The second enzyme of the de novo purine biosynthesis pathway, PurD, is involved in leaf-colour development and the cell division process in rice.
Keywords: Cell division, chloroplast development, de novo purine biosynthesis, phosphoribosylamine-glycine ligase, pigment metabolism, rice
Abstract
The de novo synthesis of purine nucleotides is crucial to all living organisms, but limited information is available for plants. In this study, we isolated a virescent-albino leaf 1 (val1) mutant of rice (Oryza sativa) that produces dynamic green-revertible albino and narrow-leaf phenotypes. In albino leaves, chloroplast development was defective, pigment contents were reduced, and cell division was impaired compared with the wild-type. Map-based cloning revealed that VAL1 encodes a phosphoribosylamine-glycine ligase (PurD), the second enzyme in the de novo purine biosynthesis pathway. Subcellular localization analysis demonstrated that VAL1 was localized in the chloroplast. Our results demonstrate that VAL1 is a crucial enzyme in the de novo purine biosynthesis pathway and is involved in regulating chloroplast development, chlorophyll metabolism, and cell division during leaf development in rice.
Introduction
Chloroplasts are semi-autonomous organelles responsible for the conversion of energy from sunlight, and for the synthesis of organic molecules by means of photosynthesis in higher plants. Chlorophyll is the primary photosynthetic pigment, and functions in the absorption and transformation of light energy into energy-storage molecules, carbohydrates, and oxygen; it is thus fundamental to the survival and reproduction of all living organisms (Fromme et al., 2003). In higher plants, defective chloroplast development and chlorophyll metabolism lead to changes in chlorophyll content and leaf colour.
Leaf-colour mutants are not only useful as markers in hybrid breeding, but can also be used to study development-related processes such as photosynthesis, chlorophyll metabolism, and chloroplast development, and thus have considerable potential for research and practical application (Du et al., 2009). In higher plants, variation in leaf colour is conspicuous, and in rice its diverse phenotypes include virescent, albino, stripe, chlorina, stay-green, zebra, and yellow variegated forms (Yoo et al., 2009). A substantial number of leaf-colour genes have been mapped, but only a few have been cloned. The two CAO homologous genes in rice, OsCAO1 and OsCAO2, encode a chlorophyll a oxygenase, which catalyses the conversion of chlorophyll a to chlorophyll b. Mutation of OsCAO1 causes a pale-green leaf phenotype (Lee et al., 2005; Yang et al., 2016). OsChlD, OsChlH, and OsChlI encode Mg-chelatase subunits that catalyse the chelation of Mg2+ into protoporphyrin IX. Mutations of these three genes all cause a yellow-green leaf phenotype to different degrees of severity (Jung et al., 2003; Deng et al., 2014; Zhang et al., 2015). YELLOW-GREEN LEAF 1 (YGL1) encodes a chlorophyll synthase that catalyses the esterification of a chlorophyllide to complete the final step of chlorophyll biosynthesis in rice. The mutant ygl1 displays yellow-green leaves in young plants owing to decreased chlorophyll accumulation (Wu et al., 2007). The above-mentioned genes all affect the development of leaf pigmentation by regulating the synthesis and degradation of chlorophyll. Defective chloroplast development can also lead to the formation of leaf-colour mutants. For example, VIRESCENT 1 (V1) encodes a chloroplast protein that regulates chloroplast RNA metabolism and promotes the establishment of the plastid genetic system in rice. VIRESCENT 2 (V2) encodes a novel type of guanylate kinase and is essential for chloroplast development in rice. The v1 and v2 mutants each develop chlorotic leaves at restrictive temperatures (Sugimoto et al., 2007). VIRESCENT 3 (V3) and STRIPE 1 (St1) respectively encode the large and small subunits of ribonucleotide reductase, which is required for chloroplast biogenesis in rice. The v3 and st1 mutants show chlorotic leaves in a growth stage-dependent manner under field conditions (Yoo et al., 2009). OsPPR1 encodes a novel pentatricopeptide repeat (PPR) protein, which is essential for chloroplast biogenesis. Antisense transgenic plants display typical chlorophyll-deficient mutant phenotypes of albinism and lethality (Gothandam et al., 2005). Thus, mutations in genes involved in chlorophyll metabolism and chloroplast development result in abnormal leaf-colour phenotypes.
The de novo synthesis of the purine nucleotide is crucial for all living organisms. The synthetic pathway is composed of 11 enzymatic steps, leading to the formation of inosinate, and provides the adenine and guanine purines necessary for almost all biochemical processes, including DNA and RNA metabolism, the biosynthetic intermediates ATP and GTP, co-enzymes, and cytokinins, as well as being a vital metabolic regulator in cell signalling (Smith and Atkins, 2002). Although purine plays an important role in plant growth and development, limited information on the synthetic pathway in plants is available. The Arabidopsis PURINE BIOSYNTHESIS 4 (PUR4) gene encodes a formylglycinamidine ribonucleotide synthase, which is the fourth enzyme in the de novo purine biosynthesis pathway. The pur4 mutation is lethal to the male gametophyte and homozygous pur4 plants cannot be obtained, which indicates that de novo purine synthesis is specifically necessary for pollen development (Berthomé et al., 2008). CIA1 encodes a Gln phosphoribosyl pyrophosphate amidotransferase 2 (ATase2), which is one of the three ATase isozymes responsible for the first committed step of de novo purine biosynthesis. The loss-of-function mutant in Arabidopsis and ATase2-RNAi transgenic tobacco (Nicotiana tabacum) plants show normal green cotyledons but small and albino/pale-green mosaic leaves and reduced chloroplast protein import, which indicates that de novo purine biosynthesis is important for chloroplast development. In addition, de novo synthesis of purine plays a crucial role in cell division (Hung et al., 2004). PHOSPHORIBOSYLAMINE-GLYCINE LIGASE (PurD) is the second enzyme in the de novo purine biosynthesis pathway, and catalyses the conversion of 5′-phosphoribosylamine (PRA) to 5′-phosphoribosyl-l-glycinamide (GAR) (Smith and Atkins, 2002). In recent years, the biological function of PurD in mycelial growth and the virulence of bacterial strains has been extensively studied. For example, strains of Gibberella zeae (Kim et al., 2007), Xanthomonas oryzae pv. oryzae (Park et al., 2007), and Brucella abortus (Truong et al., 2015) harbouring mutations in PurD show reduced virulence against plants, which indicates that purD plays a vital role in the growth and virulence of bacterial strains. However, investigation of the biological functioning of purD in plants is incomplete. Previous studies have shown that purD is a single-copy and mono-functional enzyme in Arabidopsis, and the gene shows a high degree of conserved homology to purD in prokaryotic and eukaryotic species (Schnorr et al., 1994). However, studies on the physiological characteristics and biological function of purD in plants are still required.
In this study, we isolated a virescent-albino leaf 1 (val1) mutant, which exhibits a chlorotic and narrow-leaf phenotype, decreased quantities of chlorophyll and carotenoids over the entire growth period, and abnormal chloroplast development on the albino side of the leaf. Map-based cloning revealed that VAL1 encodes a phosphoribosylamine-glycine ligase, the enzyme involved in the second step of the de novo purine biosynthesis pathway. Subcellular localization analysis demonstrated that VAL1 was localized in chloroplasts. VAL1 plays a crucial role in the expression of genes associated with chloroplast development and pigment metabolism. This is the first report to implicate involvement of the de novo purine biosynthesis pathway in the development of leaf pigmentation and in cell division, and provides novel insights into the mechanisms underlying chloroplast development, pigment accumulation, and cell division in rice.
