Abstract
The 5 human (h)KCNE β subunits each regulate various cation channels and are linked to inherited cardiac arrhythmias. Reported here are previously undiscovered protein-coding regions in exon 1 of hKCNE3 and hKCNE4 that extend their encoded extracellular domains by 44 and 51 residues, which yields full-length proteins of 147 and 221 residues, respectively. Full-length hKCNE3 and hKCNE4 transcript and protein are expressed in multiple human tissues; for hKCNE4, only the longer protein isoform is detectable. Two-electrode voltage-clamp electrophysiology revealed that, when coexpressed in Xenopus laevis oocytes with various potassium channels, the newly discovered segment preserved conversion of KCNQ1 by hKCNE3 to a constitutively open channel, but prevented its inhibition of Kv4.2 and KCNQ4. hKCNE4 slowing of Kv4.2 inactivation and positive-shifted steady-state inactivation were also preserved in the longer form. In contrast, full-length hKCNE4 inhibition of KCNQ1 was limited to 40% at +40 mV vs. 80% inhibition by the shorter form, and augmentation of KCNQ4 activity by hKCNE4 was entirely abolished by the additional segment. Among the genome databases analyzed, the longer KCNE3 is confined to primates; full-length KCNE4 is widespread in vertebrates but is notably absent from Mus musculus. Findings highlight unexpected KCNE gene diversity, raise the possibility of dynamic regulation of KCNE partner modulation via splice variation, and suggest that the longer hKCNE3 and hKCNE4 proteins should be adopted in future mechanistic and genetic screening studies.—Abbott, G. W. Novel exon 1 protein-coding regions N-terminally extend human KCNE3 and KCNE4.
Keywords: arrhythmia, KCNQ1, KCNQ4, Kv4.2, potassium channel
Ion channels are complex macromolecules that consist of multiple pore-forming and non-pore-forming subunits in vivo. Voltage-gated potassium (Kv) channels are a numerous and diverse class generated by 40 different α subunits in human tissues that can each come together to form the functional assembly, a tetramer of α subunits. The 40 α subunits are categorized into several subfamilies, and subfamily-specific heterotetramerization commonly occurs, which further diversifies the resultant complexes and the currents they generate (1). In addition, multiple different classes of non-pore-forming (β) subunits, either transmembrane or cytosolic, can coassemble with Kv α subunits to create the considerable complexity required for normal cellular functions of higher animals (2).
The most widely studied group of transmembrane Kv β subunits are the KCNE proteins, also referred to as MinK-related peptides (3, 4), with >1000 published studies on this gene family to date. KCNE proteins are so widely studied because they can exert sometimes dramatic effects on Kv channel function (5–11), are obligate partners for many Kv α subunits in vivo (5, 6, 9–17), can alter Kv channel pharmacology with respect to clinically relevant and investigational drugs (18, 19), and their reach is impressive, each modulating multiple Kv α subunits and, in some cases, other channel types (20, 21). In addition, sequence variants in each of the 5 human (h)KCNE genes are associated with one or more forms of potentially lethal cardiac arrhythmia (22–30) [for recent review, see Crump et al. (31)], and Kcne-knockout mice exhibit a wide range of sometimes dramatic phenotypes (12–17, 32–34), which suggests that additional discoveries of hKCNE-linked diseases in other tissues will follow.
KCNE proteins each consist of a single transmembrane span, an extracellular N-terminal segment, and an intracellular C-terminal segment, with known protein sequences ranging from 103 aa for the shortest (hKCNE3) to 170 aa for the longest (hKCNE4) (31). Here, I report the discovery of additional coding regions that N-terminally extend these 2 proteins by 44 and 51 residues, respectively. I present evidence that the novel longer forms are expressed in multiple human tissues, and demonstrate functional differences between short and long forms of hKCNE3 and hKCNE4.
MATERIALS AND METHODS
Genome database analyses
hKCNE3 and KCNE4 protein sequence searches on the U.S. National Center for Biotechnology Information (NCBI) database revealed putative longer forms of each protein than those currently reported (gene accession numbers are shown in Figs. 1 and 2). Expanding these searches by using the Basic Local Alignment Search Tool (BLAST) to query genomes from multiple species revealed that nucleotide and protein sequences for the predicted longer forms of each subunit were represented in multiple species, increasing confidence in the finding. Other database searches are described in Results.
Figure 1.
Gene structure and predicted protein sequence of full-length KCNE3. A) Gene structure of hKCNE3 as annotated in the NCBI database, showing 2–3 predicted exons in each of 2 different isoforms identified by their accession numbers (left). Thin, colored lines, introns; thick, colored lines: green, exons (light green, noncoding; dark green, coding); purple, entire gene; red, protein-coding exons. Black boxed region is the hKCNE3 isoform with 2 predicted exons encoding full-length hKCNE3. Upper scale is human chromosome 11 base pair numbering (GRCh38.p2 primary assembly). B) Predicted protein sequence for the novel N-terminal portion of hKCNE3. C) Predicted protein sequence for full-length KCNE3 of primates and the shorter KCNE3 predicted for M. musculus. Branched icons, consensus glycosylation sites; P, known PKC phosphorylation site. Consensus arginine amidation site indicated by open black square. Consensus sites predicted by Expasy ScanProsite (http://prosite.expasy.org/scanprosite/). Human gene numbering indicated. TM, transmembrane.
Figure 2.
Gene structure and predicted protein sequence of full-length KCNE4. A) Gene structure of hKCNE4 from NCBI databases showing 2 predicted exons. Thin, colored lines, introns; thick, colored lines: green, exons (light green, noncoding; dark green, coding); purple, entire gene; red, protein-coding exons. Upper scale is human chromosome 2 base pair numbering (GRCh38.p2 primary assembly). B) Predicted protein sequence for the novel N-terminal portion of hKCNE4. C) Predicted protein sequence for full-length KCNE4 of various vertebrates and the shorter KCNE4 predicted for M. musculus. Branched icons, consensus glycosylation sites; P, consensus PKC phosphorylation site. Consensus sites predicted by Expasy ScanProsite (http://prosite.expasy.org/scanprosite/). Human gene numbering indicated. TM, transmembrane.
RT-PCR
The majority of PCR was performed by using premade 48-tissue human cDNA arrays that were prenormalized by the manufacturer (Origene, Rockville, MD, USA) to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression. Because arrays lacked an atria-only cDNA, human atrial RNA (isolated from tissue samples kindly provided by Dr. David Christini, Weill Cornell Medical College, Institutional Review Board Protocol 1206012437) was reverse-transcribed by using a QuantiTect Reverse Transcriptase Kit (Qiagen, Valencia, CA, USA) to yield atrial cDNA. This was also used for primer annealing temperature optimization (21). The PCR primers used are given in Table 1. PCR products (either 30 or 35 cycles) were run on 1–1.5% agarose gels that contained ethidium bromide and were visualized by using a Gbox system with Gbox software (Syngene, Cambridge, United Kingdom). Each gel shown is representative of at least 2 independent PCRs and gel visualizations.
TABLE 1.
