Abstract
Akkermansia muciniphila and Faecalibacterium prausnitzii, cohabitants in the intestinal mucosa, are considered members of a healthy microbiota and reduction of both species occurs in several intestinal disorders, including inflammatory bowel disease. Little is known however about a possible link between the reduction in quantity of these species, and in which circumstances this may occur. This study aims to determine the abundances and co-occurrence of the two species in order to elucidate conditions that may compromise their presence in the gut. Loads of A. muciniphila, total F. prausnitzii and its two phylogroup (16S rRNA gene copies) were determined by quantitative polymerase chain reaction in colonic biopsies from 17 healthy controls (H), 23 patients with ulcerative colitis (UC), 31 patients with Crohn's disease (CD), 3 with irritable bowel syndrome (IBS) and 3 with colorectal cancer (CRC). Data were normalized to total bacterial 16S rRNA gene copies in the same sample. Prevalence, relative abundances and correlation analyses were performed according to type of disease and considering relevant clinical characteristics of patients such as IBD location, age of disease onset, CD behavior, current medication and activity status. Co-occurrence of both species was found in 29% of H, 65% of UC and 29% of CD. Lower levels of total F. prausnitzii and phylogroups were found in subjects with CD, compared with H subjects (P ≤ 0.044). In contrast, no differences were found with the regard to A. muciniphila abundance across different disease states, but CD patients with disease onset below 16 years of age featured a marked depletion of this species. In CD patients, correlation between A. muciniphila and total F. prausnitzii (ρ = 0.362, P = 0.045) was observed, and particularly in those with non-stricturing, non-penetrating disease behavior and under moderate immunosuppressants therapy. Altogether, this study revealed that co-occurrence of both species differs between disease status. In addition, IBD patients featured a reduction of F. prausnitzii but similar loads of A. muciniphila when compared to H subjects, with the exception of those with early onset CD. Depletion of A. muciniphila in this subgroup of subjects suggests that it could be a potential biomarker to assist in pediatric CD diagnosis.
Keywords: Akkermansia muciniphila, Faecalibacterium prausnitzii, Crohn's disease, ulcerative colitis, inflammatory bowel diseases
Introduction
Crohn's disease (CD) and ulcerative colitis (UC) are the two major types of idiopathic inflammatory bowel diseases (IBD) (Mendoza Hernández et al., 2007). Both are chronic inflammatory disorders of the gut. UC typically begins in the rectum and inflammation may extend continuously to involve the entire colon. In 20% of CD patients the disease affects the colon exclusively (Silverberg et al., 2005), but the most commonly involved areas are terminal ileum and the beginning of the colon. In CD any part of the gastrointestinal tract (from the oropharynx to the anus) may be affected in a patchy pattern (Mendoza Hernández et al., 2007). Other than location, differences in the mucosal lesions exist between these conditions. Inflammation in CD can be transmural reaching the serosa, whereas inflammation in UC patients is generally restricted to the mucosa.
The inner layer of the bowel wall is a niche of particular importance, because of the spatial proximity between epithelial cells and gut bacteria, and thus the study of human intestinal mucosa biopsies provides meaningful insights of host-bacterial interactions. Numerous studies have been prompted over the last decade aiming at deciphering the exact role of gut microbiota in IBD. Nowadays there is a wide variety of clinical and experimental studies revealing microbial implication in IBD (Sartor, 2006, 2008; Seksik et al., 2006; Manichanh et al., 2012). The most replicated finding by far has been disturbances in the intestinal microbiota composition balance, situation known as dysbiosis (Gophna et al., 2006; Manichanh et al., 2006; Martinez-Medina et al., 2006; Andoh et al., 2009; Willing et al., 2009; Sokol and Seksik, 2010; Joossens et al., 2011; Mondot et al., 2011; Machiels et al., 2013). In this state, dominating species, to whom a beneficial role to preserve gut homeostasis has been attributed, become underrepresented.
Akkermansia muciniphila inhabits mainly in the mucosa, and represents between 1 and 3% of the gut microbiota (Derrien et al., 2004, 2008). A decrease of this species has been demonstrated in feces and/or biopsies of several disorders including autism, obesity, type 2 diabetes, appendicitis, and IBD (Belzer and de Vos, 2012; Everard et al., 2013). Studies in mice models have shown that gut colonization by this species affects expression of genes involved in immune response-regulatory processes (Derrien et al., 2011) as well as in host's lipid metabolism (Lukovac et al., 2014), especially in the colon. It is of note that extracellular vesicles derived from this species have a protective function that ameliorates severity of induced colitis in mice, suggesting that it has an important role in the maintenance of intestinal homeostasis (Kang et al., 2013). A. muciniphila is essential for a healthy mucus layer in the human gut in terms of mucus production and thickness (Belzer and de Vos, 2012). This species is not only important for the host, but also for gut microbial community. Its specific capability to degrade mucus results in the release of oligosaccharides and the production of propionate and acetate (Derrien et al., 2004) as well as amino acids, important co-factors and vitamins (van Passel et al., 2011) that become available for other gut symbionts. However, significant co-occurrence of this species with other bacterial taxa present in the gut has not been revealed in feces (Lozupone et al., 2012).
In turn, Faecalibacterium prausnitzii is also an abundant intestinal microorganism with a feco-mucosal distribution, and whose relative abundance can represent between 2 and 15% of intestinal bacterial communities (Swidsinski et al., 2005; Baumgart et al., 2007; Flint et al., 2012). Several studies, of fecal and/or mucosal samples, have shown that F. prausnitzii prevalence and abundance are reduced under certain disorders such as celiac disease (Swidsinski et al., 2008; De Palma et al., 2009), obesity and type 2 diabetes (Furet et al., 2010; Graessler et al., 2012), appendicitis (Swidsinski et al., 2011), chronic diarrhea (Dörffel et al., 2012), irritable bowel syndrome (IBS) of alternating type (Rajilić-Stojanović et al., 2011), colorectal cancer (CRC) (Balamurugan et al., 2008; Lopez-Siles et al., 2016), and particularly in IBD (Sokol et al., 2008, 2009; Swidsinski et al., 2008; Willing et al., 2009; Machiels et al., 2013; Lopez-Siles et al., 2014). Low abundance of this species has been linked with active IBD (Sokol et al., 2009), and some complications such as a higher risk of post-operative recurrence (Sokol et al., 2008) or pouchitis (McLaughlin et al., 2010). Other than butyrate production (which can reduce intestinal mucosa inflammation and is the main energy source for the colonocytes), additional anti-inflammatory properties have been attributed to F. prausnitzii (Sokol et al., 2008; Miquel et al., 2013; Martín et al., 2014). Both, cell and supernatant fractions of this species, have been proven to reduce severity of acute (Sokol et al., 2008; Rossi et al., 2015), chronic (Martín et al., 2014) and low grade (Martín et al., 2015) inflammation in murine models. This has been attributed to an enhancement of intestinal barrier function related with the expression of certain tight junction proteins other than claudin (Carlsson et al., 2013). F. prausnitzii also influences gut physiology through the production of mucus O-glycans, and may help to maintain suitable proportions of different cell types of secretory linage in the intestinal epithelium, as evidenced in rodent studies (Wrzosek et al., 2013). To date, it remains unclear which conditions are likely to compromise this species in the gut. Alterations in gut pH or bile salt concentration have been suggested (Lopez-Siles et al., 2012), but a break in the ecologic relations with other gut symbionts that support its presence in the gut may also contribute, but have been little studied. Co-occurrence network analysis of gut bacteria found in feces, showed that F. prausnitzii co-occurs with several members of the C. coccoides group and Bacteroidetes (Lozupone et al., 2012). As F. prausnitzii growth is stimulated by acetate (Duncan et al., 2002), its presence in the gut may also be favored by acetate producers like A. muciniphila. However, little is known about interaction between these two species.
This work is aimed at determining the variation of mucosa-associated A. muciniphila and F. prausnitzii between healthy control subjects (H) and patients suffering from IBD, in order to elucidate in which conditions imbalances of these species take place, and if both species are affected equally. Some IBS and CRC patients have been included for comparative purposes. Prevalence and abundance of mucosa-associated A. muciniphila and F. prausnitzii have been determined in colonic samples by quantitative polymerase chain reaction (qPCR). Data have been analyzed taking into account patients' most relevant clinical characteristics. Medication at sampling was also considered in order to determine if any of the current therapies are effective in restoring these species levels to those found in H. In addition, correlation analysis of their load has also been conducted to provide supporting evidence on the effect of one population over the other, or about whether or not they are influenced by similar gut factors.
Materials and methods
Patient recruitment and characteristics
The study population was a cohort consisting of Spanish volunteers including 54 IBD (31 CD and 23 UC), three IBS, three CRC patients and 17 H (Table 1). Subjects were gender matched for all the groups. Concerning age, CD patients were younger than those in the H group (P = 0.002), whereas CRC patients were significantly older than those with IBD (P ≤ 0.028). Besides, at disease onset, CD patients were younger than UC (mean age ± SD; UC = 37.2 ± 13.3 years, CD = 28.0 ± 12.4 years; P = 0.012).
Table 1.
Sample size and clinical characteristics of subjects.
| Healthy | Irritable bowel syndrome | Colorectal cancer | IBD | P-value§ | ||
|---|---|---|---|---|---|---|
| Ulcerative colitis | Crohn's disease | |||||
| n (patients) | 17 | 3 | 3 | 23 | 31 | |
| Age (mean years ± SD) | 48.1 ± 15.8 | 35.0 ± 7.0 | 69.3 ± 16.0 | 40.5 ± 13.5 | 33.6 ± 12.2 | 0.002** |
| Male (n, %) | 10 (58.8%) | 1 (33.3%) | 3 (100%) | 13 (56.5%) | 20 (64.5%) | 0.826† |
| Active (n, %) | na | na | na | 3 (13.0%) | 13 (41.9%) | 0.036† |
| Previous surgery (n, %) | na | na | na | 1 (4.3%) | 6 (19.4%) | 0.213† |
| Smokers (n, %) | 0 (0%) | 0 (0%) | 0 (0%) | 2 (8.7%) | 5 (16.1%) | 0.410† |
| Treatment (n, %)* | 0.494† | |||||
| No treatment | na | na | na | 11 (47.8%) | 10 (32.3%) | |
| Mesalazine | na | na | na | 3 (13.0%) | 3 (9.7%) | |
| Moderate immunosuppressant | na | na | na | 4 (17.4%) | 10 (32.3%) | |
| Anti-TNFα (infliximab, adalimumab) | na | na | na | 3 (13.0%) | 6 (19.4%) | |
| CD Montreal classification | ||||||
| Age of diagnosis (n, %)* | 0.007 | |||||
| <16 yeays (A1) | na | na | na | na | 5 (16.1%) | |
| 17–40 years (A2) | na | na | na | 11 (47.8%) | 21 (67.7%) | |
| >41 years (A3) | na | na | na | 9 (39.1%) | 3 (9.7%) | |
| Location (N, %) | na | |||||
| Ileal-CD (L1) | na | na | na | na | 13 (41.9%) | |
| Colonic-CD (L2) | na | na | na | na | 7 (22.6%) | |
| Ileocolonic-CD (L3) | na | na | na | na | 10 (32.3%) | |
| Behavior (N, %)* | na | |||||
| Non-stricturing, non-penetrating (B1) | na | na | na | na | 19 (61.3%) | |
| Stricturing (B2) | na | na | na | na | 8 (25.8%) | |
| UC classification (n, %)* | na | |||||
| Ulcerative proctitis (E1) | na | na | na | 5 (21.7%) | na | |
| Distal UC (E2) | na | na | na | 11 (47.8%) | na | |
| Extensive UC or ulcerative pancolitis (E3) | na | na | na | 6 (26.1%) | na | |
IBD, Inflammatory bowel disease; TNF, tumor necrosis factor; na, not applicable.
