Abstract
Non-small cell lung cancer (NSCLC) accounts for approximately 85% of lung cancer cases, and it is one of the leading causes of cancer death in both men and women worldwide due to diagnosis in the advanced stage, rapid metastasis, and recurrence. At present, precision molecular targeted therapeutics directed toward NSCLC driven genes has made great progress and significantly improved the overall survival of patients with NSCLC, but can easily lead to acquired drug resistance. New methods are needed to develop real-time monitoring of drug efficacy and drug resistance, such as new molecular markers for more effective early detection and prediction of prognosis. Exosomes are nano-sized extracellular vesicles, containing proteins, nucleic acids and lipids, which are secreted by various cells, and they play an important role in the development of lung cancer by controlling a wide range of pathways. Tumor-derived exosomes are of great significance for guiding the targeted therapy of NSCLC and exosomes themselves can be a target for treatment. In this review, we describe the potential roles of tumor-derived exosomes and their clinical significance in NSCLC.
Keywords: NSCLC, Tumor-derived exosomes, Clinical implications, Targeted therapy
Background
Lung cancer is one of the leading causes of cancer-related death both in men and women [1] and remains the most frequently diagnosed cancer in the world [2], which is classified into two histological subtypes: non-small cell lung cancer(NSCLC)accounting for 85% and small-cell lung cancer (SCLC) accounting for the remaining 15% [3]. Only 17.7% of the patients live over 5 years after being diagnosed with lung cancer [4]. The standard treatment of NSCLC is curative surgical resection, combined with or without chemoradiotherapy for the patients with early-stage including stage I, II and a part of stage III [5, 6]. However, most patients with NSCLC are diagnosed in the advanced stage [7], which makes the cancer difficult to surgically resect and increases the rate of postoperative recurrence, while the effect of radiotherapy and chemotherapy has plateaued [8]. On the other hand, treatment of NSCLC is evolving from the use of cytotoxic chemotherapy to precision treatment based on changes in molecular and gene levels [9], which inevitably leads to drug resistance sooner or later. In the past, the golden standard of the patients’ diagnosis and gene mutations detection is tissue biopsy, which limits the assessment of lung cancer development and prognosis due to tumor heterogeneity and evolution [10]. Liquid biopsy is typically used to separate and analyze circulating free DNA and RNA from the blood of cancer patients, or other body fluids, which has potential advantages, such as real-time monitoring of treatment response, rapid and accurate identification of drug resistance genes, identification of minimal residual disease and prediction of prognosis [10, 11]. Exosomes are nano-sized extracellular vesicles, containing proteins, nucleic acids and lipids and the encapsulated contents in exosomes can escape from the degradation. Exosomes play an important role in cell-to-cell communication, tumor progression and drug resistance and have excellent prospects in liquid biopsy [12–14]. In this review, we discuss the close relationship between tumor-derived exosomes and NSCLC, and the application of tumor-derived exosomes in the targeted therapy for NSCLC.
Exosomes and tumor-derived exosomes
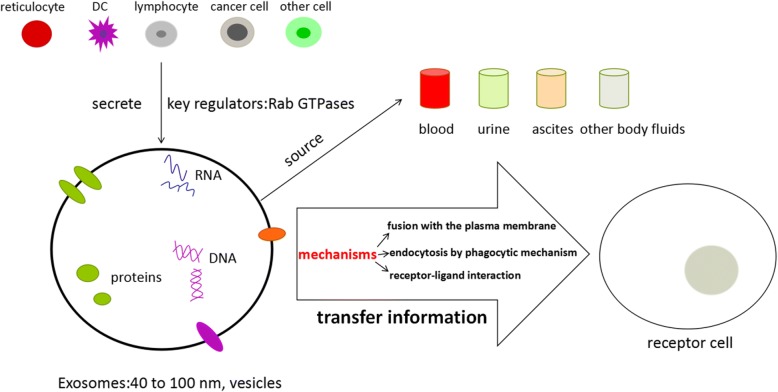
The diameter of exosomes which was first reported in the process of sheep reticulocytes maturation ranged from 40 to 100 nm, showing a characteristic cup-shaped morphology (after negative staining) or round well-delimited vesicles observed by transmission and cryo-electron microscope [15, 16]. Exosomes are small vesicles of endosomes that can be released by many cell types, such as reticulocytes [16], dendritic cells [17], lymphocytes [18, 19] and cancer cells [20]. Furthermore, exosomes have been successfully purified from many body fluids such as blood, urine, pleural effusions, ascites and broncoalveolar fluid [21]. Also, exosomes can transfer information, including DNA, RNA and proteins, to the receptor cells through fusion with the plasma membrane, endocytosis by phagocytic mechanism or receptor-ligand interaction with the cell [22] and have the heterogeneous biological behaviors due to their different secretory cell types with diverse cell status [23], thus participating in different physiological and pathological processes. Rab GTPases, including Rab27a and Rab27b, are the key regulators of exosomes secretion and Rab27 is closely related to the occurrence and the development of tumor, which indicates the role of exosomes secretion in tumor biology [24]. (These are summarized in Fig. 1).
Fig. 1.
The biology of exosomes. Exosomes are some vesicles ranged from 40 to 100 nm, which can be released by many cells and purified from many body fluids. Exosomes can transfer information to the receptor cells through fusion with the plasma membrane, endocytosis by phagocytic mechanism or receptor-ligand interaction with the cell
The immunological activities of exosomes affect the mechanisms of immunoregulation including immune activation, antigen expression regulation, intercellular communication, immunization surveillance, and immunosuppression [25]. Tumor-derived exosomes which carry tumor-associated antigens can interfere with anti-tumor immunotherapies [26]. It has been reported that about 2000 trillion exosomes can be detected in normal human blood versus 4000 trillion exosomes in the blood of cancer patients [27], which indicates that tumor cells produce more exosomes than their normal counterparts and can be a useful diagnostic biomarker [28]. Tumor-derived exosomes are believed to be important mediators of intercellular signaling and epithelial–mesenchymal transition (EMT), which transforms cancer cells into more aggressive phenotype, and contributes to the tropism of metastatic disease in specific cancer types with pre-metastatic niche [29]. Azmi et al. found tumor cells exposed to hypoxia secreted exosomes which had enhanced potential of angiogenesis and metastasis, suggesting that tumor cells adapt themselves to the hypoxic microenvironment by secreting exosomes, so as to stimulate angiogenesis or create a more favorable tumor microenvironment to promote tumor metastasis [30]. Furthermore, tumor-derived exosomes are of crucial importance in tumor growth and drug resistance, as they transfer nucleic acids and oncogenic proteins to the tumor cells, which indicates tumor-derived exosomes and their contents may be of potential value in the diagnosis, prognosis, prediction of treatment response and targeted therapy [31]. Due to the importance of tumor-derived exosomes, the methods to detect exosomes including isolation purification and analysis require further development (These methods are summarized in Table 1).
Table 1.
Detection methods of exosomes
| Contents | Methods | References |
|---|---|---|
| Isolation and purification | Ultracentrifugation Density gradients centrifugation Immunobeads Size exclusion chromatography Spin column-based methods |
[127–131] |
| Analysis of size and shape | TEM | [132] |
| Analysis of size distribution and concentration | Nanoparticle Tracking Analysis FAVS |
[133–135] |
| Analysis of proteins | Mass spectrometry, Western-blot SDS-PAGE |
[136, 137] |
| Analysis of RNA | NGS, Microarray, RT-PCR | [138–140] |
Roles of Tumor-derived exosomes in the NSCLC
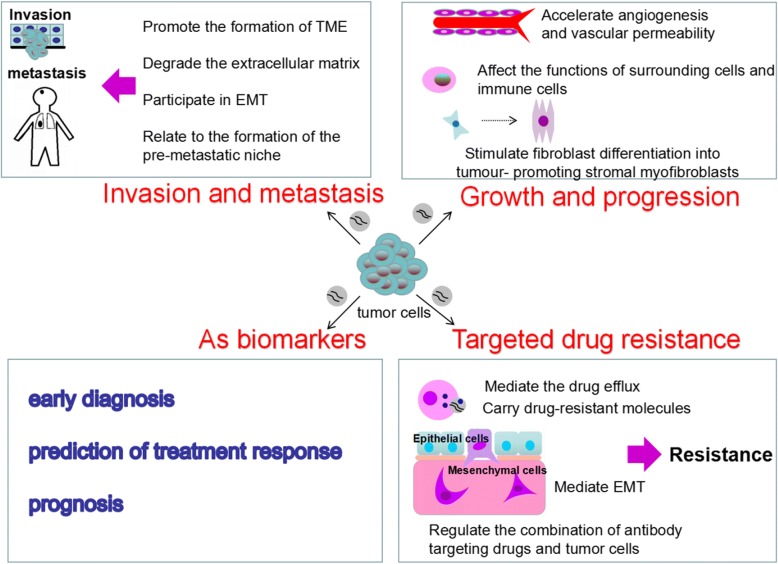
The formation and development of NSCLC is influenced by many factors and mechanisms, which is a long-term and complex process. Exosomes secreted by lung cancer cells play a vital role in this process as mediators of intercellular communication. (These roles are summarized in Fig. 2).
Fig. 2.