PCR primer details
| Primer pair | Name | Sequence, 5′–3′ | Template strand | Product size (bp) |
|---|---|---|---|---|
| KCNE3-1 | KCNE3-F | GCTGAGAGGCAGAGCTTCTA | Plus | 399 |
| KCNE3-1R | GTGTCTTGGTCTTCCACCGT | Minus | ||
| KCNE3-2 | KCNE3-F | GCTGAGAGGCAGAGCTTCTA | Plus | 503 |
| KCNE3-2R | GACGACCTCCCTTAACCGTG | Minus | ||
| KCNE3-3 | KCNE3-F | GCTGAGAGGCAGAGCTTCTA | Plus | 505 |
| KCNE3-3R | GTGACGACCTCCCTTAACCG | Minus | ||
| KCNE3-4 | KCNE3-F | GCTGAGAGGCAGAGCTTCTA | Plus | 318 |
| KCNE3-4R | CTACGCTTGTCCACTTTGCG | Minus | ||
| KCNE3-5 | KCNE3-F | GCTGAGAGGCAGAGCTTCTA | Plus | 323 |
| KCNE3-5R | GGTCACTACGCTTGTCCACT | Minus | ||
| KCNE4-1 | KCNE4-1F | GAACCCTCTTGGACTGGACG | Plus | 850 |
| KCNE4-1R | CAAAGTCAGTGTACGCGCTG | Minus | ||
| KCNE4-2 | KCNE4-2F | TCACCGCTACCTGAAAACCC | Plus | 890 |
| KCNE4-2R | AAGGTTGCTGGCTGATGGAG | Minus |
F, forward primer; R, reverse primer. The same forward primer (KCNE3-F) was used for all 5 KCNE3 primer pairs, with various reverse primers.
Western blotting
For Western blotting of human tissues, premade Western blots (Zyagen, San Diego, CA, USA) were probed, washed, and detected as described below for Chinese hamster ovary (CHO) cell samples. For CHO cell biochemistry, CHO cells were transfected with cDNAs for long KCNE4 (KCNE4L) in pCDNA3.1+ or short KCNE4 (KCNE4S) in pCINeo. Two days post-transfection, cells were lysed in 1 ml lysis buffer (for a 100-mm dish) that contained 150 mM NaCl, 50 mM Tris-HCL (pH 7.4), 20 mM NaF, 10 mM NaVO4, 1 mM PMSF (Thermo Fisher Scientific Life Sciences, Waltham, MA, USA), 1% Nonidet P-40 (Pierce, Rockland, IL, USA), 1% CHAPS [3-((3-cholamidopropyl) dimethylammonio)-1-propanesulfonate; Sigma-Aldrich, St. Louis, MO, USA], 1% Triton X-100 (Thermo Fisher Scientific Life Sciences), and 0.5% SDS (Sigma-Aldrich) with protease inhibitor cocktail (Sigma-Aldrich). Homogenate was subjected to centrifugation (4000 g, 10 min, 4°C), and aliquots of the supernatant were then mixed with sample buffer that contained 5% β-mercaptoethanol, heated at 65°C for 10 min, and loaded onto 4–12% Bis-Tris SDS-PAGE gels (Thermo Fisher Scientific Life Sciences). After electrophoresis, proteins were transferred onto PVDF membranes (Bio-Rad, Hercules, CA, USA). Membranes were incubated overnight at 4°C with 1:1000 dilutions of primary antibodies in 5% dried milk/Tris-buffered saline–Tween 20 (TBST) as follows: α-KCNE3S, in-house rabbit polyclonal antibody raised to the C-terminal domain as previously reported (14); α-KCNE3L, a custom antibody (Genscript, Piscataway, NJ, USA) raised to a peptide based on the KCNE3L-specific N-terminal segment (residues 10–23), sequence PSVRGRRVQRANGR-C; α-KCNE4S, rabbit polyclonal antibody raised to a peptide within the C-terminal domain (residues 136–150), as previously reported (35) (a kind gift from Drs. Daniel Levy and Steve A. N. Goldstein, University of California, Irvine); and α-KCNE4L, a custom antibody (Genscript) raised to a peptide based on the KCNE4L-specific N-terminal segment (residues 17–32), sequence RAQSRTEQKNPLGLD-C. Blots were then incubated for 1–2 h at room temperature with 1:5000 horseradish peroxidase–conjugated goat anti-rabbit IgG secondary antibody (Bio-Rad) in TBST with or without 1% dried milk, followed by chemiluminescent detection by using Luminata Forte chemiluminescence substrate (Millipore, Temecula, CA, USA). Blots were washed for 3 × 10 min in TBST between each step and for an additional 3–5 min in PBS before chemiluminescent detection, then analyzed with a Gbox system using Gbox software (Syngene).
Xenopus laevis oocyte expression
cRNA transcripts encoding hKCNE3L and hKCNE4L were generated by using in vitro transcription (T7 polymerase mMessage mMachine kit; Thermo Fisher Scientific Life Sciences) from synthetic genes subcloned into pCDNA3.1+ with Xenopus laevis β-globin 5′ and 3′ untranslated regions (UTRs) flanking the coding region to enhance translation and cRNA stability after vector linearization. hKCNE3S and hKCNE4S [in pGA1 (22)] and rKv4.2, hKCNQ1, and hKCNQ4 were similarly transcribed from cDNA templates that also incorporated X. laevis β-globin 5′ and 3′ UTRs. Channel subunit cRNA was quantified by spectrophotometry. Defolliculated stage V and VI X. laevis oocytes (Ecocyte Bioscience, Austin, TX, USA) were injected with ≥1 of the cRNAs above (2–10 ng for KCNE subunits, 2 ng Kv4.2, 2.5–10 ng KCNQ1, 10 ng KCNQ4/oocyte). Oocytes were incubated at 16°C in ND96 solution that contained penicillin and streptomycin, with daily washing, for 2–3 d before 2-electrode voltage-clamp (TEVC) electrophysiology.
TEVC
TEVC recordings were performed at room temperature using an OC-725C amplifier (Warner Instruments, Hamden, CT, USA) and pClamp8 software (Molecular Devices, Sunnyvale, CA, USA). Oocytes were bathed in a small-volume oocyte bath (Warner Instruments) and viewed with a dissection microscope. Bath solution was (in mM): 96 NaCl, 4 KCl, 1 MgCl2, 1 CaCl2, 10 HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid; pH 7.6] (bath chemicals were from Sigma-Aldrich). TEVC pipettes were of 1–4 MΩ resistance when filled with 3 M KCl. For quantification of current–voltage relationships, currents were recorded in response to a voltage protocol that consisted of pulses between −120 or −80 mV and 40 or 60 mV at 20-mV intervals. In the case of KCNQ1 and KCNQ4, prepulses were each followed by a tail pulse to −30 mV from a holding potential of −80 mV. For quantification of Kv4.2 steady-state inactivation, oocytes were held at −100 mV and prepulsed to voltages between −120 and 0 mV, followed by a tail pulse to +40 mV. TEVC data analysis was performed with pClamp8 and Origin 6.1 (OriginLab, Northampton, MA, USA). Values are stated as means ± sem. Normalized tail currents for Kv4.2 steady-state inactivation and KCNQ1 and KCNQ4 G/V relationships were plotted vs. prepulse voltage and fitted with a single Boltzmann function according to: g = (A1 – A2)/{1 + exp[(V1/2 – V)/Vs]} + A2, where g is the normalized tail conductance, A1 is the initial value at −∞, A2 is the final value at +∞, V1/2 is the half-maximal voltage of activation, and Vs the slope factor. Kv4.2 current inactivation curves were fitted with a standard (zero-shift) single exponential decay function with Chebyshev 4-point smoothing filter. Where appropriate, currents were compared with one another using 1-way ANOVA to assess statistical significance (P < 0.05). If multiple comparisons were performed, a post hoc Tukey’s HSD test was performed after ANOVA.