Medical treatment at the time of sampling was available in 21/23 UC, and 29/31 CD patients; Age of disease onset was available for 20/23 UC patients, and 29/31 CD patients; Disease behavior at last follow-up before the time of sampling was available in 27/31 CD patients, and none had penetrating CD (B3); Maximal disease extent at the time of sampling was available in 22/23 UC and 30/31 CD patients.
Groups were compared by appropriate statistical tests, and P-value ≤ 0.05 was considered significant, χ2 test,
ANOVA. Analyses statistically significant are highlighted in boldface.
Subjects were recruited by the Gastroenterology Services of the Hospital Universitari Dr. Josep Trueta (Girona, Spain) and the Hospital Santa Caterina (Salt, Spain) between 2006 and 2010. To avoid bias between centers, patients with IBD were diagnosed according to standard clinical, pathological, and endoscopic criteria and categorized according to the Montreal classification (Silverberg et al., 2005). IBS patients were diagnosed according to Rome III criteria (available at http://www.romecriteria.org/criteria/). The diagnosis of CRC was established by colonoscopy and biopsy, and data correlated with high risk of developing this disease was recorded. Controls consisted of subjects who underwent colonoscopy for different reasons as rectorrhagia (n = 8), colorectal cancer familial history (n = 3), and abdominal pain (n = 6), and all featured normal colonoscopy. Clinically relevant data of all the patients was collected (Table 1). Percentage of active patients was higher in CD patients than in UC (P = 0.036). Individuals included in this study were >18 years old, did not have any other intestinal disease and were not pregnant. Antibiotic treatment within 2 months before colonoscopy was an exclusion criterion.
Ethics statement
This work was approved by the Ethics Committee of Clinical Research of the Hospital Universitari Dr. Josep Trueta (Girona, Spain) and the Institut d'Assistència Sanitària of Girona (Salt, Spain) on 26th May 2006 (protocol code BACTECCU, Ref CEIC: 08/06) and 21st April 2009 (protocol code BACTODIAG, Ref CEIC: 10/08), respectively. All subjects gave written informed consent in accordance with the Declaration of Helsinki.
Sample collection and DNA extraction
Cleansing of the gastrointestinal tract using Casenglicol® was performed prior to colonoscopy, following manufacturer's guidelines. During routine endoscopy and following standard procedures, a biopsy (<25 mg) from non-affected tissue of the colon was taken for each subject. All biopsies were immediately placed in sterile tubes without any buffer. Following completion of the whole endoscopic procedure, all samples were stored at −80°C upon analysis.
To discard transient and loosely attached bacteria, biopsies were subjected to two mild ultrasound wash cycles, as reported previously (Martinez-Medina et al., 2006). Afterwards, DNA was extracted using the NucleoSpin® Tissue Kit (Macherey-Nagel GmbH & Co., Duren, Germany). The support protocol for Gram positive bacteria (consisting of pre-incubation during 1 h at 37°C with buffer T1 (20 mm Tris/HCl, 2 mM EDTA, 1% Triton X-100, pH 8) supplemented with 20 mg/ml lysozyme), and the RNAse treatment step were carried out. Genomic DNA was eluted with 10 mM Tris-HCl (pH 7.4) and stored at −80°C until use. DNA concentration and purity of the extracts were determined with a NanoDrop ND-100 spectrophotometer (NanoDrop Technologies, USA). The average purity and concentration of the DNA extracts was (mean ± SD) 1.794 ± 0.579 Abs260/280 ratio and 174.864 ± 131.829 ng/μl, respectively.
Quantification standards for qPCR
Standard DNA templates from F. prausnitzii strain S3L/3 (phylogroup I), F. prausnitzii DSM 17677 (phylogroup II) and A. muciniphila (ATCCBAA-835) were prepared as genetic constructs after PCR amplification of the whole 16S rRNA as previously reported (Lane, 1991; Weisburg et al., 1991), and subsequent insertion of this gene into a pCR®4-TOPO® cloning plasmid (Invitrogen, CA, USA) following manufacturer's guidelines.
After purification with the NucleoSpin® Plasmid kit (Macherey-Nagel GmbH & Co., Duren, Germany), plasmids were linearized with SpeI and quantified using Qubit™ Quantitation Platform (Invitrogen, Carlsbad, USA). Initial target concentration was inferred taking into consideration the theoretical molecular weight (3.58 × 106 Da) and the size of recombinant plasmid (5421 pb).
Standard curves were obtained from 10-fold serial dilutions of the titrated suspension of linearized plasmids. Strains used to construct each standard curve are indicated in Table 2. To prepare the standard curve, only dilutions within the linear dynamic range span of each reaction were used, as detailed in Table 2. Total bacteria 16S rRNA gene quantification was used to intercalibrate all the standard curves, in order to make sure that results obtained were comparable. Accordingly, plasmid preparations for A. muciniphila standard and phylogroups standards were run as unknown samples in a total bacterial qPCR. Specific intercalibration of total F. prausnitzii qPCR was not required as it uses the same standard curve template employed for total bacterial qPCR (Table 2). Quantification values obtained were compared to initial target concentration inferred from DNA concentration, and <10% variation was obtained.
Table 2.
16S rRNA-targeted primers and probes used in this study.
| Target | Primers and probe | Sequence (5′-3′)a | Final conc. (nM) | Strain used as standard | Standard curve* | qPCR conditionsc | References | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Fluorescence reporting method | Cycles | Denat. Ta(°C); t (s) | Annealing and extension Ta(°C), t (s) | Melting curved | |||||||
| Total bacteria (Eubacteria) | F_Bact 1369 | CGGTGAATACGTTCCCGG | 300 | F. prausnitzii DSM 17677 | 107-103; 89.0 ± 7.0 |
Hydrolysis probe | 40 | 95; 30 | 60; 60 | No | Furet et al., 2009 |
| R_Prok1492 | TACGGCTACCTTGTTACGACTT | 300 | |||||||||
| P_TM1389F | FAM-CTTGTACACACCGCCCGTC-TAMRA | 250 | |||||||||
| Akkermansia muciniphila | AM1-F | CAGCACGTGAAGGTGGGGAC | 300 | A. muciniphila ATCCBAA-835 | 2 × 102 - 2 × 107; 93.9 ± 4.2 |
SYBR Green | 50 | 95; 15 | 65; 30 and 72; 32 | Yes | Collado et al., 2007 |
| AM2-R | CCTTGCGGTTGGCTTCAGAT | 300 | |||||||||
| Faecalibacterium prausnitzii (total) | Fpra428F | TGTAAACTCCTGTTGTTGAGGAAGATAA | 300 | F. prausnitzii DSM 17677 | 107-103; 84.8 ± 3.2 |
Hydrolysis probe | 40 | 95; 15 | 60; 60 | No | Lopez-Siles et al., 2014 |
| Fpra583R | GCGCTCCCTTTACACCCA | 300 | |||||||||
| Fpra493PR | FAM-CAAGGAAGTGACGGCTAACTACGTGCCAG-TAMRA | 250 | |||||||||
| IACb | IAC F | TACGGATGAGGAGGACAAAGGA | 300 | DNA IAC | n.a. | Hydrolysis probe | 40 | 95; 15 | 60; 60 | No | Lopez-Siles et al., 2014 |
| IAC R | CACTTCGCTCTGATCCATTGG | 300 | |||||||||
| IAC PR | VIC®-CGCCGCTATGGGCATCGCA-TAMRA | 250 | |||||||||
| F. prausnitzii (phylogroups) | Fpra 136F | CTCAAAGAGGGGGACAACAGTT | 900 | F. prausnitzii S3L/3 (phylogroup I) and DSM 17677 (phylogroup II) | 106-10; 86.3 ± 6.1; 95.3 ± 11.9 |
Hydrolysis probe | 40 | 95; 15 | 64; 60 | No | Lopez-Siles et al., 2016 |
| Fpra 232R | GCCATCTCAAAGCGGATTG | 900 | |||||||||
| PHG1 180PR | 6FAM-TAAGCCCACGACCCGGCATCG-BHQ1 | 300 | |||||||||
| PHG2 180PR | JOE-TAAGCCCACRGCTCGGCATC-BHQ1 | 300 | |||||||||
Range span (16S rRNA gene copies), Efficiency (mean ± standard deviation); n.a., not applicable.
Probe sequences are in bold. FAM™, 6-carboxyfluorescin; VIC®, 6-carboxyrhodamine; JOE, 4′,5′-dichloro-2′,7′-dimethoxy-5(6)-carboxyfluorescein; TAMRA™, tetramethylrhodamin; BHQ1, Black Hole Quencher1.
IAC, internal amplification control; 103 copies of appropriate DNA template were added in each reaction. DNA IAC sequence (5′-3′), DNA IAC sequence: 5′-TACggATgAggAggACAAAggACgCCgCTATgggCATCgCACCAATggATCAgAgCgAAgTg-3′ (according to Lopez-Siles et al., 2014).
An amperase treatment (50°C, 2 min) and an initial denaturing step (95°C, 10 min) were performed for all the reactions. For assays based on hydrolysis probes, annealing and extension steps were performed simultaneously.
Melting curve consisted on 95°C 15 s, 60°C 1 min, 95°C 15 s, and 60°C 15 s (average temperature slope 0.58°C/s).
qPCR assays
Previously reported 16S rRNA gene-targeted primers and probes were used for total F. prausnitzii (Lopez-Siles et al., 2014), phylogroups (Lopez-Siles et al., 2016), A. muciniphila (Collado et al., 2007) and total bacterial (Furet et al., 2009) quantification through qPCR.