Material and methods
Plant material
The rice (Oryza sativa) virescent-albino leaf 1 (val1) mutant was derived from an ethylmethane sulfonate-treated population of rice ‘Jinhui 10’, which was used as the wild-type (WT) strain for phenotypic observations. The plants were grown in an experimental field at the Southwest University Rice Research Institute (Chongqing, China).
Physiological analysis
Photosynthetic pigments were extracted from ~0.1 g of newly developed mature leaves from the WT, val1, and transgenic plants in 25 ml extraction buffer (ethanol:acetone, 1:1; v/v) for 24 h at room temperature in the dark. The concentrations of chlorophyll a and b, and carotenoids were determined using a UV-1800PC spectrophotometer (Mapada Co. Ltd, China) at 663, 645, and 470 nm, respectively, and calculated according to the method of Lichtenthaler (1987). The first leaf from the top of the WT, val1, and transgenic plants was used to measure photosynthetic rate, stomatal conductance, intercellular CO2 concentration, and transpiration rate using a LI-6400 portable photosynthesis meter (LiCor, Lincoln, NE, USA) according to the manufacturer’s instructions. Data presented are the means (±SD) of three biological repeats.
Transmission electron microscopy
Leaves of the WT, val1, and transgenic plants at the tiller stage were collected and cut into 1-mm2 squares, fixed in primary fixative solution (3.5% glutaraldehyde), and then post-fixed for 2 h in 1% osmium tetroxide after washing with 0.1 mol l−1 phosphate buffer saline. Tissues were stained with uranyl acetate, dehydrated in ethanol, and embedded in Spurr’s medium prior to thin-sectioning. Samples were stained again and examined using a H-7500 TEM (Hitachi, Japan) as described previously (Dong et al., 2013).
Paraffin-sectioning and histological analysis
Leaves were collected at the tiller stage and fixed in 50% ethanol, 0.9 M glacial acetic acid, and 3.7% formaldehyde for 12 h at 4 °C. The fixed samples were dehydrated with a graded series of ethanol, infiltrated with xylene, and embedded in paraffin (Sigma). A rotary microtome (RM2245; Leica, Hamburg, Germany) was used to cut 8-μm-thick sections, which were transferred onto poly-L-lysine-coated glass slides, deparaffinized in xylene, and dehydrated through an ethanol series (Ren et al., 2018). Light microscopy was performed using an Eclipse E600 microscope (Nikon, Tokyo, Japan).
Fine-mapping of VAL1
The val1 mutant was crossed with ‘Xinong1A’ (bred by the Southwest University Rice Research Institute) and 2885 F2 plants exhibiting the mutational phenotype were selected for high-resolution mapping. Gene fine-mapping was conducted using simple sequence repeat markers obtained from the publicly available rice databases Gramene (http://www.gramene.org) and the Rice Genomic Research Program (http://rgp.dna.affrc.go.jp/E/publicdata/caps/index.html). Insertion/deletion markers were developed from comparisons of genomic sequences from ‘Xinong1A’ and ‘Jinhui 10’ in our laboratory. The sequences of primers used in the fine-mapping and candidate gene analysis are listed in Supplementary Table S1.
Vector construction for genetic complementation, RNAi, and over-expression
For the complementation test, a 7199-bp genomic fragment was cloned into the binary vector pCAMBIA1301. The resulting recombinant plasmids were introduced into val1 using the Agrobacterium tumefaciens-mediated transformation method as described previously (Zhang et al., 2017). To generate a construct for RNAi, a 328-bp fragment of VAL1 complementary DNA was amplified and inserted into the vector pTCK303 to obtain the intermediate vector. For overexpression of VAL1, the 1580-bp full-length coding sequence of VAL1 was amplified from ‘Jinhui 10’ cDNA and cloned into vector pTCK303 to generate the intermediate vector. VAL1 was driven by the rice ubiquitin promoter. The RNAi and overexpression recombinant plasmids were transformed into ‘Jinhui 10’ plants using the A. tumefaciens-mediated transformation method (Zhang et al., 2017). The primer sequences are listed in Supplementary Table S1.
RNA extraction and quantitative real-time PCR analysis
Total rice RNA was extracted from the roots, culms, leaf sheaths, leaves, and young panicles using the RNAprep Pure Plant Kit (Tiangen Co. Ltd, China). The first-strand cDNA was synthesized using the SuperScript® III Reverse Transcriptase Kit (Invitrogen). qRT-PCR analysis was performed using Novostar-SYBR Supermix (Novoprotein, Shanghai, China) in an ABI Biosystems 7500 Real-Time PCR System. At least three replicates were performed and the mean expression level was calculated. Genes used for qRT-PCR analysis were as previously described (Wang et al., 2017). The primer sequences are listed in Supplementary Table S1.
Multiple sequence alignment and phylogenetic tree construction
Protein sequences were acquired using the BLAST tool in the PHYTOZOME portal, with a 10−5 EXPECT value threshold (http://phytozome.jgi.doe.gov/pz/portal.html#!search? show=BLAST). A phylogenetic tree was constructed using MEGA 5.0 with the maximum-likelihood method based on the Jones–Taylor–Thornton matrix-based model with the lowest Bayesian information criterion scores (Tamura et al., 2011).
Nucleotide and cytokinin measurements
Samples of the WT and val1 mutant seedlings (0.1 g) were frozen in liquid nitrogen and homogenized. Nucleotides were extracted using the trichloroacetic acid (TCA) method as described previously (Hajirezaei et al., 2003). The nucleotide and cytokinin contents in the TCA extracts were measured using a HPLC system (Kontron, Eching, Germany) fitted with a Partisil-SAX anion-exchange column, and detected by absorption at OD254. Identification and quantification was completed by comparison with standards, as described previously (Hajirezaei et al., 2003).
Nucleotide treatment in vitro
Wild-type and val1 seeds were surface-sterilized after husking and grown in half-strength Murashige and Skoog medium (pH 5.8) containing different concentrations (0, 1, 5, and 10 mM) of adenine nucleotide (AMP), guanine nucleotide (GMP), or AMP and GMP. The seeds were incubated at 28 °C under a 16/8 h light/dark photoperiod. Continual observation of seedling phenotypes was undertaken for 7–10 d.
Subcellular localization
The full-length coding region of VAL1 was amplified and cloned into the expression cassette 35S-GFP-NOS (pA7) in order to generate the VAL1-GFP (green fluorescent protein) fusion vector. Both pA7-VAL1-GFP and empty pA7-GFP plasmids were transformed into rice protoplasts as described previously (Ma et al., 2017). After overnight incubation at 28 °C, GFP fluorescence was observed using a confocal laser-scanning microscopy (Olympus FLUOVIEW FV1000; Japan). The primer sequences are listed in Supplementary Table S1.
In situ hybridization
For the VAL1 and His4 probes, a gene-specific cDNA was amplified and labelled using a DIG RNA Labelling Kit (Roche). Pretreatment of sections, hybridization, and immunological detection were performed as described previously (Zhang et al., 2017). The primer sequences are listed in Supplementary Table S1.
Western blot analysis
Total proteins from 10-d-old WT and val1 seedlings were isolated using a Plant Total Protein Extraction Kit (Sangon Biotech). An equal amount of protein was separated on 10% SDS-polyacrylamide gels. The proteins were transferred to iBlot 2 PVDF Regular Stacks using the iBlot 2 Gel Transfer Device (Invitrogen), immunoblotted with various primary antibodies (Agrisera and Beijing Protein Innovation Company) and AP-conjugated Affinipure Goat Anti-Rabbit IgG (Jackson ImmunoResearch) according to the recommended dilution ratios in the iBind Cards using the iBind Western Device (Invitrogen), and detected using the NBT/BCIP substrate (Sangon Biotech).
