Abstract
Circadian clocks in adipose tissue are known to regulate adipocyte biology. Although circadian dysregulation is associated with development of obesity, the underlying mechanism has not been established. Here we report that disruption of the clock gene, brain and muscle Arnt-like 1 (Bmal1), in mice led to increased adipogenesis, adipocyte hypertrophy, and obesity, compared to wild-type (WT) mice. This is due to its cell-autonomous effect, as Bmal1 deficiency in embryonic fibroblasts, as well as stable shRNA knockdown (KD) in 3T3-L1 preadipocyte and C3H10T1/2 mesenchymal stem cells, promoted adipogenic differentiation. We demonstrate that attenuation of Bmal1 function resulted in down-regulation of genes in the canonical Wnt pathway, known to suppress adipogenesis. Promoters of these genes (Wnt10a, β-catenin, Dishevelled2, TCF3) displayed Bmal1 occupancy, indicating direct circadian regulation by Bmal1. As a result, Wnt signaling activity was attenuated by Bmal1 KD and augmented by its overexpression. Furthermore, stabilizing β-catenin through Wnt ligand or GSK-3β inhibition achieved partial restoration of blunted Wnt activity and suppression of increased adipogenesis induced by Bmal1 KD. Taken together, our study demonstrates that Bmal1 is a critical negative regulator of adipocyte development through transcriptional control of components of the canonical Wnt signaling cascade, and provides a mechanistic link between circadian disruption and obesity.—Guo, B., Chatterjee, S., Li, L., Kim, J. M., Lee, J., Yechoor, V. K., Minze, L. J., Hsueh, W., Ma, K. The clock gene, brain and muscle Arnt-like 1, regulates adipogenesis via Wnt signaling pathway.
Keywords: circadian rhythm, adipocyte development, obesity
Circadian clocks that exist in essentially all tissues and cell types of the body are integral components of many key biological processes involved in the regulation of energy homeostasis and metabolism (1–3). Both preadipocytes and adipocytes contain functional intrinsic clocks that drive not only rhythmic mRNA expression (4–6) but also secretion of various adipokines (7, 8). Though many of these processes are responsive to rhythmic input signals from the central clock (suprachiasmatic nuclei; refs. 1, 9), they also exhibit cell-autonomous properties due to regulation by their intrinsic clock (peripheral clock). Recently, it has been recognized that there is a strong association between disruption of circadian physiology and development of obesity. Epidemiological studies revealed that shift workers have higher body mass index that is positively associated with the length of their shift work (10, 11), whereas experimental settings mimicking circadian disruption with alteration of lighting schedules demonstrated detrimental effects on body weight in both human subjects (12) and animal models (13).
Although circadian disruption affects energy balance through the central clock, there is strong evidence that the peripheral clock in adipose tissue also impacts adipogenesis directly (14, 15). Brain and muscle Arnt-like 1 (Bmal1), a basic HLH transcription factor, is a central positive regulator of the core molecular clock (16). Bmal1 functions as a heterodimer with circadian locomotor output cycles kaput (CLOCK) to initiate oscillation of the molecular clock through transcriptional activation of negative factors including Period 1, 2, and 3 (Pers) and cryptochrome 1 and 2 (Crys). Pers and Crys, in turn, bind to and inhibit the activity of Bmal1/CLOCK, resulting in rhythmic oscillation of their own transcription. Temporally controlled, proteosome-mediated degradation elicits rhythms of these proteins, as well. In addition, Bmal1 promoter itself responds to the negative and positive regulation of Rev-erbα and Rev-erbβ (Rev-erbs) and retinoic acid receptor-related orphan receptors α, β, and γ (RORs), respectively, through ROR response elements (ROREs). Interestingly, in genome-wide association studies, Bmal1 and its heterodimer partner, CLOCK, were found to be associated with type 2 diabetes and obesity (17, 18), while CLOCK mutant mice are known to develop a metabolic syndrome-like phenotype (19). In addition, Per2 and Rev-erbα, components of the molecular clock and direct target genes of Bmal1, can directly regulate the master adipogenic factor peroxisome proliferator-activated receptor γ (PPARγ; refs. 14, 15).
Recently, a genome-wide chromatin immunoprecipitation sequencing (ChIP-Seq) analysis in liver by Rey et al. (20) identified Bmal1 occupancy and its associated binding sites (E-boxes or, E1–E2 elements in tandem) in many genes of the canonical Wingless-type (Wnt) signaling pathway, including the Wnt ligand (Wnt10a), its intracellular signaling components Frizzled 5 (Fzd5), Dishevelled2 (Dvl2), transcriptional activator T-cell factor 3 (TCF3), and target gene Cyclin D1. Among these, Wnt10a is involved in brown adipose differentiation (21), while both Fzd5 and Dvl2 are highly expressed in preadipocytes and down-regulated on differentiation (22). The canonical Wnt pathway, mediated by stabilization of β-catenin in the cytosol, subsequent nuclear translocation, and transcriptional activation after ligand stimulation, is a well-characterized mechanism that inhibits differentiation of adipocytes through suppression of the key adipogenic factors CCAAT/enhancer-binding protein α (CEBPα) and PPARγ (23). In addition, Wnt pathway is also known to play critical roles in cell fate determination (24), and could regulate earlier events in preadipocyte commitment to adipocyte fate (25, 26). In line with this, a recent study in epidermal stem cells by Janich et al. (27) elucidated that the molecular clock, particularly Bmal1, modulates stem cell properties through regulation of Wnt and other related developmental pathways.
Given this evidence, we investigated the hypothesis that Bmal1 could suppress adipogenesis via direct transcriptional control of components involved in the Wnt signaling cascade. We employed a genetic model of circadian disruption, the Bmal1-null (Bmal1−/−) mice, and cellular differentiation systems of C3H10T1/2 (10T1/2) and mouse embryonic fibroblast (MEF) cells. We found that Bmal1−/− mice developed obesity and attenuation of Bmal1 function promoted adipogenesis. This is mediated by down-regulation of Wnt pathway genes, many of which are direct Bmal1 targets. Our study demonstrates that Bmal1 regulation of adipogenesis via transcriptional control of the Wnt pathway could serve as an underlying molecular link between circadian disruption and obesity.
MATERIALS AND METHODS
Animals
Global Bmal1-null (Bmal1−/−) mice were obtained from Jackson Laboratory (Bar Harbor, ME, USA; ref. 28), and animals were maintained in the Methodist Hospital Research Institute mouse facility under a constant 12-h light-dark cycle, with lights on at 7:00 AM [zeitgeber time (ZT) 0]. All experiments were done following approval of the protocol by the animal care research committee of Methodist Hospital Research Institute. Mice of 6–12 wk of age were used to exclude effects of aging-related pathologies, unless otherwise indicated. Tissue samples were obtained at 5:00 PM (ZT 10), a time point with high expression of Bmal1 protein.