RESULTS
Genome databases predict N-terminally extended hKCNE3 and hKCNE4 protein products
Genomic organization of the hKCNE family was previously determined, with exon boundaries identified and maps established for all 5 genes, each containing a single protein-coding exon. Furthermore, additional exons were described in KCNE genes 1–4, but not KCNE5, that were predicted to generate 5′ UTR but not code for protein. In addition, splice variants were identified in KCNE1 and KCNE3 that generated alternative transcription start sites (2 variants per gene) and were predicted to alter the 5′ UTR but not the translated protein product (36).
Here, analyzing the NCBI protein database, 2 alternative predicted protein products were identified each for hKCNE3 and hKCNE4. This was followed by further analysis of multiple human genomes on the 1000 Genomes Browser (http://browser.1000genomes.org/Homo_sapiens/Info/Index) and the genomes of other species by using BLAST searching of NCBI databases with nucleotide and protein sequences for either form of hKCNE3 and hKCNE4. These analyses predicted that primate KCNE3 genes contained a novel translation initiation start site on exon 1 that is in-frame with the currently known hKCNE3 coding region, which resulted in an additional 44 aa N-terminal to the established start methionine encoded by part of exon 1 and the 5′ end of exon 3, with the putative exon 2 either spliced out or incorrectly identified as an exon (Fig. 1A–C). The hKCNE3 protein would thus be extended from 103 to 147 aa and of interest, bears a consensus amidation site not present in the predicted KCNE3 proteins of other primates analyzed, Cercocebus atys (Sooty mangabey) and Macaca mulatta (Rhesus macaque). In contrast to primate KCNE3 proteins, Mus musculus Kcne3 is not predicted to encode the additional N-terminal residues (Fig. 1C) because it lacks an ATG start site at the appropriate position in exon 1.
I also discovered a novel translation initiation start site on exon 1 of hKCNE4, in-frame with the known hKCNE4 coding region, which is predicted to generate an additional 51 residues at the N-terminal of hKCNE4, encoded by the 3′ end of exon 1 and the 5′ end of exon 2 (Fig. 2A, B), which extends the length of hKCNE4 from 170 to 221 aa. The longer form of KCNE4 is widely represented among vertebrates and contains an additional consensus glycosylation site in the novel N-terminal segment; however, the longer form of KCNE4 is predicted to be absent from M. musculus because mouse Kcne4 lacks an ATG start site in exon 1 (Fig. 2C).
Experimental validation of database predictions of extended hKCNE3 and hKCNE4 transcripts
RT-PCR was used to confirm existence of full-length transcripts covering the predicted novel coding region spanning both hKCNE3 exon 1 and 3 (Fig. 3A) and hKCNE4 exon 1 and 2 (Fig. 4A). Amplification of human cDNAs from a 48-tissue array prenormalized to GAPDH by the manufacturer (Origene) confirmed that hKCNE3 exon 1 and 3–spanning transcript (omitting exon 2; Fig. 3) and hKCNE4 exon 1 and 2–spanning transcript (Fig. 4) are expressed in multiple human tissues by using multiple primer pairs and confirmatory sequencing of amplicons for each. Expression patterns matched and expanded on those previously described for each gene. Thus, full-length (dual exon–spanning) hKCNE3 transcript was most readily detectable in the lower gastrointestinal tract—colon, duodenum, small intestine, and rectum—and was also detected in the atria, among other tissues (Fig. 3B). Full-length hKCNE4 transcript was most readily detected in the uterus and was also detected in the atria as well as in several other tissues (Fig. 4B). A primer pair designed to anneal to KCNE3 exons 2 and 3 failed to amplify any products on the 48-tissue array, also using 35 cycles (data not shown), potentially suggesting that hKCNE3 exon 2 RNA is absent or rarely transcribed.
Figure 3.
hKCNE3L transcript is expressed in multiple human tissues. A) Primer pairs used to verify expression of exon 1 and 3–spanning full-length hKCNE3 (hKCNE3L) transcript in human tissues, with annealing sites on exons 1 and 3 and product sizes indicated. B) Image of agarose gels showing the 399-bp hKCNE3L amplicon generated by PCR by using primer pair 1 from cDNA in human 48-tissue array. Image of agarose gel showing the various hKCNE3L amplicons with sizes as predicted (A, right), generated by using primer pairs 1–5, and human atrial cDNA. Each gel is representative of at least 2 independent PCRs and gel visualizations.
Figure 4.
hKCNE4L transcript is expressed in multiple human tissues. A) Primer pairs used to verify expression of exon 1 and 2–spanning full-length hKCNE4 (hKCNE4L) transcript in human tissues, with annealing sites on exons 1 and 2 indicated. B) Image of agarose gels showing the 850-bp hKCNE4L amplicon generated by PCR by using primer pair 1 from cDNA in human 48-tissue array (left). Image of agarose gel showing the 890-bp hKCNE4L amplicon with sizes as were predicted in panel A, generated by using primer pair 2, and human atrial cDNA (center). Image of agarose gel showing the 890-bp hKCNE4L amplicon with sizes as predicted (A), generated by using primer pair 2, and human uterus cDNA (right). Each gel is representative of at least 2 independent PCRs and gel visualizations.
Full-length predicted coding regions each contain adequate Kozak consensus sequences (AUG, and A/G in the −3 position); each also contains a G at the −6 position, which is also thought to facilitate translation initiation (37). Moreover, the previously accepted exon 2 start-site for hKCNE4 is GCCTCAATGCTG, which is considered a weaker Kozak sequence than the newly identified exon 1 start-site, because the former contains neither the G at +4 nor the A/G at −3 (Fig. 5A). Furthermore, exon 1 sequences for both hKCNE3 (Fig. 5B, C) and hKCNE4 (Fig. 5D) contain strong transcription start sites and promoter binding sites and motifs, as predicted by using the Swiss Institute of Bioinformatics, Eukaryotic Promoter Database (http://epd.vital-it.ch/). In sum, these features increased confidence that the exon 1 consensus start sites are functional, which I next tested directly in vitro and ex vivo.
Figure 5.
In silico promoter analysis of hKCNE3 and hKCNE4. A) Alignment of consensus start sites for human hKCNE3S, hKCNE3L, hKCNE4S, and hKCNE4L. ATG start sites underlined; Kozak sequence features and other features considered important for translation in bold. B) In silico analysis for hKCNE3 performed by using Swiss Institute of Bioinformatics Eukaryotic Promoter Database New (EPDnew; http://epd.vital-it.ch/). Features identified include human promoter binding region (bright blue), CpG islands (green), transcription start sites (beige), and transcription factor binding sites (black). C) In silico analysis for hKCNE4 performed by using EPDnew. Features identified include human promoter binding region (bright blue), CpG islands (dark green), transcription start clusters (blue line), histone H3K4me3 (trimethylated) binding region (bright green), and RNA polymerase II (Pol III) binding site.