Transcriptome analysis
Seedlings of the WT and val1 mutant were used for RNA extraction and transcriptome sequencing with two biological replicates. The purity and concentration of RNA were measured using a NanoPhotometer spectrophotometer (Implen, Westlake Village, CA, USA) and a Qubit® RNA Assay Kit in a Qubit® 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA). Library construction and RNA sequencing (RNA-seq) were conducted by Omicgene Biological Technology (Wuhan, China) on a HiSeq X Ten platform (Illumina, San Diego, CA, USA). RNA-seq short reads were aligned to the indica rice genome with HISAT2 (https://ccb.jhu.edu/software/hisat2/index.shtml), and the expression level of each gene was computed using StringTie (https://ccb.jhu.edu/software/stringtie/). The expression-level data were analysed using the gene set enrichment analysis (GSEA) software (version 2.07) to identify functionally related groups of genes (gene sets) with statistically significant enrichment (Subramanian et al., 2005). The gene set files used in GSEA were constructed in accordance with the genome annotation (http://www.mbkbase.org/R498). The significance of differentially expressed genes (DGEs) was determined using q-values <0.005 and |log2 (fold-change)| >1. Gene Ontology (http://www.geneontology.org/) analyses were performed by referring to GOseq (http://dx.doi.org/10.18129/B9.bioc.goseq).
Flow cytometric analysis
For nuclear staining, the shoot apex from rice seedlings was cut into small pieces in 400 µl Partec CyStain UV Precise P Lysis solution (Sysmex Partec) on ice, and then filtered with a 30-µm filter to remove cellular debris. After staining with Partec CyStain UV Precise P Staining solution (including DAPI) (Sysmex Partec), the nuclei were analysed using CyFlow Space (Sysmex Partec). A total of 30 000 events were recorded, and the data were analysed using the FCS Express software (https://www.denovosoftware.com/).
Results
The val1 mutation confers a green-revertible albino and narrow-leaf phenotype
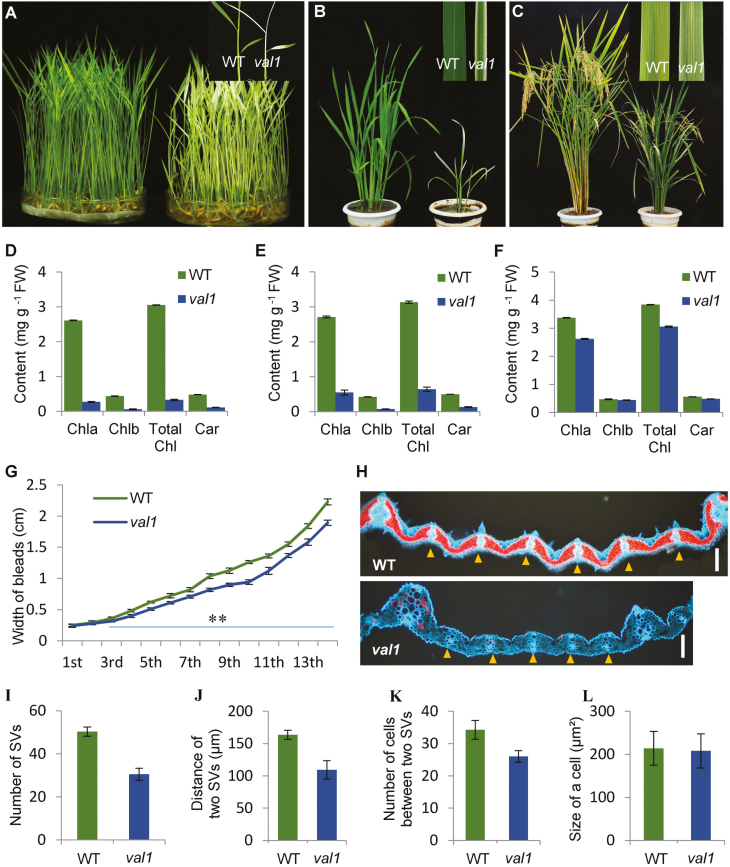
In the wild type (WT), the leaves of the plant appeared green throughout the growth period except in the later mature stage. In contrast, in the val1 mutant, new albino leaves were developed at the early seedling stage (Fig. 1A). The leaves subsequently turned green with an albino margin and displayed a mixed phenotype in which green and albino leaves were present from the later seedling stage to the tiller stage (Fig. 1B). During the heading stage, the leaves were almost gray-green in colour, and only a small number of leaves showed marginal albinism (Fig. 1C). Thus, the older the leaves, the smaller the area of albinism (Fig. 1A–C). These observations demonstrated that the phenotype of val1 was displayed in a growth stage-dependent manner. In addition, in accordance with the albino leaf phenotype of the val1 mutant, the leaf chlorophyll content was significantly reduced in the mutant compared with the WT at the seedling and tiller stages (Fig. 1D, E), and slightly reduced at the mature stage (Fig. 1F). Taken together, these observations over the full life cycle demonstrated that val1 was a dynamic green-revertible albino mutant. We also examined the expression of the phenotypes and the leaf pigment contents in relation to temperature. The val1 mutant always displayed albino leaves with lower pigment contents than those of the WT at temperatures of 20 °C (C20), 30 °C (C30), and under an alternate light/dark cycle (12 h light at 30 °C/12 h dark at 20 °C; L30 °C/D20 °C) (Supplementary Fig. S1). These results indicated that the albino-leaf phenotype of val1 in the early seedling stage was not affected by temperature, and we therefore determined that val1 was a temperature-insensitive, green-revertible albino mutant.
Fig. 1.
Phenotypic characteristics of the wild-type (WT) and the val1 mutant. (A–C) Phenotypes of the WT and val1 at the seedling stage (A), tiller stage (B), and heading stage (C). Close-up images of the leaves of the WT and val1 are shown at top-right. (D–F) Chlorophyll content of the leaves of the WT and val1 at the seedling stage (D), tiller stage (E), and heading stage (F). (G) Leaf blade widths of the WT and val1 plants for the 1st–14th leaves produced across the total growth period. (H) Transverse sections of the seventh leaf blade of the WT and val1. Yellow triangles indicate small vascular bundles. Scale bars are 100 µm. (I–L) Comparisons of the number of small vascular bundles (SV) (I), the distance between the small vascular bundles (J), and the number of cells (K) and the size of cells (L) between the small vascular bundles in the WT and val1. Data are means (±SD) of three biological replicates.
The leaf width in val1 was reduced significantly compared with the WT, and the narrow-leaf phenotype was increasingly pronounced over the course of plant development (Fig. 1A–C, G). To examine the basis for the narrow-leaf phenotype, histological analysis of transverse sections of the leaf blade showed that, compared with the WT, the number of small vascular bundles was reduced by 39.3% (Fig. 1H, I), and the distance between the small vascular bundles was reduced by 33.2% (Fig. 1H, J). The number of cells between the small vascular bundles was reduced by 24.1% (Fig. 1H, K), but the size of the cells was similar to that of the WT (Fig. 1H, L). These results indicated that the reduced leaf width may have been caused by the reduced number of vascular bundles and the reduced number of cells between the small vascular bundles. In addition, val1 plants also displayed dwarfism and smaller spikelets compared with the WT (Fig. 1A–C).