Analysis of energy homeostasis in mice
NMR analysis was carried out using an EchoMRI whole-body composition analyzer (Echo Medical Systems, Houston, TX, USA). Energy expenditure was measured by the Oxymax Comprehensive Lab Animal Monitoring System (CLAMS; Columbus Instruments, Columbus, OH, USA), which monitors O2 and CO2 gas fractions to compute Vo2 and Vco2. Locomotion activity was monitored simultaneously. Animals were individually housed and allowed to acclimate for 2 d before measurements were taken continuously for 4 d under normal light-dark cycle.
3T3-L1, 10T1/2, and MEF cell differentiation
The 3T3-L1 and 10T1/2 cell lines were obtained from the American Type Culure Collection (ATCC; Manassas, VA, USA) and maintained at subconfluence between passages in DMEM with 10% FBS and antibiotics. For induction of adipocyte differentiation, cells were grown to 2 d postconfluence and differentiated with standard differentiation medium (DMEM containing 10% FBS, 1.6 μM insulin, 1 μM dexamethasone, and 0.5 mM isobutylmethylxanthine). MEF cells were isolated as described elsewhere (29) from embryonic day 14.5 (E14.5) embryos, and the third passage was used for differentiation. For these cells, commercial adipocyte induction medium (Human Adipocyte Differentiation Medium; Cell Applications, San Diego, CA, USA) was used for the first 3 d of differentiation. RNA and protein samples were collected at indicated time points during differentiation. Wnt3a-conditioned medium was obtained from cultures of Wnt3a-producing L cells (Wnt3a-L cells) as described previously (26), and used in a 1:1 ratio for the first 3 d of adipogenic induction, with conditioned medium from L cells as control. GSK-3β inhibitor SB-2167638 (8μM; EMD Chemicals, Billerica, MA, USA) was added for the first 2 d of differentiation.
Generation of stable knockdown (KD) and overexpression cell lines
Bmal1 small hairpin RNA (shRNA) constructs in pGIPZ vector (VGM5520-99941526, VGM5520-99342254, VGM5520-99211363) and scrambled control (SC) vector (RHS4346) were purchased from Open Biosystems (Lafayette, CO, USA), and cDNA (in pcDNA3 vector) was a gift from Dr. Loning Fu (Baylor College of Medicine; ref. 30). Stable 10T1/2 cell lines expressing Bmal1 shRNA or cDNA were constructed by transient transfection using FuGene 6 reagent (Roche, Indianapolis, IN, USA) according to the manufacturer's instructions. Puromycin (for shRNA selection) or neomycin (for pcDNA3) selection was started 48 h after transfection and continued for 7–10 d. Whole transfected plates were subcultured in order to minimize the effect of confluency on differentiation efficiency.
Serum shock and circadian expression analysis
Serum shock was carried out in 10T1/2 cells using standard protocol, as described previously (31, 32). To exclude effects of cell cycle on gene expression patterns over the time course, 10T1/2 cells were grown to 100% confluency before serum shock (32). Cells were cultured in serum-free medium overnight and subjected to serum-shock synchronization by 20% FBS for 1 h. The medium was then removed and replaced with regular culture medium containing 10% serum. This was considered circadian time (CT) 0, and total RNA was collected at 1 h and every 4 h afterward for 48 h.
Oil-red-O staining, quantification, and LipidTOX green staining
Oil-red-O staining was carried out in cultured cells after fixation of cells with 0.5% Oil-red-O solution in 60% isopropanol for 1 h. Phase-contrast pictures were taken using an Olympus IX81 microscope (Olympus, Tokyo, Japan). For quantification, 1 ml of isopropyl alcohol was used for extraction, and absorbance was monitored spectrophotometrically at 510 nm, as described previously (33). HCS LipidTOX green neutral lipid staining (Invitrogen, Grand Island, NY, USA) was carried out according to the manufacturer's instructions at 1:1000 dilution.
RNA extraction and quantitative reverse transcriptase PCR analysis
Trizol reagent (Invitrogen) or RNeasy miniprep kit (Qiagen, Valencia, CA, USA) was used to extract total RNA from snap-frozen adipose tissues or cells, respectively. cDNA was generated using q-Script cDNA Supermix kit (Quanta Biosciences, Gaithersburg, MD, USA), and quantitative PCR was performed using an Roche 480 Light Cycler with Perfecta SYBR green Supermix (Quanta Biosciences). Relative expression levels were determined using the comparative Ct method and normalized to β-actin or 36B4. The specific primer sequences used are listed in Supplemental Fig. S2A.
Immunoblot analysis
Total proteins (30 μg) from tissues or cell homogenates were used for each sample on SDS-PAGE gel. Proteins were then transferred to PVDF membrane after electrophoresis, blotted using specific primary and secondary antibody, and detected by chemiluminescence (Supersignal, Pierce Biotechnology, Rockford, IL, USA). CEBPα (sc-150), PPARγ (sc-7196), and actin antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA); Bmal1 antibody (AB93806) was from Abcam (Cambridge, MA, USA), and β-catenin antibody (610153) was from BD Biosciences (San Jose, CA, USA). Assessment of cytosolic level of β-catenin was carried out as described elsewhere (34) after Wnt3a or control medium treatment for 6 h. Briefly, cells were lysed by incubating on ice for 20 min, followed by 30 strokes in a Dounce homogenizer. Membranous and cytosolic material was obtained by ultracentrifugation at 100,000 g for 40 min at 4°C in a Beckman Coulter Optima TLX-120 ultracentrifuge (Beckman Coulter, Fullerton, CA, USA). The supernatant was collected as the cytosolic fraction, and pellets were solubilized in protein lysis buffer containing 0.1% SDS and designated the membranous fraction. The cytosolic (12 μg) and total protein (4 μg) fractions were used to determine β-catenin level. Quantification of blots was performed using ImageJ software (U. S. National Institutes of Health, Bethesda, MD, USA).
TOPFlash luciferase assay
10T1/2 cells were seeded to ∼80% confluency overnight in 24-well plates. Transient transfection using FuGene 6 (Roche) was carried out in 4 replicates as described previously (35). Per well, the transfection mixture contained 150 ng of Super TOPFlash (8xTCF binding sites) or FOPFlash (with mutated-TCF binding sites) luciferase reporter (Addgene 12456 and 12457; Addgene, Cambridge, MA, USA) as described by Randall Moon and colleagues (36), together with 20 ng of Renilla luciferase (pRL-TK, Promega, Madison, WI, USA) as an internal control. At 16 h after transfection, medium was replaced by Wnt3a or control medium. Luciferase activity was measured using the Dual-Glo luciferase assay system (Promega) by Tecan Safire2 luminometer (Tecan Group Ltd., Männedorf, Switzerland) 24 h after Wnt3a addition. TOPFlash luciferase values were normalized to Renilla readings after subtraction of FOPFlash values.