Amplification reactions were performed as described elsewhere (Collado et al., 2007; Furet et al., 2009; Lopez-Siles et al., 2014, 2016) with slight modifications detailed in Table 2. Briefly, quantifications were carried out in a total volume of 20 μl reactions containing: 1 × TaqMan® Universal PCR Master Mix 2 × or SYBR®Green PCR Master Mix 2 × (Applied Biosystems, Foster City, CA, USA) as required, 300–900 nM of each primer and 250–300 nM of probe if necessary. Up to 50 ng of genomic DNA template was added in each reaction. All primers and probes used in this study as well as PCR conditions are detailed in Table 2. Total F. prausnitzii, and total bacteria primers and hydrolysis probes were purchased from Applied Biosystems (Foster City, CA, USA), whereas primers and hydrolysis probes for F. prausnitzii phylogroups and A. muciniphila were acquired from Biomers (Ulm, Germany). DNA of the internal amplification control (IAC) was synthesized by Bonsai technologies group (Alcobendas, Spain). All oligonucleotides were purified by HPLC. Plates and optical caps were provided by Applied Biosystems (Ref. 4323032 and Ref. 4306737, respectively).
Samples were run at least in duplicate in the same plate (Table S1), which was set up manually. For data analysis, the mean of the quantifications was used. Duplicates were considered valid if the standard deviation between quantification cycles (Cq) was <0.34 (i.e., a difference of <10% of the quantity was tolerated), and if not quantification was repeated. Quantification controls consisting of at least five reactions with a known number of target genes were performed to assess inter-run reproducibility. Inhibition was controlled on total F. prausnitzii quantification by adding 103 copies of an internal amplification control (IAC) template to each reaction. It was considered that there was no inhibition if the obtained Cq was <0.34 different from those obtained when quantifying the IAC alone for any of the replicates. In each run, a non-amplification control (NTC) which did not contain any DNA template (either bacterial or IAC) was also included. In all cases with hydrolysis probes, NTC resulted in undetectable Cq values whereas for SYBR Green assays NTC had Cq >35, and melting curve analysis confirmed no specific amplification.
A 7500 Real Time PCR system (Applied Biosystems, USA) was used to perform all qPCR. The thermal profile used for each assay is detailed in Table 2. In summary, it consisted of a first step at 50°C during 2 min for amperase treatment followed by a 95°C hold for 10 min to denature DNA and activate Ampli-Taq Gold polymerase; and a further 40–50 cycles consisting of a denaturation step at 95°C for 15 s, followed by an annealing and extension step at 60°C (or at 64°C for phylogroups quantification) for 1 min. When required, melting curve analysis was performed to assess whether or not fluorescence was due to specific amplification products. Data were collected and analyzed using the 7500 SDS system software version 1.4 (Applied Biosystems). Assays were performed under average PCR efficiencies of (mean ± SD) 89.9 ± 4.6% (Figure S2).
Data normalization and statistical analysis
Regarding the qualitative analyses, absence of F. prausnitzii, its phylogroups or A. muciniphila was considered if no detection was obtained during the qPCR analysis, corresponding to samples that carried these bacteria below the detection limit (i.e., 106.6, 1.10, 2.39, and 374.09 16S rRNA genes per reaction for total F. prausnitzii, phylogroup I, phylogroup II, and A. muciniphila, respectively). Pearson's χ2 test was used to compare the prevalence between groups of patients, by IBD disease location, age of disease onset and other clinically relevant data as activity, treatment and whether or not patients have had intestinal resection.
Referring to quantitative analyses, total F. prausnitzii, phylogroups and A. muciniphila 16S rRNA gene copy detected in each sample were normalized to the total bacterial 16S rRNA gene copies in the same sample. Data are given as the log10 of the ratio between 16S rRNA gene copies of the target microorganism and million of total bacterial 16S rRNA genes detected in the same sample. No further correction to adjust for differences in 16S rRNA gene operons in each species was performed, as no consensus has been achieved to date for F. prausnitzii according to rrnDB (Stoddard et al., 2015). For those samples with no detection of F. prausnitzii, phylogroup or A. muciniphila, the number of copies corresponding to their respective detection limit was used for calculations of relative abundances. Kruskal–Wallis non-parametric test was applied to assess differences in variables with more than two categories such as diagnostics, CD and UC disease location, age of disease onset, and current medication. Mann–Whitney U test was used to perform pairwise comparisons of subcategories of these variables, and FDR after multiple comparisons was assessed (Table S2). Mann–Whitney U test was also used to compare, variables with two categories such as activity (active CD and UC patients when CDAI > 150 (Best et al., 1976) and a Mayo score >3 (Pineton de Chambrun et al., 2010), respectively), and intestinal resection. In addition, ratios between 16S rRNA gene copies of either total F. prausnitzii, phylogroup I or phylogroup II and A. muciniphila were calculated and analyzed as detailed above. Spearman correlation coefficient and significance between total F. prausnitzii, or phylogroup quantities and A. muciniphila load was calculated. The same statistical method was used to analyze the correlation between the quantity of each bacterial group, and continuous clinical data such as time (in years) since disease onset. All the statistical analyses were performed using the SPSS 15.0 statistical package (LEAD Technologies, Inc.). Significance levels were established for P ≤ 0.05.
Results
Prevalence of mucosa-associated A. muciniphila and F. prausnitzii
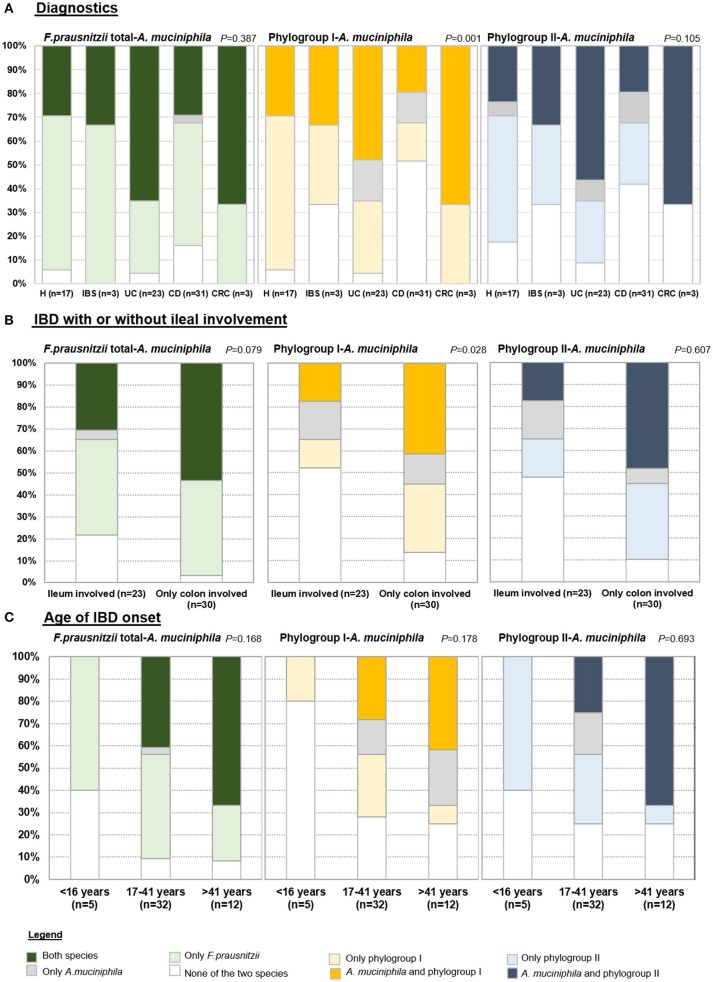
To assess co-occurrence of both species, prevalence of F. prausnitzii (total or separating by phylogroups), and A. muciniphila as calculated from positive determinations over total samples, was analyzed by condition, by IBD location and also taking into account relevant clinical data (Figure 1). Four categories of patients were established based on: detecting only F. prausnitzii, detecting only A. muciniphila, detecting both species, or none of them. Most of the subjects carried both species or F. prausnitzii alone, whilst finding A. muciniphila alone was rare and in some cases, none of the two species was found suggesting that if present, they are below the detection limit of our assays.
Figure 1.
Prevalence of total F. prausnitzii, phylogroups and A. muciniphila in (A) each group of patients, (B) in Inflammatory bowel disease (IBD) subjects according to whether or not ileum is affected, and (C) in IBD by age of disease onset. H, control subjects; IBS, irritable bowel syndrome; UC, ulcerative colitis; CD, Crohn's disease; CRC, colorectal cancer.
When analysing the cohort by diagnostics (Figure 1A), only statistically significant differences in the proportions of each category were observed for the co-occurrence of F. prausnitzii phylogroup I and A. muciniphila (P < 0.001). As a particularity of CD, none of the two species was detected in 51.61% of subjects of this group, which was a higher proportion than in the other groups of subjects (ranging from 4.35% to 33.3%). In 64.71% of H subjects only one species (F. prausnitzii phylopgroup I) was detected, while this category was present in between 16% and 34% in the other groups of subjects. F. prausnitzii phylogroup I and A. muciniphila were found in approximately 30% of H, 33.33% of IBS and 20% of CD subjects, whereas 47.82% of UC and 66.67% of CRC patients fall into this category. Similar trends were observed for F. prausnitzii phylogroup II and A. muciniphila co-occurence analysis (P = 0.105), whereas percentages of each category were more similar between groups of subjects when analyzed in conjunction prevalence of total F. prausnitzii and A. muciniphila (P = 0.387). Of note, all subjects with IBS or CRC had at least one of the two species.
When analysing the cohort by IBD disease location, no significant differences within UC and within CD subtypes were achieved (Figure S1). However, a trend toward different proportions was observed. Half of subjects with distal UC (E2) were characterized by presenting only A. muciniphila. All patients with UC and also those with C-CD carried at least one species, whereas lower prevalences were found in CD patients with ileal involvement. Particularly, none of the two species were detected in 10–31% of CD patients with ileal disease location, whereas in 23.1% of I-CD and 40% of IC-CD, both species co-occurred. As the frequencies observed in C-CD patients resembled in some cases those in UC, we analyzed IBD subjects by grouping those with (either I-CD or IC-CD) or without (i.e., C-CD or UC) ileum involved (Figure 1B). Interestingly, when analyzing co-occurrence of F. prausnitzii phylogroup I and A. muciniphila the percentage of subjects with none or only one species detected was over 80% in those with ileal disease whereas in approximately 40% of subjects with colonic disease both species were found (P = 0.028).
