Abstract
Background
Boron (B) deficiency is an agricultural problem that causes significant losses of crop yield in many areas of the world. However, systematic analysis of BOR family genes for B transport in rapeseed is still lacking. We aimed to identify and characterize BOR transporters in Brassica napus and the potential role of these transporters in B homeostasis in response to B deficiency.
Results
Here, we identified 20 BOR transporters from the Brassica napus genome, which were classified into six distinct groups that represent clear orthologous relationships to their family members in Arabidopsis. qRT-PCR revealed distinct expression profiles for BnBORs in different tissues and in response to external B levels. The B-efficient cultivar QY10 accumulated more B in shoots than the B-inefficient cultivar W10, and overexpression of BnaBOR1;1c could alleviate shoot B-deficiency symptoms in W10 by distributing more B from roots to shoots. Additionally, BnBOR1;1c expression was up-regulated by B deficiency, and the induction of BnBOR1;1c was more intense in QY10. Moreover, two conserved InDels were found in the promoter regions of BnBOR1;1c within different B-efficient genotypes.
Conclusions
Overall, the molecular characterization of the BnBOR genes of two B-efficient cultivars and their responses to B deficiency highlights the diversity of the family members in B. napus, and BnaC4.BOR1;1c has potential as a candidate gene for improving B nutrition.
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1407-1) contains supplementary material, which is available to authorized users.
Keywords: Brassica napus, BOR transporter, B deficiency, Transcriptional profile, B efficiency, Gene function
Background
Boron (B) is an essential micronutrient for higher plants [1]. In terms of physiological functions, B is important for cell wall (CW) structure, as it crosslinks the pectic polysaccharide rhamnogalacturonan II (RG-II) and pectin assembly in the CW [2, 3]. Because of the difficulty transferring B in plants, B deficiency primarily restrains rapidly growing tissues, inhibiting apical growth in the roots and shoots, and reduces fertility [4–6]. Most importantly, B deficiency often causes dried-up floral buds, fewer pods and low seed yield or sterility in plant reproductive growth.
Among essential mineral nutrients, B has the narrowest margin in soil concentrations between deficiency and toxicity [7]. A number of genes involved in efficient B uptake, transport, partitioning and export have been identified as necessary for tolerance to B deficiency and toxicity [8]. In Arabidopsis, AtBOR1 has been identified as an efflux type transporter for xylem loading and is essential for preventing shoots from B deficiency [9, 10]. Under B-deficient conditions, although AtBOR1 mRNA accumulation is not abundant, its protein showed high abundance in the PM [11]. Furthermore, BOR1 activity is repressed through ubiquitination-mediated vacuolar trafficking to avoid B toxicity under high B conditions [12, 13]. Subsequently, homologues of AtBOR1 are assumed to play crucial roles in the resistance to B deprivation in rice (OsBOR1) [14], grapevine (Vitis vinifera L.) (VvBOR1) [15], Citrus macrophylla (CmBOR1) [16], wheat (Triticum awstivum L.) (TaBOR1) [17], tomato [18] and B. napus (BnBOR1;1c) [19]. OsBOR1 is required for B uptake and xylem loading under B deficiency conditions [14]. Recently, the B exporter BOR2 was found to differ from BOR1 in Arabidopsis; it is required for the effective crosslinking of the pectin polysaccharide RG-II and root cell elongation under B limitation [20]. Additionally, AtBOR4, an Arabidopsis borate efflux transporter, is significant for the directional export of B from roots to soils to prevent the overaccumulation of B in the xylem and improve the tolerance to excess B [21]. Bot1 contributes to the high B tolerance of Sahara, a barley landrace, by exclusion of B from the roots [22]. To date, the boric acid/borate transporter activity of BORs in monocotyledon and dicotyledon plants have been demonstrated and identified; these plants have a conserved sorting motif and the boric acid channel showed homology to the mammalian Slc4 bicarbonate (HCO3−) family [23, 24].
Allotetraploid rapeseed (Brassica napus L., AnAnCnCn, 2n = 38, 840 Mb), which originated from a natural hybridization between Brassica rapa (ArAr, 2n = 20, 312 Mb) and Brassica oleracea (CoCo, 2n = 18, 540 Mb) approximately 7500–12,500 years ago, is extremely sensitive to B deficiency and shows a notable reduction in seed yield and quality with a low B supply [25–27]. In recent years, because borate rock is a depletable and non-renewable mineral resource, numerous effective measures have been taken to address this problem, including the application of borate fertilizers to soils with low B abundance. A molecular understanding of B deficiency responses in plants is pivotal for developing crop varieties with high B use efficiency under low B conditions. Such understanding is rapidly progressing for BOR1 function in Brassica napus. Six B transporter genes (BnBOR1s) homologous to AtBOR1 were identified and divided into three groups in B. napus. Each group was comprised of two members, one of which originated from B. rapa (BnBOR1;1a, BnBOR1;2a, and BnBOR1;3a) and the other from B. oleracea (BnBOR1;1c, BnBOR1;2c, and BnBOR1;3c) [28]. Unlike the non-transcriptional regulation of AtBOR1, BnaC4.BOR1;1c (BnBOR1;1c) is highly expressed not only in roots but also in shoot nodal regions and flowers in response to B limitation, and is critical for the development and fertility of inflorescences in rapeseed [19].
Here, we report the systematic analysis of the gene structure, phylogeny, motif composition, chromosomal localization and expression patterns of BOR genes in B. napus under different B conditions. Furthermore, transgenic BnaC4.BOR1;1c knockdown and overexpression lines in the B. napus cultivars QY10 and Westar 10 were established as examples to study the functional relationship between B transporters and B efficiency.
Methods
Identification and physicochemical parameter analysis of BOR genes in B. napus
All BOR genes were identified in B. napus based on their homology similarity to the 7 BOR protein sequences in Arabidopsis from the TAIR10 database (http://www.arabidopsis.org/index.jsp) using the BLAST search program in the CNS-Genoscope database (http://www.genoscope.cns.fr/brassicanapus/) [27]. The exon-intron structures of the BnBOR family members were investigated based on coding sequence alignments with corresponding genomic sequences, and the diagram was drawn using the online Gene Structure Display Server (GSDS; http://gsds.cbi.pku.edu.cn/) [28]. The physicochemical parameters, including molecular weight (MW) and isoelectronic point (pI), for each BnBOR protein were calculated using the compute pI/MW tool in ExPASy (http://www.expasy.org/tools/). GRAVY (grand average of hydropathicity) values were calculated using the PROTPARAM tool (http://web.expasy.org/protparam/).
Chromosome localization
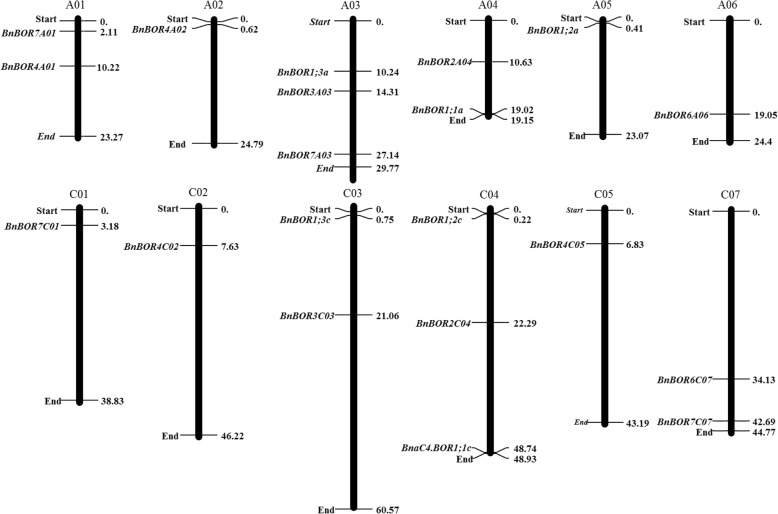
To determine the physical locations of the BnBOR genes, the starting and ending positions of all BnBOR genes on each chromosome were obtained from the Brassica database (BRAD; http://www.genoscope.cns.fr/brassicanapus/). The MapInspect software was used to draw the gene chromosome location diagrams. The 20 BnBOR gene members were distributed non-randomly on 12 B. napus chromosomes between chromosomes A and C. Chromosome A03 and C04 contained the most BnBOR genes with three, whereas chromosomes A02, A05, A06, C01, C02 and C05 contained one gene each.