Roles of Tumor-derived exosomes in the NSCLC. Tumor-derived exosomes play a vital role in tumor growth and progression, invasion and metastasis, targeted drug resistance, and can also be used as biomarkers for early diagnosis, prediction of treatment response and prognosis
Tumor-derived exosomes in the growth and progression of NSCLC
It has been shown that cancer-associated fibroblasts (CAFs) in the tumor microenvironment can secrete exosomes containing lipids, amino acids, and TCA-cycle intermediates which can promote tumor growth under nutrient-stressed or nutrient-deprivation conditions [32]. Angiogenesis is also vital for tumor growth since tumor vessels are the important sources of nutrient substances in the tumor cells [33]. Tumor-derived exosomes can accelerate angiogenesis and tumor growth in a TGFβ1 (transforming growth factor β1)-dependent pathway to stimulate fibroblasts to differentiate into tumor-promoting stromal myofibroblasts [34]. Exosomal miR-23a from hypoxic lung cancer cells can increase angiogenesis and vascular permeability by targeting tight junction protein ZO-1(zonula occludens-1) and prolyl hydroxylase [35]. STAT3-regulated exosomal miR-21 enhances the level of vascular endothelial growth factor (VEGF), thereby promoting tumor angiogenesis and inducing malignant transformation of bronchial epithelial cells [36]. Exosomal miR-210 from lung cancer regulates the level of tyrosine receptor kinase A3 (ephrin A3) in matrix cells and promotes tumor angiogenesis to maintain the growth of tumor cells [37]. All of these indicated that tumor-derived exosomes can promote the growth and progression of NSCLC by angiogenesis. Lung cancer cell-derived exosomes can also affect the progression of lung cancer by affecting the physiological functions of other cells in the surrounding tissues and microenvironment. Mesenchymal stem cell (MSC) is an important one among these cells, which can be transformed into a pro-inflammatory phenotype via NFκB-TLR signaling pathway regulated by lung cancer cell-derived exosomes, thus promoting the growth of lung cancer [38]. Furthermore, lung cancer cell-derived exosomes can indirectly regulate the progression of tumor by affecting the function of immune cells in the tumor microenvironment. Tumor-derived exosomes can help with immune escape by transferring specific proteins and RNA into the receptor cells [39] and promote tumor progression by reprogramming functions of immune cells [26]. Lung cancer cell-derived exosomes containing the epidermal growth factor receptor (EGFR) can induce tolerogenic dendritic cells and tumor antigen-specific regulatory T cells (Treg) which can inhibit the function of CD positive T cells with anti-tumor function and promote the growth of lung cancer [40, 41]. Lung cancer cell-derived exosomes containing HSP72 (heat shock protein 72) can activate the STAT3 signaling pathway to mediate the immunosuppressive effect of MDSCs (myeloid-derived suppressor cells) and thereby suppress T cell activation [42, 43]. In conclusion, these studies revealed that tumor-derived exosomes could play an important role in the growth and progression of NSCLC with different contents and provide a novel future for the therapy of NSCLC, which should be further researched.
Tumor-derived exosomes in the invasion and metastasis of NSCLC
Metastasis is one of the major causes of death in lung cancer patients, which is related to a variety of mechanisms and involves multiple steps [44]. Tumor-derived exosomes, as carriers of information transmission, promote metastasis through their direct or indirect roles [13]. Exosomes can promote the formation of the lung cancer microenvironment to increase the invasiveness of tumor cells [45]. Due to the instability of oncogenes, hypoxia, acidosis and inflammatory immune response can promote tumor cells to release more exosomes to form tumor microenvironment [46, 47] which is beneficial to the rapid growth of tumor cells and enhances their ability of invasion. Tumor-derived exosomes are associated with invadopodia that initiates invasion through degradation of the extracellular matrix [48]. Exosomal contents can also promote metastasis and transfer metastatic potential to recipient cells [13]. EMT is the disappearance of the epithelial-like characteristics and gains the phenotype of stromal cells, [49] which is an important process before the metastasis of the tumor cells and also a complex process, including cytoskeleton changes, down-regulation of the expression of adherens junction molecule E-cadherin and so on [50]. It has been reported that several proteins and miRNAs are involved in the EMT [51–54]. Tumor-derived exosomes have also been reported to be associated with the formation of the pre-metastatic niche [13] that forms at the site of the future metastasis and promotes the growth of disseminated tumor cells [55]. The main sites of NSCLC metastasis are brain, adrenal gland, bone and the liver [44]. Different types of metastatic cancer cells have significant differences in organ tropism [56] which is associated with tumor-derived exosomal integrins [57]. (The major findings of tumor-derived exosomes associated with invasion and metastasis in NSCLC are summarized in Table 2).
Table 2.
Major findings of tumor-derived exosomes in NSCLC
| Contents | Findings | References |
|---|---|---|
| Invasion | exosomal TGF-β and IL-10 may enhance migration ability in vitro under hypoxia | [47] |
| exosomal vimentin may induce EMT | [51] | |
| exosomal Rab3D which activates AKT/GSK3β signaling may induce EMT | [52] | |
| exosomal miR-23a may affect the changes of EMT related phenotype | [53] | |
| Exosomal miR-302b can inhibit the proliferation and migration of lung cancer cells through the TGFβRII/ERK pathway | [54] | |
| Drug resistance | exosomal VEGF and TGF2β may result in drug resistance | [61, 62] |
| exosomal proteins and phospholipids components are involved in gefitinib resistance | [63, 64] | |
| exosomes may antagonize the chemotherapeutic effect of cisplatin by upregulating autophagy | [65] | |
| exosomes can regulate the combination of antibody targeting drugs and tumor cells | [71, 72] |
Tumor-derived exosomes in the targeted drug resistance
Resistance to chemotherapy, radiotherapy and targeted therapy remains a major obstacle to cancer treatment [30]. Drug resistance is a multifaceted problem. It has already been reported that resistance of targeted therapy and platinum based chemotherapy is associated with microRNAs [58–60]. Here the authors discuss mechanisms associated with exosomes. (1) Tumor-derived exosomes mediate EMT by transferring related tissue factors (such as VEGF, TGF2β), and thus the tumor cells can resist apoptosis, which usually result in drug resistance [61, 62]. (2) Tumor cells and stromal cells in the tumor microenvironment can secrete exosomes carrying drug-resistant molecules (miRNAs, proteins) which are transferred to enhance the tolerance of the tumor cells to the drug through the interaction of exosomes in the TME (tumor microenvironment) [63–65]. (3) Tumor-derived exosomes can also mediate the drug efflux by transferring multi-drug resistant (MDR) protein encapsulated drug, thus influencing the drug efficacy. In the development of many malignant tumors, a special transporter system which is MDR-associated ATP-binding cassette transporter (MDR-ABC) is activated to mediate the drug from the intracellular to the extracellular [66]. MDR-ABC is a type of intracellular transporter proteins, which has the same transmembrane domain and is usually located on the membrane of exosomes and multivesicular bodies (MVBs). The chemotherapeutic or targeted therapy drug and its metabolites in the cell can be transferred into the inner body which can aggregate to form MVBs. When the MVBs are fused with the cell membrane, the inner body is released into the extracellular matrix in the form of exosomes and completes the drug efflux [67–71]. (4) Tumor-derived exosomes can also influence the drug effect by regulating the combination of antibody targeting drugs and tumor cells to enhance the drug resistance of tumor cells [71, 72]. All of these mechanisms associated with exosomes can induce the targeted therapy drug resistance. (The major findings of tumor-derived exosomes associated with drug resistance in NSCLC are summarized in Table 2).
Tumor-derived exosomes as markers in prognosis of NSCLC
Increasingly, studies suggest that exosomal miRNAs and proteins are positively associated with the stage and degree of tumor progression [73], indicating that exosomal components can be used as prognostic markers to improve treatment selection for the patients with NSCLC [74]. For example, Sandfeld-Paulsen et al. investigated exosomes from plasma of 276 non-small-cell lung cancer patients, which were phenotyped by using the Extracellular Vesicle Array. As a result, they found that NY-ESO-1, PLAP, EGFR, Alix and EpCam were correlated to overall survival (OS), which indicated that exosomal membrane-bound proteins were strong prognostic biomarkers in NSCLC [74]. In addition, Liu et al. found that elevated levels of exosomal miR-10b-5p, miR-23b-3p and miR-21-5p were associated with poor overall survival by analyzing 84 plasma exosomal miRNAs in lung adenocarcinoma patients and healthy controls, which suggested that these exosomal miRNAs may also be used as prognostic biomarkers of NSCLC [75]. The downregulation of the miRNA-146-5p indicated a poor progression free survival (PFS) compared to patients with higher exosomal miRNA [76]. All of these indicate that exosomes can be used as non-invasive prognostic biomarkers.
Clinical implications of tumor-derived exosomes in the targeted therapy of NSCLC
Liquid biopsy samples are becoming more frequently used in blood or other body fluids as biomarkers for NSCLC early diagnosis, treatment guidance and drug resistance monitoring [77]. The specificity of the cell types and the accessibility from the body fluids make the exosomes important candidates for the diagnosis and target therapy of tumors [78].