When analyzing prevalence taking into account clinical data of the patients, no significant differences were observed by activity, medication or intestinal resection neither in CD nor in UC. No differences were found within CD patients according to disease behavior. Interestingly, A. muciniphila was not detected in any of the CD patients diagnosed with disease onset below 16 years of age (Figure 1C). It remains to be stablished if this is a common issue with UC patients, as none with early disease onset was included in our cohort.
Abundances of mucosa-associated A. muciniphila and F. prausnitzii
A. muciniphila, total F. prausnitzii and its phylogroups load was compared amongst patients with different intestinal conditions (Table 3). Total F. prausnitzii was less abundant in CRC and IBD patients as compared to H subjects. However, statistically significant differences were achieved only for CRC (P = 0.028) and CD patients [P = 0.021, but not sustained after FDR assessment (Table S2)], probably because the reduction is lower for UC patients and the high variability between subjects. In CD patients, those with ileal involvement presented the lowest levels of this bacterium (P = 0.050), whereas IC-CD patients and C-CD were similar to UC (Table 3). Slight differences in average load were also found within UC patients although these differences were not statistically supported. Patients with ulcerative proctitis (E1) and extensive UC (E3) presented F. prausnitzii loads similar to H subjects, whereas those with E2 had abundances between CD patients and H subjects.
Table 3.
Abundances of mucosa-associated F. prausnitzii, its phylogroups and A. muciniphila in controls (H), irritable bowel syndrome (IBS), colorectal cancer (CRC), Ulcerative Colitis (UC), and Crohn's disease (CD) patients.
| n patients | F. prausnitzii*§ | Phylogroup I*§ | Phylogroup II*§ | A. muciniphila | |
|---|---|---|---|---|---|
| H | 17 | 5.07 ±0.70a | 3.00 ±0.81a | 2.32 ±1.69ab | 3.49 ±0.81 |
| IBS | 3 | 5.50 ±0.16ab | 2.16 ±1.54ab | 2.00 ±1.33ab | 3.63 ±0.95 |
| CRC | 3 | 4.24 ±0.34b | 1.12 ±0.91bc | 2.06 ±1.62ab | 2.84 ±1.35 |
| UC | 23 | 4.82 ±0.67ab | 2.16 ±1.26ac | 2.79 ±1.07a | 3.07 ±1.14 |
| UC location | |||||
| Ulcerative proctitis (E1) | 5 | 5.01 ± 0.20 | 3.07 ± 0.32 | 3.44 ± 0.51 | 2.77 ± 1.36 |
| Distal UC (E2) | 11 | 4.77 ± 0.61 | 2.41 ± 1.11 | 2.74 ± 0.99 | 3.13 ± 1.24 |
| Extensive UC or ulcerative pancolitis (E3) | 6 | 5.03 ± 0.76 | 1.18 ± 1.36 | 2.69 ± 1.23 | 3.11 ± 0.98 |
| CD | 31 | 4.22 ±1.31b | 1.13 ±1.64b | 1.47 ±1.37b | 3.19 ±1.35 |
| CD location | |||||
| Ileal-CD (L1) | 13 | 3.52 ± 1.33• | 0.17 ± 1.10• | 1.18 ± 1.55 | 2.87 ± 1.28 |
| Colonic-CD (L2) | 7 | 5.00 ± 0.93■ | 1.73 ± 1.94■ | 2.12 ± 1.36 | 3.33 ± 1.23 |
| Ileocolonic-CD (L3) | 10 | 4.47 ± 1.18•■ | 1.87 ± 1.60■ | 1.28 ± 1.09 | 3.42 ± 1.61 |
| p-value group of subjects | 0.024 | 0.002 | 0.015 | 0.540 | |
| p-value UC location | 0.568 | 0.110 | 0.432 | 0.961 | |
| p-value CD location | 0.050 | 0.025 | 0.349 | 0.538 | |
Mean log10 (16S rRNA gene copies/million bacterial 16S rRNA gene copies) ± standard deviations.
Statistics was calculated separately for each variable (column). Only for those analyses statistically significant (P-value in bold), pairwise comparisons were conducted, and groups of patients with similar abundances are indicated with the same superscript (a, b). Disease locations of UC and CD patients have been analyzed as independent groups. Similarly, patients' subtypes with similar abundances are indicated with the same superscript (•■). In both cases, groups not sharing superscript are those with statistically different median abundance values (P-value < 0.05).
F. prausnitzii phylogroup I load was reduced in all groups of patients in comparison to H subjects. This reduction was particularly noticeable in CD and CRC patients (P < 0.008), while in UC and IBS patients, it was observed as well, but less apparent. When analyzing data by disease location, I-CD patients showed the most marked reduction of phylogroup I counts in comparison to other CD locations (P = 0.025). Values did not differ significantly in UC patients when analyzed by location. However, loads in E2 and E3 subjects resembled that of CD patients, while for E1 subjects, their profiles were closer to that observed in H subjects.
With respect to F. prausnitzii phylogroup II, its abundance was significantly reduced in CD patients when compared to UC subjects (P = 0.015) while similar loads were observed between all the other groups of subjects (Table 3). Although no differences by IBD location were found, loads tend to be lower in those with ileal involvement (either I-CD or IC-CD, P = 0.069).
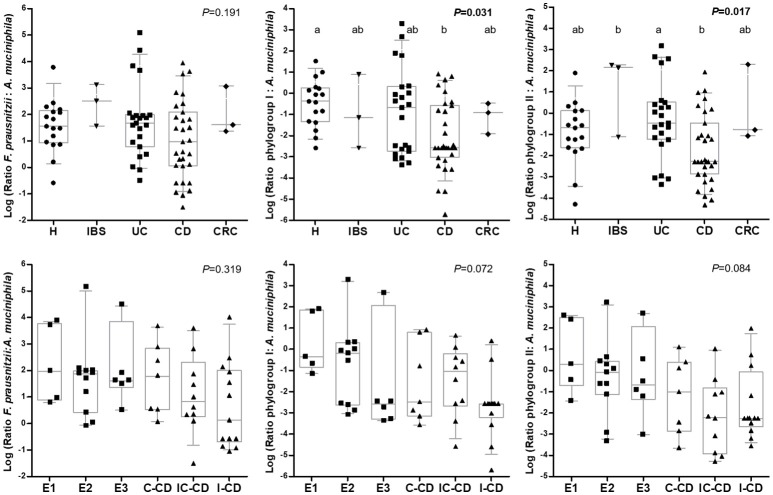
Interestingly, A. muciniphila load was similar between all the groups of subjects and also no differences were observed between IBD locations (P > 0.540; Table 3). In all groups of subjects, total F. prausnitzii counts outnumbered A. muciniphila, but there was a high variability between subjects, even within each condition. No difference in the ratio of total F. prausnitzii: A. muciniphila was found by group of subjects, neither when analyzing by IBD subtypes according to disease location. In contrast, when calculating these ratios by F. prausnitzii phylogroups, significant differences were found between conditions (Figure 2). CD patients, featured lower phylogroup I:A. muciniphila ratios than H (P = 0.031), and also lower phylogroup II: A. muciniphila ratios compared to UC (P = 0.017). When analyzing IBD groups by disease location, no significant differences were observed, probably due to the high dispersion of data and to the fact that when separating by location the number of patients included within each category is reduced. Nonetheless, subjects with a larger disease extension, or with ileum involvement, tended to feature lower values of both ratios. These differences in ratios are due to differences in F. prausnitzii (or phylogroup) load, as A. muciniphila load was similar across all subjects.
Figure 2.
Box and whiskers plot of the ratio total F. prausnitzii: A. muciniphila, F. prausnitzii phylogroup I: A. muciniphila, and F. prausnitzii phylogroup II: A. muciniphila: (from left to right) by group of subjects (up) and by Inflammatory Bowel Disease subtype (down). Data are represented as log10 of each ratio. The median is represented by the horizontal line in each box. Boxes cover the 25 and 75% quantiles, and bars show the 10 and 90% quantiles. Individual data are also shown. Homogeneous subgroups in each plot are indicated with the same superscript. H, control subjects; IBS, irritable bowel syndrome; UC, ulcerative colitis; CD, Croh's disease; CRC, colorectal cancer; E1, ulcerative proctitis; E2, ulcerative left-sided colitis; E3, ulcerative pancolitis; IC-CD, ileocolonic CD, I-CD, ileal CD; C-CD, colonic CD. A. muciniphila, 16S rRNA gene Akkermansia muciniphila; F. prausnitzii, 16S rRNA gene total Faecalibacterium prausnitzii; Phylogroup I, 16S rRNA gene F. prausnitzii phylogroup I; Phylogroup II, 16S rRNA gene F. prausnitzii phylogroup II.
A. muciniphila and F. prausnitzii abundances in relation to patients clinical and treatment data
No differences in A. muciniphila, counts were observed in either UC or CD patients according to activity status. Nevertheless, those patients with active UC featured the lowest load. F. prausnitzii and the abundance of the phylogroups did not differ between active and inactive UC patients (Table 4). CD patients with active disease feature lower levels of total F. prausnitzii and phylogroups in comparison to patients in remission, but differences did not achieve statistical significance either.
Table 4.
F. prausnitzii, its phylogroups and A. muciniphila abundance in inflammatory bowel disease patients by disease activity status.
| Diagnostics§ | n | F. prausnitzii* | P-value | Phylogroup I* | P-value | Phylogroup II* | P-value | A. muciniphila* | P-value |
|---|---|---|---|---|---|---|---|---|---|
| UC | |||||||||
| Active | 19 | 4.85 ± 0.71 | 0.464 | 2.12 ± 1.35 | 0.787 | 2.76 ± 1.11 | 0.523 | 3.04 ± 1.06 | 0.651 |
| Inactive | 3 | 4.61 ± 0.52 | 2.39 ± 0.97 | 3.18 ± 1.04 | 2.56 ± 1.29 | ||||
| CD | |||||||||
| Active | 18 | 4.10 ± 1.42 | 0.650 | 0.86 ± 1.45 | 0.373 | 1.36 ± 1.54 | 0.514 | 3.06 ± 1.28 | 0.514 |
| Inactive | 13 | 4.39 ± 1.19 | 1.52 ± 1.87 | 1.63 ± 1.15 | 3.36 ± 1.48 | ||||
Resection in CD patients was not a determining factor for A. muciniphila loads, either (Table 5). Instead, F. prausnitzii abundance was lower in those CD patients that underwent intestinal resection, with significant statistical differences for phylogroup II. Precisely, resected subjects had 10 times less phylogroup II than those without intestinal surgery (P = 0.018) whereas the phylogroup I load was only slightly lower in resected than non-resected patients.
Table 5.
