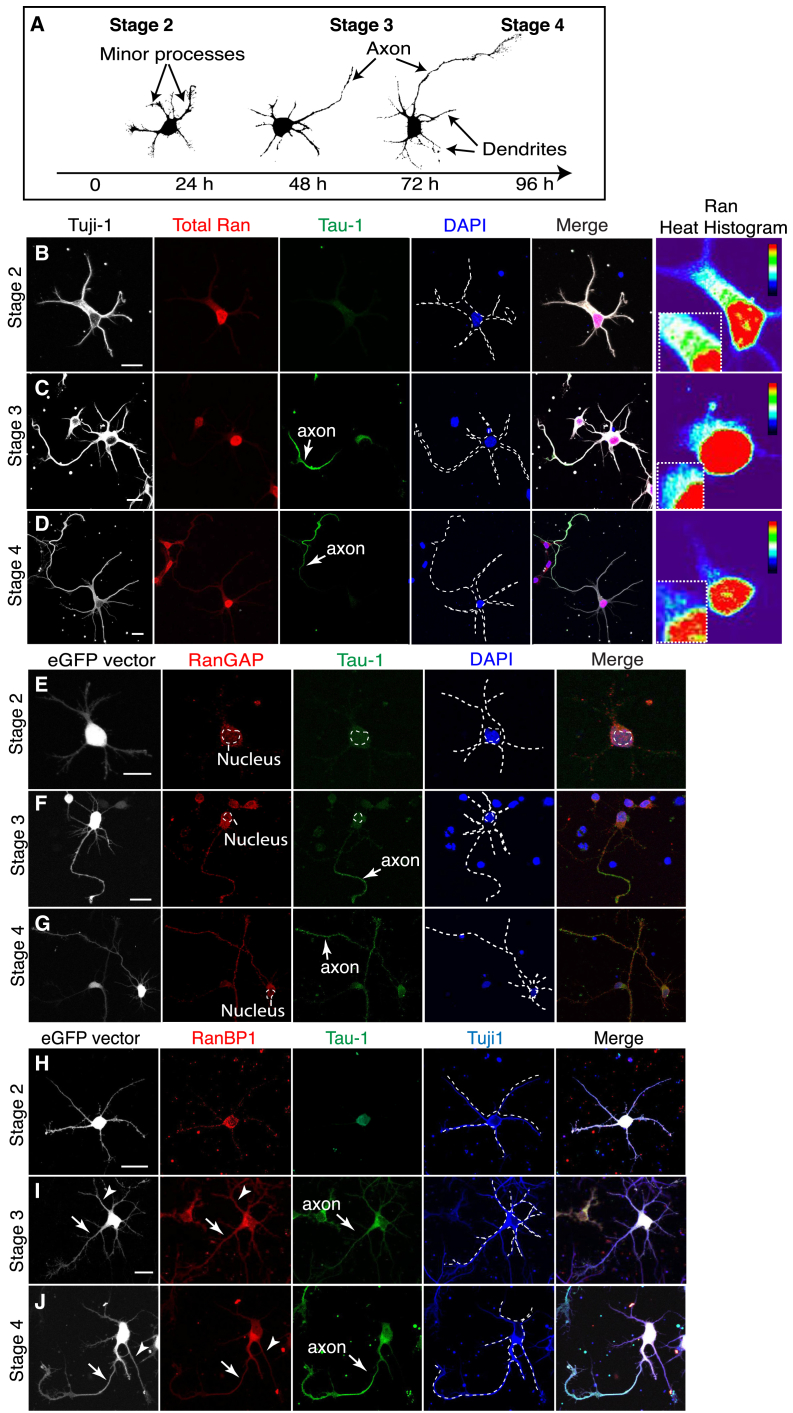
Next, we investigated whether regulated Ran-dependent nucleocytoplasmic transport promotes axon growth in vertebrate neurons using neonatal (postnatal day 0 [P0]) rat cortical neurons. Cortical neurons differentiate in vitro following four defined morphological stages (Figure 1A). During the first 12 to 24 hr, cells extend short processes (stages 1 and 2). After 36 hr, one process extends rapidly and acquires axon-specific markers (stage 3). This is followed by further axon elongation and dendrite development between 72 and 96 hr (stage 4) in culture (Dotti et al., 1988).
Figure 1.
Subcellular Localization of Ran, RanGAP, and RanBP1 in Cortical Neurons
(A) Successive stages of rat cortical neuron polarization in vitro. Neurons transfected with the control eGFP-reporter plasmid were imaged at different stages of development, and z stacks of individual cells were skeletonized using ImageJ (NIH).
(B–D) Rat cortical neurons stained for Tuji-1, Ran, and Tau-1 at Stage 2 (B), Stage 3 (C), and Stage 4 (D). The right column shows a heat histogram of Ran staining intensity. Scale bar, 10 μm.
(E–G) Neurons stained for eGFP, RanGAP, Tau-1, and DAPI at Stage 2 (E), Stage 3 (F), and Stage 4 (G). Scale bar, 10 μm.
(H–J) Neurons stained for eGFP, Tuji-1, Tau-1, and RanBP1 at Stage 2 (H), Stage 3 (I), and Stage 4 (J) using a previously validated RanBP1 antibody (Yudin et al., 2008). A white arrow points to RanBP1 staining in the axon and the arrowhead points to RanBP1 staining in a dendritic process.
We studied the expression pattern of the Ran pathway proteins during neuron polarization. In polarized neurons (stages 3 and 4), Ran was enriched in the nucleus and detected at lower levels in the cell body, dendrites, and the axon (Figures 1B–1D). RanGAP was distributed evenly throughout the cytoplasm, dendrites, and the axon (Figures 1E–1G). Using an antibody previously validated in peripheral sensory neurons (Yudin et al., 2008), we found that RanBP1 was localized in the cytosol, axon, and dendrites (Figures 1H–1J). However, we also found that a fraction of RanBP1 associated with the trans-Golgi network (TGN) using two independent antibodies. The specificity of these two antibodies was confirmed in cortical neurons using small interfering RNA (siRNA) (Figures S2A–S2F). Consistent with RanBP1 being associated with the TGN, scattering of the Golgi apparatus using brefeldin A (BFA) led to the scattering of RanBP1 (Figures S2G and S2H). To further investigate the link between the Golgi apparatus and RanBP1, we performed subcellular fractionation of rat brain cortices. A first fractionation based on sedimentation velocity was used to enrich for Golgi markers, followed by equilibrium sedimentation of the pooled peak fractions to further resolve the Golgi-enriched peak. Western blotting using antibodies targeting a number of Golgi proteins showed that a fraction of RanBP1 co-fractionates with the Golgi compartments (Figure S2I). Our results show that RanBP1 is enriched in the cytosol, including the Golgi apparatus, proximal to the nascent axon.