ChIP analysis
10T1/2 cells were fixed by formaldehyde, lysed, and sonicated to shear the chromatin. Immunoprecipitation was performed with 5 μg Bmal1 antibody (AB93806; Abcam) or control rabbit IgG plus protein A/G beads. The immunoprecipitated chromatin fragments were treated with proteinase K and purified using the Qiaquick PCR purification kit (Qiagen). Real-time PCR using Perfecta SYBR Green Supermix (Quanta Biosciences) was carried out with specific primers flanking the identified E-box sites according to ChIP-Seq annotation (20). The sequences are listed in Supplemental Fig. S2B. Data are expressed as fold enrichment over IgG after normalization to 1% input.
Statistical analysis
Data are expressed as means ± se. The growth-curve experiments and comparisons of circadian gene expression after serum shock in different groups were analyzed by 1-way analysis of variance (ANOVA). Other experiments were assessed by Student's t test, and values of P ≤ 0.05 were considered statistically significant.
RESULTS
Bmal1−/− mice develop obesity
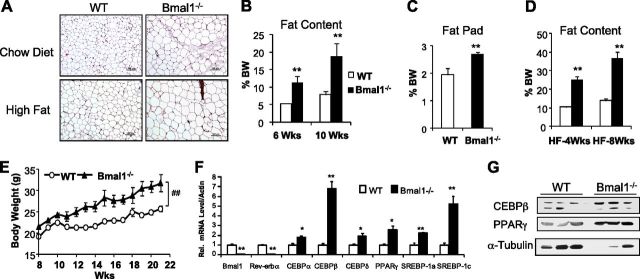
Loss of the core clock gene, Bmal1, resulted in complete loss of circadian rhythmicity under free-run condition (28). We therefore used global Bmal1−/− mice to study the effect of circadian disruption on obesity. Since these mice develop an aging phenotype starting at ∼26 wk of age (37), we limited our study to mice of 2–3 mo of age before the onset of aging-related pathologies (except growth curves that monitored mice up to 5 mo). We found that Bmal1−/− mice developed obesity on both chow and high-fat diet with significant adipocyte hypertrophy (Fig. 1A), compared to wild-type (WT) controls. Loss of Bmal1 led to ∼2-fold increase in total body fat content (Fig. 1B) which was evident at 6 wk of age and continued to increase at 10 wk, although body weight did not differ significantly from the controls. Direct measurement of the perigonadal fat pad (Fig. 1C) confirmed the finding by NMR in mice fed the chow diet. This markedly higher body fat composition was further exacerbated by the high-fat diet (45%) with the fat content in Bmal1−/− mice reaching ∼36.64%, as compared to 13.83% in the controls, after 8 wk (Fig. 1D). The body weight of mice fed the high-fat diet (Fig. 1E) was correspondingly higher throughout the 14-wk period (P<0.01; 1-way ANOVA). Up-regulation of key adipogenic factors, including CEBPα, CEBPβ (∼7-fold), and PPARγ (∼3-fold), as well as lipogenic factors sterol regulatory element binding proteins (SREBPs) 1a and 1c (Fig. 1F), indicated increased adipogenesis and lipogenesis in white adipose tissue in the absence of Bmal1, which was also evident by immunoblot analysis (Fig. 1G). Bmal1 and its direct target gene, Rev-erbα, were nearly undetectable, as expected.
Figure 1.
Bmal1−/− mice develop obesity when fed regular chow or a high-fat diet. A) Hematoxylin and eosin staining of white adipose tissue at 10 wk of age. B, C) Total body fat content of mice fed regular chow, as determined by NMR (B) and direct measurement of perigonadal white fat pad weight (C). D) Total body fat content analysis of mice fed a high-fat diet. E) Growth curve of mice fed a high-fat diet (n=7–10/group). ##P < 0.05; 1-way ANOVA. F, G) Gene expression level of adipogenic factors in white adipose tissue, as determined by quantitative RT-PCR (F; n=5) and Western blot analysis (G; n=3). *P < 0.05, **P < 0.01 vs. WT.
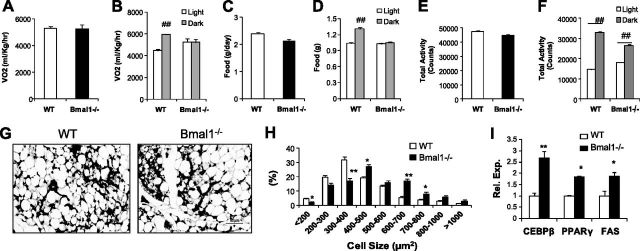
We next performed indirect calorimetry in mice fed the chow diet to assess whether alterations in energy balance contribute to the metabolic phenotype (Fig. 2A–D). Though the light-dark cycle variation that was apparent in WT controls was abolished in the Bmal1−/− mice due to loss of central clock activity rhythm (28), total oxygen consumption and food intake were comparable. Similarly, total activity levels were not altered (Fig. 2E, F), though with reduced diurnal partitioning. To test whether Bmal1 may directly modulate adipocyte development, we examined adipose tissue in 7-d-old neonates, which was unlikely to be affected by potential alterations of energy balance. Even at this early stage of postnatal adipose development, there was modest adipocyte hypertrophy (Fig. 2G) with a shift in size distribution to larger cells (Fig. 2H) accompanied by higher transcript levels of adipogenic genes (Fig. 2I) in Bmal1−/− pups.
Figure 2.
CLAMS analysis and adipose development in 7-d-old neonates. A–F) Total (A, C, E) and light vs. dark cycle (B, D, F) oxygen consumption (A, B), food intake (C, D) and activity level (E, F) in WT and Bmal1−/− mice fed a chow diet (n=5–6/group). ##P < 0.01 vs. light cycle. G–I) Histology (G), quantification of adipocyte cell size distribution (H), and gene expression analysis (I) of subcutaneous white adipose tissue (n=4–5) in 7-d-old neonates. Adipocyte cell size was quantified from 3 representative fields (×200) from each mouse. *P < 0.05, **P < 0.01 vs. WT.
