Abstract
Parainfluenza virus type 3 (PIV-3) can infect a wide variety of mammals including humans, domestic animals, and wild animals. In the present study, bovine parainfluenza virus type 3 (BPIV-3) was isolated from nasal swabs of Egyptian cattle presenting with clinical signs of mild pneumonia. The virus was isolated in Madin-Darby bovine kidney (MDBK) cells and confirmed by reverse transcription-polymerase chain reaction (RT-PCR). The complete genome of Egyptian BPIV-3 strain was sequenced by using next generation (Illumina) sequencing. The new isolate classified with genotype A of BPIV-3 and was closely related to the Chinese NM09 strain (JQ063064). Subsequently in 2015–16, a molecular surveillance study was undertaken by collecting and testing samples from cattle and buffaloes with respiratory tract infections. The survey revealed a higher rate of BPIV-3 infection in cattle than in buffaloes. The infection was inversely proportional to the age of the animals and to warm weather. This report should form a basis for further molecular studies on animal viruses in Egypt.
Keywords: BPIV-3, Egypt, Cattle, Phylogenetic analysis, Parainfluenza
1. Introduction
Bovine parainfluenza virus type 3 (BPIV-3) is a member of respirovirus genus in the family Paramyxoviridae [1]. The virus is pleomorphic and enveloped and has a diameter of 150–300 nm [2]. The non-segmented negative-stranded RNA genome contains approximately 15,000 nucleotides (nt) organized to encode six structural proteins including nucleocapsid protein, phosphoprotein, matrix protein, fusion protein, hemagglutinin-neuraminidase protein, and large polymerase protein [3]. The HN (hemagglutinin-neuraminidase) and F (fusion) proteins are surface glycoproteins found in all parainfluenza viruses (PIVs). There are significant differences in the number of HN glycosylation sites among PIVs, and even among strains within a single type, which may be part of a strategy to escape immune detection [4]. The F protein is integral to virus-host interactions and helps in fusion of membranes, which allows the viral nucleocapsid to enter and infect a host cell [5]. The M protein interacts with the surface glycoproteins (HN and F) and directs their insertion and aggregation at specific cell membrane sites [6]. The L, P and N proteins are closely associated with the viral RNA [7].
Parainfluenza virus type 3 (PIV-3) can infect a wide variety of mammals including humans, domestic animals, and wild animals [8], [9]. Cross-species infections have also been reported e.g., BPIV-3 in humans and sheep and ovine PIV-3 in cattle [10], [11]. The three genotypes of BPIV-3 are named as A (BPIV-3a), B (BPIV-3b) and C (BPIV-3c) [12] of which genotype A, which was first recognized in the USA, is the most common. Genotypes B and C were detected for the first time in Australia and China, respectively [12], [13]. It was previously thought that genotype C was circulating only in China but recently it has been reported from both Korea and Argentina [13], [14].
The BPIV-3 is one of the causes of bovine respiratory disease complex (BRDC) [15]. This virus may cause tissue damage and immunosuppression resulting in severe bronchopneumonia due to secondary bacterial infections, especially when animals are under stressful conditions [16]. The virus was first isolated in the United States in 1959 from nasal discharge of cattle with shipping fever and was initially named as myxovirus SF-41 [1]. In Egypt, the virus was first isolated in 1963 from lung tissues of imported Somali cattle with respiratory illness at the Suez quarantine station [17]. Since then, the virus has been involved in several outbreaks in Egypt and neighboring countries either alone or with other viruses e.g., infectious bovine rhinotracheitis virus (IBRV), bovine respiratory syncytial virus (BRSV) and/or bovine viral diarrhea virus (BVDV) [18], [19].
In 2014, we observed sporadic cases of pneumonia in bovine calves in Sharkia province in Egypt. Inoculation of nasal swab suspensions in MDBK (Madin-Darby bovine kidney) cells followed by RT-PCR revealed the presence of BPIV-3. No other virus e.g., IBRV, BVDV, BRSV or bovine enterovirus (BEV) was identified. In this study, we report on the complete genome sequence of BPIV-3 using Illumina sequencing. The Egyptian BPIV-3 strain was classified as a member of genotype A based on its nucleotide and amino acid (aa) sequences. The study was extended to investigate the presence of BPIV-3 in cattle and buffaloes in 2015–16. The investigation included examination of nasal swabs of animals suffering from respiratory manifestation and pneumonia and those admitted to the clinic at Faculty of Veterinary Medicine, Zagazig University, Zagazig, Egypt.