Tumor-derived exosomes as biomarkers for NSCLC diagnosis and targeted therapy guidance
The exosomes secreted by lung cancer cells, which enrich various proteins, such as EGFR, KRAS, claudins and RAB-family proteins, and promote the development of lung cancer, are effective biomarkers for early diagnosis of lung cancer [79] and the basis of targeted therapy. For example, Birgitte et al. used EV array to detect exosomal proteins in NSCLC tissues and normal tissues, and found that markers CD151, CD171 and tetraspanin 8 were higher in patients with cancer of all histological subtypes than patients without cancer [80]. Huang and colleagues carried out immunostaining analysis of exosomes of NSCLC tissues and chronic pneumonia tissues, and found that 80% of the NSCLC specimens were EGFR positive on the surface of the exosomes, while only 2% of the chronic pneumonic tissues were EGFR positive, which suggested that exosomal EGFR protein could be used as a biomarker for differential diagnosis of lung cancer [41] and indicated that gene detection could be further carried out to provide a molecular basis for targeted therapy. Recently, the translocation ALK-EML4 has also been identified inside the exosomes [81], which is a biomarker for response to first generation ALK-TKIs [82]. Vykoukal and colleagues found that the expression of 108 proteins in vesicle preparations of lung adenocarcinoma plasmas was significantly different compared to controls, of which 43 were identified in EVs from lung adenocarcinoma cell lines [83].
MicroRNAs (MiRNAs) are a class of small noncoding RNAs (the length of 18–25 nucleotides), post-transcriptional regulation molecules expressed in many organisms [84]. The expression of miRNAs, like other cancer-associated genes expression, can be altered by chromosomal amplification/deletion, transcription factor activation [85]. It has been found that the miRNAs expression profiles of NSCLC exosomes which are often detected by miRNA-microarray are different from normal tissues. Zhao et al. [86] analyzed the expression of the plasma exosomal miRNAs in the tumor tissues of 150 patients with non-small cell lung cancer, and found that plasma levels of the exosomal hsa-miRNAs (hsa-miR-25, hsa-miR-122, hsa-miR-195, hsa-miR-21 and hsa-miR-125b) were associated with EGFR mutation, which could help to determine whether or not to use targeted therapy drugs and provide a new way to detect NSCLC gene mutations. The miRNAs for adenocarcinoma diagnosis showed an AUC value of 0.936, with a sensitivity of 80.65% and a specificity of 91.67% [87]. Two studies showed that upregulation of miR-1246 and miR-208-a was associated with resistance to radiotherapy and high proliferation of the tumor by targeting the genes p21 and DR5 respectively, which could not only lead to a prognostic biomarker, but also to a new target against NSCLC [88]. It has been possible to detect the rearrangement of EML4-ALK in NSCLC patients by analyzing the exosomal miRNAs in blood and has been proved that the anaplastic lymphoma kinase (ALK)-EML4 translocation inside the exosomes with a specificity of 100% and a sensitivity of 64% [89].
In summary, exosomes are becoming increasingly important in the diagnosis of NSCLC as biomarkers and the molecular basis of NSCLC targeted therapy.
Tumor-derived exosomes as biomarkers for NSCLC targeted therapy resistance
With the increasing understanding of molecular biology and genetics of tumors, the research and clinical application of targeted therapy has become a hot topic, which can improve prognosis and also guide therapeutic decision, thus reducing morbidity and mortality. On average, there are more than 300 mutations in each lung cancer, but only a few of these genes can promote or “drive” the lung tumorigenesis [90], mainly including EGFR_(epidermal growth factor receptor), ALK_(anaplastic lymphoma kinase), c-met and so on [91–93]. EGFR, also called ErbB1 or HER1, is involved in the signal transduction pathway of cell proliferation and apoptosis, which is a part of the ErbB family of transmembrane receptor tyrosine kinases [94]. The EGFR mutations, first reported in 2004 [95], usually contain in-frame duplications/insertions in exon 20, in-frame deletions (around amino acid residues 747 to 750) in exon 19 and single missense mutations in exon 21_(L858R mutation) [96], and reveal a potential responsiveness of NSCLC to the tyrosine kinase inhibitors [97]. EGFR TKIs improve time to progression, response rates and overall survival, but acquired resistance to EGFR-TKIs is inevitable [98], and thereby tools to predict the risk of drug resistance are necessary to improve the clinical treatment choices. Tumor-derived exosomes can be used as biomarkers to evaluate therapeutic effect of targeted therapy by liquid biopsy. T790 M mutation is found in patients treated with EGFR-TKIs [99], accounting for about 50–60% with acquired resistance to erlotinib or gefitinib [99, 100]. The third generation of EGFR-TKIs includes osimertinib which overcomes T790 M-mediated resistance to EGFR-TKIs [101], but it’s still unable to avoid the targeted therapy drug resistance, and some of the patients develop a new drug resistance mutation C797S [102]. It has been reported that exosomal RNA is used to detect EGFR T790 M and activating EGFR mutations, and the sensitivity is 90% and 98%, respectively [103]. So it’s possible to identify targeted therapy drug resistance in the tumor-derived exosomes. In addition, Choi et al. [63]. identified PC9R cells with rich EV proteins were resistant to gefitinib due to EGFR T790 M mutation by Nano-LC–MS/MS analysis and would help to develop new diagnostic strategies to predict and assess the drug resistance of gefitinib. Chromosome rearrangements of ALK are detected in the NSCLC of 3–7%, which indicates a response to first generation ALK-TKIs (such as crizotinib). However, most patients develop resistance to this therapy [104]. L1196 M and G1269A are identified as secondary mutation, accounting for resistance to ALK-TKIs [104, 105]. This acquired drug resistance may be predicted by detecting exosomes.
The potential value of tumor-derived exosomes in the targeted therapy of NSCLC
Studies have found that research based on exosomes and related components can provide new inspiration for the precision treatment of NSCLC.
Exosomes have been developed as drug delivery vehicles for a variety of drugs, such as small molecular drugs, nucleic acid proteins and other drugs for cancer treatment, with low immunogenicity and toxicity [106, 107]. Lai et al. used electroporation or lipofection to transfect the drugs of interest directly into the exosomes or transfer the genes that encode for the RNA/protein of interest into exosome-secreting cells [108]. Mendt and colleagues established a standard operating procedure to generate engineered exosomes which had the ability to target oncogenic Kras (iExosomes), and was confirmed to suppress oncogenic Kras and increase the survival of mouse models with pancreatic cancer, which laid the foundation for exosomes in the targeted therapy of NSCLC [109]. Exosomes can be targetable to specific tissues, resistant to metabolic processes and membrane-permeable [110], which have a wide application prospect in the targeted therapy of NSCLC.
Exosomes play an important role in tumor development, which indicates reducing the contents of exosomes can help the targeted therapy of NSCLC. Exosomes may also serve as a direct target for NSCLC. Data shows that the prevention of exosomes production can inhibit tumorigenesis and a series of methods have been suggested to reduce the contents of exosomes [110]: (1) Blood purification: Aethlon Medical designed a blood filtration therapy which could capture a large number of antibodies and other similar substances, such as aptamers, protein ligands, and exosomes to realize the new treatment of NSCLC [111]. Other studies have shown that the use of hollow fiber filtration technology to remove exosomes from the patient’s blood can minimize the immune tolerance caused by the exosomes [112]. (2) Change the local pH of the tumor: In addition to blood purification, proton pump inhibitors (PPIs) can also improve the low pH of cells by PPI pretreatment in vivo to reduce the contents of tumor-derived exosomes in the plasma, so PPI may likely be an effective method to inhibit the secretion of exosomes in NSCLC [113]. (3) Drug usage: For example, Fabbri et al. [114]. found that the use of GW4869, a neutral sphingomyelase inhibitor (regulates ceramide biosynthesis, promotes exosomes inward budding), could inhibit the production of exosomes in mice and reduce the metastasis of lung cancer. Some studies directly target exosomes as a drug target. For example, amiloride can inhibit the synthesis and secretion of exosomes [115], and diannexin can impede the absorption of exosomes by receptor cells [116]. (4) Interfere with signal pathway: some studies have shown that interference with the signal pathway associated with production or secretion of tumor-derived exosomes can inhibit the exosomes secretion. Ostrowski and colleagues found that knockout Rab27 or its effector (SYTL4 and EXPH5) could inhibit the secretion of exosomes in HeLa cells [24]. It may be useful for inhibiting the secretion of exosomes in NSCLC. It has already been proved that ISGylation as a novel ubiquitin-like modifier can control exosomes production [117] and syndecan-syntenin-ALIX plays a key role in the biogenesis of exosomes [118], which can also be targeted in NSCLC.
The diversity of exosomes components and functions associated with NSCLC provides multiple potential therapeutic targets for the treatment of NSCLC. Yang et al. found that the promotion of the expression of let-7 in exosomes was a potential target for the treatment of NSCLC because of its high tumor suppressor effect, great clinical significance and tissue specificity [119]. Zhang et al. [120]. identified that exosomes separated from H460 cells with restoration of LKB1 (liver kinase B1) had a higher ability in lung cancer cell migration, which could be a novel target. Nao et al. [121]. developed a new strategy of antibody therapy with anti-CD9 or anti-CD63 to target on tumor-derived exosomes and inhibit metastasis of breast cancer in mouse models, which laid the foundation for targeted therapy of NSCLC. Lung cancer cells can secrete survivin through exosomes to inhibit the apoptosis of lung cancer cells and promote the growth of lung cancer cells, so the use of survivin gene negative mutant (Survivin-D53A) can promote the apoptosis of NSCLC cells and may become a potential gene therapy drug [122–124]. Previously described exosomal miR-302b also provided a potential target for the treatment of NSCLC [54]. The exosomes membrane can contain specific tumor antigen through gene transformation, which has a certain target function and can be used for the treatment of NSCLC [125, 126].