BnBOR member gene structure and conserved motif analysis
Conserved motif structures encoded by the BnBOR family genes were identified by the Multiple Expectation Maximization for Motif Elicitation (MEME) program version 4.11.2 (http://meme-suite.org/tools/meme) [29]. The parameter settings included output motifs (10), minimum motif width (10), and maximum motif width (100). The MEME motifs were annotated using the Pfam (http://pfam.xfam.org/search) and NCBI databases.
Phylogenetic analysis
Multiple sequence alignment of all the predicted BOR genes from B. napus and Arabidopsis was performed using the NCBI BLASTP (Target type: Proteome) and ClustalW2 programs with default parameters. The 37 BOR amino acid sequences (Supplemental data for the amino acids) from two taxonomic families containing core conserved BORs were downloaded using the Phytozome 11 online software from the Joint Genome Institute. These 37 BOR amino acid sequences were from four plant species including Brassica napus, Arabidopsis, Oryza sativa and Zea mays. The evolutionary history was inferred using the Neighbour-Joining method [30], and the numbers displayed in the phylogenetic tree represent bootstrap values that were estimated (with 500 replicates) to assess the relative support for each branch using MEGA6.0 software.
Plant materials and treatments
Plump seeds from the B-efficient cultivar QY10, Ningyou 7 and B-inefficient cultivar W10, Bakow were used for hydroponic culture experiments in an illuminated growth room at 24/22 °C (day/night) under a photoperiod 14/10 h (light/dark) with a light density of 300–320 μmol m− 2 s− 1. The samples were surface-sterilized for 15 min using 0.5% NaClO (w/v) and rinsed completely with sterilized ultrapure water (> 18.25 MΩ·cm). The seeds were then sown on moistened gauze after being soaked in deionized water for 1 d. After 5 d of germination, the uniform seedlings were transplanted into 10-L black plastic containers filled with Hoagland and Arnon solution with 0.25 μM B (low B) and 25 μM B (high B) for 7 d or 10 d. The nutrient solution was replaced every 3 days. The rapeseed seedlings were first grown in one quarter-strength solution, afterwards progressing to one-half-strength and eventually full-strength.
RNA extraction, reverse transcription and real-time quantitative PCR
Total RNA was extracted from plant fresh samples independently using the RNAiso™ Plus reagent (Takara Bio, Otsu, Shiga, Japan) according to the manufacturer’s recommendations. RNA samples were treated with RNase-free DNase I (Invitrogen, Grand Island, NY, USA). Subsequently, first strand cDNA was synthesized using the PrimeScript™ RT Master Mix (Takara, Tokyo, Japan) according to the manufacturer’s protocol. The specific primers for the BnBOR genes were designed with Primer-NCBI (http://www.ncbi.nlm.nih.gov/tools/primersblast/index.cgi?LINK=BlastHome) and are listed in Additional file 1: Table S1. Real-time fluorescence quantitative PCR (RT-qPCR) to detect the expression of the target genes was performed on a CFX96™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) with the SYBR Green Real-Time PCR Master Mix Kit (TOYOBO, Japan). The PCR conditions were as follows: 95 °C for 5 min, followed by 40 cycles of 95 °C for 10 s, 60 °C for 15 s and 72 °C for 20 s. The reference gene BnaActin was used as an internal control, and the fold change was analysed via the 2-ΔΔCt method [31]. The gene-specific primers are listed in Additional file 1: Table S1. The primer specificity and PCR products were confirmed by sequencing and checked on the NCBI website (https://www.ncbi.nlm.nih.gov/) or rapeseed database using Blast (Http://www.genoscope.cns.fr/brassicanapus). The primer efficiency was checked according to Zhang et al. [19]
Characterization of cis-elements in the BnaC4.BOR1;1c promoter region in B. napus
The 2000-bp upstream sequence relative to the translation start codon in BnaC4.BOR1;1c was downloaded from CNS-Genoscope. The BnaC4.BOR1;1c promoter was analysed to determine the cis-regulatory elements using the plant cis-element database PlantCARE [32].
Vector construction and plant transformation
To obtain the BnaC4.BOR1;1c overexpression (OE) lines, a full-length BnaC4.BOR1;1c gene coding sequence was amplified with Pfu DNA polymerase (Promega, Madison, WT) using gene-specific primers (Additional file 1: Table S1). The amplicon was ligated into the pGEMT easy vector (Promega, Madison, WI) and confirmed by DNA sequencing. Then, the coding sequence was subsequently cloned into the XbaІ and XhoІ sites in the pBinGlyRed3 vector with the 2 × 35S promoter using DNA fusion technology. The vector was introduced into the Agrobacterium tumefaciens strain GV3101 by electroporation. W10 were transformed according to the method in [33]. Putative transformants (T0) were transferred to soil for growth. The genomic DNA was isolated from young leaves and used to determine the positive plants by PCR using vector-specific primers. Seedlings from the T1 generation were examined for 3:1 segregation, and the seedlings with an OE construction (either heterozygous or homozygous) were kept to obtain the T2 generation. Expression of BnaC4.BOR1;1c in homozygous OE lines was determined by quantitative RT-PCR to detect the efficiency using specific primer pairs. Three homozygous lines were selected for further analysis. The QY10, W10 and BnaC4.BOR1;1c OE lines were grown hydroponically for 10 d under normal (25 μM) and low (0.25 μM) B stress as described above.
Transmission electron microscopy
For transmission electron microscopy (TEM) analysis, juvenile leaves (approximately 1 mm2) from the fresh seedlings were sampled and immediately fixed in 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4, containing 2% sucrose for 2 h. Post-fixation was performed in 1% osmium tetroxide in the same buffer for 1.5 h. The samples were then dehydrated using an ethanol series (30%, 50%, 70%, 80%, 90%, 95% and 100% [v/v] ethanol) and propylene oxide for dehydration and embedded in epoxy resin. Ultra-thin 0.5–1.0 μm sections were cut with an ultramicrotome (Leica UC6/FC6, Germany), and a transmission electron microscope (HITACHI, H7650, Japan) was used to examine uranyl acetate-stained sections at 40 kV–120 kV.
Boron measurement
The samples (root and shoot) were separated and dried at 105 °C for 30 min and then to a constant weight at 65 °C. The dried samples were ground into fine powders using a carnelian mortar, and B was lixiviated with 10 mL of 1 M HCl on a 250-rpm shaker for 2 h. The B concentration was measured using an inductively coupled plasma mass spectrometer (ICP-MS; Perkin Elmer, ELAN DRC-e, USA). B accumulation was calculated as the B concentration × dry weight.
Statistical analysis
Each graphical plot represents the results from multiple independent experiments (n ≥ 3), and the values are the means ± SD. Statistical significance between two genotypes or two organs in the same B condition was used independent Student’s t-test. LSD test was used for multiple comparisons at the p < 0.05 level among the different B treatments and genotypes in the same organ. Data were analyzed in SPSS 18.0 software and p value < 0.05 was considered statistically significant.
Results
Genome-wide identification of BnBOR gene family members in B. napus
A total of 20 BOR genes (BnBOR1s-BnBOR7s) were identified in the B. napus genome based on their homology to the 7 BOR protein sequences in Arabidopsis from the TAIR10 database using the BLAST search program in the CNS-Genoscope database. The systematic analysis of the BnBORs is shown in Table 1. Large variations in the encoding amino acid (aa) length of these genes were found, with lengths ranging from 660 aa (BnBOR7A03) to 738 aa (BnBOR3A03), and the molecular weights range from 74.67 to 81.24 kDa. ExPASy analysis revealed that these protein sequences have stable isoelectric points (pI) ranging from 6.91 to 9.26, and most of the sequence have similar parameters. Almost all BOR proteins have relatively high isoelectric points (pI > 7), except for BnBOR4A02 and BnBOR4C05. GRAVY values are defined as the sum of the hydropathy values of all amino acids divided by the protein length. All of the BnBORs are hydrophilic, with values ranging from 0.162 to 0.266. Additionally, TargetP and WoLF PSORT were used to predict the subcellular location of the 20 BnBOR proteins, which were similar to BnaC4.BOR1;1c located in the cell PM, implying the functions of the BnBOR family members. These results for the parameter analysis indicate that BnBOR family members have stable essential characteristics, physicochemical properties and an abundant chromosomal distribution.