Attenuation of Bmal1 function promotes adipogenic differentiation
Our analysis indicates that overall energy balance does not explain the increased adiposity of Bmal1−/− mice, and suggests that it could be a result of increased adipogenesis. We therefore directly tested the cell-autonomous function of Bmal1 in adipogenesis using three different cell models that are capable of differentiating into mature adipocytes: the 3T3-L1 preadipocytes, 10T1/2 mesenchymal stem cells, and MEFs. We generated stable Bmal1-KD (shBmal1) cells using shRNA in 3T3-L1 and 10T1/2 cells and used one of three constructs, which achieved ∼50% knockdown efficiency in Bmal1 mRNA (Fig. 3A) and effective reduction in protein level (Fig. 3B) in subsequent experiments. The known Bmal1 target gene, Rev-erbα, was down-regulated similarly to Bmal1. We used Oil-red-O staining, which stains for neutral lipids, to assess lipid accumulation as a result of adipogenic differentiation. Interestingly, compared to SC, KD of Bmal1 in 3T3-L1 led to increased lipid staining by Oil-red-O in early stage of differentiation (D3), indicative of enhanced adipogenesis (Fig. 3C), though the staining was weak due to very low numbers of differentiated cells at this stage. Consistent with this, expression of key adipogenic factors (CEBPα, CEBPβ, PPARγ, SREBP-1a) and adipocyte marker genes (FABP4 and FAS) were significantly up-regulated (Fig. 3D) at early time point of d 1. CEBPβ remained elevated in KD cells throughout the differentiation time course. However, at later time points (d 3; Fig. 3D), these genes were suppressed, and the Bmal1-KD cells accumulated less lipids (Supplemental Fig. S1).
Figure 3.
Stable Bmal1 KD promoted adipogenesis during early differentiation of 3T3-L1 preadipocytes. A) mRNA level of Bmal1 and Rev-erbα in SC and KD (shBmal1) cells, as determined by quantitative RT-PCR (n=3). B) Western blot analysis of Bmal1 protein. C) Oil-red-O staining at d 3 of differentiation. D) Gene expression analysis during differentiation, as determined by RT-qPCR (n=3). *P < 0.05, **P < 0.01 vs. SC.
These findings suggest that Bmal1 may have distinct roles during specific stages of adipocyte development and may suppress early adipocyte differentiation. We thus used mesenchymal precursor 10T1/2 and MEF cells to test its role in the preadipocyte commitment step of adipogenesis. Consistent with its effects on early differentiation of 3T3-L1, attenuation of Bmal1 levels enhanced differentiation in both cell types. As shown in Fig. 4, Bmal1 KD in 10T1/2 led to increased Oil-red-O staining (Fig. 4A, B) and CEBPβ and PPARγ protein levels (Fig. 4D), as well as mRNA expression of adipogenic markers FABP4, CEBPβ, and PPARγ (Fig. 4E). Transcript levels of Bmal1 and its direct target gene, D-element binding protein (DBP), were reduced to ∼50% of that of the SC, with substantially lower protein expression (Fig. 4C), as well. The MEF cells from both Bmal1−/− and heterozygote mice exhibited augmented adipogenic conversion, as assessed by lipid staining and quantification (Fig. 5A, B), compared to the WT controls. Elevated mRNA levels of FABP4, CEBPβ, and PPARγ further corroborated this result (Fig. 5C). Strikingly, adipogenic markers were markedly induced in Bmal1−/− MEFs even before adipogenic induction (d 0; Fig. 5C), suggesting spontaneous differentiation of these cells with complete deficiency of Bmal1. Bmal1 protein (Fig. 5D) was absent from homozygote Bmal1−/− MEF cells, and its target gene, DBP, was down-regulated (Fig. 5C). Thus, our results from 3 distinct cell types that are capable of adipogenic conversion clearly demonstrated that attenuation of Bmal1 function promotes adipogenesis, possibly through both early and terminal differentiation events.
Figure 4.
Stable Bmal1 KD promoted adipogenic differentiation of 10T1/2 cells. A, B) Oil-red-O staining of whole plate (A) and representative microscopic images (×100; B) at d 8 of adipogenic differentiation.C, D) Immunoblot analysis of Bmal1 (C) and adipogenic factors (D) during differentiation. E) Quantitative RT-PCR analysis of adipogenic factors and circadian genes during differentiation (n=3). *P < 0.05, **P < 0.01 vs. SC; Student's t test.
Figure 5.
Enhanced adipogenic differentiation in MEF cells of Bmal1−/− mice. A) Oil-red-O staining of MEF cells (top panels) and quantification (bottom panels). B) Oil-red-O staining (top panels) and phase-contrast images (bottom panels). C) Quantitative PCR analysis of adipogenic marker gene expression at d 0, 3, and 6. D) Bmal1 protein level, as determined by immunoblot analysis (n=3). HET, heterozygote; KO, knockout (homozygote). *P < 0.05, **P < 0.01 vs WT.
Bmal1 regulates genes in the Wnt signaling pathway via transcriptional activation
ChIP-seq analysis in mouse liver (20) revealed Bmal1 occupancy of many genes in the canonical Wnt pathway, including Wnt10a, β-catenin, Fzd5, Dvl2, and TCF3. As Wnt signaling is known to suppress adipogenesis (23, 29, 39), we tested whether the inhibitory effect of Bmal1 on adipogenesis is mediated by direct transcriptional control of genes of this pathway. We first examined whether Bmal1 affects expression of these candidate genes. Interestingly, mRNA levels of these components were regulated during differentiation of 10T1/2 (Fig. 6A) or MEF cells (Fig. 6B). Genes such as Axin2, Wnt10a, Dvl2, and TCF3 exhibited an early peak of expression at 4 h on adipogenic induction, followed by a second peak at d 3. In line with our hypothesis, we found that both Bmal1 KD (10T1/2 cells) and deficiency (MEFs) resulted in marked down-regulation of the putative Bmal1 targets (Wnt10a, Fzd5, Dvl2, and TCF3), under basal conditions and during most of the differentiation time points. In Bmal1−/− and heterozygote MEFs, these genes were most substantially suppressed at d 3 of differentiation when Wnt components were at their peak of expression (Fig. 6B). Notably, β-catenin levels were significantly lower, suggesting that this critical intracellular effector step of Wnt pathway is subjected to Bmal1 transcriptional control. Effects of these steps on Wnt pathway signaling appeared to be synergistic, as expression of Axin 2, a target gene of Wnt signaling, is ∼10% of that of the control at its peak in both cell types. Canonical Wnt signaling ultimately is transduced by β-catenin stabilization and accumulation in the cytosol and subsequent gene activation, following its nuclear translocation. We thus analyzed β-catenin abundance in the cytosol to assess Wnt activity. As shown in Fig. 6C, in MEF cells with lack of Bmal1, cytosolic β-catenin was markedly reduced to ∼40% of the control level (cytosolic to total ratio) with concomitant reduction in total cellular β-catenin expression.
Figure 6.