Nucleocytoplasmic Transport Regulates Axon Specification
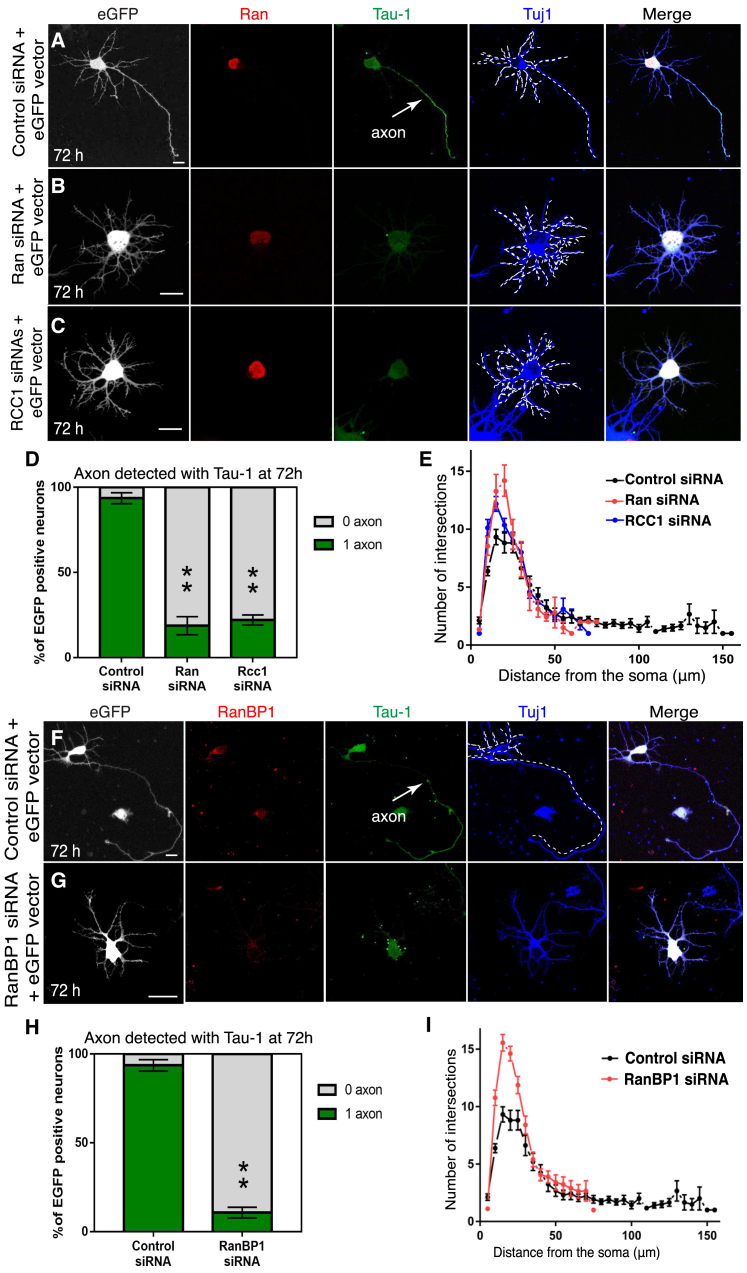
To study whether nucleocytoplasmic transport affected neuron polarity, we inhibited ran expression using siRNA. Most of the cortical neurons (∼80%) transfected with ran siRNA failed to specify their axon after 72 hr and showed short and hyperbranched neurites, whereas neurons transfected with scrambled siRNA (scrambled siRNA/GFP) differentiated normally (Figures 2A, 2B, 2D, 2E, S3A, and S3B). A similar phenotype was observed when the expression of the RanGEF RCC1 (Figures 2C–2E) and RanBP1 (Figures 2F–2I and S3C–S3E) was reduced.
Figure 2.
The Ran Pathway Is Required for Rat Cortical Neuron Polarization
(A) 72 hr cortical neuron co-transfected with a control siRNA and an eGFP reporter plasmid.
(B) 72 hr cortical neuron co-transfected with ran siRNAs and an eGFP-reporter plasmid (scale bar, 10 μm).
(C) 72 hr cortical neuron co-transfected with rcc1 siRNAs and an eGFP-reporter plasmid (scale bar, 10 μm).
(D) Neuron polarity was scored at 72 hr after transfection using the axonal marker Tau-1. The mean percentage of neurons with 0 or 1 axon over three independent replicates was determined for each condition: control siRNA (n = 31 neurons), ran siRNA (n = 32 neurons), and rcc1 siRNA (n = 28 neurons). After converting percentages to arcsin values, two-tailed unpaired t tests were performed, comparing control siRNA to ran siRNA (p = 0.0019) and Rcc1 siRNA (p = 0.0014).
(E) Sholl analysis of ran siRNAs neurons (n = 32 neurons), rcc1 siRNAs neurons (n = 28 neurons), and control scrambled siRNA neurons (n = 31 neurons) was performed 72 hr after transfection. A statistically significant increase in intersection numbers was observed for ran knockdown between 10 and 20 μm from the cell body (at 10 and 15 μm ∗p < 0.05; at 20 μm ∗∗p < 0.01) and for rcc1 knockdown between 10 and 15 μm from the cell body (at 10 and 15 μm ∗p < 0.05). No intersections were detected beyond 60 μm from the soma in the ran or rcc1 knockdown neurons, whereas in the wild-type at least one intersection could be readily detected 90 μm from the soma. Error bars represent SEMs. Statistics were calculated by unpaired two-tailed Student’s t test.
(F and G) 72 hr cortical neurons co-transfected with (F) control siRNA and an eGFP-reporter plasmid or with (G) siRNAs targeting ranBP1 and an eGFP-reporter plasmid and stained for RanBP1, Tau-1, and Tuj1. Ninety percent of the ranBP1 knockdown neurons failed to polarize by 72 hr (n = 38 neurons analyzed from at least three separate transfection experiments). In contrast, 94% of control neurons are polarized (n = 31 neurons, from at least three separate transfection experiments).
(H) The percentage of 72 hr neurons with 0 or 1 axon over three independent replicates was determined for each condition: control siRNA (n = 31 neurons) and ranBP1 siRNA (n = 38 neurons). After converting percentages to arcsin values, a two-tailed unpaired t test was performed, comparing control siRNA to ranBP1 siRNA (p = 0.00084).
(I) Sholl analysis of ranBP1 siRNAs neurons (n = 38) and control siRNA neurons (n = 31 neurons) was performed 72 hr after transfection. A statistically significant increase in the number of intersections was observed between 5 and 25 μm from the cell body (between 5 and 20 μm ∗∗∗p < 0.001; at 25 μm ∗p < 0.05, two-tailed unpaired Student’s t test). No intersections were detected beyond 75 μm from the soma in the ranBP1 knockdown neurons, whereas in the case of wild-type neurons, at least one intersection could be readily detected 90 μm from the soma. Error bars represent SEMs.
To complement our analysis of the ranBP1 phenotype, we examined formation of the axonal initial segment (AIS) using ankyrin-G antibodies. Reducing the expression of RanBP1 using siRNA abolished the AIS in a significant proportion of neurons when compared to control neurons. Only 20% of ranBP1-siRNA neurons had one AIS compared to 90% of neurons treated with scrambled siRNA (Figures S4A–S4C). This result indicates that the inhibition of RanBP1 expression largely prevents axon specification and morphogenesis in mammalian cortical neurons. To rule out potential effects on cell viability, RanBP1 levels were maintained low for 15 days using repeated rounds of siRNA transfections. Under these conditions, only 10% of RanBP1-siRNA neurons presented one axon compared to 80% of scrambled RNAi neurons with one axon (Figures S4D–S4F). Taken together, these data indicate that the Ran pathway and RanBP1 regulate axonogenesis in cortical neurons.
Next, to investigate whether Ran, RCC1, and RanBP1 link nucleocytoplasmic transport to the establishment of neuronal polarity, we asked whether hRanBP1[E37K], a mutated form of human-RanBP1 that does not bind to Ran-GTP (Petersen et al., 2000), could rescue the ranBP1 siRNA-mediated axonogenesis defects. As a control, we used hRanBP1::GFP, which localizes in the cytosol and rescues neuronal polarity in 72% of transfected cells (Figures S5A, S5B, and S5D). In contrast, expression of hRanBP1[E37K]::GFP, which also localizes in the cytosol (Figure S5C), exacerbated the axonless phenotype and failed to rescue axonogenesis in ranBP1-deficient neurons (Figure S5D). These experiments confirm that the Ran-dependent nucleocytoplasmic transport pathway regulates polarity in rat cortical neurons. Our results show that in these cells, RanBP1 is a key regulator of axonogenesis and dendritic arborization.
RanBP1 Regulates the Nuclear Export of Par4/LKB1
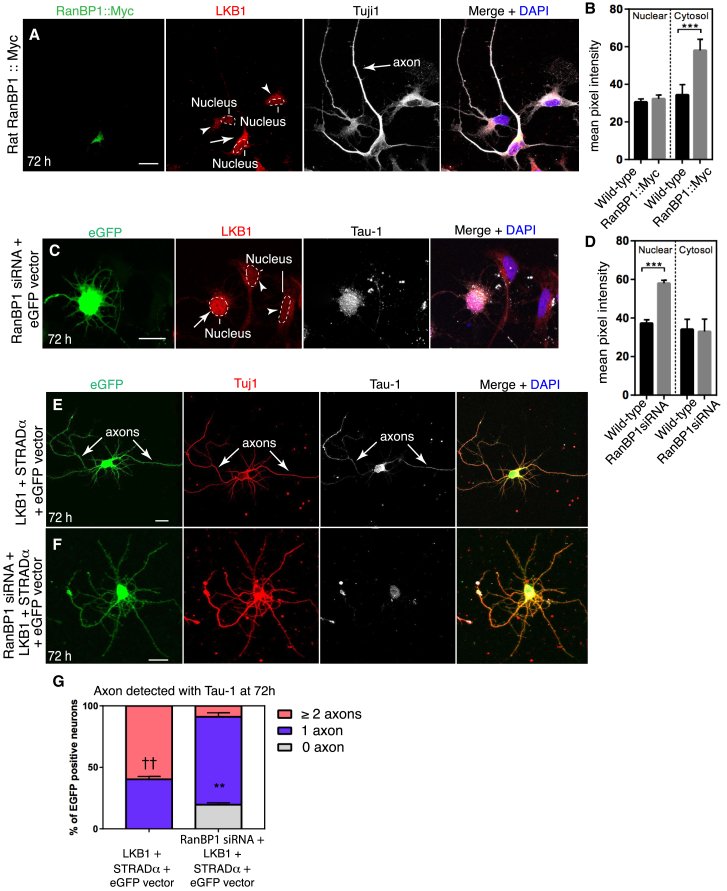
A potential cargo of the Ran pathway that could mediate RanBP1 function during axonogenesis is LKB1 (Barnes et al., 2007, Shelly et al., 2007). Because we found that RanBP1 is localized proximal to the nascent axon, we hypothesized that it may promote axon specification by unloading LKB1 from the nuclear export complex at this location. To test this hypothesis, we overexpressed RanBP1 in cortical neurons and analyzed the subcellular distribution of LKB1. Expression of RanBP1 led to substantial accumulation of LKB1 in the cytosol, relative to controls (Figures 3A and 3B). Conversely, compared to wild-type neurons, inhibition of RanBP1 levels using siRNA led to decreased cytoplasmic accumulation and increased nuclear levels of LKB1 (Figures 3C and 3D). Thus, we concluded that RanBP1 regulates the distribution of LKB1 between the nucleus and cytosol during neuron polarization.