Molecular cloning and identification of VAL1
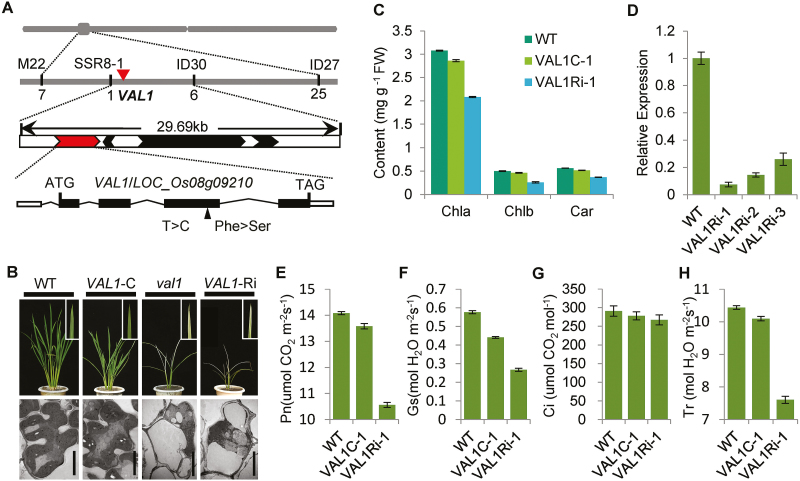
The VAL1 gene was previously mapped to a region of about 171 kb on chromosome 8 (Ma, 2014). In the current study, the location of VAL1 was narrowed to within a physical distance of 29.69 kb between the simple sequence repeat marker SSR8-1 and the insertion/deletion marker ID30. This interval includes four annotated genes (http://www.gramene.org/). Sequencing analysis identified a single-nucleotide substitution from T to C within the LOC_Os08g09210 gene, causing an amino acid mutation of Phe-358 to Ser-358 in the val1 mutant (Fig. 2A). To confirm whether the mutation of LOC_Os08g09210 resulted in the mutant phenotype, we performed a complementation experiment by transforming a 7198-bp WT DNA fragment containing LOC_Os08g09210 into the val1 mutant. The mutant phenotype was completely rescued in the transgenic plants (Fig. 2B). The contents of chlorophyll and carotenoids and the photosynthetic rate in the transgenic plants were almost identical to those of the WT (Fig. 2C, E–H). In addition, the chloroplast ultrastructure as observed by TEM was similar to that of the WT, displaying well-developed lamellar structures and equipped with normal thylakoid membranes and stacked grana (Fig. 2B). We performed RNAi to silence VAL1 in the WT plants. In the transgenic plants, the quantity of VAL1 transcripts was greatly reduced (Fig. 2D), and a dynamic green phenotype similar to that of the val1 mutant was observed (Fig. 2B). The contents of chlorophyll and carotenoids and the photosynthetic rate in the transgenic plants were significantly reduced compared with the WT (Fig. 2C, E–H). Observation by TEM showed that the mesophyll cells were almost empty and lacked obvious intact organelles, and the chloroplasts were completely degraded, similar to those of val1 (Fig. 2B). Taken together, these results confirmed that LOC_Os08g09210 corresponded to the VAL1 gene.
Fig. 2.
Molecular identity of VAL1. (A) Map-based cloning of the VAL1 gene. (B) Phenotypes of the wild-type (WT), complemented (-C), val1, and VAL1-RNAi plants. Scale bars are 5 µm. (C) Chlorophyll contents of the leaves of the WT, complemented, and VAL1-RNAi plants at the tiller stage. (D) Expression analysis of VAL1 in the leaves of the WT and the three RNAi lines by qRT-PCR. (E–H) Photosynthetic rate (E), stomatal conductance (F), intercellular CO2 concentration (G), and transpiration rate (H) of the WT, complemented, and VAL1-RNAi plants at the heading stage. Data are means (±SD) of three biological replicates.
Expression pattern of VAL1 and subcellular localisation of the VAL1 protein
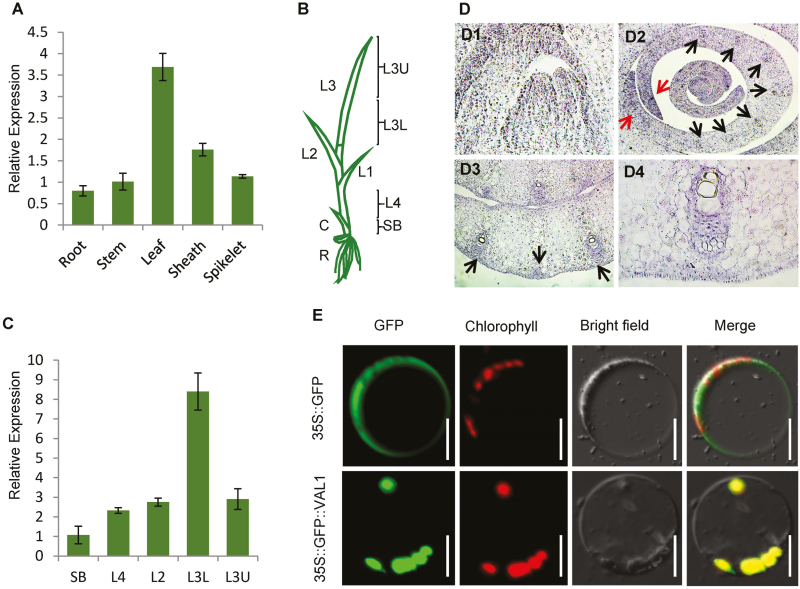
To determine the expression pattern of VAL1, quantitative reverse transcription-PCR (qRT-PCR) analysis was performed. VAL1 was expressed in a variety of tissues, including the roots, stems, leaves, leaf sheaths, and panicles, with higher levels in the leaves (with higher chlorophyll content) than in the other tissues examined (Fig. 3A). Previous studies have shown that the leaves of rice emerge from the sheath in an ordered manner. Generally, when the third leaf has fully emerged from the sheath, the shoot includes the fourth to seventh immature leaves. The leaf cells in the upper half of the third leaf (L3U) and basal (lower) half of the third leaf (L3L) contain mature chloroplasts, whereas those in the shoot base (SB) and fourth leaf above the shoot base (L4) contain proplastids and early-developing immature chloroplasts (Fig. 3B; Yoo et al., 2009). In our present study, qRT-PCR analysis showed that VAL1 was universally expressed in the SB, L4, L2, L3L, and L3U samples, with the highest levels detected in L3L (Fig. 3C).
Fig. 3.
Expression patterns of VAL1 and subcellular localization of the VAL1 protein. (A) Expression pattern of VAL1 in different tissues as indicated by real-time PCR. (B, C) Expression patterns of VAL1 in the leaf as indicated by real-time PCR. (D) Expression patterns of VAL1 by in situ hybridization. Black arrows indicate the procambium cells and vascular bundles. Red arrows indicate the margin of the P3 primordium. (E) Analysis of the subcellular localization of the VAL1 protein in rice protoplasts. Scale bars are 50 µm. SB, shoot base; L4, fourth leaf; L2, second leaf; L3L, basal half of the third leaf; L3U, upper half of the third leaf. Data are means (±SD) of three biological replicates.
For a more detailed analysis of the expression pattern of VAL1, transverse and longitudinal sections of the shoot apical meristem (SAM) were examined for VAL1 signals by in situ hybridization (Fig. 3D). First, strong signals were observed in the SAM (Fig. 3D1). Next, scattered strong signals were detected in the P2 primordia (Fig. 3D2). In the P3 primordia, VAL1 signals were concentrated at the margin of the P3 primordium and the procambium cells (Fig. 3D2), which may predict the sites of differentiation of procambium cells from fundamental cells. In the P4 primordia, VAL1 signals were primarily restricted to the vascular bundles, with no preference for the xylem or phloem (Fig. 3D3, D4). These results suggested that VAL1 may be involved in leaf development.
To examine the subcellular localization of the VAL1 protein, constructs producing the GFP and the GFP-VAL1 ORF fusion protein were transiently expressed in rice. In cells that expressed GFP alone, green fluorescence was detected consistently throughout the cell except in the vacuole in protoplasts, and the GFP-VAL1 ORF fusion protein was localized in the chloroplast (Fig. 3E). These results indicated that VAL1 was localized in the chloroplast.