2. Materials and methods
2.1. Samples and virus isolation
In 2014, nasal swabs were collected from 12 sporadic cases of calf pneumonia from the Abo-Kebeer area of Sharkia province in Egypt and were tested for the presence of BRDC viruses (IBRV, BVDV, BRSV and BPIV-3) and BEV. Briefly, the swabs were squeezed in phosphate buffered saline (PBS, pH 7.4) solution followed by centrifugation at 2500g for 10 min. The supernatants were filtrated through 0.22 µm filters and then inoculated in MDBK cells for virus isolation. The inoculated cells were incubated at 37 °C for 2 h before adding modified Eagle’s minimum essential medium (MEM) supplemented with 1.5 mg/mL trypsin (Fisher Scientific, Loughborough, UK), 4% fetal bovine serum (Fisher Scientific), 100 μg/mL streptomycin (Sigma-Aldrich, Dorset, UK), 100 U/mL penicillin (Sigma-Aldrich, Dorset, UK), and 2.5 µg/mL fungizone (Fisher Scientific, Loughborough, UK). The inoculated cells were observed daily under a light microscope for the appearance of virus-induced cytopathic effects (CPE). When CPE appeared in 80% of the cell monolayer, the virus was harvested by three cycles of freezing and thawing, followed by centrifugation at 1200g for 15 min to remove cellular debris.
2.2. Polymerase chain reaction (PCR) for the detection of IBRV
Viral DNA was extracted from cell culture supernatants using DNeasy blood and tissue kit (Qiagen, Valencia, CA). Extracted DNA was subjected to PCR using HotStarTaq master mix kit (Qiagen, Valencia, CA) and IBR gB primer set (Table 1). Briefly, the amplification reaction was performed in a 25 μL reaction mixture under the following conditions: an initial denaturation step of 15 min at 95 °C, followed by 35 cycles of 1 min at 94 °C (denaturation); 1 min for annealing at 51 °C; 1 min at 72 °C (extension) and one final extension step of 10 min at 72 °C [20].
Table 1.
Primers used in PCR and RT- PCR reactions.
| Virusa | Primer sequence | Annealing temperature | Amplicon size |
|---|---|---|---|
| BHV-1 GB | F GTA CAC GTT CAA GGC CTA CA R TCG TCT CGC AGC ATT TC |
51 °C | 668 bp |
| BVD | F CAT GCC CTT AGT AGG AC R CTC CAT GTG CCA TGT ACA G |
48 °C | 400 bp |
| BRSV | F CAT CAA TCC AAA GCA CCA CAC TGT C R GCT AGT TCT GTG GTG GAT TGT TGT C |
55 °C | 381 bp |
| BPI-3 | F AGT GAT CTA GAT GATGAT CCA R GTT ATT GAT CCA ATT GCT GT |
47 °C | 328 bp |
| BEV | F ATG GAC AAG AGG TAY GTC GTC GT R GGG CAC ACT CCG GAT TTT CTC C |
55 °C | 450 bp |
BHV-1, bovine herpesvirus type 1 or infectious bovine rhinotracheitis virus; BVDV, bovine viral diarrhea virus; BRSV, bovine respiratory syncytial virus; BPIV-3, bovine parainfluenza 3 virus; BEV, bovine enterovirus.
2.3. Reverse transcription-polymerase chain reaction (RT-PCR)
RNA was extracted from cell culture supernatants using QIAamp Viral RNA Mini kit (Qiagen, Valencia, CA). The extracted RNA was subjected to RT-PCR using one-step RT-PCR kit (Qiagen, Valencia, CA). The primer sets used for the detection of BVDV, BRSV, BPIV-3 and BEV are shown in Table 1. The amplification protocol used 25 µL reaction mixture incubated in a thermocycler at 50 °C for 30 min in RT step and initial denaturation at 95 °C for 15 min. This was followed by 35 cycles consisting of 94 °C for 1 min for denaturation followed by annealing for 1 min at appropriate temperature for each primer set (Table 1) and then elongation at 72 °C for 1 min followed by final extension cycle at 72 °C for 10 min. The products were analyzed in ethidium bromide-stained 1.2% agarose gel. A single band of expected product size confirmed the presence of the target virus.