In summary, studies on the treatment of lung cancer-related exosomes will provide a new idea for exploring precision treatment of NSCLC. (These potential values are summarized in Table 3).
Table 3.
Tumor-derived exosomes in the targeted therapy of NSCLC
| Potential value | Methods | References |
|---|---|---|
| A tool for drug delivery | Use electroporation or lipofection | [106–110] |
| A direct target for reducing the content | Blood purification: ADAPT, hollow fiber filtration technology Change the local pH of the tumor: PPIs Drug usage: GW4869, amiloride, diannexin Interfere with signal pathway: knockout Rab27, SYTL4 ,EXPH5, ISGylation and syndecan-syntenin-ALIX |
[24, 110–118] |
| Other targets | let-7, LKB1, CD9, CD63, surviving, miR-302b | [54, 119–126] |
Conclusion
In 2006, WHO officially identified lung cancer as a chronic controlled disease whose occurrence and development was a multidirectional, multistep complex network process. Liquid biopsy for tumor-derived exosomes has the advantages of noninvasive and real-time monitoring, which provides a new reference for precision medical and individualized treatment, and develops a new method for early diagnosis, prognosis evaluation, drug delivery and targeted therapy. At present, the application of exosomes in the diagnosis and treatment of NSCLC is still in their initial stage. In the future, further study in exosomes, including biogenesis, secretion, interaction with target cells and the roles of exosomal components may improve applications to medical treatment and improve the survival rate of NSCLC patients. A variety of problems remain to be overcame: (1) The specific mechanism of exosomes as an important part of the tumor microenvironment in the evolution of the NSCLC is not yet clear; (2) The sensitivity and specificity of exosomes application in the diagnosis and treatment of NSCLC still need improvement; (3) The acquisition of high purity exosomes is still an issue due to technical limitation and high cost; (4) The quantification, purification and preservation of exosomes has not yet been standardized; (5) The side effects of exosomes used in the targeted therapy cannot be completely determined. All of these problems restrict the application of exosomes in NSCLC. These reviews synthetically expound the multi-faceted nature of exosomes and potential value in the targeted therapy of NSCLC.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural Science Foundations of China (No: 81773218, 81472773, 81703009 and 81272566).
Availability of data and materials
Not applicable.
Abbreviations
- ADAPT
Adaptive dialysis-like affinity platform technology
- AKT
Protein Kinase B
- ALK
Anaplastic lymphoma kinase
- CAFs
Cancer-associated fibroblasts
- EGFR
Epidermal growth factor receptor
- EMT
Epithelial–mesenchymal-transition
- ERK
Extracellular signal-regulated kinase
- GSK3β
Glycogen synthase kinase 3β
- GTPase
Guanosine triphosphatase
- HSP72
Heat shock protein 72
- IL-10
Interleukin-10
- KRAS
Kirsten rat sarcoma viral oncogene
- LKB1
Liver kinase B1
- MDR
Multi-drug resistant
- MDR-ABC
MDR-associated ATP-binding cassette transporter
- MDSCs
Myeloid-derived suppressor cells
- MSCs
Mesenchymal stem cells
- MVBs
Multivesicular bodies
- NF-kB
Nuclear factor kappa B
- NSCLC
Non-small cell lung cancer
- OS
Overall survival
- PFS
Progression free survival
- PPIs
Proton pump inhibitors
- SCLC
Small-cell lung cancer
- STAT3
Signal transducers and activators of transcription 3
- TD-exosomes
Tumor-derived exosomes
- TGF-β
Transforming growth factor β
- TGF-β1
Transforming growth factor β1
- TKIs
Tyrosine kinase inhibitors
- TLR
Toll-like receptor
- TME
Tumor microenvironment
- Treg
Regulatory T cell
- VEGF
Vascular endothelial growth factor
- ZO-1
Zonula occludens-1
Authors’ contributions
All authors contributed to the conception and design of the manuscript, collection and assembly of data, data analysis and interpretation, manuscript writing and final approval.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hongmei Zheng, Email: 940302784@qq.com.
Yuting Zhan, Email: 326797103@qq.com.
Sile Liu, Email: 349162439@qq.com.
Junmi Lu, Email: 1136980635@qq.com.
Jiadi Luo, Email: 838014743@qq.com.
Juan Feng, Email: 445312788@qq.com.
Songqing Fan, Phone: 86.731. 85292029, Email: songqingfan@csu.edu.cn.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Fan H, Shao ZY, Xiao YY, Xie ZH, Chen W, Xie H, Qin GY, Zhao NQ. Incidence and survival of non-small cell lung cancer in Shanghai_a population-based cohort study. BMJ Open. 2015;5(12):e009419. doi: 10.1136/bmjopen-2015-009419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oser MG, Niederst MJ, Sequist LV, Engelman JA. Transformation from non-small-cell lung cancer to small-cell lung cancer_ molecular drivers and cells of origin. Lancet Oncol. 2015;16(4):e165–e172. doi: 10.1016/S1470-2045(14)71180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howlader N, Noone AM, Krapcho M. SEER Cancer Statistics Review, 1975–2013, based on November 2015 SEER data submission, posted to the SEER web site, April 2016. Bethesda: National Cancer Institute; 2016. Available at: http://seer.cancer.gov/csr/1975_2013/. Accessed 3 Mar 2017.
- 5.Ettinger DS, Wood DE, Akerley W, Bazhenova LA, Borghaei H, Camidge DR, Cheney RT, Chirieac LR, D'Amico TA, Demmy TL, Dilling TJ, Govindan R, Grannis FW, Jr, Horn L, Jahan TM, Komaki R, Kris MG, Krug LM, Lackner RP, Lanuti M, Lilenbaum R, Lin J, Loo BW, Jr, Martins R, Otterson GA, Patel JD, Pisters KM, Reckamp K, Riely GJ, Rohren E, Schild S, Shapiro TA, Swanson SJ, Tauer K, Yang SC, Gregory K, Hughes M. Non-small cell lung cancer, version 1.2015. J Natl Compr Cancer Netw. 2014;12(12):1738–1761. doi: 10.6004/jnccn.2014.0176. [DOI] [PubMed] [Google Scholar]
- 6.Lee SW, Choi EK, Chung WK, Shin KH, Ahn SD, Kim JH, Kim SW, Suh C, Lee JS, Kim WS, Kim DS, Kim DK, Park SI, Sohn KH. Postoperative adjuvant chemotherapy and radiotherapy for stage II and III non-small cell lung cancer (NSCLC) Lung Cancer. 2002;37(1):65–71. doi: 10.1016/S0169-5002(02)00026-0. [DOI] [PubMed] [Google Scholar]
- 7.Walters S, Maringe C, Coleman MP, Peake MD, Butler J, Young N, Bergström S, Hanna L. Lung cancer survival and stage at diagnosis in Australia, Canada, Denmark, Norway, Sweden and the UK_ a population-based study, 2004–2007. Thorax. 2013;68(6):551–564. doi: 10.1136/thoraxjnl-2012-202297. [DOI] [PubMed] [Google Scholar]
- 8.Kumarakulasinghe NB, van Zanwijk N, Soo RA. Molecular targeted therapy in the treatment of advanced stagenon-small cell lung cancer (NSCLC) Respirology. 2015;20(3):370–378. doi: 10.1111/resp.12490. [DOI] [PubMed] [Google Scholar]
- 9.Devarakonda S, Morgensztern D, Govindan R. Genomic alterations in lung adenocarcinoma. Lancet Oncol. 2015;16(7):e342–e351. doi: 10.1016/S1470-2045(15)00077-7. [DOI] [PubMed] [Google Scholar]
- 10.Levy B, Hu ZI, Cordova KN, Close S, Lee K, Becker D. Clinical Utility of Liquid Diagnostic Platforms in Non-Small Cell Lung Cancer. Oncologist. 2016;21(9):1121–1130. doi: 10.1634/theoncologist.2016-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molina-Vila MA, Mayo-de-Las-Casas C, Giménez-Capitán A, Jordana-Ariza N, Garzón M, Balada A, Villatoro S, Teixidó C, García-Peláez B, Aguado C, Catalán MJ, Campos R, Pérez-Rosado A, Bertran-Alamillo J. Liquid Biopsy in Non-Small Cell Lung Cancer. Front Med (Lausanne) 2016;3:69. doi: 10.3389/fmed.2016.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO, Skog J. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milane L, Singh A, Mattheolabakis G, Suresh M, Amiji MM. Exosome mediated communication within the tumor microenvironment. J Control Release. 2015;219:278–294. doi: 10.1016/j.jconrel.2015.06.029. [DOI] [PubMed] [Google Scholar]