Table 1.
Gene sequence characteristics of 20 BnBORs and their protein physicochemical parameters
| BnaBORs | Mapping | ID | Name | Description | Physicochemical parameters | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Length (bp) |
Size (Aa) |
Intron | Exon | Weight (kDa) |
pI | GRAVY | ||||
| BnaBOR1s | A04 | GSBRNA2T00003348001 | BnaA04g26910D | 2106 | 701 | 9 | 10 | 78.24 | 8.68 | 0.195 |
| C04 | GSBRNA2T00038178001 | BnaC04g51480D | 2106 | 701 | 9 | 10 | 78.31 | 8.77 | 0.183 | |
| A05 | GSBRNA2T00132977001 | BnaA05g00740D | 2115 | 704 | 9 | 10 | 78.6 | 8.86 | 0.194 | |
| C04 | GSBRNA2T00064725001 | BnaC04g00350D | 2115 | 704 | 9 | 10 | 78.64 | 8.86 | 0.200 | |
| A03 | GSBRNA2T00138903001 | BnaA03g21650D | 2112 | 703 | 11 | 12 | 78.46 | 8.94 | 0.177 | |
| C03 | GSBRNA2T00097768001 | BnaC03g72490D | 2112 | 703 | 11 | 12 | 78.49 | 8.98 | 0.187 | |
| BnaBOR2s | A04 | GSBRNA2T00093109001 | BnaAnng31830D | 3069 | 707 | 11 | 12 | 78.97 | 8.52 | 0.193 |
| C04 | GSBRNA2T00156837001 | BnaC04g21390D | 3069 | 706 | 11 | 12 | 78.84 | 8.2 | 0.201 | |
| BnaBOR3s | A03 | GSBRNA2T00111858001 | BnaA03g29440D | 2217 | 738 | 11 | 12 | 81.24 | 9.15 | 0.234 |
| C03 | GSBRNA2T00125304001 | BnaC03g34730D | 2214 | 737 | 11 | 12 | 81.21 | 9.26 | 0.22 | |
| BnaBOR4s | A01 | GSBRNA2T00083145001 | BnaA02g16980D | 2028 | 675 | 12 | 13 | 75.98 | 7.59 | 0.117 |
| A02 | GSBRNA2T00069284001 | BnaA02g35930D | 2052 | 683 | 12 | 13 | 76.46 | 6.91 | 0.162 | |
| C02 | GSBRNA2T00063784001 | BnaCnng08280D | 2052 | 683 | 12 | 13 | 76.41 | 6.88 | 0.172 | |
| C05 | GSBRNA2T00068220001 | BnaC05g11780D | 2028 | 675 | 12 | 13 | 75.98 | 7.59 | 0.117 | |
| BnaBOR6s | A06 | GSBRNA2T00147638001 | BnaA06g27700D | 2019 | 672 | 11 | 12 | 75.76 | 7.26 | 0.266 |
| C07 | GSBRNA2T00133614001 | BnaC07g29340D | 2016 | 671 | 11 | 12 | 75.84 | 7.94 | 0.258 | |
| BnaBOR7s | A01 | GSBRNA2T00121111001 | BnaA01g04520D | 3648 | 1215 | 23 | 24 | 134.72 | 7.82 | 0.047 |
| A03 | GSBRNA2T00077798001 | BnaA03g52100D | 1983 | 660 | 9 | 10 | 74.68 | 8.58 | 0.225 | |
| C01 | GSBRNA2T00130576001 | BnaC01g06010D | 2013 | 670 | 11 | 12 | 75.4 | 7.21 | 0.226 | |
| C07 | GSBRNA2T00099855001 | BnaC07g43840D | 1986 | 661 | 9 | 10 | 74.67 | 8.58 | 0.24 | |
Structure and conserved motif analysis of BnBOR genes
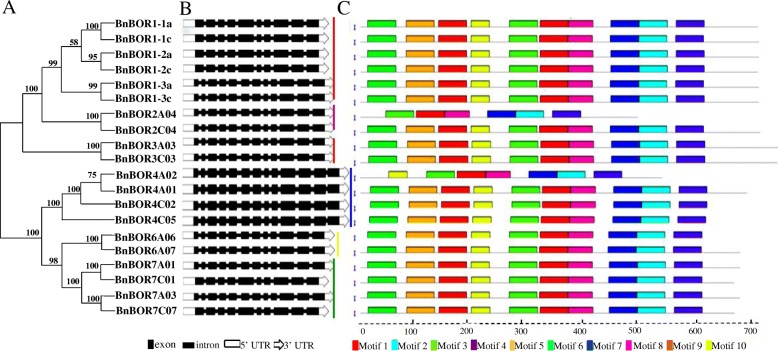
To examine the classification of the BnBORs in detail, independent classification maps were constructed with the 20 BOR family members (Fig. 1a). According to the Arabidopsis BOR1–7 family members, the BnBORs were divided into 6 subfamilies (Fig. 1a and Table 1). The BnBOR1 subfamily consists of six gene members. BnBOR2, BnBOR3 and BnBOR6 have the lowest members with only two genes in each subfamily. The other two subfamilies, BnBOR4 and BnBOR7, contain four members. Additionally, abiding with the classification criteria, no proteins homologous to BOR5 were found in B. napus. Since intron/exon organization and numbers are typical imprints of evolution within gene families, we analysed the BnBOR gene structures by comparing the gDNA sequences with their corresponding coding sequences (Fig. 1b). BnBOR1;1a, BnBOR1;1c (BnaC4.BOR1;1c), BnBOR1;2a, BnBOR1;2c, BnBOR7A03 and BnBOR7C07 contain 9 introns and 10 exons, while BnBOR4A01, BnBOR4A02, BnBOR4C02 and BnBOR4C05 consist of 12 introns and 13 exons. The other genes had 11 introns and 12 exons.
Fig. 1.
Schematic representation of the exon-intron compositions and conserved motifs in the BnBOR genes in Brassica napus. a Neighbour-joining (NJ) phylogenetic tree for BnBORs. b Exon-intron organization of BnBOR genes. The wide black boxes represent exons, the narrow black lines represent introns, the white rectangles represent the 5’-UTR and the white arrows represent the 3’-UTR. c Distribution of conserved motifs in BnBOR proteins. Different motifs are shown by different colours numbered 1–10. Some motifs are highlighted with different coloured boxes with numbers. Lines represent protein regions without a detected motif
To compare the differences in the protein structure, MEME was used to investigate the conserved motifs among the B. napus BOR proteins, revealing a total of 10 conserved motifs (designated motifs 1–10) (Fig. 1c). BnBOR2A04 possesses 6 motifs, except motifs 5, 6, 9 and 10, and BnBOR4A01 does not contain motifs 5, 6 and 9. The remaining motifs, including 1, 2, 3, 4, 7 and 8, constitute the conserved bicarbonate (HCO3−) domain, were identified by the Pfam domains and WebLogo programs, and are present in all BOR family members. The BnBORs are unevenly distributed on 12 of 19 B. napus chromosomes (Fig. 2). The majority of BnBOR genes are located on chromosome arms that are associated with high rates of recombination. Ten BnBOR genes (BnBOR7A01, BnBOR4A01, BnBOR4A02, BnBOR1;3a, BnBOR3A03, BnBOR7A03, BnBOR2A04, BnBOR1;1a, BnBOR1;2a, and BnBOR6A06) are distributed on chromosome AA, including A01, A02, A03, A04, A05, and A06, and nine BnBOR genes (BnBOR7C01, BnBOR4C02, BnBOR1;3c, BnBOR3C03, BnBOR1;2c BnBOR2C04, BnBOR1;1c, BnBOR4C05, BnBOR6C07) are distributed on C01, C02, C03, C04, C05 and C07. Interestingly, three BnBOR genes (BnBOR1;3a, BnBOR3A03a and BnBOR7A03) are distributed on A03, which had an important quantitative trait locus (QTL) for B efficiency in our previous studies [34]. These results indicate that most motifs were distributed in all BOR genes, which correlates with their functional consistency and divergence.
Fig. 2.