Inhibition of Wnt signaling pathway by attenuation of Bmal1 function. A, B) Gene expression analysis by quantitative RT-PCR in 10T1/2 cells (A) and Bmal1−/− MEF cells (B) at d 0–6; n = 3. HET, heterozygote; KO, knockout (homozygote). *P < 0.05, **P < 0.01; Student's t test. C, D) Immunoblot analysis of β-catenin protein level in Cyt fraction and total lysate in WT and Bmal1−/− MEF cells (C) and Bmal1-KD and Bmal1-overexpressing (cDNA) 10T1/2 cells with and without Wnt3a treatment (D). Right panels show quantification of cytosolic to total lysate ratio (n=3). *P < 0.05, **P < 0.01; ##P < 0.01. E) TOPFlash luciferase activity with and without Wnt3a in Bmal1-KD, Bmal1-overexpressing, and control 10T1/2 cells. Values are expressed as TOPFlash reporter activity normalized to Renilla readings after FOPFlash subtraction (n=4). **P < 0.01; ##P < 0.01.
We further tested effects of Bmal1 KD or overexpression (cDNA) and their responses to Wnt ligand in 10T1/2 cells (Fig. 6D) and found that Bmal1 KD similarly decreased basal level of β-catenin accumulation (∼60% of SC), whereas forced expression augmented this activity by ∼3-fold. Furthermore, Wnt3a-stimulated β-catenin accumulation in the KD cells was significantly blunted compared to the robust activation in SCs. In contrast, although forced expression of Bmal1 substantially increased Wnt activity under unstimulated condition, there was a complete lack of response to Wnt3a, suggesting that it may have resulted in maximal stabilization of β-catenin. As β-catenin accumulation reflects activity of only steps upstream of this event in the Wnt pathway, we further analyzed the effect of Bmal1 on the entire pathway by assessing TCF-mediated transcriptional outcome using the TOPFlash luciferase reporter that contains optimized TCF binding sites (36). As shown in Fig. 6E, Bmal1 KD led to 50% reduction of the basal Wnt activity as compared to SC (0.0221 vs. 0.0457). Furthermore, whereas Wnt3a robustly stimulated this activity (∼143-fold) in control cells, it was minimally induced in the KD cells (∼10% of SC level). On the other hand, forced expression of Bmal1 was able to further augment Wnt-stimulated TOPFlash activity. The effect of Bmal1 KD, as assessed by TCF-mediated transactivation, was substantially more pronounced than reflected by cytosolic β-catenin level (Fig. 6C, D), indicating that the identified steps affected by Bmal1 KD both upsteam and downstream of β-catenin in the Wnt pathway synergistically contributed to the dampening of Wnt activity. Together, these experiments demonstrated that Bmal1 not only regulates expression of many components in the Wnt cascade; this, in turn, leads to alterations of its signaling activity.
Bmal1 transcriptional control of Wnt genes imparts circadian expression rhythm
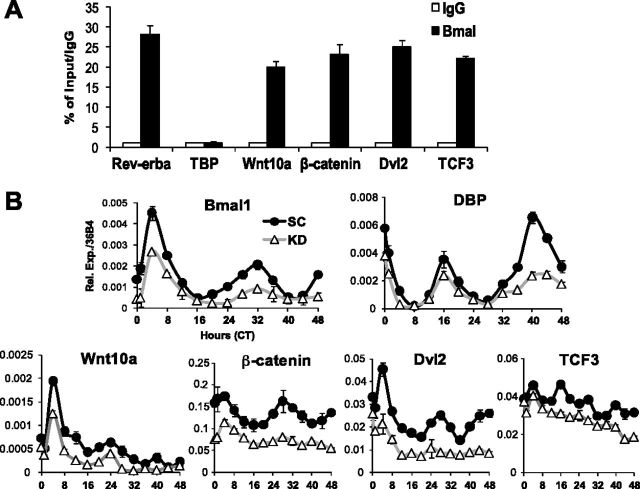
Next, we performed ChIP to assess whether the genes in Wnt pathway affected by Bmal1 level are direct transcriptional targets. We designed specific primers flanking E-boxes in their respective promoters (Supplemental Fig. S2B) according to the binding sites identified by Rey et al. (20). The known Bmal1 target gene Rev-erbα showed ∼30-fold enrichment of Bmal1 recruitment to its promoter over IgG control, whereas it was absent from the TBP promoter (Fig. 7A). This analysis identified strong Bmal1 binding to the E-boxes in Wnt10a, β-catenin, Dvl2, and TCF3 promoters, to a similar extent (∼20- to 25-fold enrichment) as compared to Rev-erbα. This provided direct evidence that these components in the Wnt pathway are target genes of Bmal1. We then tested whether Bmal1 regulation of these genes would impart a circadian expression profile, which can be elicited by serum shock, an established method to synchronize clocks in cell culture (31, 32). We performed serum shock in 10T1/2 cells and determined the expression level of the identified Bmal1 target genes in Wnt pathway over 48 h. As expected, serum shock (Fig. 7B) elicited rhythmic expression of Bmal1 and its known circadian output target gene, DBP, with a period of ∼24 h in opposite phase. Further, both their basal expression and oscillation amplitude were significantly blunted by Bmal1 KD. Notably, serum shock was able to induce the intrinsic circadian profiles of Wnt10a, β-catenin, and Dvl2 in phase with that of the Bmal1, and this oscillation pattern was blunted by Bmal1 KD. In contrast, although the expression level of TCF3 was decreased in Bmal1 KD cells, it did not display a clear rhythm, suggesting that there might be additional circadian elements involved in its transcription, or long half-life of the transcript may have masked its oscillation. These findings revealed that many steps of the Wnt pathway are under direct transcriptional control of Bmal1 and, as a result, possess intrinsic circadian oscillatory rhythms that can be elicited by serum shock, similarly to known clock genes.
Figure 7.
Components of Wnt pathway are direct target genes of Bmal1. A) ChIP assay of Bmal1 occupation in promoters containing E-boxes. Rev-erbα primers are used as positive controls and TBP primers as negative controls for the analysis. Data are expressed as percentage of input level normalized to IgG control (n=3). B) Circadian expression patterns of Bmal1 target genes in Wnt pathway induced by serum shock in 10T1/2 cells, with CT 0 as the first time point after serum shock (n=3/time point).