- 14.Sharma S, Zuñiga F, Rice GE, Perrin LC, Hooper JD, Salomon C. Tumor-derived exosomes in ovarian cancer – liquid biopsies for early detection and real-time monitoring of cancer progression. Oncotarget. 2017;8(61):104687–104703. doi: 10.18632/oncotarget.22191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simons M, Raposo G. Exosomes—vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21(4):575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 16.Pan BT, Teng K, Wu C, Adam M, Johnstone RM. Electron microscopic evidence for externalization of the transferring receptor in vesicular form in sheep reticulocytes. J Cell Biol. 1985;101(3):942–948. doi: 10.1083/jcb.101.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thery C, Regnault A, Garin J, Wolfers J, Zitvogel L, Ricciardi-Castagnoli P, Raposo G, Amigorena S. Molecular characterization of dendritic cell-derived exosomes. Selective accumulation of the heat shock protein hsc73. J Cell Biol. 1999;147(3):599–610. doi: 10.1083/jcb.147.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blanchard N, Lankar D, Faure F, Regnault A, Dumont C, Raposo G, Hivroz C. TCR activation of human T cells induces the production of exosomes bearing the TCR/CD3/zeta complex. Immunol. 2002;168(7):3235–3241. doi: 10.4049/jimmunol.168.7.3235. [DOI] [PubMed] [Google Scholar]
- 19.Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ, Geuze HJ. B lymphocytes secrete antigen-presenting vesicles. Exp Med. 1996;183(3):1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolfers J, Lozier A, Raposo G, Regnault A, Thery C, Masurier C, Flament C, Pouzieux S, Faure F, Tursz T, Angevin E, Amigorena S, Zitvogel L. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat Med. 2001;7(3):297–303. doi: 10.1038/85438. [DOI] [PubMed] [Google Scholar]
- 21.Conde-Vancells J, Rodriguez-Suarez E, Embade N, Gil D, Matthiesen R, Valle M, Elortza F, Lu SC, Mato JM, Falcon-Perez JM. Characterization and comprehensive proteome profiling of exosomes secreted by hepatocytes. Proteome Res. 2008;7(2):5157–5166. doi: 10.1021/pr8004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mittelbrunn M, Sanchez-Madrid F. Intercellular communication: diverse structures for exchange of genetic information. Nat Rev Mol Cell Biol. 2012;13:328–335. doi: 10.1038/nrm3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou J, Li XL, Chen ZR, Chng WJ. Tumor-derived exosomes in colorectal cancer progression and their clinical applications. Oncotarget. 2017;8(59):100781–100790. doi: 10.18632/oncotarget.20117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ostrowski Matias, Carmo Nuno B., Krumeich Sophie, Fanget Isabelle, Raposo Graça, Savina Ariel, Moita Catarina F., Schauer Kristine, Hume Alistair N., Freitas Rui P., Goud Bruno, Benaroch Philippe, Hacohen Nir, Fukuda Mitsunori, Desnos Claire, Seabra Miguel C., Darchen François, Amigorena Sebastian, Moita Luis F., Thery Clotilde. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nature Cell Biology. 2009;12(1):19–30. doi: 10.1038/ncb2000. [DOI] [PubMed] [Google Scholar]
- 25.Greening DW, Gopal SK, Xu R, Simpson RJ, Chen W. Exosomes and their roles in immune regulation and cancer. Semin Cell Dev Biol. 2015;40:72–81. doi: 10.1016/j.semcdb.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 26.Theresa L. Whiteside. Exosomes and tumor-mediated immune suppression. J Clin Invest. 2016;126(4):1216–1223. doi: 10.1172/JCI81136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126(4):1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frydrychowicz M, Kolecka-Bednarczyk A, Madejczyk M, Yasar S, Dworacki G. Exosomes-Structure, Biogenesis and Biological Role in Non-Small-Cell Lung Cancer. Scand J Immunol. 2015;81(1):2–10. doi: 10.1111/sji.12247. [DOI] [PubMed] [Google Scholar]
- 29.Blackwell Robert, Foreman Kimberly, Gupta Gopal. The Role of Cancer-Derived Exosomes in Tumorigenicity & Epithelial-to-Mesenchymal Transition. Cancers. 2017;9(12):105. doi: 10.3390/cancers9080105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Azmi AS, Bao B, Sarkar FH. Exosomes in Cancer Development, Metastasis and Drug Resistance: A Comprehensive Review. Cancer Metastasis Rev. 2013;32(3–4):623–642. doi: 10.1007/s10555-013-9441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X, Yuan X, Shi H, Wu L, Qian H, Xu W. Exosomes in cancer: small particle, big player. J Hematol Oncol. 2015;8:83. doi: 10.1186/s13045-015-0181-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao H, Yang L, Baddour J, Achreja A, Bernard V, Moss T, Marini JC, Tudawe T, Seviour EG, San Lucas FA, Alvarez H, Gupta S, Maiti SN, Cooper L, Peehl D, Ram PT, Maitra A, Nagrath D. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. elife. 2016;5:e10250. doi: 10.7554/eLife.10250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eldridge L, Moldobaeva A, Zhong Q, Jenkins J, Snyder M, Brown RH, Mitzner W, Wagner EM. Bronchial artery angiogenesis drives lung tumor growth. Cancer Res. 2016;76(20):5962–5969. doi: 10.1158/0008-5472.CAN-16-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Webber JP, Spary LK, Sanders AJ, Chowdhury R, Jiang WG, Steadman R, Wymant J, Jones AT, Kynaston H, Mason MD, Tabi Z, Clayton A. Differentiation of tumour-promoting stromal myofibroblasts by cancer exosomes. Oncogene. 2015;34(3):290–302. doi: 10.1038/onc.2013.560. [DOI] [PubMed] [Google Scholar]
- 35.Hsu YL, Hung JY, Chang WA, Lin YS, Pan YC, Tsai PH, Wu CY, Kuo PL. Hypoxic lung cancer-secreted exosomal miR-23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO-1. Oncogene. 2017;36(34):4929–4942. doi: 10.1038/onc.2017.105. [DOI] [PubMed] [Google Scholar]
- 36.Liu Y, Luo F, Wang B, Li H, Xu Y, Liu X, Shi L, Lu X, Xu W, Lu L, Qin Y, Xiang Q, Liu Q. STAT3-regulated exosomal miR-21 promotes angiogenesis and is involved in neoplastic processes of transformed human bronchial epithelial cells. Cancer Lett. 2016;370(1):125–135. doi: 10.1016/j.canlet.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 37.Cui H, Seubert B, Stahl E, Dietz H, Reuning U, Moreno-Leon L, Ilie M, Hofman P, Nagase H, Mari B, Krüger A. Tissue inhibitor of metalloproteinases-1 induces a pro-tumourigenic increase of miR-210 in lung adenocarcinoma cells and their exosomes. Oncogene. 2015;34(28):3640–3650. doi: 10.1038/onc.2014.300. [DOI] [PubMed] [Google Scholar]
- 38.Li X, Wang S, Zhu R, Li H, Han Q, Zhao RC. Lung tumor exosomes induce a pro-inflammatory phenotype in mesenchymal stem cells via NFκB-TLR signaling pathway. J Hematol Oncol. 2016;9:42. doi: 10.1186/s13045-016-0269-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ridder K, Sevko A, Heide J, Dams M, Rupp AK, Macas J, Starmann J, Tjwa M, Plate KH, Sültmann H, Altevogt P, Umansky V, Momma S. Extracellular vesicle-mediated transfer of functional RNA in the tumor microenvironment. Oncoimmunology. 2015;4(6):e1008371. doi: 10.1080/2162402X.2015.1008371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szajnik M, Czystowska M, Szczepanski MJ, Mandapathil M, Whiteside TL. Tumor-derived microvesicles induce, expand and up-regulate biological activities of human regulatory T cells (Treg) PLoS One. 2010;5(7):e11469. doi: 10.1371/journal.pone.0011469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang SH, Li Y, Zhang J, Rong J, Ye S. Epidermal growth factor receptor-containing exosomes induce tumor-specific regulatory T cells. Cancer Investig. 2013;31(5):330–335. doi: 10.3109/07357907.2013.789905. [DOI] [PubMed] [Google Scholar]