Chromosomal location of BOR members in Brassica napus. The gene chromosome location diagram was drawn using the MapInspect software. The size of each chromosome can be estimated from the bottom number (Mb). The 20 BnBORs are located on 12 B. napus chromosomes
Phylogenetic tree for BOR genes in monocotyledons and dicotyledons
To examine the phylogenetic relationships among the BOR proteins in monocotyledons and dicotyledons, an unrooted phylogenetic tree was constructed from the alignment of the codon nucleotide sequences using the Neighbour-Joining method (Fig. 3). As shown in Fig. 3, the BORs could be classified into 2 main groups, which is consistent with monocotyledons and dicotyledons. Compared to monocotyledons, BnBORs from dicotyledons were more closely related to AtBORs (Fig. 3; Additional file 1: Figure S1). The phylogenetic tree and comparative sequence analysis with AtBOR1 revealed that BnBOR1s share 70–90% amino acid identity with the Arabidopsis B efflux transporter BOR1. Additionally, BnBOR2s are AtBOR2 orthologs, which are close to AtBOR1. Furthermore, comparative sequence analysis of the BOR1 homologue was performed with the NCBI BLAST and ClustalW2 servers. The paralogous BnBOR proteins were highly similar to each other, with an amino acid identity/similarity ranging from 71/53% to 100/99% (Additional file 1: Figure S2). These results indicate that BnBORs may function as B efflux transporter similar to AtBORs.
Fig. 3.
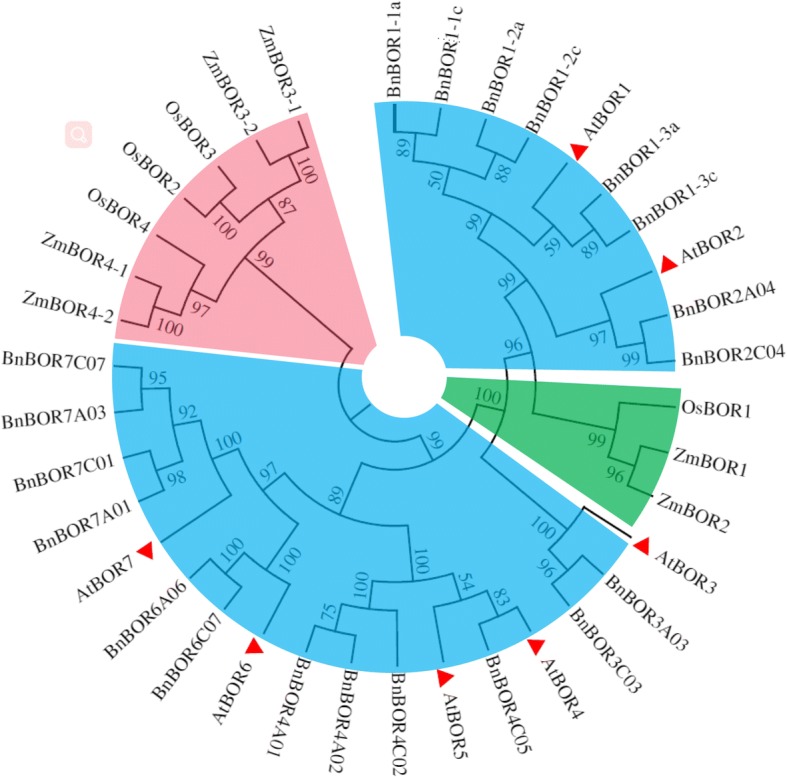
Phylogenetic tree for BORs in monocotyledons and dicotyledons. Arabidopsis BOR genes are highlighted with red triangles. The BnBOR genes are in two clades with blue colour and classified into 6 groups; ZmBOR1 and OsBOR1 were marked in pink and green, respectively
Distinct BnBOR expression profiles in different tissues and in response to boron deficiency
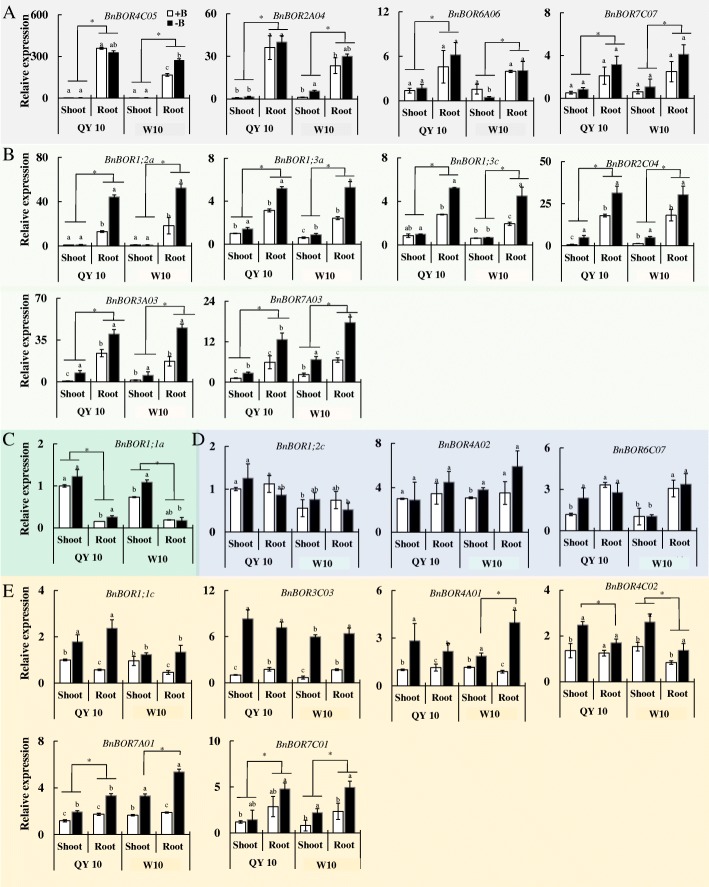
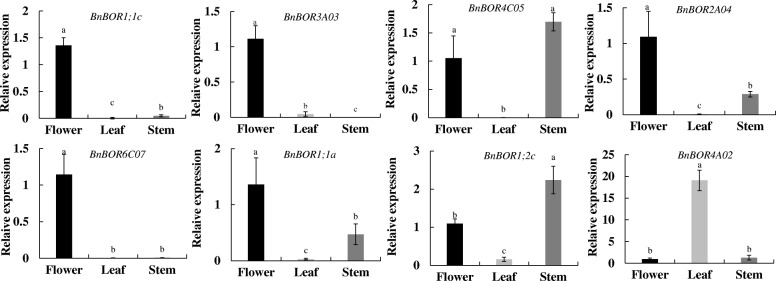
To reveal the expression profiles of BnBORs in different tissues at the seedling stage and in response to B deficiency between two cultivars, qRT-PCR was performed using gene-specific primers for 16 BnBOR genes. BnBOR members showed distinct expression patterns in Brassica napus, which could be divided into four categories based on their tissue expression and responses to B deficiency (Fig. 4). Ten genes (BnBOR4C05, BnBOR2A04, BnBOR6A06, BnBOR7C07, BnBOR1;2a, BnBOR1;3a, BnBOR1;3c, BnBOR2C04, BnBOR3A03 and BnBOR7A03) displayed much higher expression in roots than shoots, especially BnBOR4C05, BnBOR2A04 and BnBOR1;2a (Fig. 4a and b), and most of them were up-regulated by B deficiency (Fig. 4b), except BnBOR4C05, BnBOR2A04, BnBOR6A06 and BnBOR7C07 (Fig. 4a). Interestingly, we found that BnBOR1;1a was mainly expressed in shoots (Fig. 4c). The other BnBOR genes were expressed both in roots and shoots (Fig. 4d and e). Among them, the expression of BnBOR1;2c, BnBOR4A02, and BnBOR6C07 were not affected by external B status (Fig. 4d), and the others were up-regulated by B deficiency, including BnBOR1;1c, BnBOR3C03, BnBOR4A01, BnBOR4C02, BnBOR7A01 and BnBOR7C01 (Fig. 4e). Additionally, BnBOR1;1c and BnBOR1;2c showed higher expression levels in QY10 than in W10. Among them, the expression of BnBOR1;1c was up-regulated by B deficiency and the induction of BnBOR1;1c was more intense than in QY10. B. napus requires a large B supply for plant growth and reproduction, especially for the differentiation of the flowers where B is the most in demand. At reproductive stage, we found BnBOR1;1c, BnBOR3A03, BnBOR6C07 were mainly expressed in flowers and BnBOR1;1a, BnBOR4C05, BnBOR2A04, BnBOR1;2c showed higher expression both in flowers and stems (Fig. 5). Interestingly, the BnBOR4A02 was distinctly expressed in leaves (Fig. 5). The diversity in the expression patterns among these genes may reveal diverse functions of B transporters in allotetraploid B. napus.