Activation of Wnt pathway partially suppresses adipogenesis in Bmal1-KD mice
Since Wnt3a could partially restore Wnt signaling defect in Bmal1-KD cells, we tested whether this improved Wnt activity would suppress adipogenesis. At d 9 of differentiation (Fig. 8A), Bmal1 KD increased adipogenesis, as shown previously. We initially tested the time course of the Wnt3a effect. Treating cells in the first 3 d, last 3 d, or the entire length yielded similar results, with the least inhibition of adipogenesis by the first 3 d of treatment, which was used in subsequent experiments. We found that while Wnt3a completely abolished adipogenesis in the controls, as expected, it achieved only partial suppression of differentiation in the KD cells with substantial amounts of lipid accumulation. The differential response to Wnt3a was particularly evident in cell morphology (Fig. 8B). While control cells responded uniformly by reversing to original fibroblast-like appearance, there was striking lack of response in the KD cells, with the majority retaining differentiated adipocyte morphology. It is likely that a substantial population within the KD cells was partially differentiated, though not mature enough to be stained by Oil-red-O. This finding corroborated our analysis of Wnt activity (Fig. 6D, E), indicating that in addition to compromised Wnt ligand activity in Bmal1 KD, downstream components may further contribute to blunted suppression of differentiation in response to Wnt3a. Response of adipogenic genes to Wnt3a was in accord with lipid accumulation in these cells (Fig. 8C), with PPARγ still retaining ∼30% of its level, as compared to near complete inhibition in SC. We then tested whether the specific GSK-3β inhibitor, SB-216763, could mimic the effect of Wnt3a (Fig. 8D, E). As activation of Wnt potently inhibits adipogenesis, we also used LipidTOX fluorescence dye to detect weak lipid staining. As indicated by the presence of differentiated adipocytes by both staining methods (Fig. 8D), SB-216763 abrogated differentiation in the controls but failed to inhibit this process adequately in the KD cells. This response was also reflected by insufficient suppression of PPARγ and FABP4 by SB-216763 (Fig. 8E). Thus, through different means of activation of the Wnt pathway, we demonstrated that manipulation of a specific step was not sufficient to rescue the overall defects in signaling or suppress the enhanced adipogenesis induced by attenuation of Bmal1, which affects various components in this signaling cascade.
Figure 8.
Partial rescue of adipogenesis by Wnt3a and GSK-3β inhibitor in SC and Bmal1-KD 10T1/2 cells. A, B) Representative images of Oil-red-O staining (A) and phase contrast (B) of adipogenic differentiation at d 9, with and without Wnt3a treatment. C) RT-qPCR analysis of adipogenic markers at d 6 of differentiation, with and without Wnt3a treatment (first 3 d). D) Oil-red-O staining and corresponding phase-contrast images (left 2 panels) and LipidTOX Green neutral lipid staining at ×10 and ×20 view (right 2 panels) at d 9 of differentiation after GSK-3β inhibitor SB-216763 treatment. E) Expression of adipogenic markers at d 6 of differentiation after SB-216763 treatment (first 2 d). n = 3/group. **P < 0.05; Student's t test.
DISCUSSION
Circadian disruption is associated with development of obesity, and components of the molecular clock network in adipose tissue could directly modulate adipocyte development. The CLOCK mutant mice (19) exhibit obesity and a metabolic syndrome-like phenotype, and inactivation of Per2 increased adipocyte differentiation due to loss of suppression of PPARγ (14). These studies highlight the connection between disruption of molecular clock and development of obesity, which could be a common mechanism that underlies obesity in a large population. Our study demonstrated that the core clock gene, Bmal1, suppresses adipogenesis via the Wnt pathway, and points out the possibility that dysregulation of Bmal1 could be one of the underlying mechanisms contributing to the adverse metabolic consequences associated with disruption of circadian physiology.
We identified specific steps of the Wnt signaling cascade that are under Bmal1 transcriptional control, and thus, bona fide circadian-controlled genes. This is in line with two recent reports suggesting that the molecular clock, in particular Bmal1, could directly regulate this pathway (20, 27). Most of these genes identified as Bmal1 targets belong to the canonical Wnt pathway known to suppress adipogenesis (23, 40). It has been shown that the Wnt pathway suppresses not only adipocyte terminal differentiation through inhibition of CEBPα and PPARγ (23), but also preadipocyte commitment to the adipocyte fate (25, 26, 29). During development, adipocytes arise from mesenchymal precursors that first go through lineage commitment to become determined preadipocytes and subsequently undergo terminal differentiation into mature adipocytes (41, 42). 10T1/2 cells, a mesenchymal stem cell line, are developmentally more primitive than committed 3T3-L1 preadipocytes, and their differentiation does not require clonal expansion, a prerequisite for 3T3-L1 preadipocytes (43, 44). Thus, this cell type closely mimic adipocyte differentiation in vivo and has been used as a model to probe mechanisms specifically involved in the preadipocyte commitment stage of early adipocyte development (25). Our results in this cell line, and additional evidence from MEF cells, are consistent with the observed phenotype of increased adipogenesis with loss of Bmal1 in vivo.
Shimba et al. (38) initially reported that transient KD of Bmal1 in 3T3-L1 preadipocytes blunted the adipogenic response, although their recent study of Bmal1-null mice (45) revealed premature differentiation of adipocytes. Though our result of d 6 of 3T3-L1 differentiation (Supplemental Fig. S1) is in accord with that of Shimba et al. (38), we found that the early stage of adipogenesis was substantially augmented by KD of Bmal1 (Fig. 3A). This finding was further corroborated by results from 10T1/2 and MEF cells, in which attenuation of Bmal1 function led to increased adipogenic conversion. In addition, the obesity phenotype and up-regulation of adipogenic factors in Bmal1−/− mice are in line with our findings from these adipocyte cell lines, and consistent with an independent study by Lamia et al. (46) showing similarly increased adiposity in these mice. Thus, the effects seen in later stages of 3T3-L1 differentiation (Supplemental Fig. S1) could be due to its limitation as a preadipocyte model to assess the role of Bmal1 in adipogenesis. And, it is also possible that there might be certain inhibitory mechanisms of the Wnt pathway in this cell type that block Bmal1 function during the transition from d 3. In addition, the apparent differences in KD strategies (stable vs. transient) and separate sources of MEF cells might account for these results, as well. Through its direct transcriptional regulation on Wnt pathway components, our study strongly supports the notion that Bmal1 may play a more dominant role in the early stage of adipogenesis, which determines the in vivo phenotype. Elucidation of the precise mechanisms in early adipocyte commitment vs. late terminal differentiation could provide fundamental insights into regulatory pathways governing these processes, and warrants future investigation by selective inactivation of Bmal1 in distinct stages of adipocyte development.
Circadian element, as an evolutionarily conserved regulatory mechanism, has been shown to exert concerted temporal coordination over multiple steps along a specific metabolic pathway, including gluconeogenesis (3), xenobiotic metabolism (47), and bile acid homeostasis (35). Our demonstration of Bmal1 regulation of various distinct steps in the Wnt cascade could be an example of this regulatory mechanism in modulation of specific developmental pathways. This previously unknown circadian regulatory component of the Wnt pathway may impart a temporal coordination among the steps involved. Due to this regulation conferred by Bmal1 on Wnt genes, serum shock was able to elicit their rhythmic oscillation, indicating that these are bona fide circadian-controlled genes. And it is likely that impaired functions of these disparate components collectively contribute to the compromised Wnt signaling and enhanced adipogenesis observed with Bmal1 deficiency.