VAL1 encodes a phosphoribosylamine-glycine ligase
A blastp analysis was performed on the NCBI protein database using the VAL1 protein sequence. VAL1 encodes a phosphoribosylamine-glycine ligase (PurD), which belongs to the ATP-grasp superfamily. Alignment of amino acid sequences of VAL1 and other phosphoribosylamine-glycine ligase family members revealed that the sequences showed a high degree of homology. The protein structure was analysed using the PSIPRED protein prediction database (http://bioinf.cs.ucl.ac.uk/psipred/). VAL1 was predicted to include three conserved domains, designated GARS_N, GARS_A, and GRAS_C, of which the GARS_A domain contained an ATP-binding site. In addition, it was predicted by ChloroP (http://www.cbs.dtu.dk/services/ChloroP/) that VAL1 contained a chloroplast transit peptide at the N-terminus (amino acid residues 1–68; Supplementary Fig. S2).
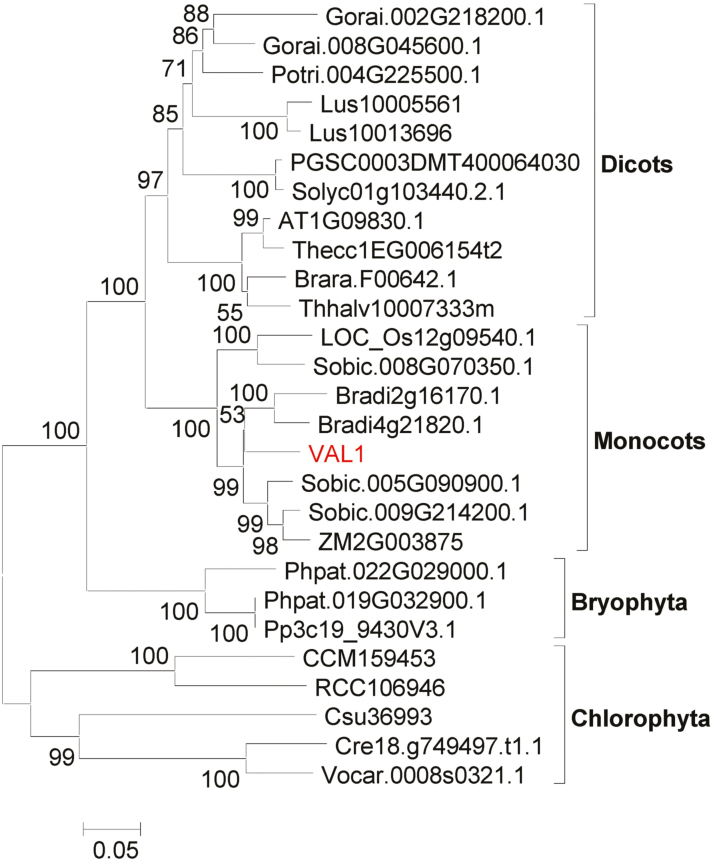
The phylogenetic tree indicated that VAL1 is widely present in many photosynthetic organisms (Fig. 4), including lower aquatic algae, terrestrial ferns, gymnosperms, and angiosperms, and it showed a high degree of homology, which suggests that VAL1 in rice may be descended from VAL1 of algae. In addition, the rice genome contains a VAL1 homolog designated LOC_Os12g09540, which is also predicted to encode a phosphoribosylamine-glycine ligase. LOC_Os12g09540 was also expressed in leaves, with the highest levels detected in L3U (Supplementary Fig. S3A, B). However, LOC_Os12g09540 and VAL1 do not belong to the same evolutionary lineage, which indicates a certain degree of differentiation in molecular function between the homologs.
Fig. 4.
Phylogenetic tree for the VAL1 protein. The tree was constructed using the maximum-likelihood method based on the Jones–Taylor–Thornton matrix-based model. Bootstrap values calculated from 500 replicates are given at the branch nodes. The VAL1 protein is highlighted in red.
Complementation of the nucleotide in vitro partially restores the phenotype
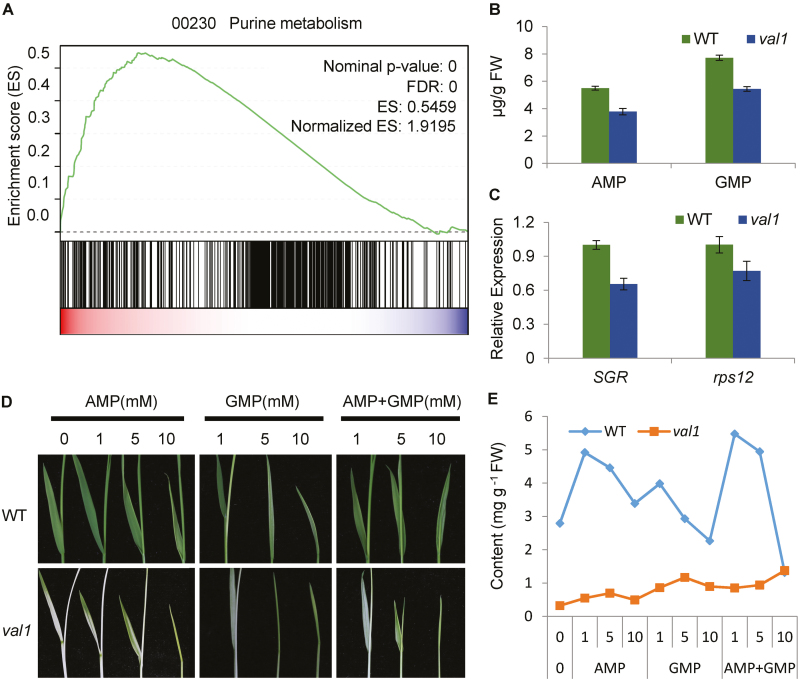
VAL1 encodes a phosphoribosylamine-glycine ligase, which is the second enzyme in the de novo purine biosynthesis pathway (Supplementary Fig. S4A). To confirm its involvement in the pathway, the transcriptomic profiles of seedlings of val1 and WT grown under normal conditions were characterized by RNA-seq with two biological replicates for each genotype. The Pearson correlation coefficient between the two replicates was greater than 0.95. Gene set enrichment analysis (GSEA) was used to examine the data. This is a statistical approach that allows identification of over-represented gene sets among differentially up- or down-regulated genes of a transcriptomic analysis (Subramanian et al., 2005). There was significant enrichment in the over-representation of genes associated with the de novo purine biosynthesis pathway (Fig. 5A, Supplementary Fig. S4B), which indicated that the mutation of VAL1 caused disruption of de novo purine biosynthesis. In addition, the contents of the de novo purine biosynthesis products AMP and GMP in val1 seedlings were lower than those of the WT (Fig. 5B), which further indicated that VAL1 was involved in the de novo purine biosynthesis pathway.
Fig. 5.
Phosphoribosylamine-glycine ligase (PurD) and de novo purine biosynthesis. (A) Gene set enrichment analysis of genes associated with the de novo purine biosynthesis pathway. The green line indicates enrichment score calculated by gene set enrichment analysis (GSEA). (B) AMP and GMP contents in the wild-type (WT) and val1 mutant. (C) Expression of rps12 and SGR in seedlings of the WT and val1. (D) Leaf morphology of the WT and val1 mutant after growth in Murashige and Skoog liquid medium supplemented with various concentrations (0, 1, 5, or 10 mM) of AMP, GMP, or AMP and GMP together for 2 weeks. (E) Chlorophyll content of plants of the WT and val1 mutant shown in (D). Data are means (±SD) of three biological replicates.