Fig. 4.
Expression profile of BnBOR genes in response to B stress in QY10 and W10. a The BnBOR genes that were mainly expressed in roots and were not regulated by the external B status. b The BnBOR genes that were mainly expressed in roots and were up-regulated by low B stress. c The BnBOR genes that were mainly expressed in shoots. d The BnBOR genes that were expressed in both roots and shoots and were not regulated by the external B status. e The BnBOR genes that were expressed in both roots and shoots and were up-regulated by low B stress. Different letters represent statistical significance among different B treatments and cultivars (p < 0.05) in the same organ. Asterisk indicates significance between shoot and root
Fig. 5.
Expression profile of BnBOR genes in different tissues of QY 10 at the reproductive-stage
Enhanced BnBOR1;1c expression alleviates B deficiency in shoots
We further examined whether the BOR protein BnBOR1;1c, which has a relatively high protein similarity to AtBOR1, is equally involved in the transfer of B from roots to shoots. To determine the biological function of BnBOR1;1c in B. napus, we performed a functional complementation test by inducing 35S::BnBOR1;1c in the B-inefficient genotype W10. Three independent lines were used for phenotypic characterization under normal and low B conditions (Fig. 6a and b). W10 displayed a range of typical B deficiency symptoms, including stunted root and shoot growth and dark green and curved leaves under low B (Fig. 6b). The BnaC4.BOR1;1c transgenic plants (OX5–2, OX9–1 and OX10–5) displayed stronger tolerance to low B stress with significantly higher shoot dry weights than W10 under low B (Fig. 6b and c). However, the primary root length was reduced in the transgenic plants under both normal B and low B conditions (Fig. 6b and d). Additionally, the transgenic plants showed significantly higher B concentrations in shoots (Fig. 6e), but the B concentration was lower in roots relative to the wild-type W10 (Fig. 6f), which resulted in a lower R/S ratio for the B concentration (Fig. 6g). These results demonstrate that BnBOR1;1c had a similar biological function with AtBOR1, which is involved in transfer of B from roots to shoots. Importantly, enhanced BnBOR1;1c expression could alleviate B deficiency in shoots by improving their B concentration.
Fig. 6.
Overexpression analysis of BnaC4.BOR1;1c in Brassica napus B-inefficient W10. a and (b) Phenotypes of QY10, W10 and overexpressing lines after 10 d of growth at normal B (NB, 25 μM B) and low B (LB 0.25 μM B), respectively. c Shoot dry weight and (d) primary root length for W10 and the overexpressing lines. The (e) shoot, (f) root and (g) root/shoot B concentrations. Each treatment was repeated three times (n = 3) and error bars denote the standard deviation (SD)
Differential physiological responses to B deficiency between two different B-efficient cultivars
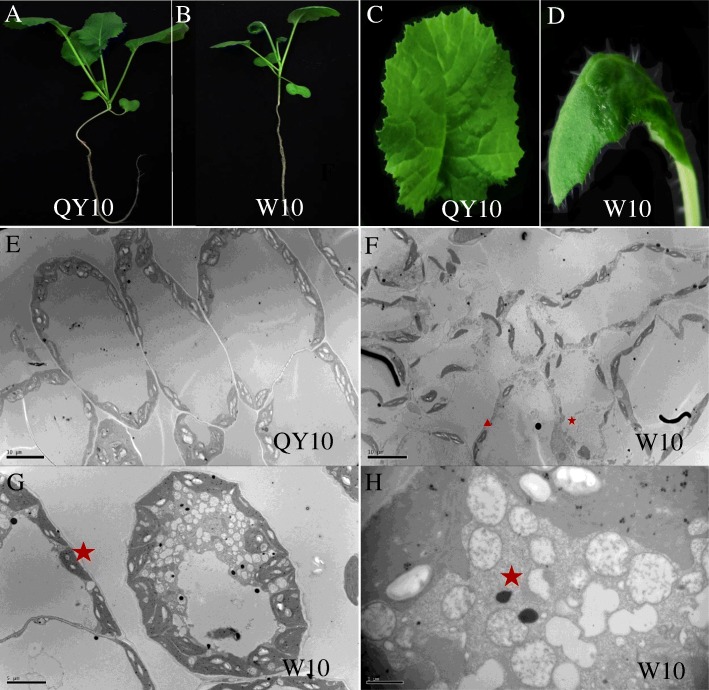
To evaluate the adaptation of QY10 and W10 to B deficiency, 0.25 μM and 25 μM B were used as normal and low B conditions in a hydroponic seedling culture system, respectively. Under low B conditions, the B-efficient cultivar QY10 showed better growth performance than the B-inefficient W10 (Fig. 7a and b). Moreover, W10 displayed crimped leaves and inhibited shoot apices, which were characteristic of B-deficiency symptoms (Fig. 7c and d). To examine the cellular changes underlying the morphologies of the B-deficiency symptoms in W10, we analysed the ultrastructure of the abnormal curved leaves using TEM. Mesophyll cells from QY10 were well organized and structurally intact (Fig. 7e), but the mesophyll cells from W10 appeared unordered and malformed (Fig. 7f). Additionally, the polymerization between cells became significantly weakened and the intercellular gap was enlarged (Fig. 7g). More importantly, we found a large number of lysosomes in W10 cytoplasms, which is viewed as a final destination for endocytic intracellular degradation (Fig. 7h). The lysosomes function as programmed cell death initiators and may lead to B deficiency phenotypes.
Fig. 7.
Phenotypes and transmission electron microscopy (TEM) analysis of the Brassica napus B-efficient cultivar QY10 and B-inefficient cultivar W10. The phenotypes of (a) QY10 and (b) W10 plants under low B (0.25 μM) conditions. The phenotypes of juvenile leaves from (c) QY10 and (d) W10. Mesophyll cells from (e) QY10 and (f-h) W10; red pentagrams mark the lysosomes
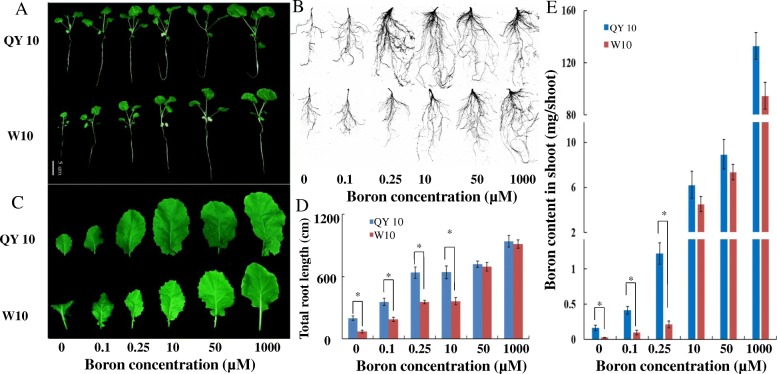
In the B gradient experiment, the growth of QY10 and W10 was facilitated by an increased supply of B nutrition (Fig. 8a). However, under B limitation conditions, QY10 grew significantly better than W10 until the B supply concentration was up to 10 μM, including more extended leaves and developed roots (Fig. 8a-d). Similarly, the shoot B content in QY10 was significantly higher than in W10 (Fig. 8e). When the B supply was increased to more than 10 μM, no significant differences in the B content were detected between the two cultivars. Overall, these results show that the B-efficient cultivar QY10 accumulated more B in shoots under low B stress.
Fig. 8.