- 42.Liu Y, Gu Y, Cao X. The exosomes in tumor immunity. Oncoimmunology. 2015;4(9):e1027472. doi: 10.1080/2162402X.2015.1027472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chalmin F, Ladoire S, Mignot G, Vincent J, Bruchard M, Remy-Martin JP, Boireau W, Rouleau A, Simon B, Lanneau D, De Thonel A, Multhoff G, Hamman A, Martin F, Chauffert B, Solary E, Zitvogel L, Garrido C, Ryffel B, Borg C, Apetoh L, Rébé C, Ghiringhelli F. Membrane-associated Hsp72 from tumor-derived exosomes mediates STAT3-dependent immunosuppressive function of mouse and human myeloid-derived suppressor cells. J Clin Invest. 2010;120(2):457–471. doi: 10.1172/JCI40483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wood SL, Pernemalm M, Crosbie PA, Whetton AD. The role of the tumor-microenvironment in lung cancer-metastasis and its relationship to potential therapeutic targets. Cancer Treat Rev. 2014;40(4):558–566. doi: 10.1016/j.ctrv.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 45.Fujita Y, Kosaka N, Araya J, Kuwano K, Ochiya T. Extracellular vesicles in lung microenvironment and pathogenesis. Trends Mol Med. 2015;21(9):533–542. doi: 10.1016/j.molmed.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 46.King HW, Michael MZ, Gleadle JM. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer. 2012;12:421. doi: 10.1186/1471-2407-12-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Yi J, Chen X, Zhang Y, Xu M, Yang Z. The regulation of cancer cell migration by lung cancer cell-derived exosomes through TGF-β and IL-10. Oncol Lett. 2016;11(2):1527–1530. doi: 10.3892/ol.2015.4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoshino D, Kirkbride KC, Costello K, Clark ES, Sinha S, Grega-Larson N, Tyska MJ, Weaver AM. Exosome secretion is enhanced by invadopodia and drives invasive behavior. Cell Rep. 2013;5(5):1159–1168. doi: 10.1016/j.celrep.2013.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vella LJ. The emerging role of exosomes in epithelial-mesenchymal-transition in cancer. Front Oncol. 2014;4:361. doi: 10.3389/fonc.2014.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramteke A, Ting H, Agarwal C, Mateen S, Somasagara R, Hussain A, Graner M, Frederick B, Agarwal R, Deep G. Exosomes secreted under hypoxia enhance invasiveness and stemness of prostate cancer cells by targeting adherens junction molecules. Mol Carcinog. 2015;54(7):554–565. doi: 10.1002/mc.22124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rahman MA, Barger JF, Lovat F, Gao M, Otterson GA, Nana-Sinkam P. Lung cancer exosomes as drivers of epithelial mesenchymal transition. Oncotarget. 2016;7(34):54852–54866. doi: 10.18632/oncotarget.10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang J, Liu W, Lu X, Fu Y, Li L, Luo Y. High expression of small GTPase Rab3D promotes cancer progression and metastasis. Oncotarget. 2015;6(13):11125–11138. doi: 10.18632/oncotarget.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim J, Kim TY, Lee MS, Mun JY, Ihm C, Kim SA. Exosome cargo reflects TGF-beta1-mediated epithelial-to-mesenchymal transition (EMT) status in A549 human lung adenocarcinoma cells. Biochem Biophys Res Commun. 2016;478(2):643–648. doi: 10.1016/j.bbrc.2016.07.124. [DOI] [PubMed] [Google Scholar]
- 54.Li J, Yu J, Zhang H, Wang B, Guo H, Bai J, Wang J, Dong Y, Zhao Y, Wang Y. Exosomes-Derived MiR-302b Suppresses Lung Cancer Cell Proliferation and Migration via TGFβRII Inhibition. Cell Physiol Biochem. 2016;38(5):1715–1726. doi: 10.1159/000443111. [DOI] [PubMed] [Google Scholar]
- 55.Alderton GK. Metastasis. Exosomes drive premetastatic niche formation. Nat Rev Cancer. 2012;12(7):447. doi: 10.1038/nrc3304. [DOI] [PubMed] [Google Scholar]
- 56.Obenauf AC, Massagué J. Surviving at a distance: organ specific metastasis. Trends Cancer. 2015;1(1):76–91. doi: 10.1016/j.trecan.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, Singh S, Williams C, Soplop N, Uryu K, Pharmer L, King T, Bojmar L, Davies AE, Ararso Y, Zhang T, Zhang H, Hernandez J, Weiss JM, Dumont-Cole VD, Kramer K, Wexler LH, Narendran A, Schwartz GK, Healey JH, Sandstrom P, Labori KJ, Kure EH, Grandgenett PM, Hollingsworth MA, de Sousa M, Kaur S, Jain M, Mallya K, Batra SK, Jarnagin WR, Brady MS, Fodstad O, Muller V, Pantel K, Minn AJ, Bissell MJ, Garcia BA, Kang Y, Rajasekhar VK, Ghajar CM, Matei I, Peinado H, Bromberg J, Lyden D. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zang H, Peng J, Wang W, Fan S. Roles of microRNAs in the resistance to platinum based chemotherapy in the non-small cell lung cancer. J Cancer. 2017;8(18):3856–3861. doi: 10.7150/jca.21267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zang H, Wang W, Fan S. The role of microRNAs in resistance to targeted treatments of non-small cell lung cancer. Cancer Chemother Pharmacol. 2017;79(2):227–231. doi: 10.1007/s00280-016-3130-7. [DOI] [PubMed] [Google Scholar]
- 60.Yu S, Cao H, Shen B, Feng J. Tumor-derived exosomes in cancer progression and treatment failure. Oncotarget. 2015;6(35):37151–37168. doi: 10.18632/oncotarget.6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Acloque H, Thiery JP, Nieto MA. The physiology and pathology of the EMT. Meeting on the epithelial-mesenchymal transition. EMBO Rep. 2008;9(4):322–326. doi: 10.1038/embor.2008.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Andarawewa KL, Erickson AC, Chou WS, Costes SV, Gascard P, Mott JD, Bissell MJ, Barcellos-Hoff MH. Ionizing Radiation Predisposes Nonmalignant Human Mammary Epithelial Cells to Undergo Transforming Growth Factor B–Induced Epithelial to Mesenchymal Transition. Cancer Res. 2007;67(18):8662–8670. doi: 10.1158/0008-5472.CAN-07-1294. [DOI] [PubMed] [Google Scholar]
- 63.Choi DY, You S, Jung JH, Lee JC, Rho JK, Lee KY, Freeman MR, Kim KP, Kim J. Extracellular vesicles shed from gefitinib-resistant nonsmall cell lung cancer regulate the tumor microenvironment. Proteomics. 2014;14(16):1845–1856. doi: 10.1002/pmic.201400008. [DOI] [PubMed] [Google Scholar]
- 64.Jung JH, Lee MY, Choi DY, Lee JW, You S, Lee KY, Kim J, Kim KP. Phospholipids of tumor extracellular vesicles stratify gefitinib-resistant nonsmall cell lung cancer cells from gefitinib-sensitive cells. Proteomics. 2015;15(4):824–835. doi: 10.1002/pmic.201400243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li XQ, Liu JT, Fan LL, Liu Y, Cheng L, Wang F, Yu HQ, Gao J, Wei W, Wang H, Sun GP. Exosomes derived from gefitinib-treated EGFR-mutant lung cancer cells alter cisplatin sensitivity via up-regulating autophagy. Oncotarget. 2016;7(17):24585–24595. doi: 10.18632/oncotarget.8358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jones PM, George AM. The ABC transporter structure and mechanism: perspectives on recent research. Cell Mol Life Sci. 2004;61(6):682–699. doi: 10.1007/s00018-003-3336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee CH. Reversing agents for ATP-binding cassette drug transporters. Methods Mol Biol. 2010;596:325–340. doi: 10.1007/978-1-60761-416-6_14. [DOI] [PubMed] [Google Scholar]
- 68.Krishna R, Mayer LD. Multidrug resistance (MDR) in cancer. Mechanisms, reversal using modulators of MDR and the role of MDR modulators in influencing the pharmacokinetics of anticancer drugs. Eur J Pharm Sci. 2000;11(4):265–283. doi: 10.1016/S0928-0987(00)00114-7. [DOI] [PubMed] [Google Scholar]
- 69.Hegedus C, Ozvegy-Laczka C, Szakács G, Sarkadi B. Interaction of ABC multidrug transporters with anticancer protein kinase inhibitors: substrates and/or inhibitors? Curr Cancer Drug Targets. 2009;9(3):252–272. doi: 10.2174/156800909788166565. [DOI] [PubMed] [Google Scholar]
- 70.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ciravolo V, Huber V, Ghedini GC, Venturelli E, Bianchi F, Campiglio M, Morelli D, Villa A, Della Mina P, Menard S, Filipazzi P, Rivoltini L, Tagliabue E, Pupa SM. Potential role of HER2-overexpressing exosomes in countering trastuzumab-based therapy. J Cell Physiol. 2012;227(2):658–667. doi: 10.1002/jcp.22773. [DOI] [PubMed] [Google Scholar]
- 72.Aung T, Chapuy B, Vogel D, Wenzel D, Oppermann M, Lahmann M, Weinhage T, Menck K, Hupfeld T, Koch R, Trümper L, Wulf GG. Exosomal evasion of humoral immunotherapy in aggressive B-cell lymphoma modulated by ATP-binding cassette transporter A3. Proc Natl Acad Sci U S A. 2011;108(37):15336–15341. doi: 10.1073/pnas.1102855108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Z, Chen JQ, Liu JL, Tian L. Exosomes in tumor microenvironment: novel transporters and biomarkers. J Transl Med. 2016;14(1):297. doi: 10.1186/s12967-016-1056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sandfeld-Paulsen B, Aggerholm-Pedersen N, Bæk R, Jakobsen KR, Meldgaard P, Folkersen BH, Rasmussen TR, Varming K, Jørgensen MM, Sorensen BS. Exosomal proteins as prognostic biomarkers in non-small cell lung cancer. Mol Oncol. 2016;10(10):1595–1602. doi: 10.1016/j.molonc.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu Q, Yu Z, Yuan S, Xie W, Li C, Hu Z, Xiang Y, Wu N, Wu L, Bai L, Li Y. Circulating exosomal microRNAs as prognostic biomarkers for non-small-cell lung cancer. Oncotarget. 2017;8(8):13048–13058. doi: 10.18632/oncotarget.14369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yuwen DL, Sheng BB, Liu J, Wenyu W, Shu YQ. MiR-146a-5p level in serum exosomes predicts therapeutic effect of cisplatin in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2017;21(11):2650–2658. [PubMed] [Google Scholar]