The decreased AMP and GMP contents in the val1 mutant may influence plastid genome replication and division. We therefore used qRT-PCR to examine the expression of SGR and rps12, which are single-copy genes in the plant nuclear and plastid genomes, as representatives of the copy numbers of nuclear and chloroplastic genomes, respectively (Sugimoto et al., 2007; Yoo et al., 2009). The expression levels of both SGR and rps12 were reduced in the val1 mutant (Fig. 5C), which indicated that nuclear and chloroplastic genome replication appeared to be arrested by limited AMP and GMP levels.
Next, we investigated whether the mutant phenotypes could be complemented biochemically by exogenous application of AMP and GMP. Application of AMP, GMP, and AMP and GMP together partially restored the green colour of the val1 leaf (Fig. 5D). Low concentrations (1 and 5 mM) of AMP, GMP, and AMP and GMP promoted accumulation of chlorophyll in WT leaves, whereas high concentrations (10 mM) repressed chlorophyll accumulation and inhibited plant growth (Fig. 5D). In the val1 mutant, chlorophyll content was elevated with increasing concentrations of AMP, GMP, and AMP and GMP (Fig. 5E). Thus, the applications partially restored the accumulation of chlorophyll in the val1 mutant. In addition, low concentrations (10 and 100 nM) of the synthetic cytokinin benzylaminopurine (6-BA) did not rescue the mutant phenotype, and a high concentration of 6-BA (1000 nM) severely inhibited plant growth (Supplementary Fig. S5).
VAL1 affects chloroplast development and pigment metabolism
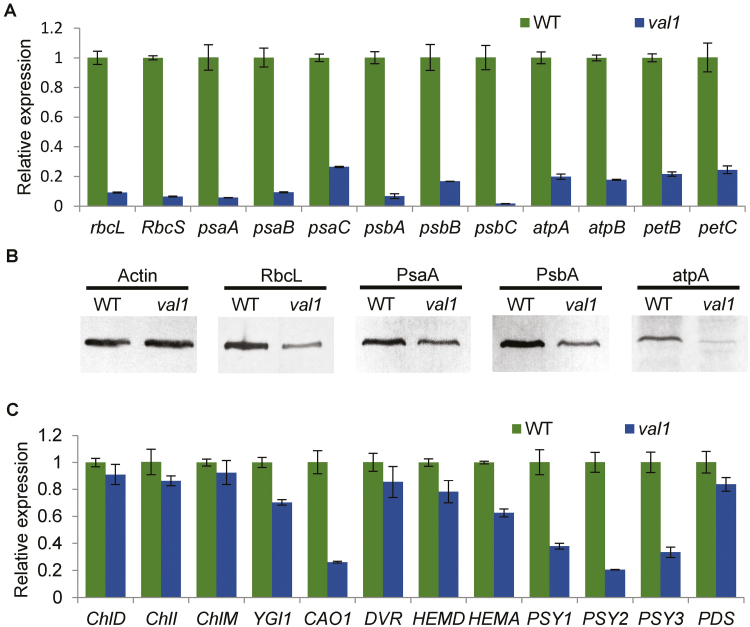
Previous research has indicated that chloroplast development in the albino portion of the leaf in val1 plants is severely defective (Ma, 2014). Thus, expression analysis of genes associated with chloroplast development and photosynthesis were compared between WT and val1 plants by qRT-PCR analysis. The transcript levels of rbcL and RbcS (encoding the large and small subunit of Rubisco, respectively), psaA, psaB, psaC, and psbA, psbB, psbC (encoding two reaction-centre polypeptides of PSI and PSII, respectively), atpA and atpB (encoding the alpha- and beta-subunits of chloroplast ATP synthase, respectively), petB and petC (encoding the cytochrome b6 subunit and the Rieske FeS centre of cytochrome b6f complex, respectively) were significantly reduced in the val1 mutant compared with the WT (Fig. 6A). The protein accumulation levels of RbcL, PsaA, PsbA, and atpA were consistent with the transcript levels (Fig. 6B). Taken together, these results indicated that the VAL1 mutation affected the expression of genes associated with chloroplast development and photosynthesis, which resulted in defective chloroplast development.
Fig. 6.
Expression of genes associated with chloroplast development, photosynthesis, and pigment metabolism in the wild-type (WT) and val1 mutant. (A) Expression analysis of genes associated with chloroplast development and photosynthesis in the WT and val1. (B) Western blots of proteins associated with chloroplast development and photosynthesis. Actin was used as an internal control. (C) Expression analysis of genes associated with pigment metabolism in the WT and val1. Data are means (±SD) of three biological replicates.
Given that the val1 mutant displayed reduced chlorophyll and carotenoid contents, the expression levels of pigment metabolism-related genes were compared between WT and val1 plants by qRT-PCR analysis. The transcript levels of pigment metabolism-related genes were reduced in the val1 mutant compared with those of the WT to different degrees. For example, the transcript levels of the chlorophyll synthesis pathway gene CAO and the carotenoid synthesis pathway genes PSY1, PSY2, and PSY3 were all significantly reduced in the val1 mutant compared with the WT. The other genes involved in the synthesis of chlorophyll and carotenoids, such as ChlD, ChlI, ChlM, YGL1, DVR, HEMA, HEMD, and PDS were also slightly reduced in the val1 mutant compared with the WT (Fig. 6C). In the heading stage, the expression of these genes related to chloroplast development, photosynthesis, and pigment metabolism were not significantly reduced in the val1 mutant compared with the WT (Supplementary Fig. S6A, B).
To obtain further insights into the function of VAL1, the transcriptomic profiles of seedings of val1 and the WT were characterized. A total of 3683 DEGs between val1 and WT were identified, including 1813 up- and 1870 down-regulated DEGs. GO enrichment analysis showed that the expression of genes involved in chloroplast development, photosynthesis, and pigment biosynthesis were changed dramatically. For chloroplast development and photosynthesis a total of 34 DEGs were identified, comprising 32 down- and two up-regulated genes; for example, the expression of rbcL, psaG,and psbR showed decreases of 53.6, 74.1, and 82.6%, respectively, in val1 (Supplementary Table S2). For pigment biosynthesis a total of 25 DEGs were identified, comprising 16 down- and nine up-regulated genes; for example, the expression of PORA and CAO1 showed decreases of 85.9% and 47.5%, respectively, in val1 (Supplementary Table S3).
VAL1 is involved in cell division in leaves
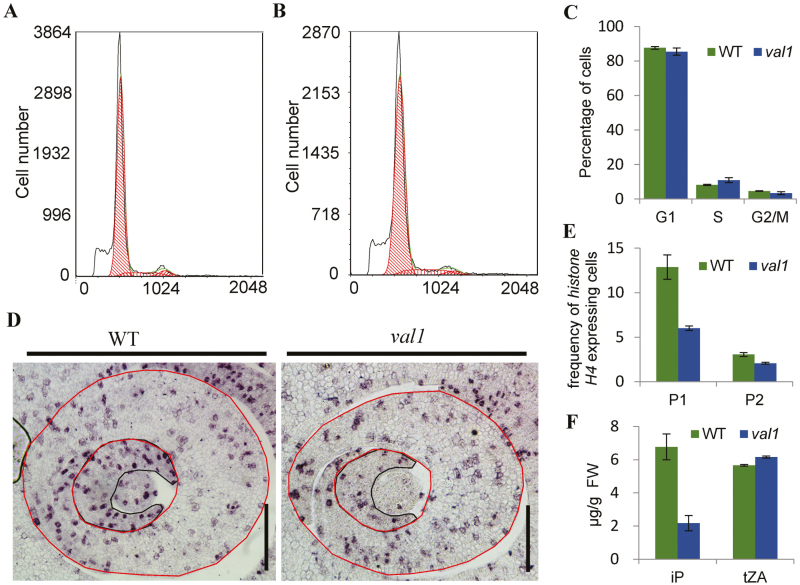
Since the number of vascular bundles and cells between the small vascular bundles was significantly reduced in the narrow leaves of val1, we speculated that this may have been caused by impaired cell proliferation of leaf primordia. To text this hypothesis, flow cytometry was used to monitor the amount of DNA in the nuclear suspensions in the shoot apex of WT and val1 plants. The results revealed a significant increase in the number of cells in the S phase and decreases in the G2/M phases of the cell cycle, implying that the cell cycle was delayed at the S phase (Fig. 7A–C). The expression patterns of histone H4 were characterized by in situ hybridization and it was found to be relatively evenly expressed throughout the P1 leaf primordium in the WT, whereas the frequency of cells that expressed histone H4 was reduced by 29.2% in the val1 mutant (Fig. 7D, E). In the P2 leaf primordium, histone H4 was mainly expressed at the margins of the primordium, and the frequency of cells that expressed histone H4 was reduced by 27.7% in the val1 mutant compared with the WT (Fig. 7D, E). These results confirmed that the decreased number of cells was caused by impaired cell proliferation during the early development of primordia. Given that de novo purine synthesis leads to the synthesis of cytokinins, we measured the contents of isopentenyladenine (iP) and trans-zeatin (tZA) in WT and val1 seedlings. The content of iP was reduced by 67.9%, whereas that of tZA was increased slightly in the val1 mutant compared with the WT (Fig. 7F). Taken together, these results supported the contention that the decreased number of cells was caused by impaired cell proliferation, which may have been due to the reduced content of cytokinins during the early development of leaf primordia in the val1 mutant.