2.4. Next generation sequencing (Illumina sequencing)
For molecular characterization, cell culture supernatants were loaded on FTA cards and shipped to the University of Minnesota Veterinary Diagnostic Laboratory (UMVDL), Saint Paul, Minnesota, USA. Viral RNA was extracted from loaded FTA cards using the QIAamp Viral RNA Mini kit (Qiagen, Valencia, CA). Equal amounts of RNA from all samples were combined to make a single pool for Illumina sequencing. The pooled RNA sample was sent to the University of Minnesota Genetic Center (UMGC) for Illumina MiSeq paired end 250 cycles. The sample passed sequencing quality assessment in which total RNA was quantified using a fluorimetric RiboGreen assay. The library was created using Illumina’s Truseq RNA sample preparation kit (Cat. # RS-122-2001) and good quality libraries were sequenced on MiSeq. The obtained sequence reads were analyzed by CLC genomic workbench 6.0.1 Trimming and sequence quality were tested followed by preparation of contigs by de novo assembly. Extracted contigs were analyzed by tBlastx analysis on NCBI.2 The ORF finder tool3 was used to find ORFs in the obtained sequences. The obtained nt sequences were translated through translator.4 Prediction of coding and non-coding regions was done based on nt and aa alignments with reference BPIV-3 (NM09; JQ063064) sequence in GenBank.
2.5. Phylogenetic analysis and genome characterization
The alignment of compatible nt sequences, calculation of P distance and tree construction were done by using MEGA 6.0 (Molecular Evolutionary Genetic Analysis). Based on nt blast analysis on NCBI (blastn), sequences representing different geographical areas, years, and species were selected for further comparison. A phylogenetic tree of aligned sequences was constructed by selecting the best fit Maximum Likelihood model in Mega 6.0 based on the lowest BIC score (Bayesian Information Criterion). The Maximum-Likelihood methods used for constructing phylogenetic tree for complete genome, HN and M genes were the nucleotide substitution model GTR + G + I, GTR + I and T92 + I models, respectively [21]. The sequence of BPIV-3/Egypt/2014 was submitted to GenBank with accession number KP757872.
2.6. Samples for molecular diagnosis of BPIV-3
Nasal samples were collected for molecular detection of BPIV-3 in clinically ill animals with varying degrees of respiratory manifestation and without history of vaccination against BPIV-3. The samples were collected with long plastic swabs. They were then placed in tubes contained 1 mL of PBS each. The swabs were squeezed in PBS, and then centrifuged to obtain the sample supernatants. The number of samples collected was 67 and 74 in 2015 and 2016, respectively. The RNA from these supernatants was extracted using QIAamp Viral RNA Mini kit (Qiagen, Valencia, CA) followed by RT-PCR for BPIV-3 as described above.
3. Results
3.1. Virus isolation
Eight of the 12 samples showed CPE on MDBK cells on the fourth day of inoculation in the form of many scattered, rounded cells. The PCR and RT-PCR of infected cell culture supernatant indicated the absence of IBRV, BVDV, BRSV and BEV. However, a single band of BPIV-3 appeared in all eight samples.
3.2. Illumina sequence analysis
Illumina sequence analysis on the pool of these eight isolates revealed the presence of BPIV-3 complete genome with a total length of 15,435 nt divided into 6 coding (N, P, M, F, HN and L) and 6 non-coding regions (Leader UTR, N-P UTR, P-M UTR, M-F UTR, F-HN UTR, HN-L UTR and Trailer UTR) (Table 2). The first gene of the coding region, the N gene (1548 nt), had 84–99.9% nt (91–100% aa) identity with the other strains of BPIV-3. The P gene was 1791 nt (597 aa) long with stop codon and had 81–100% nt (91–100% aa) identity with other strains of BPIV-3. The M gene (1056 nt) ended in stop codon with 82.2–100% nt identity and high degree of aa conservation (96–100%) with other BPIV-3 sequences. The F gene (1623 nt) had 81.2–100% nt and 86–100% aa identity with other BPIV-3 sequences. HN gene was 1719 nt (573 aa) long showing 80.1–99.8% nt and 93–99% aa identity with other BPIV-3 sequences. The L gene was the largest (6702 nt) with stop codon and had 84.3–100% nt (91–100% aa) identity with other BPIV-3 viruses (Table 3, Table 4).
Table 2.
Coding and non-coding regions of BPIV-3/Egypt/2014/KP757872.
| Genome region | Nucleotide | No. of amino acids |
|---|---|---|
| Leader UTR | 1–95 | |
| Gene N | 96–1644 | 516 |
| N UTR | 1645–1768 | |
| Gene P | 1769–3560 | 597 |
| P UTR | 3561–3719 | |
| Gene M | 3720–4776 | 352 |
| M UTR | 4777–5050 | |
| Gene F | 5051–6674 | 541 |
| F UTR | 6675–6784 | |
| Gene HN | 6785–8504 | 573 |
| HN UTR | 8505–8624 | |
| Gene L | 8625–15.327 | 2234 |
| Trailer UTR | 15.328–15.435 | |
| Total | 15.435 | 4813 |
Table 3.