- 77.Tang Y, Qiao G, Xu E, Xuan Y, Liao M, Yin G. Biomarkers for early diagnosis, prognosis, prediction, and recurrence monitoring of non-small cell lung cancer. Onco Targets Ther. 2017;10:4527–4534. doi: 10.2147/OTT.S142149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhao L, Liu W, Xiao J, Cao B. The role of exosomes and “exosomal shuttle microRNA” in tumorigenesis and drug resistance. Cancer Lett. 2015;356(2 Pt B):339–346. doi: 10.1016/j.canlet.2014.10.027. [DOI] [PubMed] [Google Scholar]
- 79.Clark DJ, Fondrie WE, Yang A, Mao L. Triple SILAC quantitative proteomic analysis reveals differential abundance of cell signaling proteins between normal and lung cancer-derived exosomes. Proteomics. 2016;133:161–169. doi: 10.1016/j.jprot.2015.12.023. [DOI] [PubMed] [Google Scholar]
- 80.Sandfeld-Paulsen B, Jakobsen KR, Bæk R, Folkersen BH, Rasmussen TR, Meldgaard P, Varming K, Jørgensen MM, Sorensen BS. Exosomal Proteins as Diagnostic Biomarkers in Lung Cancer. J Thorac Oncol. 2016;11(10):1701–10. doi: 10.1016/j.jtho.2016.05.034. [DOI] [PubMed] [Google Scholar]
- 81.Brinkmann K, Enderle D, Koestler T, Bentink S, Emenegger J, Spiel A, Mueller R, Vincent O, Skog J, Noerholm M. Abstract 545: Plasma-based diagnostics for detection of EML4-ALK fusion transcripts in NSCLC patients. Cancer Res. 2015;75:545. doi: 10.1158/1538-7445.AM2015-545. [DOI] [Google Scholar]
- 82.Solomon BJ, Mok T, Kim DW, Wu YL, Nakagawa K, Mekhail T, Felip E, Cappuzzo F, Paolini J, Usari T, Iyer S, Reisman A, Wilner KD, Tursi J, Blackhall F. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med. 2014;371(23):2167–2177. doi: 10.1056/NEJMoa1408440. [DOI] [PubMed] [Google Scholar]
- 83.Vykoukal J, Sun N, Aguilar-Bonavides C, Katayama H, Tanaka I, Fahrmann JF, Capello M, Fujimoto J, Aguilar M, Wistuba II, Taguchi A, Ostrin EJ, Hanash SM. Plasma-derived extracellular vesicle proteins as a source of biomarkers for lung adenocarcinoma. Oncotarget. 2017;8(56):95466–95480. doi: 10.18632/oncotarget.20748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cheng G. Circulating miRNAs: roles in cancer diagnosis, prognosis and therapy. Adv Drug Deliv Rev. 2015;81:75–93. doi: 10.1016/j.addr.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 85.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20(8):460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 86.Zhao Q, Cao J, Wu YC, Liu X, Han J, Huang XC, Jiang LH, Hou XX, Mao WM, Ling ZQ. Circulating miRNAs is a potential marker for gefitinib sensitivity and correlation with EGFR mutational status in human lung cancers. Am J Cancer Res. 2015; 15; 5(5):1692–705. eCollection 2015. [PMC free article] [PubMed]
- 87.Jin X, Chen Y, Chen H, Fei S, Chen D, Cai X, Liu L, Lin B, Su H, Zhao L, Su M, Pan H, Shen L, Xie D, Xie C. Evaluation of Tumor-Derived Exosomal miRNA as Potential Diagnostic Biomarkers for Early-Stage Non-Small Cell Lung Cancer Using Next-Generation Sequencing. Clin Cancer Res. 2017;23(17):5311–5319. doi: 10.1158/1078-0432.CCR-17-0577. [DOI] [PubMed] [Google Scholar]
- 88.Reclusa P, Taverna S, Pucci M, Durendez E, Calabuig S, Manca P, Serrano MJ, Sober L, Pauwels P, Russo A, Rolfo C. Exosomes as diagnostic and predictive biomarkers in lung cancer. J Thorac Dis. 2017;9(Suppl 13):S1373–S1382. doi: 10.21037/jtd.2017.10.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rolfo C, Laes JF, Reclusa P, Valentino A, Peeters M. P2.01-093 Exo-ALK Proof of Concept: Exosomal Analysis of ALK Alterations in Advanced NSCLC Patients. J Thorac Oncol. 2017;12(1):S844–S845. doi: 10.1016/j.jtho.2016.11.1145. [DOI] [Google Scholar]
- 90.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr KKW. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Steuer CE, Ramalingam SS. Targeting EGFR in lung cancer: Lessons learned and future perspectives. Mol Asp Med. 2015;45:67–73. doi: 10.1016/j.mam.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shaw AT, Engelman JA. ALK in lung cancer: past, present, and future. J Clin Oncol. 2013;31(8):1105–1111. doi: 10.1200/JCO.2012.44.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gelsomino F, Rossi G, Tiseo M. MET and Small-Cell Lung Cancer. Cancers (Basel) 2014;6(4):2100–2115. doi: 10.3390/cancers6042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7(3):169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 95.Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 96.Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba II, Fong KM, Lee H, Toyooka S, Shimizu N, Fujisawa T, Feng Z, Roth JA, Herz J, Minna JD, Gazdar AF. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97(5):339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 97.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 98.Wu SG, Shih JY. Management of acquired resistance to EGFR TKI–targeted therapy in advanced non-small cell lung cancer. Mol Cancer. 2018;17(1):38. doi: 10.1186/s12943-018-0777-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kobayashi S, Boggon TJ, Dayaram T, Jänne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352(8):786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 100.Wu SG, Liu YN, Tsai MF, Chang YL, Yu CJ, Yang PC, Yang JC, Wen YF, Shih JY. The mechanism of acquired resistance to irreversible EGFR tyrosine kinase inhibitor-afatinib in lung adenocarcinoma patients. Oncotarget. 2016;7(11):12404–12413. doi: 10.18632/oncotarget.7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cross DA, Ashton SE, Ghiorghiu S, Eberlein C, Nebhan CA, Spitzler PJ, Orme JP, Finlay MR, Ward RA, Mellor MJ, Hughes G, Rahi A, Jacobs VN, Red Brewer M, Ichihara E, Sun J, Jin H, Ballard P, Al-Kadhimi K, Rowlinson R, Klinowska T, Richmond GH, Cantarini M, Kim DW, Ranson MR, Pao W. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov. 2014;4(9):1046–1061. doi: 10.1158/2159-8290.CD-14-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Thress KS, Paweletz CP, Felip E, Cho BC, Stetson D, Dougherty B, Lai Z, Markovets A, Vivancos A. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med. 2015;21(6):560–562. doi: 10.1038/nm.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Krug AK, Enderle D, Karlovich C, et al. Improved EGFR mutation detection using combined exosomal RNA and circulating tumor DNA in NSCLC patient plasma. Ann Oncol. 2018;29(3):700–706. doi: 10.1093/annonc/mdx765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Friboulet L, Li N, Katayama R, Lee CC, Gainor JF, Crystal AS, Michellys PY. The ALK inhibitor ceritinib overcomes crizotinib resistance in non-small cell lung cancer. Cancer Discov. 2014;4(6):662–673. doi: 10.1158/2159-8290.CD-13-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Doebele RC, Pilling AB, Aisner DL, Kutateladze TG, Le AT, Weickhardt AJ, Kondo KL, Linderman DJ, Heasley LE, Franklin WA, Varella-Garcia M, Camidge DR. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin Cancer Res. 2012;18(5):1472–1482. doi: 10.1158/1078-0432.CCR-11-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Batrakova EV, Kim MS. Using exosomes, naturally-equipped nanocarriers, for drug delivery. J Control Release. 2015;219:396–405. doi: 10.1016/j.jconrel.2015.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Harding CV, Heuser JE, Stahl PD. Exosomes: looking back three decades and into the future. J Cell Biol. 2013;200(4):367–371. doi: 10.1083/jcb.201212113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lai RC, Yeo RW, Tan KH, Lim SK. Exosomes for drug delivery - a novel application for the mesenchymal stem cell. Biotechnol Adv. 2013;31:543–551. doi: 10.1016/j.biotechadv.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 109.Mendt M, Kamerkar S, Sugimoto H, McAndrews KM, Wu CC, Gagea M, Yang S, Blanko EVR, Peng Q, Ma X, Marszalek JR, Maitra A, Yee C, Rezvani K, Shpall E, LeBleu VS, Kalluri R. Generation and testing of clinical-grade exosomes for pancreatic cancer. JCI Insight. 2018;3(8). [DOI] [PMC free article] [PubMed]