Figure 3.
RanBP1 Is Required for LKB1-Dependent Axonogenesis
(A) LKB1 localization in a representative 72 hr rat cortical neuron overexpressing RanBP1::Myc, compared to non-transfected neurons (open triangles). Scale bar, 10 μm.
(B) Quantification of the LKB1 signal intensity in the nucleus and cytosol of neurons overexpressing RanBP1::Myc (n = 23 neurons), compared to neighboring non-transfected neurons (n = 29 neurons). Wild-type nuclear: 30.55 ± 1.69; RanBP1::Myc nuclear: 32.32 ± 1.94. Error bars represent SEMs (p = 0.50, two-tailed unpaired t test. Wild-type cytosol: 34.37 ± 1.72; RanBP1::Myc cytosol: 58.03 ± 1.70. Error bars represent SEMs (p < 0.0001, two-tailed unpaired t test).
(C) LKB1 localization (white arrow) in representative RanBP1 deficient 72 hr neurons compared to non-GFP neurons. Scale bar, 10 μm.
(D) Quantification of LKB1 signal intensity in the nucleus and cytosol of neurons 72 hr after siRNA-mediated knock down of ranBP1 (n = 20) compared to non-GFP neurons (n = 19). Wild-type nuclear: 37.25 ± 1.81; ranBP1-siRNA nuclear: 57.98 ± 1.61. Error bars represent SEMs (p = 0.0001, two-tailed unpaired t test). Wild-type cytosol: 34.13 ± 1.35; ranBP1-siRNA cytosol: 32.97 ± 1.71. Error bars represent SEMs (p = 0.60, two-tailed unpaired t test).
(E) Co-expression of LKB1 and STRADα together with an eGFP reporter plasmid in rat cortical neurons. White arrows indicate axons.
(F) ranBP1-siRNA neurons overexpressing LKB1 and STRADα.
(G) The mean percentage of 72 hr neurons with 0, 1, or ≥2 axons over three independent replicates was determined for each condition: overexpression of LKB1+STRADα (n = 32 neurons) and overexpression of LKB1+STRADα with knock down of ranBP1 (n = 34 neurons). After converting percentages to arcsin values, two-tailed unpaired t tests were performed. Comparing LKB1+STRADα against LKB1+STRADα for the proportion of neurons with 1 axon (p = 0.00187, ∗∗) and 2 axons (p = 0.00790, ††).
Overexpression of LKB1 and STRADα induces supernumerary axons in cultured neurons (Figure 3E) (Barnes et al., 2007, Shelly et al., 2007). To further study the relation between LKB1 and RanBP1, we asked whether RanBP1 is required for this phenotype. Further supporting the hypothesis that RanBP1 plays a key role in regulating the availability of LKB1 during neuron polarity, we found that overexpression of LKB1 and STRADα did not generate additional axons when RanBP1 expression was also decreased, and in 20% of the cases, no axon could be detected (Figures 3E–3G). These results show that RanBP1-dependent unloading of LKB1 from the nuclear export complex is a key step in regulating LKB1 function during axonogenesis.
RanBP1 Regulates Neuron Polarity through the LKB1-STRAD-STK25 Pathway
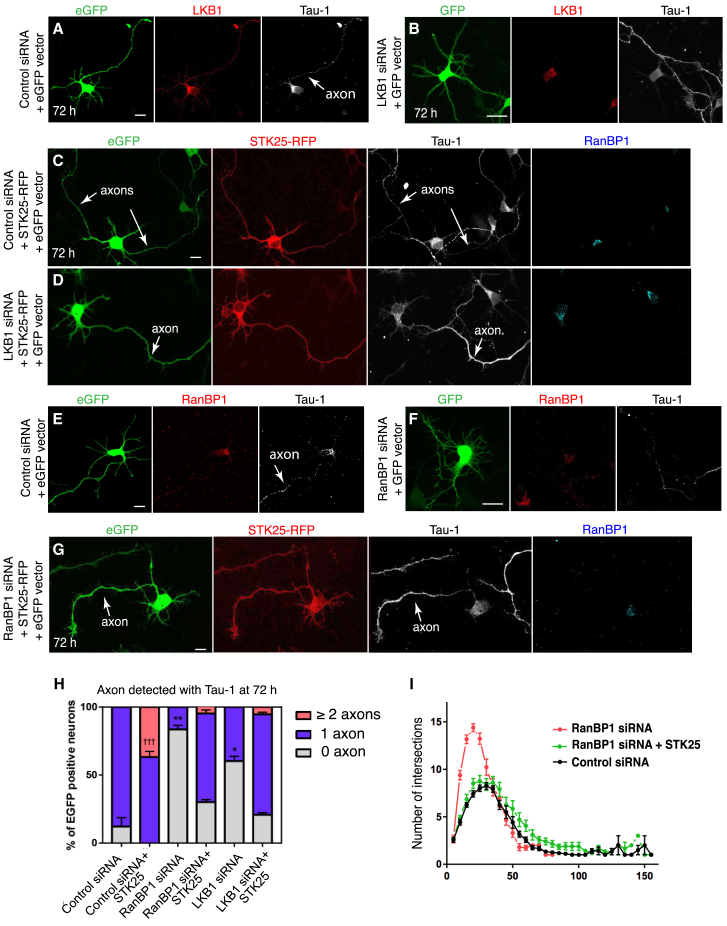
LKB1 function during axon specification is partly mediated by STK25, and overexpression of STK25 in LKB1-deficient cells can restore axon specification (Figures 4A–4D and 4H) (Matsuki et al., 2010). Our work raises the possibility that LKB1 is the main cargo that mediates RanBP1 function during neuronal polarization. Consistent with this hypothesis, overexpression of STK25 in neurons where RanBP1 expression has been decreased ameliorates the polarity phenotype, with 65% of ranBP1-siRNA(+)STK25 neurons presenting an axon, compared to 12% of neurons transfected with ranBP1-siRNA (Figures 4E–4H). In addition, the hyperbranched dendrite phenotype of RanBP1-siRNA neurons was also suppressed when expressing STK25 (Figure 4I). These findings indicate that during neuron polarization, RanBP1 controls the availability of LKB1 in the cytosol and enables axonogenesis through the STK25 pathway.
Figure 4.
LKB1 Is the Main Cargo Regulated by the Ran/RanBP1 Pathway during Axonogenesis
(A and B) Rat cortical neuron transfected with control siRNAs and the eGFP-reporter (A) and siRNAs targeting LKB1 and the eGFP-reporter plasmid (B).
(C) 72 hr cortical neuron overexpressing STK25-red fluorescent protein (RFP) and scrambled siRNA together with an eGFP-reporter plasmid.
(D) 72 hr cortical neuron overexpressing STK25-RFP and ranBP1-siRNA together with the eGFP-reporter plasmid.
(E and F) 72 hr cortical neuron transfected with scrambled siRNA and the eGFP-reporter (E) and 72 hr cortical neuron transfected with siRNAs targeting ranBP1 and the eGFP-reporter plasmid (F).
(G) 72 hr cortical neuron transfected with siRNAs ranBP1, STK25-RFP, and an eGFP-reporter plasmid.