Putative amino acid identities of BPIV-3/Egypt/2014/KP757872 compared with those of BPIV-3, SPIV3 and HPIV3.a
* Bovine PIV3, ** Swine PIV3, • Human PIV3.
GBA: GenBank accession number.
I%: Per cent identity.
Table 4.
Nucleotide identities of BPIV-3/Egypt/2014/KP757872 compared with those of BPIV-3, SPIV3 and HPIV3.a
| GenBankAccession | Strain | Complete | Leader UTR | N | N-P UTR | P | P-M | M | M-F | F | F-HN | HN | HN-L | L | Trailer UTR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| JQ063064 | BPIV-3 | 99.9 | 100 | 99.9 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 99.8 | 100 | 99.9 | 100 |
| AF178655 | 92.1 | 90.3 | 93.1 | 90.4 | 90.8 | 77.5 | 92.5 | 83.9 | 91.6 | 79.6 | 93.0 | 79.3 | 93.4 | 74.5 | |
| AF178654 | 92.1 | 90.3 | 92.9 | 88.0 | 91.5 | 79.4 | 92.0 | 82.4 | 91.7 | 80.6 | 93.0 | 78.5 | 93.3 | 72.7 | |
| AB770484 | 91.7 | 93.5 | 91.9 | 87.2 | 91.2 | 76.9 | 90.1 | 82.4 | 90.8 | 81.6 | 92.1 | 76.0 | 93.5 | 70.9 | |
| AB770485 | 91.7 | 93.5 | 91.9 | 87.2 | 91.1 | 76.9 | 90.1 | 82.4 | 90.8 | 81.6 | 92.1 | 76.0 | 93.4 | 70.9 | |
| EU277658 | 82.8 | 77.4 | 85.5 | 72.0 | 81.8 | 65.0 | 82.2 | 64.4 | 81.9 | 69.4 | 81.9 | 63.6 | 85.0 | 65.5 | |
| HQ530153 | 82.1 | 77.4 | 83.5 | 72.0 | 81.7 | 63.1 | 82.7 | 58.4 | 81.2 | 63.3 | 80.1 | 52.9 | 85.0 | 61.8 | |
| LC000638 | 82.2 | 74.2 | 83.9 | 73.6 | 81.3 | 61.9 | 83.3 | 59.6 | 81.4 | 60.2 | 80.8 | 54.5 | 84.9 | 61.8 | |
| JX969001 | 81.8 | 74.2 | 84.1 | 72.8 | 81.1 | 62.5 | 82.7 | 59.6 | 81.7 | 63.3 | 80.7 | 52.1 | 84.3 | 52.7 | |
| EU439428 | SPIV-3 | 91.4 | 93.5 | 91.8 | 86.4 | 91.3 | 73.8 | 90.1 | 82.0 | 90.9 | 78.6 | 91.4 | 77.7 | 93.0 | 72.7 |
| EU439429 | 92.0 | 90.3 | 93.1 | 90.4 | 90.6 | 78.8 | 92.5 | 83.9 | 91.6 | 79.6 | 93.0 | 78.5 | 93.0 | 76.4 | |
| EU424062 | HPIV-3 | 78.0 | 51.6 | 79.6 | 58.4 | 73.7 | 51.3 | 79.3 | 51.3 | 78.2 | 62.2 | 74.3 | 50.4 | 82.5 | 50.9 |
| KF687346 | 78.0 | 51.6 | 79.8 | 56.8 | 73.4 | 49.4 | 79.5 | 53.2 | 77.6 | 65.3 | 74.0 | 52.1 | 82.7 | 54.5 | |
| KF530245 | 78.1 | 51.6 | 79.4 | 58.4 | 73.2 | 50.6 | 80.1 | 53.6 | 77.7 | 66.3 | 74.6 | 52.1 | 82.7 | 54.5 | |
| FJ455842 | 77.8 | 51.6 | 79.5 | 57.6 | 73.4 | 52.5 | 79.8 | 54.3 | 77.9 | 67.3 | 74.0 | 51.2 | 82.0 | 52.7 | |
| AB012132 | 78.0 | 51.6 | 79.6 | 54.4 | 72.8 | 51.9 | 79.9 | 55.8 | 78.3 | 64.3 | 74.5 | 51.2 | 82.5 | 56.4 | |
* Bovine parainfluenza virus type 3 (BPIV-3); ** Swine parainfluenza virus type 3 (SPIV-3); • Human parainfluenza virus type 3 (HPIV-3).