Experimental validation of extended hKCNE3 and hKCNE4 protein expression and function
Human tissue lysates were next probed for KCNE3 expression by Western blot using 2 rabbit polyclonal antibodies. α-KCNE3-CT antibody was raised to the C-terminal domain common to both the 103-residue short form of KCNE3 (designated here KCNE3S) and the novel, 147-residue form (designated here KCNE3L). The α-KCNE3L antibody was raised to an N-terminal epitope only present in the novel longer form (KCNE3L). The short form of KCNE3 protein that is expressed in mice migrates much more slowly (∼37 kDa) in SDS-PAGE than would be predicted solely from its 103-residue length (∼14 kDa), primarily because of glycosylation (3 sites) (5, 14, 38). Deglycosylation eliminates the 37-kDa band and leaves a single band for mouse KCNE3 at 14 kDa (38). The newly discovered portion of hKCNE3 does not contain additional consensus glycosylation sites (Fig. 1B). On the basis of the native migration pattern for mouse KCNE3, I therefore predicted a migration distance for hKCNE3L of 45–55 kDa. Strikingly, in human colon—in which KCNE3 is known to be enriched on the basis of previous mouse studies (14, 38)—and esophagus, the highly predominant band detected by α-KCNE3L antibody was at a migration distance close to the 50-kDa marker, which corresponded to the expected migration distance for hKCNE3L (Fig. 6A). The α-KCNE3-CT antibody detected similar bands at 50 kDa in colon and esophagus and also detected bands at 37 kDa in the stomach, colon, and intestine. In the case of the stomach, dense bands were also visible at 30 and 24 kDa (Fig. 6B). Results suggest that in tissues in which KCNE3 is thought to be functionally important, hKCNE3L constitutes a major isoform. In addition, hKCNE3S is also potentially expressed in stomach, intestine, and colon, but was not detectable in esophagus.
Figure 6.
hKCNE3L protein expression in human tissues. A) Western blot showing detection of 50-kD band in human colon and esophagus by using anti-KCNE3L antibody. B) Western blot showing detection of 50-kD band in human colon and esophagus and faster-migrating bands in several tissues by using anti–KCNE3-CT antibody.
I next studied expression of the hKCNE4 protein in human tissues by Western blot analysis using rabbit polyclonal antibodies raised to epitopes either on the C-terminal domain (α-KCNE4-CT antibody) common to both the 170-residue short form of KCNE4 (designated here KCNE4S) and the novel, 221-residue form (designated here KCNE4L), or to an N-terminal epitope (α-KCNE4L antibody) only present in KCNE4L. Previous studies showed that KCNE4S, the previously accepted form of KCNE4, migrates at a distance corresponding to the 25 kDa marker, whereas native rat KCNE4, which I now predict to be KCNE4L, runs at a migration distance corresponding to ∼30 kDa (35). Here, the α-KCNE4L antibody detected bands at 25 kDa in several human tissue lysates separated by SDS-PAGE, a strong band at ∼30 kDa in kidney, and a weaker ∼30-kDa band in uterus. The α-KCNE4-CT antibody detected strong bands at ∼30 kDa in kidney, thymus, and uterus and only very faint bands at 25 kDa in all 5 tissues tested (Fig. 7A). Because the α-KCNE4L antibody cannot detect KCNE4S, the 25-kDa band it detects may either by an immature form or nonspecific. The presence of an ∼30-kDa band detected by one or both antibodies strongly suggested that hKCNE4L is expressed in human kidney, thymus, and uterus, and the relatively low intensity of the 25-kDa band when probing with α-KCNE4-CT antibody suggests that hKCNE4L is the predominant and, possibly, sole hKCNE4 isoform in the tissues tested.
Figure 7.
hKCNE4L protein expression in human tissues. A) Western blots showing detection of ∼30-kD band in human kidney and, faintly, in uterus by using anti-KCNE4L antibody. A 25-kD band, possibly nonspecific, is also visible in all tissues tested (top). Western blots showing detection of ∼30-kD band in human kidney, thymus, and uterus by using anti–KCNE4-CT antibody (bottom). B) Western blots showing detection using anti-KCNE4L antibody of ∼30-kD band doublet in lysates of CHO cells transfected with hKCNE4L; band was absent from lysates of CHO cells transfected with hKCNE4S and from lysates of nontransfected CHO cells (top). Western blots showing detection using anti–KCNE4-CT antibody of ∼30-kD band doublet in lysates of CHO cells transfected with hKCNE4L and of ∼25-kD band doublet in lysates of CHO cells transfected with hKCNE4S; band was absent from lysates of nontransfected CHO cells (bottom).
To further test these conclusions, CHO cells that were transfected with cDNAs that corresponded to hKCNE4S or hKCNE4L were lysed, separated by SDS-PAGE, and probed with either antibody. Cells transfected with hKCNE4S cDNA expressed a protein recognized only by the α-KCNE4-CT antibody, with 2 dense bands at 24–26 kDa and a much fainter band at 20 kDa, which may reflect nonglycosylated and monoglycosylated forms [KCNE4S has one N-glycosylation site (35)] and another form with alternative post-translational modification—hKCNE4S has a consensus PKC site (Fig. 2B) and multiple consensus myristoylation sites (not shown). CHO cells that were transfected with hKCNE4L expressed a protein recognized by either antibody, with a predominant doublet at 28–30 kDa and much fainter bands at 25 and 20 kDa (Fig. 7B). This band pattern is consistent with the mature (diglycosylated) form of hKCNE4L migrating at ∼30 kDa that I also detected in human tissues, and with 3 other forms (non- and monoglycosylated, and, possibly, one lacking additional post-translational modification as discussed above for hKCNE4S) migrating at 28, 25, and 20 kDa, which is consistent with the novel portion of hKCNE4L that contains an additional consensus glycosylation site (Fig. 2B). Of interest, it is possible that the 2 higher-weight KCNE4L bands that run as a doublet close to 30 kDa in Western blots from CHO cell lysates (Fig. 7B) are differentially detected by α-KCNE4L and α-KCNE4CT antibodies in tissue lysates, as the most prominent band detected by the latter in the human tissues analyzed seems to correspond to the heavier band of the CHO cell lysate doublet, whereas the band detected by the α-KCNE4L antibody corresponds more to the migration distance of the lighter band of the CHO cell lysate doublet (Fig. 7A, B).
Electrophysiologic analysis of functional effects of full-length hKCNE3 and hKCNE4
cRNAs that corresponded to the novel hKCNE3L and hKCNE4L transcripts were injected into X. laevis oocytes with cRNAs that encode known KCNE3 and KCNE4 Kv channel α subunit partners Kv4.2 (KCND2), Kv7.1 (KCNQ1) and Kv7.4 (KCNQ4), and functional effects were compared with those of hKCNE3S and hKCNE4S by using TEVC electrophysiology. hKCNE3S was previously found to strongly inhibit the fast-inactivating Kv4.2 channel, which generates transient outward current in musine auditory neurons (39) and human cardiac myocytes (40), whereas rat KCNE4S was previously found to slow Kv4.2 inactivation (41). Here, I recapitulated these effects with hKCNE4L, but found that hKCNE3L was ineffective at inhibiting rKv4.2 and, instead, slowed its inactivation by ∼43% (Fig. 8A–C). As previously reported for rat KCNE4S (41), both hKCNE3L and hKCNE4L right-shifted the voltage dependence of Kv4.2 steady-state inactivation (Fig. 8D, E).