(H) The mean percentage of 72 hr neurons with 0, 1, or ≥2 axons over three independent replicates were determined for each condition. The absence of axon in ranBP1 and LKB1 siRNA neurons is rescued by expressing STK25-RFP. At least 24 neurons were scored for the number of axons for each condition.
(I) Sholl analysis of siRNA control (n = 15), ranBP1-siRNAs neurons (n = 18), and ranBP1-siRNAs neurons rescued by overexpressing STK25-RFP (n = 21) was performed 72 hr after transfection. A statistically significant decrease in intersection numbers was observed for the rescue experiment between 10 and 25 μm from the cell body (at 10, 15, 20, and 25 μm ∗p < 0.001). No intersections were detected beyond 60 μm from the soma in the ranBP1 knockdown neurons, whereas in the rescued experiment and control siRNA, intersections reaching 90 μm could be readily detected. Comparing control siRNA and ranBP1 siRNASTK25(+) shows no statistically significant differences in the first 70 μm from the cell soma. Comparing control siRNA and ranBP1-siRNA shows statistically significant differences at 10, 15, 20, and 25 μm from the cell (p < 0.0001).
Error bars represent SEMs. Statistics were calculated by two-tailed unpaired Student’s t test.
RanBP1 Regulates Radial Migration of Neurons during Embryonic Cortical Development
To investigate the role of the nucleocytoplasmic transport machinery during neuronal differentiation and radial neural migration in vivo, we turned to the developing mouse cortex. We found that the Ran, RanGAP, and RanBP1 expression patterns in vivo resembled those found in cultured neurons (Figures S6A–S6C). RanBP1 and RanGAP were mostly localized to the cytosol (Figure S6A and S6B), while Ran was enriched in the nucleus relative to the cytosol (Figure S6C).
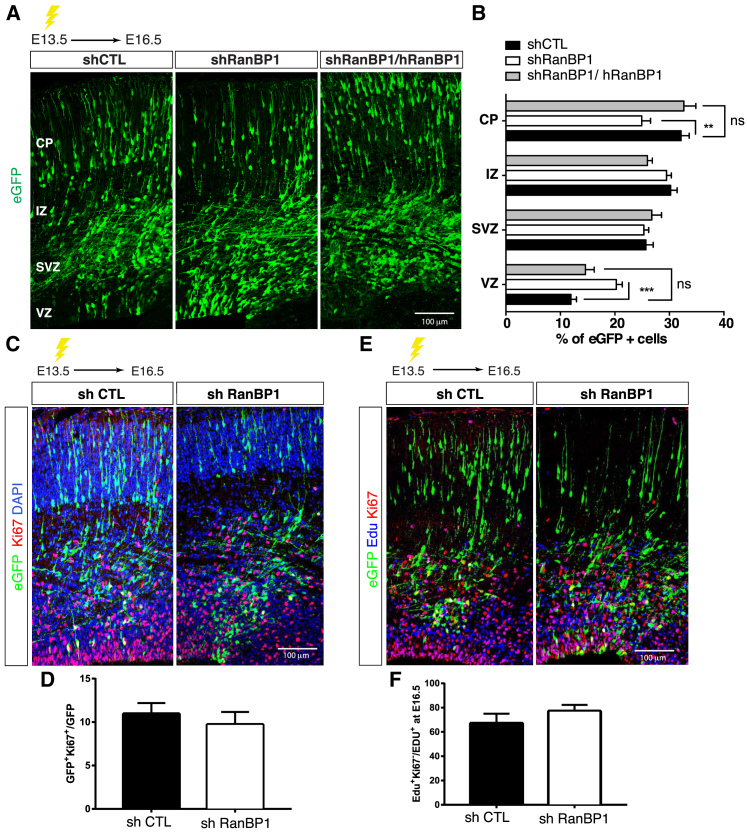
During cortex development, the establishment of neuronal polarity is a prerequisite for radial migration. To test RanBP1 function during cortex development, we generated three independent small hairpin RNA (shRNA) plasmids. All three shRNAs efficiently inhibited RanBP1 levels in cell lines and in the embryonic mouse cortex (Figures S7A–S7C). Mouse brains were electroporated at embryonic day 13.5 (E13.5), and radial migration was analyzed after 72 hr by counting the number of GFP-expressing neurons that had reached the cortical plate. In utero electroporation using the two most efficient ranBP1-shRNA constructs (shRNA2 and -3) resulted in a strong reduction in GFP+ neurons when compared to the control shRNA (small hairpin control [shCTL]), which may be due either to cell death or defects of neural progenitor proliferation. Loss of GFP+ cells was not detected using the less efficient ranBP1-shRNA construct (ranBP1-shRNA1), allowing us to investigate the role of RanBP1 during cortical radial migration. A significant proportion of neurons electroporated with ranBP1-shRNA1 accumulated within the ventricular (VZ) and SVZ and failed to reach the cortical plate (CP) (Figures 5A and 5B). This was not due to off-target effects of the ranBP1-shRNA1 because the defects were fully rescued by co-electroporation with a vector encoding shRNA-resistant human ranBP1 (Figure 5A). These findings indicate that inhibition of RanBP1 induces a significant delay in radial neural migration.
Figure 5.
RanBP1 Regulates Neuronal Migration In Vivo
(A) E13.5 mice were electroporated with shCTL, shranBP1 (shranBP1_1), or shranBP1 + hranBP1 and analyzed at E16.5. Electroporated cells are GFP+ (green).
(B) Quantification of the distribution of electroporated cells in indicated cortical areas represented as the percentage of GFP+ cells. Embryos were obtained from three independent electroporation experiments (shCTL [n = 14], shRanBP1 [n = 17], shranBP1+hranBP1 [n = 10]). Neuronal migration was analyzed using two-way ANOVA with Tukey’s post-hoc test. p values are as follows: for VZ: shCTL versus sh1 0.0003, shCTL versus sh1+Rescue 0.9757, sh1 versus sh1+Rescue 0.1526; for SVZ: shCTL versus sh1 > 0.9999, shCTL versus sh1+Rescue > 0.9999, sh1 versus sh1+Rescue 0.9999; for IZ: shCTL versus sh1 > 0.9999, shCTL versus sh1+Rescue 0.6250, sh1 versus sh1+Rescue 0.8169; for CP: shCTL versus sh1 0.0040, shCTL versus sh1+Rescue > 0.9999, sh1 versus sh1+Rescue 0.0063. CP, cortical plate; IZ, intermediate zone; SVZ, subventricular zone; VZ, ventricular zone.
(C) E13.5 embryos were subjected to in utero electroporation and analyzed at E16.5. Representative images of coronal mouse embryonic sections stained for GFP and Ki67 antibodies. Scale bar, 100 μm.
(D) Quantification of cells expressing Ki67. Data are from 3 independent experiments and 10 embryos per condition.
(E) Representative images of coronal mouse embryonic sections stained for GFP, Edu, and Ki67 antibodies.
(F) The proliferation index was measured as GFP+Ki67+/GFP+ cells. No significant difference was detected when comparing shCTL (10.96 ± 1.24) to shranBP1 (9.72 ± 1.45). The rate of cell-cycle exit was measured as the ratio of Edu+Ki67−/Edu+ cells. No significant difference was detected when comparing shCTL (67.13 ± 7.91) to shranBP1 (77.14 ± 5.18).
To investigate whether the high number of electroporated cells present in the VZ and SVZ was due to abnormal neuronal precursor cell (NPC) proliferation, we studied the expression of Ki67, a proliferation marker that is upregulated in all active phases of the cell cycle. At E16.5, the number of cells expressing Ki67 was similar in brains electroporated with ranBP1-shRNA1 and shCTL (Figures 5C and 5D). To test whether the neural migration phenotype observed in ranBP1 knockdown cortices was due to a delay of NPCs in exiting the cell cycle, E15.5 pregnant mice electroporated either with ranBP1-shRNA1 or shCTL were injected with 5-ethynyl-2′-deoxyuridine (EdU) to label proliferating NPCs. After 24 hr, embryos were collected and brain sections were co-stained for EdU and Ki67. No differences were observed in the number of EdU+, Ki67− cells between ranBP1-shRNA1 and shCTL embryos, indicating that inhibition of RanBP1 expression does not affect the exit of NPCs from the cell cycle (Figures 5E and 5F). Thus, the accumulation of neurons in the VZ and SVZ and the abnormal radial migration observed in cortices electroporated with ranBP1-shRNA1 was not due to NPC proliferation defects.