Phenotypes of the Brassica napus B-efficient cultivar QY10 and B-inefficient cultivar W10 in response to different B levels. a, b, and c The growth phenotypes of QY10 and W10 grown for 15 d at different B levels. d The total root length of the two cultivars. e The shoot B content. Each treatment was repeated three times (n = 3) and error bars denote the standard deviation (SD). Asterisk indicates significance between two genotypes
Comparison of the BnBOR1;1c promoter sequences between B-efficient and B-inefficient genotypes
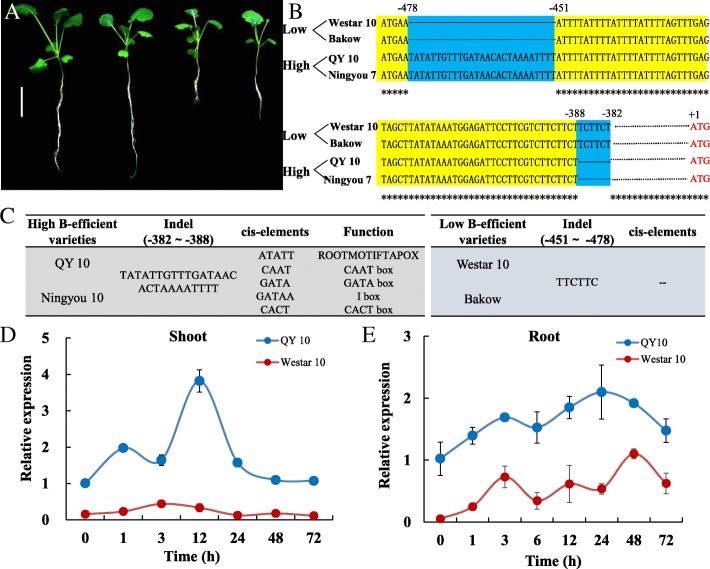
Among the BnBORs, the expression of BnBOR1;1c was up-regulated by B deficiency and the induction of BnBOR1;1c was more intense in QY10 compared to W10. We next isolated and assessed the BnBOR1;1c coding sequences and promoter from the two cultivars using CNS-Genoscope. Interestingly, the coding sequence was the same and two InDels were found in the promoter regions (Additional file 1: Figure S3). One is an insertion of 27 bp at − 478 bp to − 451 bp in the QY10 promoter relative to W10. The other is an insertion of two TTC repeats at − 388 bp to − 382 bp in W10 (Fig. 9b). Interestingly, we found that the two InDels are conserved in the B-efficient genotypes Ningyou 7 and QY10, but they are lost in the B-inefficient genotypes Bakow and Westar 10 (Fig. 9c). The B-efficient and B-inefficient cultivars were identified from 210 Brassica napus cultivars using a B efficiency coefficient (BEC), which is the ratio of the biomass or seed yield under B deficiency to that under normal B supply [35]. Under low B conditions, Ningyou 7 and QY10 grew significantly better than Bakow and Westar 10 (Fig. 9a). Furthermore, four cis-acting elements, YACT, GATA BOX, IBOXCORE, and ROOTMOTIF TAPOX І, were identified in the 27-bp insertion regions in the QY10 promoter using the PLACE website (Fig. 9c). Among these elements, the GATA sequence is a core element in the 35CaMV promoter and ROOTMOTIF TAPOX І (ATATT) is a novel binding site for WRKY transcription factors. Quantitative RT-PCR results verified that BnaC4.BOR1;1c expression levels in the roots and shoots of QY10 plants were significantly greater than those in W10 (Fig. 9d, e). The differential expression of BnaC4.BOR1;1c may contribute to the high B efficiency in QY10 shoots. Additionally, we further analysed the cis-element in the BnaC4.BOR1;1c promoter in QY10 using the online software PlantCARE and found various types of cis-elements, such as stress response-, hormone response-, and development-related elements. These results indicated that the insertion in the BnaC4.BOR1;1c promoter of QY10 contains potential cis-acting regulatory elements, which may be responsible for the higher expression levels in QY10.
Fig. 9.
Expression and promoter analysis of BnaC4.BOR1;1c in B-efficient and B-inefficient cultivars. a Phenotypic performance of B-efficient cultivars (QY 10 and Ningyou 7) and B-inefficient cultivars (W10 and Bakow) that were cultivated in a hydroponic culture under B-limitation conditions (0.25 μM) for 10 d. b Sequence alignment of BnaC4.BOR1;1c promoters from two cultivars. c A 27-bp insertion in the region from − 451 bp to − 478 bp in the QY10 promoter that harbours four important cis-acting elements predicted using PLACE. Expression of BnaC4.BOR1;1c in (d) shoots and (e) roots from QY10 and W10; the plants were grown in 10 μM boric acid for 7 d, then transplanted in 0.25 μM B to analyse the expression after 1, 3, 12, 24, 48, and 72 h of treatment
Discussion
BOR genes in B. napus and their evolution
The anion exchanger bicarbonate (HCO3−) domain related to anion transport has been widely described in bacteria, plants and animals [36]. The conserved domain in the BOR gene family has been reported in a number of plant species, but few have been reported in Brassica napus, an important oil crop cultivated worldwide. In the present study, we performed a comprehensive search for HCO3− domain-containing BOR genes throughout the B. napus genome, and a total of 20 full-length BnBORs were identified. BnBOR gene family members with high homologous conservation in their amino acid coding sequence have similar protein physicochemical parameters and structural properties, and all the members of this family are predicted to be located on the plasma membrane. In this study, these genes were divided into six distinct groups based on domain organization and phylogenetic analysis, which is highly consistent with the results from Arabidopsis BORs. As the A. thaliana genome contains 7 BOR genes, the number of BOR genes in B. napus is about three times that in A. thaliana. B. napus is a recent allopolyploid that originated by combining the intact genomes of B. oleracea and B. rapa and shows 20 BOR genes from the two immediate progenitor species B. rapa and B. oleracea. Copy number expansion of the BOR family in B. napus has primarily occurred through genome duplication events, and the B. napus genome appears to be on the more stable side. Due to the origin and evolutionary independence of the two diploids (B. oleracea and B. rapa) over the past 4.6 MYA [37], the chromosomal locations of BnBORs in the A genome are not completely conserved in homologous regions in the C genome. Tandem and segmental duplication events are the main reasons for gene expansion as organisms adapt to new and changing environments [38]. The BOR genes are unevenly distributed on 12 of 19 B. napus chromosomes, with the majority of the BnBOR genes being located on chromosome arms that are associated with high rates of recombination. The uneven and clustered distribution of BOR genes has also been found in rice and maize [14, 22, 39]. The diversification of BnBOR genes was observed from many aspects, including phylogenesis, genomic structure, as well as chromosome rearrangement. This diversity in the structure of BOR genes is likely to be triggered by gene duplication accompanied by the loss or gain of introns and exons, which presumably contributes to protein functional diversity.
Polyploidy or whole-genome duplication (WGD) plays key roles in the evolution of gene families throughout the evolutionary history of plants, and helps plants adapt to new and changing environments [38]. Divergent expression patterns in different tissue responses to B deficiency were detected among BnBOR paralogs (Fig. 4), indicating the differential roles of each member in the regulation of B nutrition in B. napus. BnBOR4C05, BnBOR2A04 and BnBOR1;2a showed root-specific expression (Fig. 4a and b) and BnBOR1;1a was mainly expressed in shoots (Fig. 4c). Additionally, BnBOR3C03 was strongly induced by B deficiency (Fig. 4e). B requirement for the differentiation of the flowers is much higher than for vegetative growth. In our previous study, we found knockdown of BnaC4.BOR1;1c caused severe inhibition of inflorescence growth only under boron limitation [19]. In this research, we found other BnBORs also showed a higher expression in flowers (Fig. 5), which may exist functional redundancy for inflorescence development by facilitating boron transport to the growing reproductive tissues. All these results reveal a more complex network of B nutrition in B. napus with complex genomes.
Genetic effects of BnBOR1;1c on B efficiency
B-deficiency-induced alterations in gene expression profiles and proteomic profile in C. sinensis and Brassica napus [34, 40–42]. The function of the BOR1 gene has been well-characterized in Arabidopsis, rice, maize and wheat [10, 14]. However, the genetic basis underlying B efficiency in Brassica napus remains poorly elucidated. The allotetraploid rapeseed is extremely sensitive to B deficiency and shows a notable reduction in seed yield and quality with a low B supply [10, 14, 43]. Based on the comparative differential physiological responses to B deficiency between genotypes, we found that the B-efficient cultivar QY10 appears better growth performance (Fig. 7) and accumulates more B in shoots under low B stress (Fig. 8). We examined transgenic rapeseed oil by inducing 35S::BnBOR1;1c in the B-inefficient genotype W10. The transgenic plants showed better growth and higher B contents in shoots under B deficiency than in wild type W10 (Fig. 6), indicating that BnBOR1;1c may function in B transport, similar to AtBOR1, while BnBOR1;1c overexpression conferred a strong tolerance to low B stress in B. napus.