Fig. 7.
Involvement of VAL1 in cell division in leaves. (A, B) Flow cytometry measurements of nuclei from the shoot apex in 10-d-old wild-type (WT) (A) and val1 plants (B). (C) Quantification of the DNA profiles of WT and val1 plants. (D) Expression of histone H4 in cross-sections of the shoot apical meristems in the WT and val1 at the tiller stage. Black and red lines represent P1 and P2 primordia, respectively. Scale bars are 100 µm. (E) Frequency of histone H4-expressing cells in the leaf primordia indicated by red circles in (D). P1, P1 promordium; P2, P2 promordium. (F) Contents of isopentenyladenine and trans-zeatin in the WT and val1 mutant seedlings. Data are means (±SD) of three biological replicates.
Application value of VAL1 in rice breeding
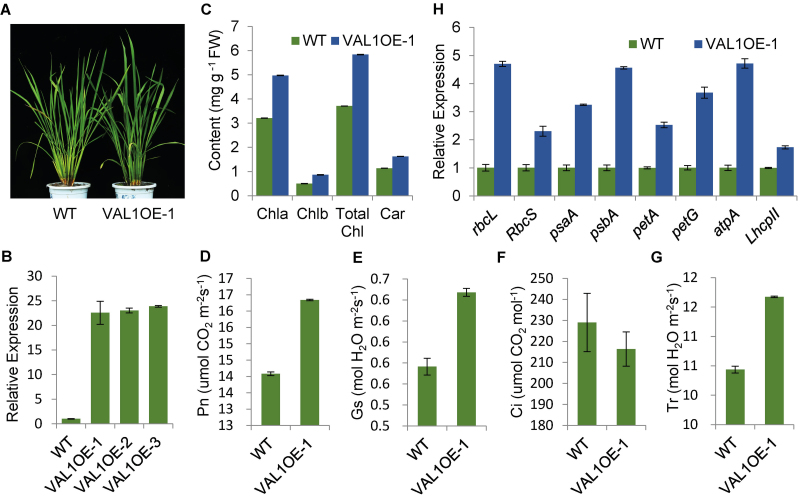
The shade of the leaf colour in rice is determined by the content of chlorophyll. Chlorophyll is the main pigment of plants for photosynthesis and plays a key role in the absorption, transmission, and conversion of light energy (Fromme et al., 2003). To assess the application value of VAL1 in rice breeding, we expressed VAL1 under the control of the ubiquitin promoter in the WT. In the tillering stage, the green leaf colour was slightly darker in transgenic plants compared with the WT (Fig. 8A). The chlorophyll and carotenoid contents were significantly increased (Fig. 8C), and the photosynthetic rate was increased in transgenic plants compared with the WT (Fig. 8 D). The qRT-PCR results showed that the expression level of VAL1 was significantly increased in transgenic plants compared with the WT (Fig. 8B). The expression levels of photosynthesis-associated genes rbcL, RbcS, psaA, psbA, petA, petG, atpA, and LhcpII, were significantly increased to different degrees in transgenic plants compared with the WT (Fig. 8H). These results indicated that VAL1 played an important role in regulation of plant photosynthesis by modulating the expression of related genes. Thus, plants with overexpressed VAL1 showed a higher photosynthetic ability, and could hence they could be utilized in breeding.
Fig. 8.
Application value of VAL1 in rice breeding. (A) Phenotypes of the wild-type (WT) and VAL1-overexpressing plants in the tiller stage. (B) Expression analysis of VAL1 in the leaf of the WT and three VAL1-overexpressing lines by real-time PCR. (C) Chlorophyll content of the leaf of the WT and VAL1-overexpressing plants in the tiller stage. (D–G) Photosynthetic rate (D), stomatal conductance (E), intercellular CO2 concentration (F), and transpiration rate (G) of the WT and VAL1-overexpressing plants at the heading stage. (H) Expression analysis of genes associated with photosynthesis in the WT and VAL1-overexpressing plants. Data are means (±SD) of three biological replicates.
Discussion
In our present study, VAL1 was shown to encode a phosphoribosylamine-glycine ligase, the second enzyme in the de novo purine biosynthesis pathway. Subcellular localization indicated that the VAL1 protein was localized in the chloroplast (Fig. 3). VAL1 is a novel regulator that regulates chloroplast development, pigment metabolism, and cell division, and thereby affects the development of leaf colour and blade width in rice.
Many genes associated with leaf colour have been cloned in rice. Metabolic pathways associated with chlorophyll synthesis and degradation, and chloroplast development have been elucidated, and mutation of genes involved in these metabolic pathways leads to changes in leaf colour. For example, mutations in the genes OsCHlH (Jung et al., 2003), OsCHlD and OsCHlI (Zhang et al., 2015), OsDVR (Wang et al., 2010), OsGluRs (Liu et al., 2007), YGL1 (Wu et al., 2007), OsCAO1 and OsCAO2 (Lee et al., 2005; Yang et al., 2016), OsPPR1 (Gothandam et al., 2005), YSA (Su et al., 2012), SGR (Jiang et al., 2007), NYC1 (Kusaba et al., 2007), V1 (Kusumi et al., 2011), V2 (Sugimoto et al., 2004), and V3 and st1 (Yoo et al., 2009) produce different types of leaf-colour mutants. In the val1 mutant, the expression levels of chlorophyll- and photosynthesis-associated genes were significantly reduced, and chloroplast development was defective (Figs 2D, 6). Thus, similar to the above-mentioned leaf-colour mutants, the green-revertible albino phenotype of the val1 mutant was caused by disruption of chlorophyll metabolism and chloroplast development. Previous research has indicated that the phenotypes of the majority of green-revertible albino leaf-colour mutants are regulated by temperature and development, or by a combination of the two. For example, the temperature-sensitive seedling-colour mutant tsc-1 displays a white, light-yellow, and normal green leaf when grown at 23.1 °C, 26.1 °C, and 30.1 °C, respectively (Dong et al., 2001). In contrast, the albino leaf phenotype of the val1 mutant at the early seedling stage was not affected by temperature, which indicated that val1 is a temperature-insensitive mutant (Supplementary Fig. S1). Developmentally regulated mutants, such as the ysa mutant, develop albino leaves before the three-leaf (L3) stage and gradually turn green and recover to the normal green leaf colour at the six-leaf stage (Su et al., 2012). The temperature-conditional and developmentally-regulated mutants v3 and st1 largely exhibit a normal green-leaf phenotype up to the L3 stage and develop chlorotic leaves until the maximum tiller stage, but recover to develop mostly green leaves after heading. At temperatures of 20 °C or 30 °C, these mutants produce chlorotic leaves, whereas under an alternate light/dark cycle (12 h light at 30 °C/12 h dark at 20 °C), these mutants develop leaves of an almost normal green colour (Yoo et al., 2009). In our present study, the val1 mutant initially produced albino new leaves, but these subsequently turned green and only a small number of leaves showed marginal albinism at later developmental stages (Fig. 1). The results indicated that val1 is a novel green-revertible albino mutant, for which the phenotype is regulated by development and is independent of temperature.