Discussion
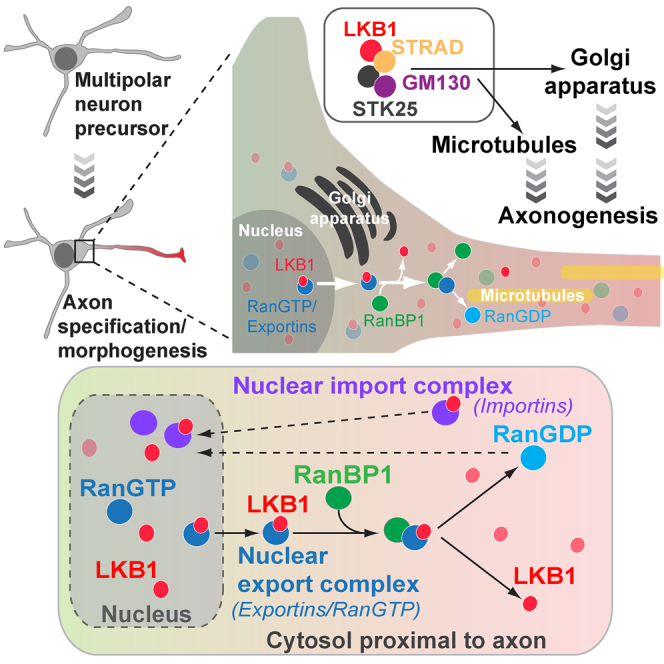
Nucleocytoplasmic transport of proteins and RNAs influences a broad range of signaling and transduction pathways in eukaryotes. Our work reveals that in neurons the nucleocytoplasmic transport pathway regulates polarity because it is required for both radial migration in vivo and axonogenesis in cultured neurons. In cultured neurons, we find that nucleocytoplasmic transport and, in particular, RanBP1 regulate the cytosolic level of the key polarity cargo, LKB1, which promotes axon specification through microtubule regulation (Barnes and Polleux, 2009) and Golgi regulation downstream of STK25-GM130 (Matsuki et al., 2010). Thus, our results indicate that regulated export and unloading of LKB1 from the nucleocytoplasmic transport machinery is a key regulatory step of axonogenesis. Furthermore, we find that the requirement for the Ran pathway during axonogenesis is conserved through evolution.
The Nucleocytoplasmic Transport Machinery Regulates Neuron Polarity
RanBP1 terminates nuclear export by promoting cargo release from the export complex and at the same time catalyzing the conversion of RanGTP into RanGDP. In cultured cortical neurons, the localization of the nucleus is strongly asymmetric, such that most of the cytosol, including organelles, is found proximal to the nascent axon. This suggests that most RanBP1-dependent cargo unloading occurs next to the nascent axon. Depending on the coefficient of diffusion of the export complexes and that of the cargoes exported to the cytosol, it is possible that in cortical neurons, the nucleocytoplasmic pathway initiates polarity by promoting the local accumulation of cargoes, including that of the polarity factor LKB1. Our finding that RanBP1 overexpression increases the amount of LKB1 detected in this region supports this hypothesis. In addition to its cytosolic localization, a fraction of RanBP1 is associated with the TGN. However, our attempts to identify the domain in RanBP1 that mediates this association were unsuccessful. Although it is possible that RanBP1 is modified in the Golgi, whether this event influences nucleocytoplasmic transport remains unclear.
Ran can be detected at the tip of developing neurites (Chen et al., 2017), and previous work performed in vertebrate sciatic nerve axons has shown that RanBP1 functions as part of a retrograde transport that signals to the nucleus to regulate the response to injury (Yudin et al., 2008). Consistent with Ran regulation taking place during neurite outgrowth and axonogenesis, we detected RanBP1 in the cytosol, neurites, and the nascent axon in cultured cortical neurons. It is therefore possible that during axonogenesis, RanBP1 functions as part of a retrograde pathway that involves LKB1. It will be interesting to test whether LKB1 is one of the cargoes that is regulated by the importin-β/α-dynein complex in response to nerve injury in peripheral neurons.
We also note that LKB1 is expressed in the primary cilium (Mick et al., 2015), raising the possibility that LKB1 is among the cargoes that mediate RanGTP/RanBP1 function during cilium initiation (Fan et al., 2011).
Next to axonogenesis, we find that RanBP1, and therefore the nucleocytoplasmic transport machinery, is required for radial migration during cortex development. LKB1 and STK25/GM130 have been shown to regulate radial migration in the developing cortex (Asada et al., 2007, Matsuki et al., 2013), and it is therefore possible that RanBP1-dependent export of this kinase mediates part of the RanBP1 radial migration phenotype. It is also possible that the availability of cargoes other than LKB1 is regulated by RanBP1 during this early step of neuron polarization.
RanBP1 Regulates Neuron Polarity through Golgi Regulations
Our work demonstrates that during the establishment of neuronal polarity, a functional cross-talk takes place between two major trafficking pathways: the nucleocytoplasmic transport pathway and trafficking associated with the Golgi apparatus. The Golgi apparatus is an important regulator of neuron morphogenesis. It is located on the basal side of the migrating neuron progenitors, when post-mitotic neurons acquire their polarity (de Anda et al., 2005, Distel et al., 2010, Umeshima et al., 2007). The Golgi then relocates to the side of the apical leading edge, where it invades the apical dendrite (de Anda et al., 2010, Zhao et al., 2015). Interfering with the localization of the Golgi during this phase compromises cortex development (Horton et al., 2005), and Golgi condensation downstream of STK25 correlates with ectopic axonogenesis (Matsuki et al., 2010). However, at present it is not clear exactly how Golgi condensation may influence axonogenesis. Nevertheless, our finding that the loss of axon and the induction of hyperbranched neurites observed in RanBP1-deficient cortical neurons can be rescued by expressing STK25 indicates that most of RanBP1 function during axonogenesis and neurite branching is mediated through LKB1. It will be interesting to test whether a function for the nucleocytoplasmic transport pathway during polarity is limited to neurons or whether it extends to other cell types.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-Ki67 | Abcam | ab15580; RRID:AB_443209 |
| Chicken polyclonal anti-GFP | Abcam | ab13970; RRID:AB_300798 |
| Rabbit polyclonal anti-RanBP1 | Sigma-Aldrich | SAB1411184 |
| Rabbit polyclonal anti-RanBP1 | Cell Signaling | 8780 |
| Rabbit polyclonal anti-RanBP1 | Abcam | ab97659; RRID:AB_10680142 |
| Chicken polyclonal anti-Tau | Abcam | ab75714; RRID:AB_1310734 |
| Mouse monoclonal anti-Tau | Millipore | mab3420; RRID:AB_94855 |
| Mouse anti-TGN38 | NovusBio | NB300-575SS |
| Mouse anti-MAP2 | Abcam | ab11267; RRID:AB_297885 |
| Rabbit anti-MAP2 | Millipore | ab5622; RRID:AB_91939 |
| Mouse anti-LKB1 | NovusBio | NBP2-14835SS |
| Rabbit anti-Ran | Sigma-Aldrich | SAB4502579; RRID:AB_10747705 |
| Mouse anti-RanGAP1 | Santa Cruz | sc-28322; RRID:AB_2176987 |
| Mouse anti-Myc | Santa Cruz | 9E10 |
| Chicken anti-Tuj1 | Abcam | ab107216 |
| mouse anti-Cathepsin D | BD Transduction Lab | 610800; RRID:AB_CVCL_G671 |
| mouse anti-mitofusin | Abcam | ab126575 |
| mouse anti-BIP | BD Transduction Lab | 610978; RRID:AB_11141234 |
| rabbit anti-Transferrin Receptor | Abcam | ab84036; RRID:AB_10673794 |
| Bacterial Strains- Plasmids | ||
| shRNA1-3 RanBP1 | pSuper GFP plasmid | Oligoengine |
| shCTL shRNA | Egan et al., 2013 | pSuper GFP plasmid |
| Human RanBP1 | pCAG-IRES-RFP | This paper |
| Kruppel-Gal4 | This paper | N/A |
| RanBP1::MYC_YQRL | This paper | CMV-GFP plasmid |
| humanRanB1::GFP_YQRL | This paper | CMV-GFP plasmid |
| human RanB1(E37K)::GFP_YQRL | This paper | CMV-GFP plasmid |
| Mouse LKB1 cDNA | Barnes et al., 2007 | pCAGGS |
| Mouse STRAD-α | Barnes et al., 2007 | pCAGGS |
| Mouse STK25 | Matsuki et al., 2010 | pCAGGS-RFP |
| RanBP1-Myc-DDK | Origene | RR201578 |
| RanBP1-GFP | Origene | RG223305 |
| Critical Commercial Assays | ||
| Rat Neurons Nucleofector kit | Lonza | VSPI-1003 |
| EndoFree plasmid purification kit | QIAGEN | 12362 |
| Lipofectamine RNAiMAX reagent | Thermo Fisher | 13778030 |
| SYBR Mesa Blue Kit | Eurogentec | N/A |
| Experimental Models: Cell Lines | ||
| PC12 cells | ATCC | CRL-1721 |
| Primary rat cortical neurons | This work | N/A |
| Primary mouse cortical neurons | This work | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse pups, E13.5 | Charles River | C57BL/6 |
| Drosophila RNAi lines | Vienna Drosophila RNAi Centre | N/A |
| Rat pups, P0 | UCL facility | N/A |
| Oligonucleotides | ||
| siRNAs RAN (rat) | Sigma | NM_053439 |
| siRNAs RanBP1 (rat) | Sigma | NM_001108324 |
| siRNA RCC1(rat) | Sigma | NM_001128189 |
| siRNA LKB1(rat) | Sigma | NM_001108069 |
| siRNA cntrl (rat) | Sigma | SIC001 |
| RAT GAPDH | Eurofins Genomics | N/A |
| For: 5′-GACATGCCGCCTGGAGAAAC-3′ | ||
| Rev: 5′-AGCCCAGGATGCCCTTTAGT-3′ | ||
| RAT RAN | Eurofins Genomics | N/A |
| For: 5′-AGGACCCATCAAGTTCAACG-3′ | ||
| Rev: 5′TTCACACACGCGTACCAGAT-3′ | ||
| RAT RanBP1: | Eurofins Genomics | N/A |
| 5′-GTGCCAACCACTACATCACG-3′ | ||
| 5′-TGCCTGATCCTGCTTTCTTT-3′ | ||
| RAT Rcc1 | Eurofins Genomics | N/A |
| 5′-CCGGGAAAGTGGAACTACAA-3′ | ||
| 5′-ACCTTTACCACCTGCGTGTC-3′ | ||
| RAT LKB1 | Eurofins Genomics |
N/A |
| 5′-CCTGCAGAGAAAACCCAGAG-3′ | ||
| 5′-GCAGCTTCAAGTTTCCCAAG-3′ | ||
| Software and Algorithms | ||
| ImageJ | NIH GOV | N/A |
| LAS AF software | Leica | N/A |
| Other | ||
| Sequencing | Eurofins Genomics | N/A |
Contact for Reagents and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Franck Pichaud (f.pichaud@ucl.ac.uk).