The nt sequences from the non-coding regions of BPIV-3/Egypt/2014/KP757872 genome differed markedly from those of the previously reported BPIV-3 sequences but were 100% identical to those of NM09 sequence (JQ063064) from China (Table 4). The P distance revealed that the coding regions of BPIV-3/Egypt/2014/KP757872 had 99.9%, 99.8% and 99.9% identity with the Chinese isolate NM09 (JQ063064) in N, HN and L genes, respectively. The nt identities for the coding regions between BPIV-3/Egypt/2014/KP757872 and the other BPIV-3 sequences were in the range of 80.7–93.4% (Table 4). The nt identities for the complete genome were 91.7–99.9% between BPIV-3/Egypt/2014/KP757872 and the other BPIV-3 genotype A, 82.8% with genotype B (EU277658), and 81.8–82.2% with genotype C (HQ530153, LC000638 and JX969001). The nt identity between BPIV-3/Egypt/2014/KP757872 and the swine strain EU439428-29 ranged from 91.4 to 92.0%. The lowest nt identity (77.8–78.1%) was observed with human strains FJ455842 and KF530245.
In comparison to the highly identical NM09 Chinese strain (JQ063064), HN gene sequence analysis revealed one synonymous nt substitution at position C 633 T and two non-synonymous substitutions at positions C 563 A, and A 761 G. The aa changes included a threonine to asparagine and a lysine to arginine at positions 188 and 254, respectively. The study sequence had eight synonymous nt substitutions (G1326A, C2196T, A2523G, C2526T, A2538G, T3639G, C3642T and T6219A) in L gene and two nt substitutions (A675G and G684A) in N gene in comparison with the Chinese isolate NM09.
3.3. Phylogenetic analysis
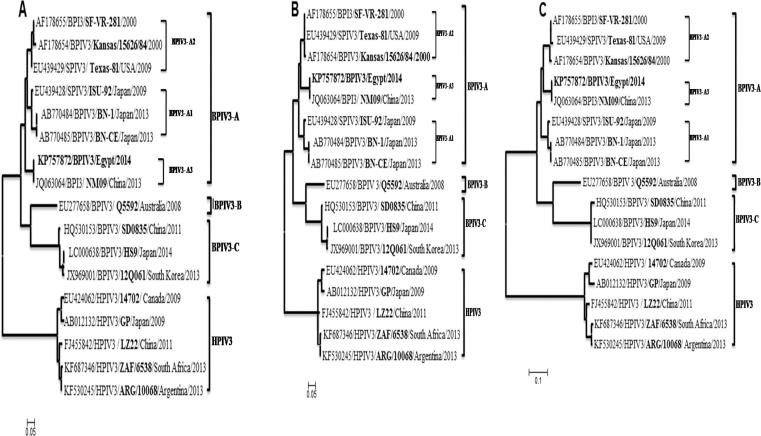
Phylogenetic reconstructions based on the alignment of the complete genome and the HN and M gene nt sequences demonstrated that the Egyptian isolate grouped with genotype A [Chinese NM09 isolate (JQ063064)] (Fig. 1). The phylogenetic tree produced from the complete nt sequence was almost identical to that produced from HN and M gene nt sequences. The full-length HN gene and M gene phylogenetic tree indicated three discernible subgenotypes in BPIV-3 genotype A: BPIV3- A1 represented by ISU_92 (EU439428), BN-1 (AB770484) and BN-CE (AB770485) viruses; BPIV3- A2 represented by Texas-81 (EU439429), Kansas (AF178654) and SF-VR-281 (AF178655) viruses; while NM09 (JQ063064) and Egyptian strain forming the third subgenotype, BPIV3- A3 (Fig. 1).
Fig. 1.
Phylogenetic analysis of bovine parainfluenza virus type 3 strains based on nucleotide sequences of the HN (a) and Matrix (b) genes, and complete genome sequence (c). The nucleotide sequences were aligned using the MEGA 6.0 software.
3.4. Molecular diagnosis
The molecular diagnosis revealed highest positivity of BPIV-3 in 2016 samples as compared to 2015 (Table 5, Table 6). The highest positivity was detected in cattle (72%) and buffalo calves (60%) in 2016. The lowest percentage was among adult cattle (20%) and adult buffaloes (12.5%) in 2015. The results revealed that the rate of infection was inversely proportional to the animal age and warm weather (Table 5, Table 6).
Table 5.