Figure 8.
Functional effects of long and short forms of hKCNE3 and hKCNE4 on rKv4.2. A) Exemplar current traces recorded from Xenopus oocytes expressing rat Kv4.2 alone (n = 36) or with hKCNE3L (E3L; n = 19), hKCNE3S (E3S; n = 16), hKCNE4L (E4L; n = 19), or hKCNE4S (E4S; n = 10). Voltage-clamp protocol (top right). B) Mean ± sem peak current–voltage relationship for currents and n values are same as in panel A. C) Mean ± sem τ of fast inactivation (single exponential fit) for currents and n values are same as in panel A. D) Exemplar current traces recorded from Xenopus oocytes expressing rat Kv4.2 alone (n = 36) or with hKCNE3L (E3L; n = 15) or KCNE4L (E4L; n = 16) by using steady-state inactivation protocol (left). Tail current time point used for quantifying available channels indicated by arrow. E) Mean ± sem for voltage dependence of steady-state inactivation (plotted as fraction of channels available at arrow in panel D vs. voltage) for currents and n values are same as in panel D. **P < 0.01 vs. Kv4.2 alone current at +40 mV; other groups P > 0.05 vs. Kv4.2 alone (B). **P < 0.01 vs. all other groups; other comparisons P > 0.05 after pairwise analysis by 1-way ANOVA followed by Tukey’s HSD test (C).
KCNE3 forms channels with KCNQ1 in the basolateral membranes of epithelia in the lower intestinal tract, and hKCNE3S converts KCNQ1 to a constitutively active K+ channel with an essentially linear current–voltage relationship across the physiologic range (10). Here, I recapitulated this effect and also found that hKCNE3S augments KCNQ1 outward current. hKCNE3L similarly converted KCNQ1 to a constitutively active K+ channel but, in contrast, concomitantly reduced outward current amplitude (Fig. 9A, B). Comparing normalized G/V relationships measured from tail currents, KCNQ1–hKCNE3L channels retained marginally more voltage-dependence, exhibiting reduced fractional current at −120 mV, compared with KCNQ1-hKCNE3S (Fig. 9C). hKCNE4S strongly inhibits KCNQ1 (42, 43), recapitulated here with inhibition of >80%—and some of the 0.5-μA current remaining is likely to be endogenous oocyte current, which is typically ∼0.2 μA under these recording conditions. In contrast, hKCNE4L inhibition of KCNQ1 was limited to only ∼40–45% (Fig. 9A–C). This lower efficacy of inhibition is unlikely to have arisen from insufficient expression of hKCNE4L, because similar fractional current inhibition at +40 mV was observed for the same quantity of hKCNE4L cRNA injected per oocyte (10 ng) with KCNQ1 expressed from 2.5 ng cRNA per oocyte (42% inhibition) vs. KCNQ1 expressed from 10 ng cRNA per oocyte (45% inhibition; mean currents shown in Fig. 9D).
Figure 9.
Functional effects of long and short forms of hKCNE3 and hKCNE4 on hKCNQ1. A) Exemplar current traces recorded from Xenopus oocytes expressing hKCNQ1 alone (2.5 ng cRNA per oocyte; n = 17) or with 10 ng cRNA per oocyte hKCNE3L (E3L; n = 11), hKCNE3S (E3S; n = 10), hKCNE4L (E4L; n = 11), or hKCNE4S (E4S; n = 24). Voltage-clamp protocol (top). Zero current level indicated by dashed line. B) Mean ± sem peak current–voltage relationship for currents and n values are same as in panel A. C) Mean ± sem normalized G/V relationship measured at beginning of tail pulse for currents and n values are same as in panel A; symbols are same as in panel B. KCNQ1-E4S values were omitted because low currents precluded accurate tail current measurements. D) Mean ± sem peak current at +40 mV for KCNQ1 (2.5 ng vs. 10 ng cRNA per oocyte) alone (n = 10–17) or with coinjection of 10 ng/oocyte KCNE4L (E4L) cRNA (n = 11). ***P < 0.0005 (B). ***P < 0.00001 (D).
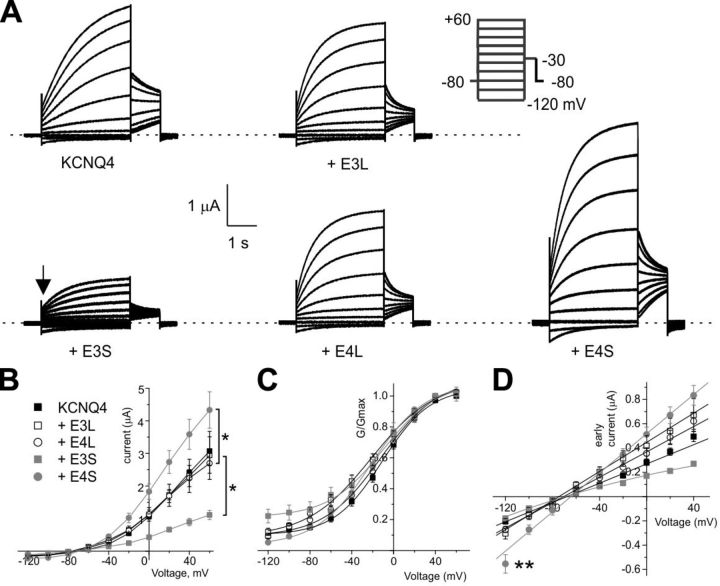
hKCNE3S was previously found to inhibit KCNQ4 activity (10), whereas hKCNE4S was found to augment KCNQ4 current (44, 45). These effects were recapitulated in the present study (Fig. 10A, B). In contrast, hKCNE3L and hKCNE4L did not affect KCNQ4 current magnitude (Fig. 10B). hKCNE3S-KCNQ4 currents exhibited relatively higher fractional current at hyperpolarized potentials as measured in the tail current, but this most likely arises from linear leak contributing relatively more to the tail because of the low current amplitude for this subunit combination (Fig. 10B, C). Of note, hKCNE4S–KCNQ4 channels produced a prominent early current at −120 and −100 mV, measured immediately after (see arrow in Fig. 10A) switching from holding potential (−80 mV). Aside from hKCNE3S–KCNQ4 channels, the early current–voltage relationship for KCNQ4 alone or with the KCNEs tested was essentially linear (Fig. 10D), in contrast to the outward-rectifying late current measured at the end of the test pulse (Fig. 10B). The early current deactivates during the first second when the oocyte is held at −120 mV, but it is evidence that KCNQ4 channels, especially those modulated by hKCNE4S, are active at the holding potential and could therefore influence the resting membrane potential of cell types in which they are expressed.
Figure 10.