Drosophila
For the F1 recessive RNAi screen in the Bolwig’s organ, RNAi lines [Vienna Drosophila RNAi Centre (VDRC)] (Dietzl et al., 2007), were expressed using a Kruppel-Gal4 driver (a generous gift from Fernando Casares, Sevilla, Spain). The Bolwig’s nerve was scored using the artificial neuronal 3 × P3-GFP transgene (Berghammer et al., 1999).
Mice
All experiments were conducted with University College London ethical committee approval. E13.5 timed-pregnant mice were handled and electroporated as previously described in (Nitarska et al., 2016). Pregnant mice were sacrificed 72h following surgery.
Primary cultures of rat cortical neurons
All experiments were conducted with University College London ethical committee approval. Cells were isolated form P0 cortices and electroporated before plating using the Nucleofector kit (Lonza). Briefly, 1 × 106 cells were resuspended in 100 μL of Nucleofector solution containing 3 μg of an expression vector or a pool of three gene-specific siRNAs (10 nM of each siRNA, 30 nM total), or a combination of expression vectors plus siRNAs. siRNAs targeting ran (NM_053439), ranBP1 (NM_001108324), rcc1 (NM_001128189) and LKB1 (NM_001108069), and a siRNA negative control (MISSION® siRNA Universal Negative Control, SIC001) (Sigma).
Method Details
Immunological Methods
Cells were washed (PBS), fixed (PBS, 3% paraformaldehyde; 15 min), washed (PBS), permeabilized in PBS-T (PBS plus 0.3% Triton X-100; 20 min), blocked (PBS-T plus 5% goat serum; 15 min) and incubated with primary antibodies overnight at 4°C and appropriate secondaries antibodies (Jackson ImmunoResearch, dilution 1:200; 1 h, room temperature). The primary antibodies used were: rabbit anti-RanBP1 (1:500, SAB1411184, Sigma-Aldrich), rabbit polyclonal anti-RanBP1 (#8780, Cell Signaling), rabbit polyclonal anti-RanBP1 (1:4000, ab97659 Abcam), chicken anti-Tau (1:1000, ab75714 Abcam), mouse anti-Tau (1:1000, mab3420, Millipore), mouse anti-TGN38 (1:100, NB300-575SS, Novusbio), mouse anti-MAP2 (1:1000, ab11267, Abcam), rabbit anti-MAP2 (1:1000, ab5622 Millipore), mouse anti-LKB1 (1:100, NBP2-14835SS, Novusbio), rabbit anti-Ran (GTP+GDP) (1:1000, SAB4502579, Sigma-Aldrich), mouse anti-RanGAP1 (1:200, sc-28322, Santa Cruz Biotechnology), mouse anti-Myc (1:500, 9E10, Santa Cruz), chicken anti-GFP (1:1000, ab13970, Abcam) and chicken anti-Tuj1 (1:1000, ab107216, Abcam).
Tissue preparation and Immunostaining
Embryonic brains were processed as previously described in (Nitarska et al., 2016). The following primary antibodies were used: rabbit anti-Ki67 (Abcam ab15580), chicken anti-GFP (Abcam ab13970), rabbit anti-RanBP1 (1:500, SAB1411184, Sigma-Aldrich).
Radial neural migration analysis
Radial migration analysis of embryos electroporated in utero with the indicated GFP vectors was performed as described previously (Hand et al., 2005) using ImageJ and excel macro.
Cell cycle index analysis
Cell cycle exit index was determined using EdU/Ki67 immunolabelling. For EdU incorporation E15.5 timed pregnant females received an intraperitoneal injection of EdU (Invitrogen) at a dose of 20 mg/kg body weight. After 24h, coronal sections were stained for EdU incorporation using the Click-iT EdU cocktail and immunolabelled with the Ki67 antibody. All imaging and image analysis were performed blind. Quantification of labeled cells within the cortical wall was performed by dividing the images into bins 100 μm wide. The bins spanned the entire coronal section from the ventricular surface to the pia. EdU+/Ki67- cells and total number of EdU+ cells were counted per each bin. Cell cycle exit index was calculated as the percentage of EdU+/Ki67- cells over the total number of EdU+ cells.
Molecular Biology
Rat RanBP1-Myc-DDK (RR201578) and human RanBP1-GFP (RG223305) expression plasmids were purchased from Origene. A genomic rescue transgene was generated that consists of a 1kb ranBP1promoter cloned upstream of the RanBP1::GFP fusion protein. Rat RanBP1-Myc-DDK-YQRL, and human RanBP1-GFP-YQRL were generated by directly ligating annealed oligos into the vector backbones using the PmeI restriction site. The RanBP1(E37K)-GFP-YQRL mutant was generated by site-directed mutagenesis (Quick Change Mutagenesis Kit, ThermoFisher). Mouse Par4/LKB1 and mouse STRADα plasmids were a kind gift of Franck Polleux, Columbia University, USA. For in utero electroporation experiments the oligo hairpins were ligated into pSUPER GFP plasmid (Oligoengine) using BglII/HindIII restriction sites. The shRNA sequences targeting mouse RanBP1 transcripts were as follows: shRNA1 5′-CGTGCAAAGCTGTTCCGGTTTGTTCAAGAGACAAACCGGAACAGCTTTGCAC3′, shRNA2 5′-CCCTACCCTTTAAGGTTTGTTTTTCAAGAGAAAACAAACC
TTAAAGGGTAGG-3′, shRNA3 5′-GGACCATCCGCCTTCTTATGATTCAAGA
GATCATAAGAAGGCGGATGGTCC-3′, shCTL shRNA sequence 5′-GCGTACGGGGAAACTTCGA-3′ was described previously (Egan et al., 2013).
Complete hRanBP1 coding sequence was subcloned into pCAG-IRES-RFP vector using NotI and EcoRI.