F. prausnitzii, its phylogroups and A. muciniphila abundance in inflammatory bowel disease patients depending on whether or not they have had intestinal resection during the course of the disease.
| Diagnostics§ | n | F. prausnitzii* | P-value | Phylogroup I* | P-value | Phylogroup II* | P-value | A. muciniphila* | P-value |
|---|---|---|---|---|---|---|---|---|---|
| UC | |||||||||
| Non-resected | 19 | 4.73 ± 0.68 | na | 1.97 ± 1.21 | na | 2.79 ± 1.01 | 0.544 | 3.05 ± 1.14 | na |
| Resected | 1 | 4.91 | 3.45 | 2.68 | 4.11 | ||||
| CD | |||||||||
| Non-resected | 21 | 4.46 ± 1.33 | 0.239 | 1.33 ± 1.84 | 0.842 | 1.85 ± 1.40 | 0.018 | 3.29 ± 1.44 | 0.476 |
| Resected | 6 | 3.89 ± 1.04 | 1.07 ± 1.33 | 0.59 ± 0.44 | 3.46 ± 0.45 | ||||
Median (log10 16S rRNA gene copies/million bacterial 16S rRNA gene copies) ± standard deviations; na, not applicable. Analyses statistically significant are highlighted in boldface.
UC, ulcerative colitis; CD, Crohn's disease.
The A. muciniphila load was lower in CD patients who presented with the disease below 16 years of age (Table 6). This group of patients also featured very low quantities of F. prausnitzii phylogroup I although statistical significance was not achieved. No differences in these bacterial loads were observed between groups of UC patients with different age of disease onset. We also analyzed disease duration, but no statistically significant correlation was found between any of the bacterial loads and time of disease duration (data not shown).
Table 6.
F. prausnitzii, its phylogroups and A. muciniphila abundances in inflammatory bowel disease patients by age of disease onset.
| Diagnostics§ | n | F. prausnitzii* | P-value | Phylogroup I* | P-value | Phylogroup II* | P-value | A. muciniphila*# | P-value |
|---|---|---|---|---|---|---|---|---|---|
| UC | |||||||||
| 17–40 years (A2) | 11 | 4.87 ± 0.47 | 0.676 | 2.52 ± 1.09 | 0.171 | 3.12 ± 0.88 | 0.305 | 2.93 ± 1.19 | 0.569 |
| >41 years (A3) | 9 | 4.64 ± 0.91 | 1.65 ± 1.42 | 2.58 ± 1.10 | 2.88 ± 1.07 | ||||
| CD | |||||||||
| <16 years (A1) | 5 | 3.62 ± 1.59 | 0.004 ± 0.44 | 1.65 ± 1.71 | 1.76 ± 0.73a | ||||
| 17–40 years (A2) | 21 | 4.26 ± 1.36 | 0.562 | 1.40 ± 1.87 | 0.112 | 1.30 ± 1.34 | 0.547 | 3.31 ± 1.16b | 0.030 |
| >41 years (A3) | 3 | 4.67 ± 0.71 | 0.97 ± 0.48 | 2.26 ± 1.47 | 4.20 ± 2.15b | ||||
Median (log10 16S rRNA gene copies/million bacterial 16S rRNA gene copies) ± standard deviations.
UC, ulcerative colitis; CD, Crohn's disease.
Statistics was calculated separately for each variable (column). Groups of patients with similar abundances of A. muciniphila are indicated with the same superscript (a, b) whereas groups not sharing superscript are those with statistically different median abundance values (P < 0.05). Analyses statistically significant are highlighted in boldface.
Finally, data were analyzed by taking into account the medication of the patients at the time of sampling (Table 7). No differences in A. muciniphila, F. prausnitzii or in phylogroups abundances were observed between medications for any IBD. However, those UC patients that received anti-tumor necrosis factor had the lowest levels of A. muciniphila. In contrast, those CD patients receiving moderate immunosupressants had lower F. prausnitzii loads than patients without treatment or receiving therapies such as mesalazine or anti-tumor necrosis factor.
Table 7.
F. prausnitzii, its phylogroups and A. muciniphila abundances in inflammatory bowel disease by medication at sampling.
| Diagnostics§ | n | F. prausnitzii* | P-value | Phylogroup I* | P-value | Phylogroup II* | P-value | A. muciniphila | P-value |
|---|---|---|---|---|---|---|---|---|---|
| UC | |||||||||
| No treatment | 11 | 4.81 ± 0.72 | 1.94 ± 1.35 | 2.74 ± 1.13 | 3.10 ± 1.11 | ||||
| Mesalazine | 3 | 4.87 ± 0.40 | 0.783 | 2.30 ± 0.97 | 0.578 | 3.20 ± 1.01 | 0.639 | 3.53 ± 1.82 | 0.387 |
| Mod. Immsup | 4 | 4.95 ± 0.61 | 2.97 ± 0.33 | 3.18 ± 1.54 | 3.06 ± 0.51 | ||||
| Anti-TNF | 3 | 4.35 ± 0.96 | 1.53 ± 1.85 | 2.37 ± 1.03 | 2.10 ± 1.52 | ||||
| CD | |||||||||
| No treatment | 10 | 4.30 ± 1.51 | 1.04 ± 1.96 | 1.67 ± 1.61 | 2.69 ± 1.08 | ||||
| Mesalazine | 3 | 5.00 ± 0.41 | 0.537 | 1.30 ± 1.67 | 0.975 | 2.24 ± 1.89 | 0.719 | 3.90 ± 2.20 | 0.125 |
| Mod. Immsup | 10 | 3.84 ± 1.21 | 0.99 ± 1.50 | 1.15 ± 1.09 | 3.72 ± 0.97 | ||||
| Anti-TNF | 6 | 4.31 ± 1.68 | 1.42 ± 1.76 | 1.60 ± 1.42 | 2.72 ± 1.78 | ||||
Median (log10 16S rRNA gene copies/million bacterial 16S rRNA gene copies) ± standard deviations.
UC, ulcerative colitis; CD, Crohn's disease; Mod. Immsup, moderate immunosuppresants; Anti-TNF, Anti-tumor necrosis factor.
Correlation between A. muciniphila and F. prausnitzii abundances
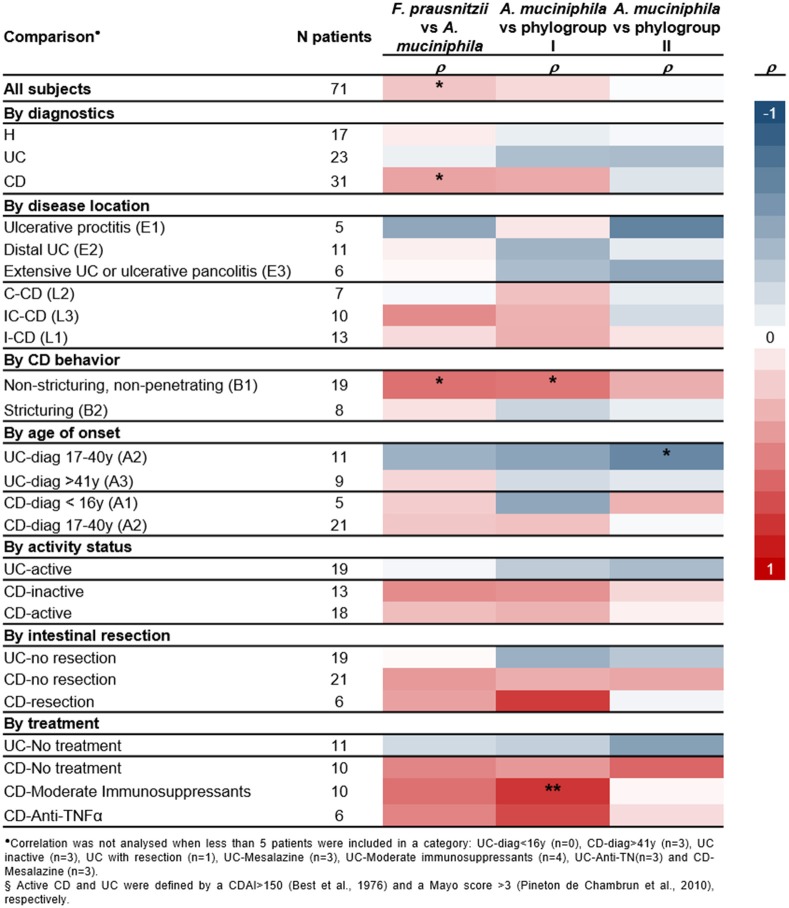
Correlation between A. muciniphila and F. prausnitzii numbers was analyzed to provide supporting evidence for a direct/indirect effect of one population over the other or about a putative common factor affecting both species populations in a given condition (Figure 3).
Figure 3.
Heatmap of spearman correlation cofficients between A. muciniphila, total F. prausnitzii and its phylogroups abundances, splitting up patients by diagnostics, inflammatory bowel disease subtypes, and main clinical characteristics. Significant correlations are indicated *P < 0.05, **P < 0.01.
No correlation between these two species was found in H or UC patients (Figure S3). Therefore, in these conditions, factors of the gut environment may be differentially impacting on each species. In contrast, positive correlation of both species load was observed in CD subjects (Figure 3). Analysis by phylogroups indicated that A. muciniphila quantity tended to positively correlate with phylogroup I in CD patients (P = 0.060), whereas no significant correlation was observed with phylogroup II (Figure S3).
Moreover, no significant correlation between the two species was observed, when splitting subjects by activity status or whether or not they have had intestinal resection. Of note, a positive correlation between A. muciniphila and F. prausnitzii (particularly phylogroup I) was observed in CD patients with non-stricturing, non-penetrating disease (B1) and in those under moderate immunosuppressants (ρ ≥ 0.539, P ≤ 0.017). In contrast, in UC patients with disease onset between 17 and 40 years of age, a negative correlation between A. muciniphila and F. prausnitzii phylogroup II was observed (ρ = −0.673, P = 0.023).
Discussion
A. muciniphila and F. prausnitzii are two symbiotic and numerically abundant members of the gut microbiota, and both have been associated with dysbiosis in several disease conditions, including IBD. Their niche is close to the intestinal mucosa, and therefore it can be hypothesized that they may play a key role in cross-talk with the host. Both species are considered to have a part in a well-functioning gut and thus are considered as promising next generation probiotics (Neef and Sanz, 2013; Martín et al., 2017). In the present study we have analyzed the prevalence and abundance of mucosa associated A. muciniphila, total F. prausnitzii and phylogroup in H and IBD subjects, taking into account the diversity of disease locations and the clinical features of patients. Some IBS and CRC have been included as well, but only for illustrative purposes given the low number of patients engaged. The abundance of both species has been previously reported to be reduced in several intestinal disorders (Belzer and de Vos, 2012; Miquel et al., 2013), but here for the first time we correlate the load of both species. Through analysis of clinical data, we consider which particular conditions this underrepresentation is favored, and whether or not the imbalance of one species is linked to changes in the abundance of the other.