Functional effects of long and short forms of hKCNE3 and hKCNE4 on KCNQ4. A) Exemplar current traces recorded from Xenopus oocytes expressing hKCNQ4 alone (n = 35), or with hKCNE3L (E3L; n = 16), hKCNE3S (E3S; n = 12), hKCNE4L (E4L; n = 38), or hKCNE4S (E4S; n = 12). Voltage-clamp protocol (top right). Zero current level indicated by dashed line. B) Means ± sem peak current–voltage relationship for currents and n values are same as in panel A. C) Mean ± sem normalized G/V relationship measured at beginning of tail pulse for currents and n values are same as in panel A. D) Mean ± sem early current (recorded at position shown by arrow in panel A) for currents and n values are same as in panel A; symbols are same as in panels B and C. *P < 0.05. ***P < 0.0001 vs. early current at −120 mV for KCNQ4 alone, other groups not significantly different from KCNQ1 alone.
DISCUSSION
The KCNE proteins are the most highly studied regulatory β subunits of Kv channels and they have also been shown to modulate the function of hyperpolarization-activated, monovalent cation-nonselective channels that provide pacemaking activity in the heart and brain (46). KCNE2 has even recently been shown to regulate l-type calcium channels (47). As a family, KCNE proteins are ubiquitously expressed, with coverage between them of most, if not all, tissues and in both excitable and nonexcitable cells. Each KCNE isoform is expressed in multiple human tissues and can regulate multiple types of Kv channel α subunits, at least in vitro. Indeed, proteins such as KCNE2 can be viewed as master regulators of cellular excitability because they can modulate so many channel types and multiple facets of channel biology, for example, gating, trafficking, localization, and even α subunit composition (48).
When we originally cloned KCNE4 by BLAST searching using residues important for KCNE1 function, we discovered the mouse Kcne4 gene but not the human isoform (22). Others then cloned the hKCNE4 gene on the basis of the published mouse sequence (49) and, understandably, did not expect that the human isoform would be so much longer than that of the mouse because of an additional coding exon. In the case of KCNE3, we and others uncovered only the short form of the human gene, again missing the exon 1–encoded portion (10, 22). Evolutionarily, it is extremely interesting that M. musculus lacks the start site required to generate protein from KCNE4 exon 1, particularly when other lower animals, such as reptilians, are predicted to translate the longer form (although I have not experimentally confirmed this), and the prairie vole, which exhibits similar heart rate to the house mouse, is also predicted to express KCNE4L. Strikingly, the chinchilla KCNE4 sequence even lacks the accepted start methionine for KCNE4S, according to the NCBI sequence, which suggests it can only express KCNE4L (Fig. 2B). In the case of KCNE3, the longer form may be a primate or higher mammal adaptation; I have thus far only found the shorter form in genome databases covering nonprimates (Fig. 1 and data not shown), but these searches are by no means exhaustive. As more α-KCNE channel functional pairings are compared between short and long forms of KCNE3 and KCNE4, the picture may emerge of how the relative modulatory effects of each isoform shape aspects of physiology unique to, for example, primates vs. lower mammals.
KCNE3 is also quite promiscuous and is expressed in numerous tissues, predominantly the lower gastrointestinal tract but also in human atria (Fig. 3). Native Western blots offer tantalizing evidence that both KCNE3S and KCNE3L might be expressed in vivo, perhaps depending on splice variation. By using primers directed toward amplifying the region between hKCNE3 exons 1 and 3, I could only detect transcripts that lacked exon 2, but it is possible that hKCNE3S is encoded by transcripts that are generated by either exons 2 and 3, or exon 3 alone (i.e., with exons 1 and/or 2 spliced out), which would not have been detected by using my PCR strategy. The short form of KCNE3 has been reported to modulate KCNQ1, KCNQ4, KCNQ5, hERG, Kv2.1, Kv3.1, Kv3.2, Kv3.4, Kv4.2, Kv4.3, and Kv12.2 (21).
Here, in an initial functional screen, the additional KCNE3 residues found in primates are shown to modify its effects on rKv4.2, which essentially swaps inhibition for slowed inactivation (Fig. 8). The longer form of hKCNE3 strongly modulated KCNQ1 voltage dependence, but was unable to inhibit KCNQ4 in contrast to hKCNE3S (Figs. 9 and 10). Thus, long and short forms of hKCNE3 differ in both their α subunit specificity and in the specific effects they can have on a particular α subunit that they both modulate. Analogously, hKCNE4L retained the ability, like hKCNE4S, to slow rKv4.2 inactivation, but was less inhibitory to KCNQ1 and lost the capacity to augment KCNQ4 current (Figs. 8–10). KCNE4 is also quite widely expressed (Fig. 4) and its previously reported modulatory capacity includes regulation of Kv4.2 and Kv4.3 inactivation kinetics as well as inhibition of Kv1.1 (KCNA1), Kv1.3 (KCNA3), KCNQ1, and even the Ca2+-activated BK potassium channel (35, 41, 42, 50). In contrast, KCNE4 augments Kv1.5 (KCNA5) activity in CHO cells and in native mouse myocytes (32) and also augments, and left-shifts the voltage dependence of activation of, KCNQ4 in vitro and in rat mesenteric artery smooth muscle cells (45). Future studies will be directed toward a fuller comparison of the functional effects of the long forms vs. the short forms of hKCNE3 and hKCNE4 upon their full repertoire of α subunit partners, including the possibility that longer forms may be able to regulate α subunits that the short forms cannot.
Sequence variants in hKCNE3 and hKCNE4 genes are associated with inherited cardiac arrhythmias. In particular, it will be of interest to determine whether the functional effects of nonsynonymous point mutations altering the sequence of the KCNE3 extracellular segment, which are associated with Brugada syndrome/Long QT syndrome (hKCNE4S-T4A) and atrial fibrillation (hKCNE4S-V17M and R53H) (24–26), are altered by the novel segment introduced nearby into the extended N-terminal region of hKCNE3L. In the case of hKCNE4, a single variant in the intracellular segment (KCNE4S-E145D) is associated with increased incidence of atrial fibrillation (29, 30), the functional implications of which could also conceivably be affected by the longer extracellular region of hKCNE4L, if it, for example, mediates altered partnering capabilities or allosteric effects. Discovery of the new portions of hKCNE3 and hKCNE4 also may suggest re-evaluation of previous genetic screens in cases in which exon 1 was not examined or in which exon 1 nucleotide sequence variants were identified but not published because they were not considered to lie within the coding region.
Finally, discovery of extended forms of hKCNE3 and hKCNE4 proteins opens up the possibility of variable isoforms of these 2 proteins in vivo, depending on splice variation, which would add yet another level of complexity to this fascinating gene family, and is something to be examined more fully in future work. Previously discovered KCNE1 and KCNE3 splice variants were not considered to alter protein sequence but, rather, the UTRs (36). A thorough examination of KCNE1, KCNE2, and KCNE5 in genome databases during the present analysis did not reveal predicted longer forms of these proteins, but to be absolutely certain, a comprehensive analysis of all possible tissues will be required.
Acknowledgments
The author is grateful for helpful discussions with Dr. Alex Mercer, Daniel Neverisky (University of California, Irvine) and Dr. Nancy Carrasco (Yale University, New Haven, CT, USA). The author thanks Dr. David Christini and Lala Tanmoy Das (Weill Cornell Medical College, New York, NY, USA) for invaluable tissue samples. This work was supported by the U.S. National Institutes of Health (National Institute of Diabetes and Digestive and Kidney Diseases Grant DK41544, and National Institute of General Medical Sciences Grant GM115189).