Age-related prevalence of BPIV-3 in cattle and buffaloes.
| Year | Cattle |
Buffaloes |
Total | |||
|---|---|---|---|---|---|---|
| Adults | Calves | Adults | Calves | |||
| 2015 | No. tested | 10 | 34 | 8 | 15 | 67 |
| No. positive | 2 | 11 | 1 | 6 | 20 | |
| Percentage | 20 | 32.3 | 12.5 | 40 | 29.8 | |
| 2016 | No. tested | 15 | 25 | 14 | 20 | 74 |
| No. positive | 7 | 18 | 5 | 12 | 42 | |
| Percentage | 46.6 | 72 | 35.7 | 60 | 56.7 | |
Table 6.
Weather-related prevalence of BPIV-3 in cattle and buffaloes.
| Year | Hot weather (April-July) | Cold weather (December-March) | Total | |
|---|---|---|---|---|
| 2015 | No. tested | 37 | 30 | 67 |
| No. positive | 6 | 14 | 20 | |
| Percentage | 16.2 | 46.6 | 29.8 | |
| 2016 | No. tested | 29 | 45 | 74 |
| No. positive | 5 | 37 | 42 | |
| Percentage | 17.2 | 82.2 | 56.7 | |
4. Discussion
BPIV-3 is one of the most important viral respiratory pathogens of young and adult cattle associated with BRDC [22]. In Egypt, the virus usually causes sporadic outbreaks in different parts of the country either alone or in association with other bacterial or viral pathogens [18], [19], [20], [21], [22], [23]. Although the virus was first detected in Egypt in 1963, no sequence analysis data are available. To our knowledge, this is the first report on complete genome sequence of BPIV-3 in Egypt. The complete genome of our isolate was 15,435 nt in length, which was 63 bases shorter than the longest previously identified BPIV-3 strain Q5592 genome (15,498 nt) [8]. Like other BPIV-3, all genes ended in stop codon and were separated by non-translated short sequences between the genes [2]. Instead of variation among the short intergenic sequences, we obtained 100% identity with a Chinese strain (JQ063064) in all non-coding parts.
Based on phylogenetic analysis, the Egyptian (BPIV-3/Egypt/2014/KP757872) sequence grouped together with genotype A cluster of BPIV-3 sequences. It was closely related to genotype A NM09 sequence (JQ063064) from China; both forming a unique subgenotype [24]. The higher aa identity (92–100% aa) among different genotypes based on M gene and identical tree topology based on the M gene and complete genome nt sequences support the use of the M gene as an appropriate target for the generation of informative phylogenetic reconstructions [8]. Unfortunately, only a small number of complete BPIV-3 genome sequences have been reported and none of them is from Egypt. It is, therefore, difficult to accurately detect any genetic variations in the BPIV-3 strains causing disease outbreaks in the country since its first isolation in 1963 [17].
Egypt imports beef from Brazil, India, Australia, New Zealand and the United States and live cattle for immediate slaughter from Brazil, Australia, Sudan and Ethiopia [25]. It is surprising, therefore, that the Egyptian strain was closest to the Chinese A strain, which was identified recently in 2012. It is difficult to pinpoint the source of this strain since there is no live animal trade between China and Egypt. Understanding of distribution pathway and geographic location/genetic variation relationship requires sequence analysis of a large number of BPIV-3 isolates from a broader range of countries.
The BPIV-3 has been reported from water buffaloes, sheep, goat and camels [9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23]. All of these animals are reared in Egypt either in mixed farms with cattle or individually without proper vaccination. The molecular relationship among BPIV-3 of different species is rarely studied. Future work should be targeted this area to define relationships, if any, among BPIV-3 from different animal species. We further conclude that BPIV-3 should be considered as one of the important causes of respiratory manifestation among young calves in Egypt and that all animals imported into Egypt should be screened for this infection.
The severity and gradation of clinical signs that appeared in BPIV-3 infected animals during examination can be explained by a variety of causative agent that can share in the infection e.g., Mycoplasma, Pasteurella multocida, BRSV, IBRV or BVDV [26]. A high percentage of infected animals was found in cold weather (December-March), which is not surprising because PIVs are stable at cold temperatures [7]. Low percentage of samples was positive during hot weather (April-July) when the temperatures can reach up to 40 °C in which the virus cannot survive [27]. A higher percentage in young animals rather than in adults may be linked to pollution, malnutrition and/or lack of maternal immunity. The environmental pollution with dust and with ammonia and hydrogen sulfide gases from manure in the closed rearing system may serve as irritants and predispose to respiratory disease [28]. The BPI-3 infection was high in cattle than in buffaloes, which may be due to some genetic and/or host factor that make the cattle more sensitive. Also, cattle respond less favorably to stress than do buffaloes [29].