Western blot and quantitative real-time PCR
PC12 cells were electroporated and harvested 48h after seeding to be analyzed by either western blot or qRT-PCR. Cells were lysed with ice-cold extraction buffer (50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 5 mM EDTA, 0.1% NP-40). Samples were centrifuged at 20,000 g for 10 min at 4°C and protein content determined by the BCA protein assay kit. The total quantity of protein (40 μg) was subjected to gel electrophoresis on a 10% SDS-PAGE, immunoblotted and probed for rabbit anti-Ran (GTP+GDP) (1:1000, SAB4502579, Sigma-Aldrich) or rabbit anti-RanBP1 (1:1000, SAB1411184, Sigma-Aldrich), together with mouse anti-GFP (1:1000, ab1218, Abcam). Membranes were incubated with the appropriate anti-mouse (1:10000, IR-680, LI-COR Biosciences) and anti-rabbit (1:10000, IR-800, LI-COR Biosciences) antibodies. Protein signal was detected by Odyssey infrared imaging system LI-COR Biosciences (LI-COR).
Total RNA was extracted using Trizol reagent (Invitrogen), and 1 μg of total RNA was used for cDNA synthesis using SuperScript RT II (Invitrogen). Real-time qPCR was performed using SYBR Mesa Blue Kit (Eurogentec). The following primers were used: GAPDH 5′-GACATGCCGCCTGGAGAAAC-3′ (forward) and 5′-AGCCCAGGATGCCCTTTAGT-3′ (reverse); Ran 5′-AGGACCCATCAAGTTCAACG-3′ (forward); 5′TTCACACACGCGTACCAGAT-3′ (reverse); RanBP1 5′-GTGCCAACCACTACATCACG-3′ (forward); 5′-TGCCTGATCCTGCTTTCTTT-3′ (reverse); Rcc1 5′-CCGGGAAAGTGGAACTACAA-3′ (forward); 5′-ACCTTTACCACCTGCGTGTC-3′ (reverse). LKB1 5′-CCTGCAGAGAAAACCCAGAG-3′ (forward); 5′-GCAGCTTCAAGTTTCCCAAG-3′ (reverse). Statistical significance was calculated using an unpaired t test with significance at p < 0.05.
Biochemical Analysis
Brains obtained from rat pups (P0) were processed on ice with a Dounce homogenizer in 1 mL ice-cold homogenization buffer (HB: 250 mM sucrose; 20 mM HEPES-HCl, pH 7.4; 1 mM EDTA; 5 mM MgCl2), supplemented with 0.2% protease inhibitors (Sigma-Aldrich) and centrifuged at 900 g for 5 min at 4°C. The supernatant was recovered and the pellet re-suspended in 500 ul HB, re-homogenized and centrifuged as described above. The supernatants were pooled to yield a post-nuclear supernatant (PNS). PNSs were fractionated by sequential centrifugation through two sucrose gradients, essentially as previously described (Ferraro et al., 2007).
Brefeldin A treatments
Brefeldin A was obtained from Sigma. Primary rat neurons were treated with brefeldin A (1 μg/ml for 90 min) to induce Golgi dispersion. Cells were then fixed and processed for immunofluorescence.
Quantification and Statistical Analysis
Axon quantification
The mean percentages of 4 stage neurons with 0, 1 or multiple axons were determined for each condition. After converting percentages to arcsin values, two-tailed, unpaired, t tests were performed comparing knockdown/mutants to control.
Morphological characteristics of neurons
Sholl analysis was used to characterize the morphology of imaged neurons. Statistics were calculated by unpaired, two-tailed Student’s t test.
Western blot quantification
Densitometry analysis of signal intensity in western blotting was performed with a Student’s t test.
Neuronal migration
Neuronal migration was analyzed using two way Anova with Tukey’s post test.
Neuronal progenitor proliferation index was measured as GFP+Ki67+ / GFP+ cells and the rate of cell cycle exit was measured as the ratio of Edu+Ki67- / Edu+ cells.
IF Colocalization
to analyze the colocalization of the proteins of interest and cellular compartments, the Pearson’s correlation coefficients were used. The Pearson correlation is +1 in the case of a perfect direct (increasing) linear relationship (correlation), and −1 in the case of a perfect decreasing (inverse) linear relationship (anticorrelation). The Person correlation values were calculated by the pixel intensity- spatial correlation analysis method using confocal images and ImageJ processing program.
Real Time PCR
Statistical significance was calculated by comparing the normalized expression ratios to 1 and using one sample Student’s t test.
All analyses were performed using data from at least three biological replicates. In all cases, a p value of ≤ 0.05 was considered to be significant.
Acknowledgments
We thank Franck Polleux, Maya Shelly, and Sven C.D. van Ijzendoorn for providing reagents. We thank the Pichaud Lab members for discussion and support. F.F. is supported by a Medical Research Council grant (MC-UU-12018/2 to Dan Cutler). This work, including support to C.M. and T.K., was funded by a Medical Research Council grant to F.P. (MC_UU_12018/3). K.M. is the recipient of a Wellcome Trust PhD studentship. A.R. is the recipient of the Wellcome Trust Investigator Award 103717/14/Z.
Author Contributions
F.P. supervised the work and conceived the project together with C.M. C.M. performed and analyzed all of the experiments using cultured cortical neurons. J.N. performed and analyzed the in vivo experiments in mice with the help of C.M. T.K. contributed to the RNAi screen in Drosophila by screening half of the RNAi lines and assisted in culturing rat cortical neurons. F.F. designed and performed the subcellular fractionation experiments and advised on the Golgi dispersion experiments. K.M. contributed to troubleshooting the RanBP1 antibodies. A.R. advised on the in vivo experiments in mice. F.P. wrote the manuscript together with C.M. and with the help of A.R.
Declaration of Interests
The authors declare no competing interests.
Published: September 4, 2018
Footnotes
Supplemental Information includes seven figures and one table and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.07.107.
Supplemental Information
References
- Asada N., Sanada K., Fukada Y. LKB1 regulates neuronal migration and neuronal differentiation in the developing neocortex through centrosomal positioning. J. Neurosci. 2007;27:11769–11775. doi: 10.1523/JNEUROSCI.1938-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas A.F., Boudeau J., Sapkota G.P., Smit L., Medema R., Morrice N.A., Alessi D.R., Clevers H.C. Activation of the tumour suppressor kinase LKB1 by the STE20-like pseudokinase STRAD. EMBO J. 2003;22:3062–3072. doi: 10.1093/emboj/cdg292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes A.P., Polleux F. Establishment of axon-dendrite polarity in developing neurons. Annu. Rev. Neurosci. 2009;32:347–381. doi: 10.1146/annurev.neuro.31.060407.125536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes A.P., Lilley B.N., Pan Y.A., Plummer L.J., Powell A.W., Raines A.N., Sanes J.R., Polleux F. LKB1 and SAD kinases define a pathway required for the polarization of cortical neurons. Cell. 2007;129:549–563. doi: 10.1016/j.cell.2007.03.025. [DOI] [PubMed] [Google Scholar]
- Berghammer A.J., Klingler M., Wimmer E.A. A universal marker for transgenic insects. Nature. 1999;402:370–371. doi: 10.1038/46463. [DOI] [PubMed] [Google Scholar]
- Bischoff F.R., Görlich D. RanBP1 is crucial for the release of RanGTP from importin beta-related nuclear transport factors. FEBS Lett. 1997;419:249–254. doi: 10.1016/s0014-5793(97)01467-1. [DOI] [PubMed] [Google Scholar]
- Bischoff F.R., Ponstingl H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature. 1991;354:80–82. doi: 10.1038/354080a0. [DOI] [PubMed] [Google Scholar]
- Bischoff F.R., Ponstingl H. Catalysis of guanine nucleotide exchange of Ran by RCC1 and stimulation of hydrolysis of Ran-bound GTP by Ran-GAP1. Methods Enzymol. 1995;257:135–144. doi: 10.1016/s0076-6879(95)57019-5. [DOI] [PubMed] [Google Scholar]
- Calderon de Anda F., Gärtner A., Tsai L.H., Dotti C.G. Pyramidal neuron polarity axis is defined at the bipolar stage. J. Cell Sci. 2008;121:178–185. doi: 10.1242/jcs.023143. [DOI] [PubMed] [Google Scholar]
- Chen W.S., Chen Y.J., Huang Y.A., Hsieh B.Y., Chiu H.C., Kao P.Y., Chao C.Y., Hwang E. Ran-dependent TPX2 activation promotes acentrosomal microtubule nucleation in neurons. Sci. Rep. 2017;7:42297. doi: 10.1038/srep42297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Anda F.C., Pollarolo G., Da Silva J.S., Camoletto P.G., Feiguin F., Dotti C.G. Centrosome localization determines neuronal polarity. Nature. 2005;436:704–708. doi: 10.1038/nature03811. [DOI] [PubMed] [Google Scholar]