Glossary
- BLAST
Basic Local Alignment Search Tool
- CHO
Chinese hamster ovary
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- hKCNE
human KCNE
- KCNE3/4L
long KCNE3/4
- KCNE3/4S
short KCNE3/4
- Kv
voltage-gated potassium
- NCBI
U.S. National Center for Biotechnology Information
- TBST
Tris-buffered saline–Tween 20
- TEVC
2-electrode voltage-clamp
- UTR
untranslated region
REFERENCES
- 1.Isacoff E. Y., Jan Y. N., Jan L. Y. (1990) Evidence for the formation of heteromultimeric potassium channels in Xenopus oocytes. Nature , 530–534 [DOI] [PubMed] [Google Scholar]
- 2.Abbott G. W., Xu X., Roepke T. K. (2007) Impact of ancillary subunits on ventricular repolarization. J. Electrocardiol. , S42–S46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abbott G. W., Goldstein S. A. (1998) A superfamily of small potassium channel subunits: form and function of the MinK-related peptides (MiRPs). Q. Rev. Biophys. , 357–398 [DOI] [PubMed] [Google Scholar]
- 4.McCrossan Z. A., Abbott G. W. (2004) The MinK-related peptides. Neuropharmacology , 787–821 [DOI] [PubMed] [Google Scholar]
- 5.Abbott G. W., Butler M. H., Bendahhou S., Dalakas M. C., Ptacek L. J., Goldstein S. A. (2001) MiRP2 forms potassium channels in skeletal muscle with Kv3.4 and is associated with periodic paralysis. Cell , 217–231 [DOI] [PubMed] [Google Scholar]
- 6.Barhanin J., Lesage F., Guillemare E., Fink M., Lazdunski M., Romey G. (1996) K(V)LQT1 and lsK (minK) proteins associate to form the I(Ks) cardiac potassium current. Nature , 78–80 [DOI] [PubMed] [Google Scholar]
- 7.Kanda V. A., Lewis A., Xu X., Abbott G. W. (2011) KCNE1 and KCNE2 provide a checkpoint governing voltage-gated potassium channel α-subunit composition. Biophys. J. , 1364–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanda V. A., Lewis A., Xu X., Abbott G. W. (2011) KCNE1 and KCNE2 inhibit forward trafficking of homomeric N-type voltage-gated potassium channels. Biophys. J. , 1354–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanguinetti M. C., Curran M. E., Zou A., Shen J., Spector P. S., Atkinson D. L., Keating M. T. (1996) Coassembly of K(V)LQT1 and minK (IsK) proteins to form cardiac I(Ks) potassium channel. Nature , 80–83 [DOI] [PubMed] [Google Scholar]
- 10.Schroeder B. C., Waldegger S., Fehr S., Bleich M., Warth R., Greger R., Jentsch T. J. (2000) A constitutively open potassium channel formed by KCNQ1 and KCNE3. Nature , 196–199 [DOI] [PubMed] [Google Scholar]
- 11.Tinel N., Diochot S., Borsotto M., Lazdunski M., Barhanin J. (2000) KCNE2 confers background current characteristics to the cardiac KCNQ1 potassium channel. EMBO J. , 6326–6330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roepke T. K., Anantharam A., Kirchhoff P., Busque S. M., Young J. B., Geibel J. P., Lerner D. J., Abbott G. W. (2006) The KCNE2 potassium channel ancillary subunit is essential for gastric acid secretion. J. Biol. Chem. , 23740–23747 [DOI] [PubMed] [Google Scholar]
- 13.Roepke T. K., Kanda V. A., Purtell K., King E. C., Lerner D. J., Abbott G. W. (2011) KCNE2 forms potassium channels with KCNA3 and KCNQ1 in the choroid plexus epithelium. FASEB J. , 4264–4273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roepke T. K., King E. C., Purtell K., Kanda V. A., Lerner D. J., Abbott G. W. (2011) Genetic dissection reveals unexpected influence of beta subunits on KCNQ1 K+ channel polarized trafficking in vivo. FASEB J. , 727–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roepke T. K., King E. C., Reyna-Neyra A., Paroder M., Purtell K., Koba W., Fine E., Lerner D. J., Carrasco N., Abbott G. W. (2009) Kcne2 deletion uncovers its crucial role in thyroid hormone biosynthesis. Nat. Med. , 1186–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roepke T. K., Kontogeorgis A., Ovanez C., Xu X., Young J. B., Purtell K., Goldstein P. A., Christini D. J., Peters N. S., Akar F. G., Gutstein D. E., Lerner D. J., Abbott G. W. (2008) Targeted deletion of kcne2 impairs ventricular repolarization via disruption of I(K,slow1) and I(to,f). FASEB J. , 3648–3660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roepke T. K., Purtell K., King E. C., La Perle K. M., Lerner D. J., Abbott G. W. (2010) Targeted deletion of Kcne2 causes gastritis cystica profunda and gastric neoplasia. PLoS One , e11451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abbott G. W., Roepke T. K. (2008) Pharmacogenetics of drug-induced arrhythmias. Expert Rev. Clin. Pharmacol. , 93–104 [DOI] [PubMed] [Google Scholar]
- 19.Panaghie G., Abbott G. W. (2006) The impact of ancillary subunits on small-molecule interactions with voltage-gated potassium channels. Curr. Pharm. Des. , 2285–2302 [DOI] [PubMed] [Google Scholar]
- 20.Abbott G. W. (2015) The KCNE2 K⁺ channel regulatory subunit: Ubiquitous influence, complex pathobiology. Gene , 162–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abbott G. W. (2016) KCNE1 and KCNE3: The yin and yang of voltage-gated K+ channel regulation. Gene , 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abbott G. W., Sesti F., Splawski I., Buck M. E., Lehmann M. H., Timothy K. W., Keating M. T., Goldstein S. A. (1999) MiRP1 forms IKr potassium channels with HERG and is associated with cardiac arrhythmia. Cell , 175–187 [DOI] [PubMed] [Google Scholar]
- 23.Gordon E., Panaghie G., Deng L., Bee K. J., Roepke T. K., Krogh-Madsen T., Christini D. J., Ostrer H., Basson C. T., Chung W., Abbott G. W. (2008) A KCNE2 mutation in a patient with cardiac arrhythmia induced by auditory stimuli and serum electrolyte imbalance. Cardiovasc. Res. , 98–106 [DOI] [PubMed] [Google Scholar]
- 24.Lundby A., Ravn L. S., Svendsen J. H., Hauns S., Olesen S. P., Schmitt N. (2008) KCNE3 mutation V17M identified in a patient with lone atrial fibrillation. Cell. Physiol. Biochem. , 47–54 [DOI] [PubMed] [Google Scholar]
- 25.Nakajima T., Wu J., Kaneko Y., Ashihara T., Ohno S., Irie T., Ding W. G., Matsuura H., Kurabayashi M., Horie M. (2012) KCNE3 T4A as the genetic basis of Brugada-pattern electrocardiogram. Circ. J. , 2763–2772 [DOI] [PubMed] [Google Scholar]