5. Conclusions
BPIV-3 was isolated from Egyptian cattle and sequenced by using next generation (Illumina). The newly identified strain classified with genotype A of BPIV-3 and was closely related to the Chinese NM09 strain (JQ063064). Subsequently in 2015–16, a molecular surveillance study was undertaken. The survey revealed a higher rate of BPIV-3 infection in cattle than in buffaloes. The infection was inversely proportional to the age of the animals and to warm weather. The control of BPI-3 infection in Egypt is under the purview of the Egyptian Ministry of Agriculture and can be achieved by organizing veterinary convoys, which can provide the rural areas with obligatory BPIV-3 vaccine for all susceptible animals. It should be responsible to provide vaccines in veterinary health units scattered in small villages all over the country. The Ministry should also generate and distribute brochures about infectious diseases including BPI-3 for increasing farmer’s attention about importance of vaccination. Finally, further studies are needed to investigate the prevalence and role of different types of BPIV-3 viruses in Egypt.
Competing interests
None of the authors has a financial or personal relationship with other people or organization that could inappropriately influence or bias the content of the paper.
Acknowledgements
The authors thank the cattle farmers in Egypt for their co-operation.
Footnotes
Peer review under responsibility of Faculty of Veterinary Medicine, Cairo University.
See: http://www.clcbio.com/products/clc-genomics-workbench/
See: https://blast.ncbi.nlm.nih.gov/Blast.cgi
See: http://www.ncbi.nlm.nih.gov/gorf/orfig.cgi
See: http://www.fr33.net/translator.php
References
- 1.Adams M.J., Lefkowitz E.J., King A.M.Q., Harrach B., Harrison R.L., Knowles N.J. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2016) Arch Virol. 2016;161:2921–2949. doi: 10.1007/s00705-016-2977-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vainionpaa R., Hyypia T. Biology of parainfluenza viruses. Clin Microbiol Rev. 1994;7:265–275. doi: 10.1128/cmr.7.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang L.F., Collins P.L., Fouchier R.A.M., Kurath G., Lamb R.A., Randall R.E. Family Paramyxoviridae. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Virus Taxonomy: Classification and Nomenclature of Viruses. Ninth Report. Elsevier; Waltham: 2012. pp. 672–685. [Google Scholar]
- 4.Henrickson K.J., Savatski L.L. Antigenic structure, function, and evolution of the hemagglutinin-neuraminidase protein of human parainfluenza virus type 1. J Infect Dis. 1997;176:867–875. doi: 10.1086/516533. [DOI] [PubMed] [Google Scholar]
- 5.Parks G.D., Lamb R.A. Defective assembly and intracellular transport of mutant paramyxovirus hemagglutinin-neuraminidase proteins containing altered cytoplasmic domains. J Virol. 1990;64:3605–3616. doi: 10.1128/jvi.64.8.3605-3616.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coronel E.C., Takimoto T., Murti G., Varich N., Portner A. Nucleocapsid incorporation into Parainfluenza virus is regulated by specific interaction with matrix protein. J Virol. 2001;75:1117–1123. doi: 10.1128/JVI.75.3.1117-1123.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henrickson K.J. Parainfluenza viruses. Clin Microbiol Rev. 2003;16:242–264. doi: 10.1128/CMR.16.2.242-264.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horwood P.F., Gravel J.L., Mahony T.J. Identification of two distinct bovine parainfluenza virus type 3 genotypes. J Gen Virol. 2008;89:1643–1648. doi: 10.1099/vir.0.2008/000026-0. [DOI] [PubMed] [Google Scholar]
- 9.Intisar K.S., Ali Y.H., Taha K.M., Musa M.Z., Khalafalla A.I. Isolation of some respiratory viruses from camels. Int J Livest Prod. 2014;5:177–181. [Google Scholar]
- 10.Stevenson R.G., Hore D.E. Comparative pathology of lambs and calves infected with parainfluenza virus type 3. J Comp Pathol. 1970;80:613–618. doi: 10.1016/0021-9975(70)90060-5. [DOI] [PubMed] [Google Scholar]
- 11.Ben-Ishai Z., Naftali V., Avram A., Yatziv S. Human infection by a bovine strain of parainfluenza virus type 3. J Med Virol. 1980;6:165–168. doi: 10.1002/jmv.1890060209. [DOI] [PubMed] [Google Scholar]