- de Anda F.C., Meletis K., Ge X., Rei D., Tsai L.H. Centrosome motility is essential for initial axon formation in the neocortex. J. Neurosci. 2010;30:10391–10406. doi: 10.1523/JNEUROSCI.0381-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K.C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Distel M., Hocking J.C., Volkmann K., Köster R.W. The centrosome neither persistently leads migration nor determines the site of axonogenesis in migrating neurons in vivo. J. Cell Biol. 2010;191:875–890. doi: 10.1083/jcb.201004154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman J., Macara I.G. STRADalpha regulates LKB1 localization by blocking access to importin-alpha, and by association with Crm1 and exportin-7. Mol. Biol. Cell. 2008;19:1614–1626. doi: 10.1091/mbc.E07-05-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotti C.G., Sullivan C.A., Banker G.A. The establishment of polarity by hippocampal neurons in culture. J. Neurosci. 1988;8:1454–1468. doi: 10.1523/JNEUROSCI.08-04-01454.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan C.M., Nyman U., Skotte J., Streubel G., Turner S., O’Connell D.J., Rraklli V., Dolan M.J., Chadderton N., Hansen K. CHD5 is required for neurogenesis and has a dual role in facilitating gene expression and polycomb gene repression. Dev. Cell. 2013;26:223–236. doi: 10.1016/j.devcel.2013.07.008. [DOI] [PubMed] [Google Scholar]
- Fan S., Whiteman E.L., Hurd T.W., McIntyre J.C., Dishinger J.F., Liu C.J., Martens J.R., Verhey K.J., Sajjan U., Margolis B. Induction of Ran GTP drives ciliogenesis. Mol. Biol. Cell. 2011;22:4539–4548. doi: 10.1091/mbc.E11-03-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro F., Ma X.M., Sobota J.A., Eipper B.A., Mains R.E. Kalirin/Trio Rho guanine nucleotide exchange factors regulate a novel step in secretory granule maturation. Mol. Biol. Cell. 2007;18:4813–4825. doi: 10.1091/mbc.E07-05-0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gärtner A., Fornasiero E.F., Munck S., Vennekens K., Seuntjens E., Huttner W.B., Valtorta F., Dotti C.G. N-cadherin specifies first asymmetry in developing neurons. EMBO J. 2012;31:1893–1903. doi: 10.1038/emboj.2012.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand R., Bortone D., Mattar P., Nguyen L., Heng J.I.-T., Guerrier S., Boutt E., Peters E., Barnes A.P., Parras C. Phosphorylation of Neurogenin2 specifies the migration properties and the dendritic morphology of pyramidal neurons in the neocortex. Neuron. 2005;48:45–62. doi: 10.1016/j.neuron.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Hatanaka Y., Murakami F. In vitro analysis of the origin, migratory behavior, and maturation of cortical pyramidal cells. J. Comp. Neurol. 2002;454:1–14. doi: 10.1002/cne.10421. [DOI] [PubMed] [Google Scholar]
- Horton A.C., Rácz B., Monson E.E., Lin A.L., Weinberg R.J., Ehlers M.D. Polarized secretory trafficking directs cargo for asymmetric dendrite growth and morphogenesis. Neuron. 2005;48:757–771. doi: 10.1016/j.neuron.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Kalab P., Pu R.T., Dasso M. The ran GTPase regulates mitotic spindle assembly. Curr. Biol. 1999;9:481–484. doi: 10.1016/s0960-9822(99)80213-9. [DOI] [PubMed] [Google Scholar]
- Kuhlmann J., Macara I., Wittinghofer A. Dynamic and equilibrium studies on the interaction of Ran with its effector, RanBP1. Biochemistry. 1997;36:12027–12035. doi: 10.1021/bi970524k. [DOI] [PubMed] [Google Scholar]
- Lai K.O., Zhao Y., Ch’ng T.H., Martin K.C. Importin-mediated retrograde transport of CREB2 from distal processes to the nucleus in neurons. Proc. Natl. Acad. Sci. USA. 2008;105:17175–17180. doi: 10.1073/pnas.0803906105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lounsbury K.M., Macara I.G. Ran-binding protein 1 (RanBP1) forms a ternary complex with Ran and karyopherin beta and reduces Ran GTPase-activating protein (RanGAP) inhibition by karyopherin beta. J. Biol. Chem. 1997;272:551–555. doi: 10.1074/jbc.272.1.551. [DOI] [PubMed] [Google Scholar]
- Matsuki T., Matthews R.T., Cooper J.A., van der Brug M.P., Cookson M.R., Hardy J.A., Olson E.C., Howell B.W. Reelin and stk25 have opposing roles in neuronal polarization and dendritic Golgi deployment. Cell. 2010;143:826–836. doi: 10.1016/j.cell.2010.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuki T., Chen J., Howell B.W. Acute inactivation of the serine-threonine kinase Stk25 disrupts neuronal migration. Neural Dev. 2013;8:21. doi: 10.1186/1749-8104-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mick D.U., Rodrigues R.B., Leib R.D., Adams C.M., Chien A.S., Gygi S.P., Nachury M.V. Proteomics of primary cilia by proximity labeling. Dev. Cell. 2015;35:497–512. doi: 10.1016/j.devcel.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemergut M.E., Mizzen C.A., Stukenberg T., Allis C.D., Macara I.G. Chromatin docking and exchange activity enhancement of RCC1 by histones H2A and H2B. Science. 2001;292:1540–1543. doi: 10.1126/science.292.5521.1540. [DOI] [PubMed] [Google Scholar]
- Nitarska J., Smith J.G., Sherlock W.T., Hillege M.M., Nott A., Barshop W.D., Vashisht A.A., Wohlschlegel J.A., Mitter R., Riccio A. A functional switch of NuRD chromatin remodeling complex subunits regulates mouse cortical development. Cell Rep. 2016;17:1683–1698. doi: 10.1016/j.celrep.2016.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor S.C., Martínez-Cerdeño V., Ivic L., Kriegstein A.R. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Petersen C., Orem N., Trueheart J., Thorner J.W., Macara I.G. Random mutagenesis and functional analysis of the Ran-binding protein, RanBP1. J. Biol. Chem. 2000;275:4081–4091. doi: 10.1074/jbc.275.6.4081. [DOI] [PubMed] [Google Scholar]
- Pollarolo G., Schulz J.G., Munck S., Dotti C.G. Cytokinesis remnants define first neuronal asymmetry in vivo. Nat. Neurosci. 2011;14:1525–1533. doi: 10.1038/nn.2976. [DOI] [PubMed] [Google Scholar]
- Seewald M.J., Kraemer A., Farkasovsky M., Körner C., Wittinghofer A., Vetter I.R. Biochemical characterization of the Ran-RanBP1-RanGAP system: are RanBP proteins and the acidic tail of RanGAP required for the Ran-RanGAP GTPase reaction? Mol. Cell. Biol. 2003;23:8124–8136. doi: 10.1128/MCB.23.22.8124-8136.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelly M., Cancedda L., Heilshorn S., Sumbre G., Poo M.M. LKB1/STRAD promotes axon initiation during neuronal polarization. Cell. 2007;129:565–577. doi: 10.1016/j.cell.2007.04.012. [DOI] [PubMed] [Google Scholar]
- Shoukimas G.M., Hinds J.W. The development of the cerebral cortex in the embryonic mouse: an electron microscopic serial section analysis. J. Comp. Neurol. 1978;179:795–830. doi: 10.1002/cne.901790407. [DOI] [PubMed] [Google Scholar]
- Umeshima H., Hirano T., Kengaku M. Microtubule-based nuclear movement occurs independently of centrosome positioning in migrating neurons. Proc. Natl. Acad. Sci. USA. 2007;104:16182–16187. doi: 10.1073/pnas.0708047104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudin D., Hanz S., Yoo S., Iavnilovitch E., Willis D., Gradus T., Vuppalanchi D., Segal-Ruder Y., Ben-Yaakov K., Hieda M. Localized regulation of axonal RanGTPase controls retrograde injury signaling in peripheral nerve. Neuron. 2008;59:241–252. doi: 10.1016/j.neuron.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.S., Arnaoutov A., Dasso M. RanBP1 governs spindle assembly by defining mitotic Ran-GTP production. Dev. Cell. 2014;31:393–404. doi: 10.1016/j.devcel.2014.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Li J., Ma Y., Wang J., Pan W., Gao K., Zhang Z., Lu T., Ruan Y., Yue W. Ezh2 is involved in radial neuronal migration through regulating Reelin expression in cerebral cortex. Sci. Rep. 2015;5:15484. doi: 10.1038/srep15484. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.