- 26.Ohno S., Toyoda F., Zankov D. P., Yoshida H., Makiyama T., Tsuji K., Honda T., Obayashi K., Ueyama H., Shimizu W., Miyamoto Y., Kamakura S., Matsuura H., Kita T., Horie M. (2009) Novel KCNE3 mutation reduces repolarizing potassium current and associated with long QT syndrome. Hum. Mutat. , 557–563 [DOI] [PubMed] [Google Scholar]
- 27.Ohno S., Zankov D. P., Ding W. G., Itoh H., Makiyama T., Doi T., Shizuta S., Hattori T., Miyamoto A., Naiki N., Hancox J. C., Matsuura H., Horie M. (2011) KCNE5 (KCNE1L) variants are novel modulators of Brugada syndrome and idiopathic ventricular fibrillation. Circ. Arrhythm. Electrophysiol. , 352–361 [DOI] [PubMed] [Google Scholar]
- 28.Splawski I., Tristani-Firouzi M., Lehmann M. H., Sanguinetti M. C., Keating M. T. (1997) Mutations in the hminK gene cause long QT syndrome and suppress IKs function. Nat. Genet. , 338–340 [DOI] [PubMed] [Google Scholar]
- 29.Zeng Z., Tan C., Teng S., Chen J., Su S., Zhou X., Wang F., Zhang S., Gu D., Makielski J. C., Pu J. (2007) The single nucleotide polymorphisms of I(Ks) potassium channel genes and their association with atrial fibrillation in a Chinese population. Cardiology , 97–103 [DOI] [PubMed] [Google Scholar]
- 30.Zeng Z. Y., Pu J. L., Tan C., Teng S. Y., Chen J. H., Su S. Y., Zhou X. Y., Zhang S., Li Y. S., Wang F. Z., Gu D. F. (2005) [The association of single nucleotide polymorphism of slow delayed rectifier K+ channel genes with atrial fibrillation in Han nationality Chinese]. Zhonghua Xin Xue Guan Bing Za Zhi , 987–991 [PubMed] [Google Scholar]
- 31.Crump S. M., Abbott G. W. (2014) Arrhythmogenic KCNE gene variants: current knowledge and future challenges. Front. Genet. , 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crump S. M., Hu Z., Kant R., Levy D. I., Goldstein S. A., Abbott G. W. (2016) Kcne4 deletion sex- and age-specifically impairs cardiac repolarization in mice. FASEB J. , 360–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu Z., Crump S. M., Anand M., Kant R., Levi R., Abbott G. W. (2014) Kcne3 deletion initiates extracardiac arrhythmogenesis in mice. FASEB J. , 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu Z., Kant R., Anand M., King E. C., Krogh-Madsen T., Christini D. J., Abbott G. W. (2014) Kcne2 deletion creates a multisystem syndrome predisposing to sudden cardiac death. Circ. Cardiovasc. Genet. , 33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levy D. I., Wanderling S., Biemesderfer D., Goldstein S. A. (2008) MiRP3 acts as an accessory subunit with the BK potassium channel. Am. J. Physiol. Renal Physiol. , F380–F387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lundquist A. L., Turner C. L., Ballester L. Y., George A. L. Jr (2006) Expression and transcriptional control of human KCNE genes. Genomics , 119–128 [DOI] [PubMed] [Google Scholar]
- 37.De Angioletti M., Lacerra G., Sabato V., Carestia C. (2004) Beta+45 G --> C: a novel silent beta-thalassaemia mutation, the first in the Kozak sequence. Br. J. Haematol. , 224–231 [DOI] [PubMed] [Google Scholar]
- 38.Preston P., Wartosch L., Günzel D., Fromm M., Kongsuphol P., Ousingsawat J., Kunzelmann K., Barhanin J., Warth R., Jentsch T. J. (2010) Disruption of the K+ channel beta-subunit KCNE3 reveals an important role in intestinal and tracheal Cl− transport. J. Biol. Chem. , 7165–7175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang W., Kim H. J., Lee J. H., Wong V., Sihn C. R., Lv P., Perez Flores M. C., Mousavi-Nik A., Doyle K. J., Xu Y., Yamoah E. N. (2014) Functional significance of K+ channel β-subunit KCNE3 in auditory neurons. J. Biol. Chem. , 16802–16813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niwa N., Nerbonne J. M. (2010) Molecular determinants of cardiac transient outward potassium current (I(to)) expression and regulation. J. Mol. Cell. Cardiol. , 12–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levy D. I., Cepaitis E., Wanderling S., Toth P. T., Archer S. L., Goldstein S. A. (2010) The membrane protein MiRP3 regulates Kv4.2 channels in a KChIP-dependent manner. J. Physiol. , 2657–2668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grunnet M., Jespersen T., Rasmussen H. B., Ljungstrøm T., Jorgensen N. K., Olesen S. P., Klaerke D. A. (2002) KCNE4 is an inhibitory subunit to the KCNQ1 channel. J. Physiol. , 119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grunnet M., Olesen S. P., Klaerke D. A., Jespersen T. (2005) hKCNE4 inhibits the hKCNQ1 potassium current without affecting the activation kinetics. Biochem. Biophys. Res. Commun. , 1146–1153 [DOI] [PubMed] [Google Scholar]
- 44.Strutz-Seebohm N., Seebohm G., Fedorenko O., Baltaev R., Engel J., Knirsch M., Lang F. (2006) Functional coassembly of KCNQ4 with KCNE-beta- subunits in Xenopus oocytes. Cell. Physiol. Biochem. , 57–66 [DOI] [PubMed] [Google Scholar]
- 45.Jepps T. A., Carr G., Lundegaard P. R., Olesen S. P., Greenwood I. A. (2015) Fundamental role for the KCNE4 ancillary subunit in Kv7.4 regulation of arterial tone. J. Physiol. , 5325–5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu H., Wu J., Potapova I., Wymore R. T., Holmes B., Zuckerman J., Pan Z., Wang H., Shi W., Robinson R. B., El-Maghrabi M. R., Benjamin W., Dixon J., McKinnon D., Cohen I. S., Wymore R. (2001) MinK-related peptide 1: A beta subunit for the HCN ion channel subunit family enhances expression and speeds activation. Circ. Res. , E84–E87 [DOI] [PubMed] [Google Scholar]
- 47.Liu W., Deng J., Wang G., Zhang C., Luo X., Yan D., Su Q., Liu J. (2014) KCNE2 modulates cardiac L-type Ca channel. J. Mol. Cell. Cardiol. , 208–218 [DOI] [PubMed] [Google Scholar]
- 48.Abbott G. W. (2012) KCNE2 and the K (+) channel: the tail wagging the dog. Channels (Austin) , 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teng S., Ma L., Zhen Y., Lin C., Bähring R., Vardanyan V., Pongs O., Hui R. (2003) Novel gene hKCNE4 slows the activation of the KCNQ1 channel. Biochem. Biophys. Res. Commun. , 808–813 [DOI] [PubMed] [Google Scholar]
- 50.Grunnet M., Rasmussen H. B., Hay-Schmidt A., Rosenstierne M., Klaerke D. A., Olesen S. P., Jespersen T. (2003) KCNE4 is an inhibitory subunit to Kv1.1 and Kv1.3 potassium channels. Biophys. J. , 1525–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]