- 12.Zhu Y.M., Shi H.F., Gao Y.R., Xin J.Q., Liu N.H., Xiang W.H. Isolation and genetic characterization of bovine parainfluenza virus type 3 from cattle in China. Vet Microbiol. 2011;149:446–451. doi: 10.1016/j.vetmic.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 13.Oem J., Lee E., Lee K., Kim S., Lee M., Hyun B. Molecular characterization of a Korean bovine parainfluenza virus type 3 isolate. Vet Microbiol. 2013;162:224–227. doi: 10.1016/j.vetmic.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 14.Maidana S.S., Lomonaco P.M., Combessies G., Craig M.I., Diodati J., Rodriguez D. Isolation and characterization of bovine parainfluenza virus type 3 from water buffaloes (Bubalus bubalis) in Argentina. BMC Vet Res. 2012;8:83–91. doi: 10.1186/1746-6148-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Snowder G.D., Van Vleck L.D., Cundiff L.V., Bennett G.L. Bovine respiratory disease in feedlot cattle: environmental, genetic, and economic factors. J Anim Sci. 2006;84:1999–2008. doi: 10.2527/jas.2006-046. [DOI] [PubMed] [Google Scholar]
- 16.Haanes E.J., Guimond P., Wardley R. The bovine parainfluenza virus type-3 (BPIV-3) hemagglutinin/neuraminidase glycoprotein expressed in baculovirus protects calves against experimental BPIV-3 challenge. Vaccine. 1997;15:730–738. doi: 10.1016/s0264-410x(96)00231-9. [DOI] [PubMed] [Google Scholar]
- 17.Singh K.V., El-Cicy I.F. Studies with bovine parainfluenza- 3 virus in U.A.R. (Egypt) Can J Comp Med Vet Sci. 1967;31:70–79. [PMC free article] [PubMed] [Google Scholar]
- 18.Aly N.M., Shehab G.G., Abd el-Rahim I.H. Bovine viral diarrhoea, bovine herpesvirus and parainfluenza-3 virus infection in three cattle herds in Egypt in 2000. Rev Sci Technol. 2003;22:879–892. doi: 10.20506/rst.22.3.1440. [DOI] [PubMed] [Google Scholar]
- 19.Zaher K.S., Syame S.M., Elhewairy H.M., Marie H.S.H. Investigation of bovine respiratory disease complex in Egypt with emphasis on some viral and bacterial pathogens. Life Sci. 2014;11:56–62. [Google Scholar]
- 20.Sobhy N.M., Mor S.K., Mohammed M.E.M., Bastawecy I.M., Fakhry H.M., Youssef C.R.B. Comparative molecular characterization of bovine herpesvirus-1 strains from Egypt and the United States. Life Sci. 2014;11:493–499. [Google Scholar]
- 21.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong X., Zhu Y., Cai H., Lv C., Gao Y., Yu Z. Studies on the pathogenesis of a Chinese strain of bovine parainfluenza virus type 3 infection in Balb/c mice. Vet Microbiol. 2012;158:199–204. doi: 10.1016/j.vetmic.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 23.Gafer A.M., Hussein H.A., Reda I.M. Isolation and characterization of PI-3 virus from sheep and goats. Int J Virol. 2009;5:28–35. [Google Scholar]
- 24.Wen Y., Shi X., Wang F., Wang W., Zhang S., Li G. Phylogenetic analysis of the bovine parainfluenza virus type 3 from cattle herds revealing the existence of a genotype A strain in China. Virus Genes. 2012;45:542–547. doi: 10.1007/s11262-012-0810-1. [DOI] [PubMed] [Google Scholar]
- 25.Hamza M. Egypt livestock and products annual. United states, Foreign agriculture service: USDA; September 11, 2014. Available from: http://www.fas.usda.gov/data/egypt-livestock-and-products-annual-0. [Accessed 20 February 2015].
- 26.Hotchkiss E.J., Dagleish M.P., Willoughby K., McKendrick I., Finlayson J., Zadoks R.N. Prevalence of Pasteurella multocida and other respiratory pathogens in the nasal tract of Scottish calves. Vet Rec. 2010;167:555–560. doi: 10.1136/vr.c4827. [DOI] [PubMed] [Google Scholar]
- 27.Sobsey M.D., Meschke J.S. World Health Organization; Geneva, Switzerland: 2003. Virus survival in the environment with special attention to survival in sewage droplets and other environmental media of fecal or respiratory origin; p. 70. [Google Scholar]
- 28.Callan R.J., Garry F.B. Biosecurity and bovine respiratory disease. Vet Clin North Am Food Anim Pract. 2002;18:57–77. doi: 10.1016/S0749-0720(02)00004-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valarcher JF, Hägglund S. Viral respiratory infections in cattle. In: Proceedings of the 24th World Buiatrics Congress – Nice, France, 2006; 384–97.