Our data show that the A. muciniphila load in the mucosa of H subjects is slightly higher (2.0- to 4.5-fold, respectively) than in IBD and CRC patients, but is not statistically significant. An increase in A. muciniphila abundance in CRC patients compared to controls has been previously found in stools (Weir et al., 2013) but not in mucosal biopsies (Mira-Pascual et al., 2015), and the analysis of our limited cohort is in line with this finding. Previous studies have reported a significant decrease of this species in IBD subjects (Png et al., 2010). Methodological differences may explain the inconsistency with our findings, as we exclusively focused on colonic samples. In biopsy samples, Png and collaborators observed a reduction of this species in IBD patients that ranged between 2.9- and 3.9-fold when compared to controls, which is similar to the reduction observed in our subjects. In that study (Png et al., 2010), differences were observed depending on whether or not the tissue was affected, with the depletion being more conspicuous in inflamed tissue, and without reaching significant differences between non-inflamed tissue of CD and controls, which is in line with our results as we used non-affected tissue. In addition, we have explored differences taking into account disease location, activity or intestinal resection, but no association between A. muciniphila load and these variables has been revealed. Intriguingly, CD patients who presented with disease below 16 years of age had a striking reduction of this species compared to those with disease onset at a later age. A. muciniphila has been reported to colonize the gut in early infancy, and loads in infants 1 year old are similar to that found in adults (Collado et al., 2007). Therefore, it seems likely that this depletion is not a general phenomenon that occurs in IBD or age-driven, but due to particular features of pediatric IBD that are sustained throughout the disease. In line with this, discrepancies between dysbiosis signatures in adult and infant IBD patients have been previously reported (Hansen et al., 2012) and it remains to be explored through prospective studies if early disease onset results in long term microbial signatures. Another future application of this finding could be to explore the usefulness of A. muciniphila depletion as a biomarker to assist in pediatric IBD diagnosis.
Regarding mucosa-associated F. prausnitzii loads we have observed a marked reduction in CRC and CD patients, especially in those with ileal involvement, affecting both phylogroups of this species. Although less prominent, UC patients also featured lower F. prausnitzii abundance than H subjects. Our study is in agreement with previous reports which found F. prausnitzii to be reduced in CRC and IBD adults (Swidsinski et al., 2005, 2008; Martinez-Medina et al., 2006; Frank et al., 2007; Sokol et al., 2008, 2009; Willing et al., 2009; McLaughlin et al., 2010; Vermeiren et al., 2012; Kabeerdoss et al., 2013; Machiels et al., 2013; Miquel et al., 2013; Lopez-Siles et al., 2014, 2016). Besides, lower abundance of both F. prausnitzii phylogroups has been previously reported concerning CD patients (Jia et al., 2010; Lopez-Siles et al., 2016), which is in line with our findings. Moreover, because we have observed differences between IBD subtypes, our results support the hypothesis that patients with ileal disease location constitute a differentiated pathological entity (Willing et al., 2009). We have corroborated that the reduction of F. prausnitzii numbers compared to H subjects takes place in both active and inactive IBD patients (Willing et al., 2009), with active CD patients featuring the lowest levels of phylogroup I. Also in agreement with previous studies (Sokol et al., 2008) lower numbers of F. prausnitzii were detected in resected CD patients, but in our study, statistically significant differences were only achieved for phylogroup II, probably because the depletion was more striking. It remains unknown why there are shifts in particular subgroups of this species. To date, several articles convey the point that the genus Faecalibacterium hosts a complex diversity (Lopez-Siles et al., 2015; Benevides et al., 2017; Martín et al., 2017). This diversity has been shown mainly through phylogenetic methods, but phenotypical diversity also exists. Supporting this point, studies characterizing several strains of this species isolated from different origins have failed to find phenotypic traits that consistently distinguish members from one or other subtype (Lopez-Siles et al., 2012; Foditsch et al., 2014; Martín et al., 2017). However, the effect of host factors differentially influencing F. prausnitzii subpopulations has been poorly explored which may explain our results. Another hypothesis could be that subtypes of F. prausnitzii interact in a different manner with other members of the microbiome, which has also been scarcely studied to date.
We have explored co-occurrence and correlation between A. muciniphila and F. prausnitzii in H and IBD patients. We considered that both species may have a syntrophic relationship, thus we hypothesize that the depletion or enrichment of one would imply the same effect on the other. In particular, A. muciniphila mucolytic activity could release oligosaccharides, co-factors, vitamins, and short chain fatty acids, including acetate that juxtaposed species could use for growth. Indeed, F. prausnitzii has been proven to be able to use some oligosaccharides derived from mucus and its growth is stimulated by acetate and requires presence of vitamins in the medium (Duncan et al., 2002; Lopez-Siles et al., 2012). These compounds, can be provided by A. muciniphila, although not exclusively, and therefore establish cross-feeding interactions. A recent study based on co-culture experiments demonstrated this trophic interaction (Belzer et al., 2017). However, in most of the cases studied here, we did not find a correlation between F. prausnitzii and A. muciniphila abundances, and the two species co-occurred only in 41.5% of subjects engaged in the study. This may be because F. prausnitzii does not depend exclusively on by-products synthesized by A. muciniphila. In agreement with that, other studies have reported that F. prausnitzii can benefit from the presence of a variety of acetate-producing species (Wrzosek et al., 2013; Rios-Covian et al., 2015). It would be interesting to determine whether this species (or other mucus-inhabiting species) increase in patients in which A. muciniphila diminishes, and thus may be partially replacing its role concerning acetate production.
Nonetheless, we observed that positive correlation between the two species happens in CD patients, and particularly for those with B1 behavior or under immunosuppressant therapy. The fact that there is a positive correlation of the two species indicates that their abundance varies in a similar way in this particular condition. The most likely scenario is that in CD the two bacteria are similarly affected by host and gut environmental factors. To support this hypothesis, both species share the characteristic that their growth is severely compromised at pH < 5.5 (Derrien et al., 2004; Lopez-Siles et al., 2012) and in turn, acidic stools have been reported for IBD patients (Nugent et al., 2001; Barkas et al., 2013). Notably, in those cases of correlation between both species, only members of phylogroup I were involved. It would be interesting to perform co-culture studies with different F. prausnitzii strains, and monitor their growth under different conditions in order to determine more accurately their relationship.
In our cohort, A. muciniphila was not detected in 57.1% of all subjects but this seems to be related to the fact that this species has a lower relative abundance in the gut, rather than to a higher sensitivity to gut disease. In contrast, A. muciniphila was more frequently found in UC patients. This could be partially explained by the fact that a higher proportion of loose mucus has been found in UC patients (Antoni et al., 2014), which is the likely niche for A. muciniphila. Another hypothesis that can not be ruled out is that some factor of UC patients favors the presence of A. muciniphila. In our limited cohort, we have also observed higher prevalence of this species in CRC group compared to controls, which is in line with previous findings (Mira-Pascual et al., 2015). However, our data points out that this higher presence does not imply an increase in the abundance at the mucosal level. In contrast, almost 90% of subjects were F. prausnitzii carriers and thus the fact that it is a second-liner in the mucosa and the fact that this species can rely on other members of the gut microbiota for cross-feeding may explain its higher ubiquity and abundance compared to A. muciniphila.
Finally, our study revealed that CD patients are characterized by a low F. prausnitzii: A. muciniphila, ratio affecting both phylogroups. This indicates that compared to H and UC, these patients have an altered proportion of beneficial microorganisms in the mucosa. Although our study does not allow us to decipher if this imbalance is a cause or a consequence of the disease, it can be an aggravator because the two species have been linked to be key for mucus integrity (Derrien et al., 2004; Wrzosek et al., 2013) and gut homeostasis. A significant depletion of both species has also been reported in children with atopic disease (Candela et al., 2012), and therapeutic strategies to restore these species needs to be explored, particularly for disorders that have in common to feature chronic inflammation. In addition, two recent studies have linked the two species with response to immunotherapy treatment (Gopalakrishnan et al., 2017; Routy et al., 2017), thus pointing out another situation in which it is relevant to have these bacteria. Further studies to assess implications in IBD treatment response would be interesting, as immunomodulators are among the usual therapies prescribed to IBD patients. Finally, further confirmation of our results in a larger cohort would be required given that we have engaged a limited number of subjects, and thus it would provide robustness to those findings not sustained after FRD assessment.
Conclusions
IBD patients are characterized by a reduction of F. prausnitzii and a slight underrepresentation of A. muciniphila in the colonic mucosa, regardless of disease activity status. While differences in F. prausnitzii load have been observed for I-CD patients, early onset CD is characterized by a lack of A. muciniphila, but further prospective studies are required to assess if this feature is sustained long term. Positive correlation between the two species was found in CD patients, and further studies are required to elucidate which common factors alter both populations in particular gut disorders.
Author contributions
XA, SD, LG-G, ML-S, and MM-M study concept and design. XA, NE-C, ML-S, and MS-M acquisition of data. ML-S and MM-M interpretation of data and statistical analysis. ML-S drafting the manuscript. XA, SD, NE-C, LG-G, MM-M, and MS-M critical revision of the manuscript for important intellectual content. LG-G and MM-M obtained funding. All authors have approved the final version of the manuscript and agree to be accountable for all aspects of the work, ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Conflict of interest statement
XA is a consultant for AbbVie, Janssen and Takeda, and has received honoraria for lectures, including services on speakers bureaus from AbbVie, MS-D, Janssen, Takeda, Shire, Zambon and Ferring. XA, LG-G, ML-S, and MM-M, have filed a European patent for a “Method for the detection, follow up and/or classification of intestinal diseases” (application number EP15382427). The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We appreciate the generosity of the patients who freely gave their time and samples to make this study possible, and the theater staff of all centers for their dedication and careful sample collection. Thanks are due to MD. David Busquets and Ms. Romà Surís for assistance with samples collection and analyses, respectively. We are thankful to Ms. Natàlia Adell for statistical help.
Footnotes
Funding. This work was funded by the Universitat de Girona projects MPCUdG2016-009 and GdRCompetUdG2017, and the Spanish Ministry of Education and Science through projects SAF2006-00414, SAF2010-15896 and SAF2013-43284-P, being the last co-funded by the European Regional Development. Dr. Sylvia H. Duncan acknowledges support from the Scottish Government Research and Environment Science and Analytical Services Division (RESAS).