We also found that BnBOR1;1c expression was much higher in both roots and shoots in the B-efficient cultivar QY10 (Fig. 6 and Fig. 9d and e). Interestingly, the coding sequence for BnaC4.BOR1;1c was the same as in QY10 and the B-inefficient cultivar W10, though two InDels were found in the promoter regions between them (Fig. 9b). Furthermore, we identified four significant cis-acting elements among the 27-bp InDels in QY10, including YACT, GATA box, I box, and ROOTMOTIF TAPOX І. These elements are related to mesophyll expression and activating reporter gene expression and involved in light-responsive and root-specific elements. Gowik [44] reported that the YACT motif is a key component in Mem1 (mesophyll expression module 1) found in the promoter of the phosphoenolpyruvate carboxylase (ppcA1) in the C4 plant Flaveria trinervia. The ROOTMOTIF TAPOX was identified for the first time in the rolD promoter in Agrobacterium rhizogenes and has been proven to be responsible for the expression of the gus gene in the roots of transgenic tobacco plants [45]. In fact, the cis-elements present upstream of the target genes can regulate gene expression and alter gene function. Ye et al. reported that an InDel in the Sl-ALMT9 promoter contributes to fruit malate accumulation and enhances aluminium tolerance in tomato [46]. Additionally, InDel analysis in other B. napus genotypes revealed that the 27-bp InDels are conserved between B-efficient (QY10 and Ningyou 7) and B-inefficient (Westar 10 and Bakow) types (Fig. 9b and c). Thus, these InDels can be used for molecular marker-assisted selection of B. napus to improve plant B nutrition. However, the biological roles of these InDels in the BnBOR1;1c promoter require further study.
Conclusions
In conclusion, 20 BnBOR genes were identified in the Brassica napus genome and named according to their phylogenetic relationships. The diversity of expression patterns among these genes may reveal diverse functions in allotetraploid B. napus. By investigating the differential physiological responses to B deficiency, the B-efficient cultivar QY10 showed better growth performance and a higher dry weight and B concentration than the B-inefficient cultivar W10 under B limitation. Among the 20 identified BnBOR genes, the expression of BnaC4.BOR1;1c was much higher in both the roots and shoots of QY10. Moreover, two InDels were found in the promoter regions of the two cultivars, and these InDels are conserved between the B-efficient and B-inefficient genotypes. Importantly, overexpressing BnaC4.BOR1;1c could alleviate B-deficiency in the B-inefficient cultivar W 10. All these results provide comprehensive insights into the BOR family members in B. napus and highlight the diversity of the family members, which could be valuable for further biological function studies focused on BnBOR genes.
Additional file
Figure S1. Phylogenetic tree for BORs in Brassica napus. The BOR phylogenetic tree was generated by MEGA 6.0 with the Neighbour-joining (NJ) method and 1000 replicates bootstraps and based on the amino acid sequences of the 20 BnBOR genes and 7 AtBOR genes. The AtBORs are marked by red diamonds. Figure S2. Identity/similarity matrix for the BnBOR proteins. Amino acid identity and similarity are indicated by the first and second number. Figure S3. BLAST analysis of the promoter region from BnaC4.BOR1;1c in different Brassica napus genotypes. Table S1. Sequences of the primers used for PCR. Table S2. Putative cis-elements in the BnaC4.BOR1;1c promoter region in B. napus. (DOCX 219 kb)
Acknowledgments
Funding
This work was funded by the National Key Research and Development Program of China (Grant No. 2016YFD0100700) and the National Natural Science Foundation of China (Grant No. 31772380) and Fundamental Research Funds for the Central Universities (Project No. 2662017QD039). The funding bodies were not contributed to the design and experiments of the study.
Availability of data and materials
The datasets generated and analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- B
Boron
- BEC
B efficiency coefficient
- BnBOR1s
Boron transporter genes in Brassica napus
- qRT-PCR
Quantitative real-time PCR
- QTL
quantitative trait locus
- QY10
Qingyou 10
- RG-II
pectic polysaccharide rhamnogalacturonan II
- TEM
Transmission Electron Microscopy
- W10
Westar 10
- WGD
whole-genome duplication
Authors’ contributions
FX, HC, and QZ designed the study. QZ and HC prepared the genetic constructs and transformation. HC carried out the nutrient solution experiments. QZ and MH measured the boron concentration. QZ, SW and LS carried out the bioinformatics analysis. QZ and HC drafted the manuscript. All the authors critically revised this manuscript. All the authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Haifei Chen, Email: chenhaifei@webmail.hzau.edu.cn.
Quan Zhang, Email: quanzhang@webmail.hzau.edu.cn.
Mingliang He, Email: bghy@webmail.hzau.edu.cn.
Sheliang Wang, Email: sheliangwang2017@mail.hzau.edu.cn.
Lei Shi, Email: leish@mail.hzau.edu.cn.
Fangsen Xu, Phone: +86-27-87282225, Email: fangsenxu@mail.hzau.edu.cn.
References
- 1.Warington K. The effect of boric acid and borax on the broad bean and certain other plants. Ann Bot. 1923;37:457–466. [Google Scholar]
- 2.O'Neill MA, Ishii T, Albersheim P, Darvill AG. Rhamnogalacturonan II: structure and function of a borate cross-linked cell wall pectic polysaccharide. Plant Bio. 2004;55:109–139. doi: 10.1146/annurev.arplant.55.031903.141750. [DOI] [PubMed] [Google Scholar]
- 3.Hänsch R, Mendel RR. Physiological functions of mineral micronutrients (Cu, Zn, Mn, Fe, Ni, Mo, B, Cl) Curr Opin Plant Bio. 2009;12:259–266. doi: 10.1016/j.pbi.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 4.Brown PH, Shelp BJ. Boron mobility in plants. Plant Soil. 1997;193:85–101. doi: 10.1023/A:1004211925160. [DOI] [Google Scholar]
- 5.Dell B, Huang L. Physiological response of plants to low boron. Plant Soil. 1997;193:103–120. doi: 10.1023/A:1004264009230. [DOI] [Google Scholar]
- 6.Lordkaew S, Dell B, Jamjod S, Rerkasem B. Boron deficiency in maize. Plant Soil. 2011;342:207–220. doi: 10.1007/s11104-010-0685-7. [DOI] [Google Scholar]
- 7.Goldberg S. Reactions of boron with soils. Plant Soil. 1997;193:35–48. doi: 10.1023/A:1004203723343. [DOI] [Google Scholar]
- 8.Miwa K, Fujiwara T. Boron transport in plants: co-ordinated regulation of transporters. Ann Bot. 2010;105:1103–1108. doi: 10.1093/aob/mcq044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noguchi K, Fujiwara T, Chino M, Matsunaga T, Watanabe-Oda H. Absorption and distribution of boron in Arabidopsis thaliana. Springer Netherlands. 1997;76:197–201. [Google Scholar]
- 10.Takano J, Noguchi K, Yasumori M, Kobayashi M, Gajdos Z, Miwa K, et al. Arabidopsis boron transporter for xylem loading. Nature. 2002;420:337–340. doi: 10.1038/nature01139. [DOI] [PubMed] [Google Scholar]
- 11.Takano J, Miwa K, Yuan L, von Wiren N, Fujiwara T. Endocytosis and degradation of BOR1, a boron transporter of Arabidopsis thaliana, regulated by boron availability. Proc Natl Acad Sci U S A. 2005;102:12276–12281. doi: 10.1073/pnas.0502060102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takano J, Tanaka M, Toyoda A, Miwa K, Kasai K, Fuji K, et al. Polar localization and degradation of Arabidopsis boron transporters through distinct trafficking pathways. Proc Natl Acad Sci U S A. 2010;107:5220–5225. doi: 10.1073/pnas.0910744107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kasai K, Takano J, Miwa K, Toyoda A, Fujiwara T. High boron-induced ubiquitination regulates vacuolar sorting of the bor1 borate transporter in Arabidopsis thaliana. J Biol Chem. 2011;286:6175. doi: 10.1074/jbc.M110.184929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakagawa Y, Hanaoka H, Kobayashi M, Miyoshi K, Miwa K, Fujiwara T. Cell-type specificity of the expression of OsBOR1, a rice efflux boron transporter gene, is regulated in response to boron availability for efficient boron uptake and xylem loading. Plant Cell. 2007;19:2624–2635. doi: 10.1105/tpc.106.049015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pérezcastro R, Kasai K, Gainzacortés F, Ruizlara S, Casaretto JA, Peñacortés H. VvBOR1, the grapevine ortholog of AtBOR1, encodes an efflux boron transporter that is differentially expressed throughout reproductive development of Vitis Vinifera L. Plant Cell Physiol. 2012;53:485–494. doi: 10.1093/pcp/pcs001. [DOI] [PubMed] [Google Scholar]