- 110.Tickner JA, Urquhart AJ, Stephenson SA, Richard DJ, O'Byrne KJ. Functions and therapeutic roles of exosomes in cancer. Front Oncol. 2014;4:127. doi: 10.3389/fonc.2014.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Marleau AM, Chen CS, Joyce JA, Tullis RH. Exosome removal as a therapeutic adjuvant in cancer. J Transl Med. 2012;10(1):134. doi: 10.1186/1479-5876-10-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ichim TE, Zhong Z, Kaushal S, Zheng X, Ren X, Hao X, Joyce JA, Hanley HH, Riordan NH, Koropatnick J, Bogin V, Minev BR, Min WP, Tullis RH. Exosomes as a tumor immune escape mechanism: possible therapeutic implications. J Transl Med. 2008;6:37. doi: 10.1186/1479-5876-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Federici C, Petrucci F, Caimi S, Cesolini A, Logozzi M, Borghi M, D'Ilio S, Lugini L, Violante N, Azzarito T, Majorani C, Brambilla D, Fais S. Exosome release and low PH belong to a framework of resistance of human melanoma cells to cisplatin. PLoS One. 2014;9(2):e88193. doi: 10.1371/journal.pone.0088193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, Zanesi N, Crawford M, Ozer GH, Wernicke D, Alder H, Caligiuri MA, Nana-Sinkam P, Perrotti D, Croce CM. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109(31):E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Savina A, Furlán M, Vidal M, Colombo MI. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J Biol Chem. 2003;278(22):20083–20090. doi: 10.1074/jbc.M301642200. [DOI] [PubMed] [Google Scholar]
- 116.Combes V, Latham SL, Wen B, Allison AC, Grau GE. Diannexin down-modulates tnf-induced endothelial microparticle release by blocking membrane budding process. Int J Innov Med Health Sci. 2016;7:1–11. doi: 10.20530/ijimhs_7_1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Villarroya-Beltri C, Baixauli F, Mittelbrunn M, Fernández-Delgado I, Torralba D, Moreno-Gonzalo O, Baldanta S, Enrich C, Guerra S, Sánchez-Madrid F. ISGylation controls exosome secretion by promoting lysosomal degradation of MVB proteins. Nat Commun. 2016;7:13588. doi: 10.1038/ncomms13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, Zimmermann P, David G. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14(7):677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- 119.Yang G, Zhang W, Yu C, Ren J, An Z. MicroRNA let-7: Regulation, single nucleotide polymorphism, and therapy in lung cancer. J Cancer Res Ther. 2015;11(Suppl 1):C1–C6. doi: 10.4103/0973-1482.163830. [DOI] [PubMed] [Google Scholar]
- 120.Zhang C, Xiao X, Chen M, Aldharee H, Chen Y, Long W. Liver kinase B1 restoration promotes exosome secretion and motility of lung cancer cells. Oncol Rep. 2018;39(1):376–382. doi: 10.3892/or.2016.4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nishida-Aok N, Tominaga N, Takeshita F, Sonoda H, Yoshioka Y, Ochiya T. Disruption of Circulating Extracellular Vesicles as a Novel Therapeutic Strategy against Cancer Metastasis. Mol Ther. 2017;25(1):181–191. doi: 10.1016/j.ymthe.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhu DE, Höti N, Song Z, Jin L, Wu Z, Wu Q, Wu M. Suppression of tumor growth using a recombinant adenoviral vector carrying the dominant-negative mutant gene Survivin-D53A in a nude mice model. Cancer Gene Ther. 2006;13(8):762–770. doi: 10.1038/sj.cgt.7700952. [DOI] [PubMed] [Google Scholar]
- 123.Galbo PM, Jr CMJ, Figel S, Maguire O, Qiu J, Wiltsie L, Minderman H, Fenstermaker RA. Circulating CD9+/GFAP+/survivin+ exosomes in malignant glioma patients following survivin vaccination. Oncotarget. 2017;8(70):114722–114735. doi: 10.18632/oncotarget.21773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.H T, Liu S, Zhang J, Zhang S, Cheng L, Li C, Zhang X, Dail L, Fan P, Dai L, Yan N, Wang R, Wei Y, Deng H. Enhancement of cisplatin sensitivity in lung cancer xenografts by liposome-mediated delivery of the plasmid expressing small hairpin RNA targeting Survivin. J Biomed Nanotechnol. 2012;8(4):633–641. doi: 10.1166/jbn.2012.1419. [DOI] [PubMed] [Google Scholar]
- 125.L A-E, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29(4):341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 126.Ohno SI, Takanashi M, Sudo K, Ueda S, Ishikawa A, Matsuyama N, Fujita K, Mizutani T, Ohgi T, Ochiya T, Gotoh N, Kuroda M. Systemically Injected Exosomes Targeted to EGFR Deliver Antitumor MicroRNA to Breast Cancer Cells. Mol Ther. 2013;21:185. doi: 10.1038/mt.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zeringer E, Barta T, Li M, Vlassov AV. Strategies for Isolation of Exosomes. Cold Spring Harb Protoc. 2015;2015(4):319–323. doi: 10.1101/pdb.top074476. [DOI] [PubMed] [Google Scholar]
- 128.Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006; Chapter 3: Unit 3.22. [DOI] [PubMed]
- 129.Momen-Heravi F, Balaj L, Alian S, Mantel PY, Halleck AE, Trachtenberg AJ, Soria CE, Oquin S, Bonebreak CM, Saracoglu E, Skog J, Kuo WP. Current methods for the isolation of extracellular vesicles. Biol Chem. 2013;394(10):1253–1262. doi: 10.1515/hsz-2013-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Enderle D, Spiel A, Coticchia CM, Berghoff E, Mueller R, Schlumpberger M, Sprenger-Haussels M, Shaffer JM, Lader E, Skog J, Noerholm M. Characterization of RNA from Exosomes and Other Extracellular Vesicles Isolated by a Novel Spin Column-Based Method. PLoS One. 2015;10(8):e0136133. doi: 10.1371/journal.pone.0136133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Baranyai T, Herczeg K, Onódi Z, Voszka I, Módos K, Marton N, Nagy G, Mäger I, Wood MJ, El Andaloussi S, Pálinkás Z, Kumar V, Nagy P, Kittel Á, Buzás EI, Ferdinandy P, Giricz Z. Isolation of Exosomes from Blood Plasma: Qualitative and Quantitative Comparison of Ultracentrifugation and Size Exclusion Chromatography Methods. PLoS One. 2015;10(12):e0145686. doi: 10.1371/journal.pone.0145686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Muller L, Hong CS, Stolz DB, Watkins SC, Whiteside TL. Isolation of biologically-active exosomes from human plasma. J Immunol Methods. 2014;41:55–65. doi: 10.1016/j.jim.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gardiner Chris, Ferreira Yannick J., Dragovic Rebecca A., Redman Christopher W.G., Sargent Ian L. Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. Journal of Extracellular Vesicles. 2013;2(1):19671. doi: 10.3402/jev.v2i0.19671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Coscia C, Parolini I, Sanchez M, Biffoni M, Boussadia Z, Zanetti C, Fiani ML, Sargiacomo M. Generation, Quantification, and Tracing of Metabolically Labeled Fluorescent Exosomes. Methods Mol Biol. 2016;1448:217–235. doi: 10.1007/978-1-4939-3753-0_16. [DOI] [PubMed] [Google Scholar]
- 135.Higginbotham JN, Zhang Q, Jeppesen DK, Scott AM, Manning HC, Ochieng J, Franklin JL, Coffey RJ. Identification and characterization of EGF receptor in individual exosomes by fluorescence-activated vesicle sorting. J Extracell Vesicles. 2016;5:29254. doi: 10.3402/jev.v5.29254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kreimer S, Belov AM, Ghiran I, Murthy SK, Frank DA, Ivanov AR. Mass-spectrometry-based molecular characterization of extracellular vesicles: lipidomics and proteomics. J Proteome Res. 2015;14(6):2367–2384. doi: 10.1021/pr501279t. [DOI] [PubMed] [Google Scholar]
- 137.Bastos-Amador P, Royo F, Gonzalez E, Conde-Vancells J, Palomo-Diez L, Borras FE, Falcon-Perez JM. Proteomic analysis of microvesicles from plasma of healthy donors reveals high individual variability. J Proteome. 2012;75(12):3574–3584. doi: 10.1016/j.jprot.2012.03.054. [DOI] [PubMed] [Google Scholar]
- 138.Selmaj I, Cichalewska M, Namiecinska M, Galazka G, Horzelski W, Selmaj KW, Mycko MP. Global exosome transcriptome profiling reveals biomarkers for multiple sclerosis. Ann Neurol. 2017;81(5):703–717. doi: 10.1002/ana.24931. [DOI] [PubMed] [Google Scholar]
- 139.Vallabhajosyula P, Korutla L, Habertheuer A, Yu M, Rostami S, Yuan CX, Reddy S, Liu C, Korutla V, Koeberlein B, Trofe-Clark J, Rickels MR, Naji A. Tissue-specific exosome biomarkers for noninvasively monitoring immunologic rejection of transplanted tissue. J Clin Invest. 2017;127(4):1375–1391. doi: 10.1172/JCI87993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chen Y, Buyel JJ, Hanssen MJ, Siegel F, Pan R, Naumann J, Schell M, van der Lans A, Schlein C, Froehlich H, Heeren J, Virtanen KA, van Marken Lichtenbelt W, Pfeifer A. Exosomal microRNA miR-92a concentration in serum reflects human brown fat activity. Nat Commun. 2016;7:11420. doi: 10.1038/ncomms11420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.