In summary, our study uncovered a novel cell-autonomous function of the core clock gene, Bmal1, in suppressing adipogenesis through direct transcriptional regulation of genes of the Wnt signaling pathway. Furthermore, it is conceivable that altered Bmal1 function as a result of disrupted circadian physiology may contribute to the prevalence of associated adverse metabolic consequences. Further elucidation of this mechanism may reveal the full effect of this conserved temporal regulatory component in possibly many other cell types during embryogenesis and adult stem cell development.
Supplementary Material
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
Acknowledgments
The authors thank The Methodist Hospital Research Institute (TMHRI) for funding support and the Center for Diabetes Research for technical assistance.
The authors also thank Dr. David D. Moore (Baylor College of Medicine, Houston, TX, USA) and Dr. Tuo Deng (TMHRI, Houston, TX, USA) for insightful discussions and for reagents.
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
- 10T1/2
- C3H10T1/2
- Bmal1
- brain and muscle Arnt-like 1
- CEBP
- CCAAT/enhancer-binding protein
- ChIP
- chromatin immunoprecipitation
- ChIP-Seq
- chromatin immunoprecipitation sequencing
- CLOCK
- circadian locomotor output cycles kaput
- CT
- circadian time
- DBP
- D-element binding protein
- Dvl2
- Dishevelled 2
- Fzd5
- Frizzled 5
- KD
- knockdown
- MEF
- mouse embryonic fibroblast
- Per1–3
- Period 1–3
- PPARγ
- peroxisome proliferator-activated receptor γ
- ROR
- retinoic acid receptor-related orphan receptor
- SC
- scrambled control
- shRNA
- small hairpin RNA
- SREBP
- sterol regulatory element binding protein
- TCF3
- T-cell factor 3
- Wnt
- Wingless-type
- WT
- wild type
- ZT
- zeitgeber time
REFERENCES
- 1.Bass J., Takahashi J. S.. Circadian integration of metabolism and energetics. Science (New York) , 1349–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green C. B., Takahashi J. S., Bass J. (2008) The meter of metabolism. Cell , 728–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Panda S., Antoch M. P., Miller B. H., Su A. I., Schook A. B., Straume M., Schultz P. G., Kay S. A., Takahashi J. S., Hogenesch J. B. (2002) Coordinated transcription of key pathways in the mouse by the circadian clock. Cell , 307–320 [DOI] [PubMed] [Google Scholar]
- 4.Otway D. T., Frost G., Johnston J. D. (2009) Circadian rhythmicity in murine pre-adipocyte and adipocyte cells. Chronobiol. Int. , 1340–1354 [DOI] [PubMed] [Google Scholar]
- 5.Wu X., Zvonic S., Floyd Z. E., Kilroy G., Goh B. C., Hernandez T. L., Eckel R. H., Mynatt R. L., Gimble J. M. (2007) Induction of circadian gene expression in human subcutaneous adipose-derived stem cells. Obesity (Silver Spring, Md.) , 2560–2570 [DOI] [PubMed] [Google Scholar]
- 6.Gimble J. M., Sutton G. M., Ptitsyn A. A., Floyd Z. E., Bunnell B. A.. Circadian rhythms in adipose tissue: an update. Curr. Opin. Clin. Nutr. Metab. Care , 554–561 [DOI] [PubMed] [Google Scholar]
- 7.Gavrila A., Peng C. K., Chan J. L., Mietus J. E., Goldberger A. L., Mantzoros C. S. (2003) Diurnal and ultradian dynamics of serum adiponectin in healthy men: comparison with leptin, circulating soluble leptin receptor, and cortisol patterns. J. Clin. Endocrinol. Metab. , 2838–2843 [DOI] [PubMed] [Google Scholar]
- 8.Kalsbeek A., Fliers E., Romijn J. A., La Fleur S. E., Wortel J., Bakker O., Endert E., Buijs R. M. (2001) The suprachiasmatic nucleus generates the diurnal changes in plasma leptin levels. Endocrinology , 2677–2685 [DOI] [PubMed] [Google Scholar]
- 9.Schibler U., Sassone-Corsi P. (2002) A web of circadian pacemakers. Cell , 919–922 [DOI] [PubMed] [Google Scholar]
- 10.Pan A., Schernhammer E. S., Sun Q., Hu F. B.. Rotating night shift work and risk of type 2 diabetes: two prospective cohort studies in women. PLoS Med. , e1001141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parkes K. R. (2002) Shift work and age as interactive predictors of body mass index among offshore workers. Scand. J. Work Environ. Health , 64–71 [DOI] [PubMed] [Google Scholar]
- 12.Scheer F. A., Hilton M. F., Mantzoros C. S., Shea S. A. (2009) Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc. Natl. Acad. Sci. U. S. A. , 4453–4458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karatsoreos I. N., Bhagat S., Bloss E. B., Morrison J. H., McEwen B. S.. Disruption of circadian clocks has ramifications for metabolism, brain, and behavior. Proc. Natl. Acad. Sci. U. S. A. , 1657–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grimaldi B., Bellet M. M., Katada S., Astarita G., Hirayama J., Amin R. H., Granneman J. G., Piomelli D., Leff T., Sassone-Corsi P.. PER2 controls lipid metabolism by direct regulation of PPARgamma. Cell Metab. , 509–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J., Lazar M. A. (2008) Bifunctional role of Rev-erbalpha in adipocyte differentiation. Mol. Cell. Biol. , 2213–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reppert S. M., Weaver D. R. (2002) Coordination of circadian timing in mammals. Nature , 935–941 [DOI] [PubMed] [Google Scholar]
- 17.Sookoian S., Gemma C., Gianotti T. F., Burgueno A., Castano G., Pirola C. J. (2008) Genetic variants of Clock transcription factor are associated with individual susceptibility to obesity. Am. J. Clin. Nutr. , 1606–1615 [DOI] [PubMed] [Google Scholar]
- 18.Woon P. Y., Kaisaki P. J., Braganca J., Bihoreau M. T., Levy J. C., Farrall M., Gauguier D. (2007) Aryl hydrocarbon receptor nuclear translocator-like (BMAL1) is associated with susceptibility to hypertension and type 2 diabetes. Proc. Natl. Acad. Sci. U. S. A. , 14412–14417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turek F. W., Joshu C., Kohsaka A., Lin E., Ivanova G., McDearmon E., Laposky A., Losee-Olson S., Easton A., Jensen D. R., Eckel R. H., Takahashi J. S., Bass J. (2005) Obesity and metabolic syndrome in circadian clock mutant mice. Science (New York) , 1043–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rey G., Cesbron F., Rougemont J., Reinke H., Brunner M., Naef F.. Genome-wide and phase-specific DNA-binding rhythms of Bmal1 control circadian output functions in mouse liver. PLoS Biol. , e1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tseng Y. H., Kriauciunas K. M., Kokkotou E., Kahn C. R. (2004) Differential roles of insulin receptor substrates in brown adipocyte differentiation. Mol. Cell. Biol. , 1918–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bennett C. N., Ross S. E., Longo K. A., Bajnok L., Hemati N., Johnson K. W., Harrison S. D., MacDougald O. A. (2002) Regulation of Wnt signaling during adipogenesis. J. Biol. Chem. , 30998–31004 [DOI] [PubMed] [Google Scholar]