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2018.00281/full#supplementary-material
References
- Andoh A., Tsujikawa T., Sasaki M., Mitsuyama K., Suzuki Y., Matsui T., et al. (2009). Faecal microbiota profile of Crohn's disease determined by terminal restriction fragment length polymorphism analysis. Aliment. Pharmacol. Ther. 29, 75–82. 10.1111/j.1365-2036.2008.03860.x [DOI] [PubMed] [Google Scholar]
- Antoni L., Nuding S., Wehkamp J., Stange E. F. (2014). Intestinal barrier in inflammatory bowel disease. World J. Gastroenterol. 20, 1165–1179. 10.3748/wjg.v20.i5.1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balamurugan R., Rajendiran E., George S., Samuel G. V., Ramakrishna B. S. (2008). Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J. Gastroenterol. Hepatol. 23, 1298–1303. 10.1111/j.1440-1746.2008.05490.x [DOI] [PubMed] [Google Scholar]
- Barkas F., Liberopoulos E., Kei A., Elisaf M. (2013). Electrolyte and acid-base disorders in inflammatory bowel disease. Ann. Gastroenterol. 26, 23–28. [PMC free article] [PubMed] [Google Scholar]
- Baumgart M., Dogan B., Rishniw M., Weitzman G., Bosworth B., Yantiss R., et al. (2007). Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum. ISME J. 1, 403–418. 10.1038/ismej.2007.52 [DOI] [PubMed] [Google Scholar]
- Belzer C., Chia L. W., Aalvink S., Chamlagain B., Piironen V., Knol J., et al. (2017). Microbial metabolic networks at the mucus layer lead to diet-independent butyrate and vitamin B12 production by intestinal symbionts. MBio 8:e00770–17. 10.1128/mBio.00770-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belzer C., de Vos W. M. (2012). Microbes inside—from diversity to function: the case of Akkermansia. ISME J. 6:1449. 10.1038/ismej.2012.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benevides L., Burman S., Martin R., Robert V., Thomas M., Miquel S., et al. (2017). New Insights into the diversity of the genus Faecalibacterium. Front. Microbiol. 8:1790. 10.3389/fmicb.2017.01790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best W. R., Becktel J. M., Singleton J. W., Kern F., Jr. (1976). Development of a Crohn's disease activity index. National Cooperative Crohn's disease study. Gastroenterology 70, 439–444. 10.1016/S0016-5085(76)80163-1 [DOI] [PubMed] [Google Scholar]
- Candela M., Rampelli S., Turroni S., Severgnini M., Consolandi C., De Bellis G., et al. (2012). Unbalance of intestinal microbiota in atopic children. BMC Microbiol. 12:95. 10.1186/1471-2180-12-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson A. H., Yakymenko O., Olivier I., Håkansson F., Postma E., Keita A. V., et al. (2013). Faecalibacterium prausnitzii supernatant improves intestinal barrier function in mice DSS colitis. Scand. J. Gastroenterol. 48, 1136–1144. 10.3109/00365521.2013.828773 [DOI] [PubMed] [Google Scholar]
- Collado M. C., Derrien M., Isolauri E., de Vos W. M., Salminen S. (2007). Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl. Environ. Microbiol. 73, 7767–7770. 10.1128/AEM.01477-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Palma G., Nadal I., Collado M. C., Sanz Y. (2009). Effects of a gluten-free diet on gut microbiota and immune function in healthy adult human subjects. Br. J. Nutr. 102, 1154–1160. 10.1017/S0007114509371767 [DOI] [PubMed] [Google Scholar]
- Derrien M., Collado M. C., Ben-Amor K., Salminen S., de Vos W. M. (2008). The mucin degrader Akkermansia muciniphila is an abundant resident of the human intestinal tract. Appl. Environ. Microbiol. 74, 1646–1648. 10.1128/AEM.01226-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M., Van Baarlen P., Hooiveld G., Norin E., Müller M., de Vos W. M. (2011). Modulation of mucosal immune response, tolerance, and proliferation in mice colonized by the mucin-degrader Akkermansia muciniphila. Front. Microbiol. 2:166. 10.3389/fmicb.2011.00166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M., Vaughan E. E., Plugge C. M., de Vos W. M. (2004). Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 54, 1469–1476. 10.1099/ijs.0.02873-054/5/1469 [DOI] [PubMed] [Google Scholar]
- Dörffel Y., Swidsinski A., Loening-Baucke V., Wiedenmann B., Pavel M. (2012). Common biostructure of the colonic microbiota in neuroendocrine tumors and Crohn's disease and the effect of therapy. Inflamm. Bowel Dis. 18, 1663–1671. 10.1002/ibd.21923 [DOI] [PubMed] [Google Scholar]
- Duncan S. H., Hold G. L., Harmsen H. J., Stewart C. S., Flint H. J. (2002). Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 52, 2141–2146. 10.1099/00207713-52-6-2141 [DOI] [PubMed] [Google Scholar]
- Everard A., Belzer C., Geurts L., Ouwerkerk J. P., Druart C., Bindels L. B., et al. (2013). Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. U.S.A. 110, 9066–9071. 10.1073/pnas.1219451110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint H. J., Scott K. P., Louis P., Duncan S. H. (2012). The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 9, 577–589. 10.1038/nrgastro.2012.156 [DOI] [PubMed] [Google Scholar]
- Foditsch C., Santos T. M., Teixeira A. G., Pereira R. V., Dias J. M., Gaeta N., et al. (2014). Isolation and characterization of Faecalibacterium prausnitzii from calves and piglets. PLoS ONE 9:e116465. 10.1371/journal.pone.0116465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D. N., St Amand A. L., Feldman R. A., Boedeker E. C., Harpaz N., Pace N. R. (2007). Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U.S.A. 104, 13780–13785. 10.1073/pnas.0706625104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furet J.-P., Firmesse O., Gourmelon M., Bridonneau C., Tap J., Mondot S., et al. (2009). Comparative assessment of human and farm animal faecal microbiota using real-time quantitative PCR. FEMS Microbiol. Ecol. 68, 351–362. 10.1111/j.1574-6941.2009.00671.x [DOI] [PubMed] [Google Scholar]
- Furet J. P., Kong L. C., Tap J., Poitou C., Basdevant A., Bouillot J. L., et al. (2010). Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss: links with metabolic and low-grade inflammation markers. Diabetes 59, 3049–3057. 10.2337/db10-0253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan V., Spencer C. N., Nezi L., Reuben A., Andrews M. C., Karpinets T. V., et al. (2017). Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science 359, 97–103. 10.1126/science.aan4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gophna U., Sommerfeld K., Gophna S., Doolittle W. F., Veldhuyzen van Zanten S. J. (2006). Differences between tissue-associated intestinal microfloras of patients with Crohn's disease and ulcerative colitis. J. Clin. Microbiol. 44, 4136–4141. 10.1128/JCM.01004-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graessler J., Qin Y., Zhong H., Zhang J., Licinio J., Wong M. L., et al. (2012). Metagenomic sequencing of the human gut microbiome before and after bariatric surgery in obese patients with type 2 diabetes: correlation with inflammatory and metabolic parameters. Pharmacogenomics J. 13, 514–522. 10.1038/tpj.2012.43 [DOI] [PubMed] [Google Scholar]
- Hansen R., Russell R. K., Reiff C., Louis P., McIntosh F., Berry S. H., et al. (2012). Microbiota of de-novo pediatric IBD: increased Faecalibacterium prausnitzii and reduced bacterial diversity in Crohn's but not in ulcerative colitis. Am. J. Gastroenterol. 107, 1913–1922. 10.1038/ajg.2012.335 [DOI] [PubMed] [Google Scholar]
- Jia W., Whitehead R. N., Griffiths L., Dawson C., Waring R. H., Ramsden D. B., et al. (2010). Is the abundance of Faecalibacterium prausnitzii relevant to Crohn's disease? FEMS Microbiol. Lett. 310, 138–144. 10.1111/j.1574-6968.2010.02057.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joossens M., Huys G., Cnockaert M., De Preter V., Verbeke K., Rutgeerts P., et al. (2011). Dysbiosis of the faecal microbiota in patients with Crohn's disease and their unaffected relatives. Gut 60, 631–637. 10.1136/gut.2010.223263 [DOI] [PubMed] [Google Scholar]
- Kabeerdoss J., Sankaran V., Pugazhendhi S., Ramakrishna B. S. (2013). Clostridium leptum group bacteria abundance and diversity in the fecal microbiota of patients with inflammatory bowel disease: a case-control study in India. BMC Gastroenterol. 13:20. 10.1186/1471-230X-13-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C.-S., Ban M., Choi E.-J., Moon H.-G., Jeon J.-S., Kim D.-K., et al. (2013). Extracellular vesicles derived from gut microbiota, especially Akkermansia muciniphila, protect the progression of dextran sulfate sodium-induced colitis. PLoS ONE 8:e76520. 10.1371/journal.pone.0076520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics. eds Stackebrandt E., Goodfellow M. (New York, NY: John Wiley and Sons; ), 115–175. [Google Scholar]
- Lopez-Siles M., Khan T. M., Duncan S. H., Harmsen H. J., Garcia-Gil L. J., Flint H. J. (2012). Cultured representatives of two major phylogroups of human colonic Faecalibacterium prausnitzii can utilize pectin, uronic acids, and host-derived substrates for growth. Appl. Environ. Microbiol. 78, 420–428. 10.1128/AEM.06858-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Siles M., Martinez-Medina M., Abellà C., Busquets D., Sabat-Mir M., Duncan S. H., et al. (2015). Mucosa-associated Faecalibacterium prausnitzii phylotype richness is reduced in patients with inflammatory bowel disease. Appl. Environ. Microbiol. 81, 7582–7592. 10.1128/AEM.02006-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Siles M., Martinez-Medina M., Busquets D., Sabat-Mir M., Duncan S. H., Flint H. J., et al. (2014). Mucosa-associated Faecalibacterium prausnitzii and Escherichia coli co-abundance can distinguish irritable bowel syndrome and inflammatory bowel disease phenotypes. Int. J. Med. Microbiol. 304, 464–475. 10.1016/j.ijmm.2014.02.009 [DOI] [PubMed] [Google Scholar]
- Lopez-Siles M., Martinez-Medina M., Surís-Valls R., Aldeguer X., Sabat-Mir M., Duncan S. H., et al. (2016). Changes in the abundance of Faecalibacterium prausnitzii phylogroups I and II in the intestinal mucosa of inflammatory bowel disease and patients with colorectal cancer. Inflamm. Bowel Dis. 22, 28–41. 10.1097/MIB.0000000000000590 [DOI] [PubMed] [Google Scholar]