- 16.Cañon P, Aquea F, Arce-Johnson P. Functional characterization of citrus macrophylla BOR1 as a boron transporter. Physiol Plantarum. 2013;149:329. doi: 10.1111/ppl.12037. [DOI] [PubMed] [Google Scholar]
- 17.Leaungthitikanchana S, Fujibe T, Tanaka M, Wang S, Sotta N, Takano J. Differential expression of three bor1 genes corresponding to different genomes in response to boron conditions in hexaploid wheat (Triticum Aestivum L.) Plant Cell Physiol. 2013;54:1056. doi: 10.1093/pcp/pct059. [DOI] [PubMed] [Google Scholar]
- 18.Uraguchi S, Kato Y, Hanaoka H, Miwa K, Fujiwara T. Generation of boron-deficiency-tolerant tomato by overexpressing an Arabidopsis thaliana borate transporter AtBOR1. Front Plant Sci. 2014;5:125. doi: 10.3389/fpls.2014.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Q, Chen H, He M, Zhao Z, Cai H, Ding G. The boron transporter BnaC4.BOR1;1c is critical for inflorescence development and fertility under boron limitation in Brassica napus. Plant Cell Environ. 2017;40:1819–1833. doi: 10.1111/pce.12987. [DOI] [PubMed] [Google Scholar]
- 20.Miwa K, Wakuta S, Takada S, Ide K, Takano J, Naito S, et al. Roles of BOR2, a boron exporter, in cross linking of rhamnogalacturonan II and root elongation under boron limitation in Arabidopsis. Plant Physiol. 2013;163:1699–1709. doi: 10.1104/pp.113.225995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miwa K, Takano J, Omori H, Seki M, Shinozaki K, Fujiwara T. Plants tolerant of high boron levels. Science. 2007;318:1417. doi: 10.1126/science.1146634. [DOI] [PubMed] [Google Scholar]
- 22.Hayes JE, Reid RJ. Boron tolerance in barley is mediated by efflux of boron from the roots. Plant Physiol. 2004;136:3376–3382. doi: 10.1104/pp.103.037028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takano J, Miwa K, Fujiwara T. Boron transport mechanisms: collaboration of channels and transporters. Trends Plant Sci. 2008;13:451–457. doi: 10.1016/j.tplants.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 24.Parker MD, Boron WF. The divergence, actions, roles, and relatives of sodium-coupled bicarbonate transporters. Physiol Rev. 2013;93:803–959. doi: 10.1152/physrev.00023.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rathke GW, Christen O, Diepenbrock W. Effects of nitrogen source and rate on productivity and quality of winter oilseed rape (Brassica napus L.) grown in different crop rotations. Field Crops Res. 2005;94:103–113. doi: 10.1016/j.fcr.2004.11.010. [DOI] [Google Scholar]
- 26.Chalhoub B, Denoeud F, Liu S, Parkin IA, Tang H. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 2014;345:950–953. doi: 10.1126/science.1253435. [DOI] [PubMed] [Google Scholar]
- 27.Sun J, Shi L, Zhang CY, Xu F. Cloning and characterization of boron transporters in Brassica napus. Mol Biol Rep. 2012;39:1963–1973. doi: 10.1007/s11033-011-0930-z. [DOI] [PubMed] [Google Scholar]
- 28.Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bailey TL, Boden M, Buske FA, Firth M, Grant CE, Clementi L. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Y, Wang H, Gilmer S, Whitwill S, Keller W, Fowke LC. Control of petal and pollen development by the plant cyclin-dependent kinase inhibitor ICK1 in transgenic Brassica plants. Planta. 2002;215:248–257. doi: 10.1007/s00425-002-0752-2. [DOI] [PubMed] [Google Scholar]
- 34.Hua Y, Zhang D, Zhou T, He M, Ding G, Shi L, et al. Transcriptomics-assisted quantitative trait locus fine mapping for the rapid identification of a nodulin 26-like intrinsic protein gene regulating boron efficiency in allotetraploid rapeseed. Plant Cell Environ. 2016;39:1601–1618. doi: 10.1111/pce.12731. [DOI] [PubMed] [Google Scholar]
- 35.Yang L, Zhang Q, Dou J, Li L, Guo L, Shi L, et al. Characteristics of root boron nutrition confer high boron efficiency in Brassica napus cultivars. Plant Soil. 2013;371:95–104. doi: 10.1007/s11104-013-1669-1. [DOI] [Google Scholar]
- 36.Romero MF, Chen AP, Parker MD, Boron WF. The SLC4 family of bicarbonate (HCO3−) transporters. Mol Asp Med. 2013;34:159–182. doi: 10.1016/j.mam.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu S, Liu Y, Yang X, Tong C, Edwards D, Parkin IAP. The Brassica Oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat Commun. 2014;5:3930. doi: 10.1038/ncomms4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bowers JE, Chapman BA, Rong J, Paterson AH. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- 39.Chatterjee M, Tabi Z, Galli M, Malcomber S, Buck A, Muszynski M, Gallavotti A. The boron efflux transporter ROTTEN EAR is required for maize inflorescence development and fertility. Plant Cell. 2014;26:2962–2977. doi: 10.1105/tpc.114.125963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marschner H. Marschner’s mineral nutrition of higher. Plants. 2012;89
- 41.Lu YB, Qi YP, Lee J, Guo P, Ye X, Jia MY, Chen LS. Long-term boron-deficiency-responsive genes revealed by cDNA-AFLP differ between Citrus sinensis roots and leaves. Front Plant Sci. 2015;6:585. doi: 10.3389/fpls.2015.00585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang LT, Lu YB, Zhang Y, Guo P, Chen LS. Proteomic profile of Citrus grandis roots under long-term boron-deficiency revealed by iTRAQ. Trees. 2016;30:1057–1071. doi: 10.1007/s00468-015-1344-7. [DOI] [Google Scholar]
- 43.Yang LT, Qi YP, Lu YB, Guo P, Sang W, Feng H, Chen LS. iTRAQ protein profile analysis of Citrus sinensis roots in response to long-term boron-deficiency. J Proteome. 2013;93:179–206. doi: 10.1016/j.jprot.2013.04.025. [DOI] [PubMed] [Google Scholar]
- 44.Udo G, Westhoff P. Cis-regulatory elements for mesophyll specific gene expression in the C-4 plant Flaveria trinervia, the promoter of the C-4 phosphoenolpyruvate carboxylase gene. Plant Cell. 2004;16:1077–1090. doi: 10.1105/tpc.019729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elmayan T, Tepfer M. Evaluation in tobacco of the organ specificity and strength of the rolD promoter, domain a of the 35S promoter and the 35S2 promoter. Transgenic Res. 1995;4:388. doi: 10.1007/BF01973757. [DOI] [PubMed] [Google Scholar]
- 46.Ye J, Wang X, Hu T, Zhang F, Wang B, Li C, Zhang Y. An InDel in the promoter of Al-ACTIVATED MALATE TRANSPORTER9 selected during tomato domestication determines fruit malate contents and aluminum tolerance. Plant Cell. 2017;29:2249–2268. doi: 10.1105/tpc.17.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Phylogenetic tree for BORs in Brassica napus. The BOR phylogenetic tree was generated by MEGA 6.0 with the Neighbour-joining (NJ) method and 1000 replicates bootstraps and based on the amino acid sequences of the 20 BnBOR genes and 7 AtBOR genes. The AtBORs are marked by red diamonds. Figure S2. Identity/similarity matrix for the BnBOR proteins. Amino acid identity and similarity are indicated by the first and second number. Figure S3. BLAST analysis of the promoter region from BnaC4.BOR1;1c in different Brassica napus genotypes. Table S1. Sequences of the primers used for PCR. Table S2. Putative cis-elements in the BnaC4.BOR1;1c promoter region in B. napus. (DOCX 219 kb)
Data Availability Statement
The datasets generated and analysed during the current study are available from the corresponding author on reasonable request.