- 23.Ross S. E., Hemati N., Longo K. A., Bennett C. N., Lucas P. C., Erickson R. L., MacDougald O. A. (2000) Inhibition of adipogenesis by Wnt signaling. Science (New York) , 950–953 [DOI] [PubMed] [Google Scholar]
- 24.Clevers H. (2006) Wnt/beta-catenin signaling in development and disease. Cell , 469–480 [DOI] [PubMed] [Google Scholar]
- 25.Bowers R. R., Lane M. D. (2008) Wnt signaling and adipocyte lineage commitment. Cell Cycle , 1191–1196 [DOI] [PubMed] [Google Scholar]
- 26.Jackson A., Vayssiere B., Garcia T., Newell W., Baron R., Roman-Roman S., Rawadi G. (2005) Gene array analysis of Wnt-regulated genes in C3H10T1/2 cells. Bone , 585–598 [DOI] [PubMed] [Google Scholar]
- 27.Janich P., Pascual G., Merlos-Suarez A., Batlle E., Ripperger J., Albrecht U., Obrietan K., Di Croce L., Benitah S. A. (2011) The circadian molecular clock creates epidermal stem cell heterogeneity. Nature , 209–214 [DOI] [PubMed] [Google Scholar]
- 28.Bunger M. K., Wilsbacher L. D., Moran S. M., Clendenin C., Radcliffe L. A., Hogenesch J. B., Simon M. C., Takahashi J. S., Bradfield C. A. (2000) Mop3 is an essential component of the master circadian pacemaker in mammals. Cell , 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawai M., Mushiake S., Bessho K., Murakami M., Namba N., Kokubu C., Michigami T., Ozono K. (2007) Wnt/Lrp/beta-catenin signaling suppresses adipogenesis by inhibiting mutual activation of PPARgamma and C/EBPalpha. Biochem. Biophys. Res. Commun. , 276–282 [DOI] [PubMed] [Google Scholar]
- 30.Fu L., Pelicano H., Liu J., Huang P., Lee C. (2002) The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell , 41–50 [DOI] [PubMed] [Google Scholar]
- 31.Balsalobre A., Damiola F., Schibler U. (1998) A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell , 929–937 [DOI] [PubMed] [Google Scholar]
- 32.Chalmers J. A., Martino T. A., Tata N., Ralph M. R., Sole M. J., Belsham D. D. (2008) Vascular circadian rhythms in a mouse vascular smooth muscle cell line (Movas-1). Am. J. Physiol. , R1529–1538 [DOI] [PubMed] [Google Scholar]
- 33.Ramirez-Zacarias J. L., Castro-Munozledo F., Kuri-Harcuch W. (1992) Quantitation of adipose conversion and triglycerides by staining intracytoplasmic lipids with Oil red O. Histochemistry , 493–497 [DOI] [PubMed] [Google Scholar]
- 34.Young C. S., Kitamura M., Hardy S., Kitajewski J. (1998) Wnt-1 induces growth, cytosolic beta-catenin, and Tcf/Lef transcriptional activation in Rat-1 fibroblasts. Mol. Cell. Biol. , 2474–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma K., Xiao R., Tseng H. T., Shan L., Fu L., Moore D. D. (2009) Circadian dysregulation disrupts bile acid homeostasis. PLoS One , e6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Veeman M. T., Slusarski D. C., Kaykas A., Louie S. H., Moon R. T. (2003) Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr. Biol. , 680–685 [DOI] [PubMed] [Google Scholar]
- 37.Kondratov R. V., Kondratova A. A., Gorbacheva V. Y., Vykhovanets O. V., Antoch M. P. (2006) Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. , 1868–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimba S., Ishii N., Ohta Y., Ohno T., Watabe Y., Hayashi M., Wada T., Aoyagi T., Tezuka M. (2005) Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proc. Natl. Acad. Sci. U. S. A. , 12071–12076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Longo K. A., Wright W. S., Kang S., Gerin I., Chiang S. H., Lucas P. C., Opp M. R., MacDougald O. A. (2004) Wnt10b inhibits development of white and brown adipose tissues. J. Biol. Chem. , 35503–35509 [DOI] [PubMed] [Google Scholar]
- 40.Ross S. E., Erickson R. L., Gerin I., DeRose P. M., Bajnok L., Longo K. A., Misek D. E., Kuick R., Hanash S. M., Atkins K. B., Andresen S. M., Nebb H. I., Madsen L., Kristiansen K., MacDougald O. A. (2002) Microarray analyses during adipogenesis: understanding the effects of Wnt signaling on adipogenesis and the roles of liver X receptor alpha in adipocyte metabolism. Mol. Cell. Biol. , 5989–5999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Christodoulides C., Lagathu C., Sethi J. K., Vidal-Puig A. (2009) Adipogenesis and WNT signalling. Trends Endocrinol. Metab. , 16–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bowers R. R., Lane M. D. (2007) A role for bone morphogenetic protein-4 in adipocyte development. Cell Cycle , 385–389 [DOI] [PubMed] [Google Scholar]
- 43.Cho Y. C., Jefcoate C. R. (2004) PPARgamma1 synthesis and adipogenesis in C3H10T1/2 cells depends on S-phase progression, but does not require mitotic clonal expansion. J. Cell. Biochem. , 336–353 [DOI] [PubMed] [Google Scholar]
- 44.Jefcoate C. R., Wang S., Liu X. (2008) Methods that resolve different contributions of clonal expansion to adipogenesis in 3T3-L1 and C3H10T1/2 cells. Methods Mol. Biol. , 173–193 [DOI] [PubMed] [Google Scholar]
- 45.Shimba S., Ogawa T., Hitosugi S., Ichihashi Y., Nakadaira Y., Kobayashi M., Tezuka M., Kosuge Y., Ishige K., Ito Y., Komiyama K., Okamatsu-Ogura Y., Kimura K., Saito M.. Deficient of a clock gene, brain and muscle Arnt-like protein-1 (BMAL1), induces dyslipidemia and ectopic fat formation. PLoS One , e25231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lamia K. A., Storch K. F., Weitz C. J. (2008) Physiological significance of a peripheral tissue circadian clock. Proc. Natl. Acad. Sci. U. S. A. , 15172–15177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gachon F., Olela F. F., Schaad O., Descombes P., Schibler U. (2006) The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. , 25–36 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.