- Lozupone C., Faust K., Raes J., Faith J. J., Frank D. N., Zaneveld J., et al. (2012). Identifying genomic and metabolic features that can underlie early successional and opportunistic lifestyles of human gut symbionts. Genome Res. 22, 1974–1984. 10.1101/gr.138198.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukovac S., Belzer C., Pellis L., Keijser B. J., de Vos W. M., Montijn R. C., et al. (2014). Differential modulation by Akkermansia muciniphila and Faecalibacterium prausnitzii of host peripheral lipid metabolism and histone acetylation in mouse gut organoids. MBio 5, e01438–e01414. 10.1128/mBio.01438-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machiels K., Joossens M., Sabino J., De Preter V., Arijs I., Eeckhaut V., et al. (2013). A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 63, 1275–1283. 10.1136/gutjnl-2013-304833 [DOI] [PubMed] [Google Scholar]
- Manichanh C., Borruel N., Casellas F., Guarner F. (2012). The gut microbiota in IBD. Nat. Rev. Gastroenterol. Hepatol. 9, 599–608. 10.1038/nrgastro.2012.152 [DOI] [PubMed] [Google Scholar]
- Manichanh C., Rigottier-Gois L., Bonnaud E., Gloux K., Pelletier E., Frangeul L., et al. (2006). Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut 55, 205–211. 10.1136/gut.2005.073817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín R., Chain F., Miquel S., Lu J., Gratadoux J. J., Sokol H., et al. (2014). The commensal bacterium Faecalibacterium prausnitzii is protective in DNBS-induced chronic moderate and severe colitis models. Inflamm. Bowel Dis. 20, 417–430. 10.1097/01.MIB.0000440815.76627.64 [DOI] [PubMed] [Google Scholar]
- Martín R., Miquel S., Benevides L., Bridonneau C., Robert V., Hudault S., et al. (2017). Functional characterization of novel Faecalibacterium prausnitzii strains isolated from healthy volunteers: a step forward in the use of F. prausnitzii as a Next-Generation Probiotic. Front. Microbiol. 8:1226. 10.3389/fmicb.2017.01226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín R., Miquel S., Chain F., Natividad J. M., Jury J., Lu J., et al. (2015). Faecalibacterium prausnitzii prevents physiological damages in a chronic low-grade inflammation murine model. BMC Microbiol. 15:67. 10.1186/s12866-015-0400-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Medina M., Aldeguer X., Gonzalez-Huix F., Acero D., Garcia-Gil L. J. (2006). Abnormal microbiota composition in the ileocolonic mucosa of Crohn's disease patients as revealed by polymerase chain reaction-denaturing gradient gel electrophoresis. Inflamm. Bowel Dis. 12, 1136–1145. 10.1097/01.mib.0000235828.09305.0c [DOI] [PubMed] [Google Scholar]
- McLaughlin S. D., Clark S. K., Tekkis P. P., Nicholls R. J., Ciclitira P. J. (2010). The bacterial pathogenesis and treatment of pouchitis. Therap. Adv. Gastroenterol. 3, 335–348. 10.1177/1756283X10370611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza Hernández J. L., Lana Soto R., Díaz-Rubio M. (2007). Definiciones y Manifestaciones Clínicas Generales Madrid: Arán Ediciones S.L. [Google Scholar]
- Miquel S., Martín R., Rossi O., Bermúdez-Humarán L., Chatel J., Sokol H., et al. (2013). Faecalibacterium prausnitzii and human intestinal health. Curr. Opin. Microbiol. 16, 255–261. 10.1016/j.mib.2013.06.003 [DOI] [PubMed] [Google Scholar]
- Mira-Pascual L., Cabrera-Rubio R., Ocon S., Costales P., Parra A., Suarez A., et al. (2015). Microbial mucosal colonic shifts associated with the development of colorectal cancer reveal the presence of different bacterial and archaeal biomarkers. J. Gastroenterol. 50, 167–179. 10.1007/s00535-014-0963-x [DOI] [PubMed] [Google Scholar]
- Mondot S., Kang S., Furet J. P., Aguirre de Carcer D., McSweeney C., Morrison M., et al. (2011). Highlighting new phylogenetic specificities of Crohn's disease microbiota. Inflamm. Bowel Dis. 17, 185–192. 10.1002/ibd.21436 [DOI] [PubMed] [Google Scholar]
- Neef A., Sanz Y. (2013). Future for probiotic science in functional food and dietary supplement development. Curr. Opin. Clin. Nutr. Metab. Care 16, 679–687. 10.1097/MCO.0b013e328365c258 [DOI] [PubMed] [Google Scholar]
- Nugent S. G., Kumar D., Rampton D. S., Evans D. F. (2001). Intestinal luminal pH in inflammatory bowel disease: possible determinants and implications for therapy with aminosalicylates and other drugs. Gut 48, 571–577. 10.1136/gut.48.4.571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineton de Chambrun G., Peyrin-Biroulet L., Lémann M., Colombel J. F. (2010). Clinical implications of mucosal healing for the management of IBD. Nat. Rev. Gastroenterol. Hepatol. 7, 15–29. 10.1038/nrgastro.2009.203 [DOI] [PubMed] [Google Scholar]
- Png C. W., Lindén S. K., Gilshenan K. S., Zoetendal E. G., McSweeney C. S., Sly L. I., et al. (2010). Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am. J. Gastroenterol. 105, 2420–2428. 10.1038/ajg.2010.281 [DOI] [PubMed] [Google Scholar]
- Rajilić-Stojanović M., Biagi E., Heilig H. G., Kajander K., Kekkonen R. A., Tims S., et al. (2011). Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology 141, 1792–1801. 10.1053/j.gastro.2011.07.043 [DOI] [PubMed] [Google Scholar]
- Rios-Covian D., Gueimonde M., Duncan S. H., Flint H. J., de los Reyes-Gavilan C. G. (2015). Enhanced butyrate formation by cross-feeding between Faecalibacterium prausnitzii and Bifidobacterium adolescentis. FEMS Microbiol. Lett. 362:fnv176. 10.1093/femsle/fnv176 [DOI] [PubMed] [Google Scholar]
- Rossi O., Khan M. T., Schwarzer M., Hudcovic T., Srutkova D., Duncan S. H., et al. (2015). Faecalibacterium prausnitzii strain HTF-F and its extracellular polymeric matrix attenuate clinical parameters in DSS-induced colitis. PLoS ONE 10:e0123013. 10.1371/journal.pone.0123013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routy B., Le Chatelier E., Derosa L., Duong C. P. M., Alou M. T., Daillère R., et al. (2017). Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97. 10.1126/science.aan3706 [DOI] [PubMed] [Google Scholar]
- Sartor R. B. (2006). Mechanisms of disease: pathogenesis of Crohn's disease and ulcerative colitis. Nat. Clin. Pract. Gastroenterol. Hepatol. 3, 390–407. 10.1038/ncpgasthep0528 [DOI] [PubMed] [Google Scholar]
- Sartor R. B. (2008). Microbial influences in inflammatory bowel diseases. Gastroenterology 134, 577–594. 10.1053/j.gastro.2007.11.059 [DOI] [PubMed] [Google Scholar]
- Seksik P., Sokol H., Lepage P., Vasquez N., Manichanh C., Mangin I., et al. (2006). Review article: the role of bacteria in onset and perpetuation of inflammatory bowel disease. Aliment. Pharmacol. Ther. 24, 11–18. 10.1111/j.1365-2036.2006.03053.x [DOI] [PubMed] [Google Scholar]
- Silverberg M. S., Satsangi J., Ahmad T., Arnott I. D., Bernstein C. N., Brant S. R., et al. (2005). Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: report of a working party of the 2005 Montreal World Congress of Gastroenterology. Can. J. Gastroenterol. 19, 5A–36A. 10.1155/2005/269076 [DOI] [PubMed] [Google Scholar]
- Sokol H., Pigneur B., Watterlot L., Lakhdari O., Bermúdez-Humarán L. G., Gratadoux J. J., et al. (2008). Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U.S.A. 105, 16731–16736. 10.1073/pnas.0804812105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H., Seksik P. (2010). The intestinal microbiota in inflammatory bowel diseases: time to connect with the host. Curr. Opin. Gastroenterol. 26, 327–331. 10.1097/MOG.0b013e328339536 [DOI] [PubMed] [Google Scholar]
- Sokol H., Seksik P., Furet J. P., Firmesse O., Nion-Larmurier I., Beaugerie L., et al. (2009). Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm. Bowel Dis. 15, 1183–1189. 10.1002/ibd.20903 [DOI] [PubMed] [Google Scholar]
- Stoddard S. F., Smith B. J., Hein R., Roller B. R., Schmidt T. M. (2015). rrnDB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res. 43, D593–D598. 10.1093/nar/gku1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidsinski A., Dörffel Y., Loening-Baucke V., Theissig F., Rückert J. C., Ismail M., et al. (2011). Acute appendicitis is characterised by local invasion with Fusobacterium nucleatum/necrophorum. Gut 60, 34–40. 10.1136/gut.2009.191320 [DOI] [PubMed] [Google Scholar]
- Swidsinski A., Loening-Baucke V., Lochs H., Hale L. P. (2005). Spatial organization of bacterial flora in normal and inflamed intestine: a fluorescence in situ hybridization study in mice. World. J. Gastroenterol. 11, 1131–1140. 10.3748/wjg.v11.i8.1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidsinski A., Loening-Baucke V., Vaneechoutte M., Doerffel Y. (2008). Active Crohn's disease and ulcerative colitis can be specifically diagnosed and monitored based on the biostructure of the fecal flora. Inflamm. Bowel Dis. 14, 147–161. 10.1002/ibd.20330 [DOI] [PubMed] [Google Scholar]
- van Passel M. W., Kant R., Zoetendal E. G., Plugge C. M., Derrien M., Malfatti S. A., et al. (2011). The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PLoS ONE 6:e16876. 10.1371/journal.pone.0016876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeiren J., Van den Abbeele P., Laukens D., Vigsnaes L. K., De Vos M., Boon N., et al. (2012). Decreased colonization of fecal Clostridium coccoides/Eubacterium rectale species from ulcerative colitis patients in an in vitro dynamic gut model with mucin environment. FEMS Microbiol. Ecol. 79, 685–696. 10.1111/j.1574-6941.2011.01252.x [DOI] [PubMed] [Google Scholar]
- Weir T. L., Manter D. K., Sheflin A. M., Barnett B. A., Heuberger A. L., Ryan E. P. (2013). Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS ONE 8:e70803. 10.1371/journal.pone.0070803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg W. G., Barns S. M., Pelletier D. A., Lane D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing B., Halfvarson J., Dicksved J., Rosenquist M., Järnerot G., Engstrand L., et al. (2009). Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn's disease. Inflamm. Bowel Dis. 15, 653–660. 10.1002/ibd.20783 [DOI] [PubMed] [Google Scholar]
- Wrzosek L., Miquel S., Noordine M. L., Bouet S., Joncquel Chevalier-Curt M., Robert V., et al. (2013). Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol. 11:61. 10.1186/1741-7007-11-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.