Nucleotide metabolism is a crucial process in all living organisms (Smith and Atkins, 2002). Although the de novo biosynthesis of purine nucleotides is essential for plant growth and development, little information is available on the molecular mechanism of the purine biosynthesis pathway in plants. The Arabidopsis gene CIA1 encodes a Gln phosphoribosyl pyrophosphate amidotransferase 2 (ATase2), which is one of three ATase isozymes responsible for the first step of de novo purine biosynthesis. The cia1 mutant displays normal green cotyledons, together with small and albino/pale-green mosaic leaves that contain slightly smaller cells but only half the cell number of the WT, which indicates that de novo purine biosynthesis is important for chloroplast biogenesis and cell division (Hung et al., 2004). Our present study showed that VAL1 encodes a phosphoribosylamine-glycine ligase, the second enzyme in the de novo purine biosynthesis pathway. In the val1 mutant, chloroplast development was severely defective and chlorophyll metabolism was disrupted, which resulted in the dynamic green-revertible albino leaf phenotype. Taken together, the findings indicated that VAL1 is a crucial enzyme that regulates the de novo purine biosynthesis pathway, which may be involved in the development of leaf colour in rice. In addition, the val1 mutant developed narrow leaves, caused by diminished cell division owing to the reduced content of cytokinins in the leaf (Fig. 7), and led to a reduced number of vascular bundles and fewer cells between the small vascular bundles (Fig. 1). Thus, the de novo purine biosynthesis pathway may also be involved in the process of cell division in rice. The location of the pathway within plant cells is presently still in dispute, although the subcellular localization of multiple enzymes provides convincing evidence (Smith and Atkins, 2002). Our current study showed that VAL1 contained a chloroplast transit peptide and was localized in the chloroplast (Fig. 3), which indicated that at least part of the de novo purine biosynthesis occurs in the chloroplasts in rice.
It is worth considering why the mutation of VAL1 caused a dynamic green-revertible albino phenotype. One hypothesis is that VAL1 paralogs might perform the same or similar functions at later growth stages, and eventually offset the loss caused by mutation in the VAL1 gene. This explanation was confirmed by phylogenetic analysis. There was one VAL1-like gene, LOC_Os12g09540, which showed high similarity to VAL1 at the amino acid level (Fig. 4; Supplementary Fig. S2), and LOC_Os12g09540 was also highly expressed in leaves, with the highest levels detected in L3U (Fig. S3A, B). One reasonable interpretation is that there is a threshold level of purine nucleotides that needs to be met for normal chloroplast biogenesis and chlorophyll metabolism. Under rapid growth conditions, purine nucleotides are channelled into cell division, the purine nucleotide level in the mutant drops below the threshold, and chloroplasts fail to develop normally, causing the mutant to display albino leaves. When the division and growth rate of leaf cells are slower, purine nucleotides have time to accumulate in the mutant cells, eventually reaching the threshold level, and the mutant leaves subsequently turn green in the later stages. In val1 seedlings, the contents of the de novo purine biosynthesis products adenine nucleotide (AMP) and guanine nucleotide (GMP) were lower than those of the WT seedlings (Fig. 5B), which indicated that the purine nucleotide level in the mutant dropped below the threshold. Furthermore, in the val1 mutant, application of AMP, GMP, and AMP and GMP together partially restored the accumulation of chlorophyll in the val1 mutant (Fig. 5D, E). Taken together, these results indicated that the dynamic green-revertible albino process may be regulated by the threshold level of purine nucleotides. In addition, purine nucleotides are maintained in intracellular pools through a combination of de novo synthesis and salvage pathways (Zrenner et al., 2006). Previous studies have demonstrated that the strong demand for nucleotides in growing and dividing cells is met by de novo synthesis, whereas non-growing cells may be able to maintain their pools of nucleotides by salvaging (Ashihara and Crozier, 1999; Zrenner et al., 2006). VAL1 achieved the highest expression levels in the WT leaves at the L3 stage, and accumulated more in L3L but decreased dramatically in L3U (Fig. 3C). These observations suggested that VAL1 might play an important role at the early leaf development stage, with a relatively minor role in the mature leaf. Thus, another reasonable interpretation is that the de novo purine biosynthesis process regulates chloroplast development and chlorophyll metabolism at the early leaf development stage, and the salvage pathway regulates chloroplast development and chlorophyll metabolism in mature leaves.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. The val1 mutant is a temperature-insensitive green-revertible albino mutant.
Fig. S2. Protein sequence alignment of VAL1.
Fig. S3. Expression patterns of LOC_Os12g09540.
Fig. S4. VAL1 participates in the de novo purine biosynthesis pathway.
Fig. S5. Exogenous application of 6-BA to the wild-type and val1 mutant.
Fig. S6. Expression of genes associated with chloroplast development, photosynthesis, and pigment metabolism in the wild-type and val1 mutant in the heading stage.
Table S1. Primers used in the study.
Table S2. DEGs annotated within chloroplast development and photosynthesis.
Table S3. DEGs annotated within pigment biosynthesis.
Supplementary Material
Acknowledgements
This work was supported by the National Key Research and Development Project (2017YFD0100201), and the Project of Chongqing Science & Technology Commission Grants CSTCCXLJRC201713 and CSTC2016SHMS-ZTZX0017.
References
- Ashihara H, Crozier A. 1999. Biosynthesis and metabolism of caffeine and related purine alkaloids in plants. Advances in Botanical Research 30, 117–205. [Google Scholar]
- Berthomé R, Thomasset M, Maene M, Bourgeois N, Froger N, Budar F. 2008. pur4 mutations are lethal to the male, but not the female, gametophyte and affect sporophyte development in Arabidopsis. Plant Physiology 147, 650–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng XJ, Zhang HQ, Wang Y, He F, Liu JL, Xiao X, Shu ZF, Li W, Wang GH, Wang GL. 2014. Mapped clone and functional analysis of leaf-color gene Ygl7 in a rice hybrid (Oryza sativa L. ssp. indica). PLoS ONE 9, e99564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Fei GL, Wu CY, et al. 2013. A rice virescent-yellow leaf mutant reveals new insights into the role and assembly of plastid caseinolytic protease in higher plants. Plant Physiology 162, 1867–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong YJ, Dong WQ, Shi SY, Jin QS. 2001. Identification and genetic analysis of a thermo-sensitive seedling-colour mutant in rice (Oryza sativa L.). Breeding Science 51, 1–4. [Google Scholar]
- Du P, Ling YH, Sang XC, Zhao FM, Xie R, Yang ZL, He GH. 2009. Gene mapping related to yellow green leaf in a mutant line in rice (Oryza sadva L.). Genes & Genomics 31, 165–171. [Google Scholar]
- Fromme P, Melkozernov A, Jordan P, Krauss N. 2003. Structure and function of photosystem I: interaction with its soluble electron carriers and external antenna systems. FEBS Letters 555, 40–44. [DOI] [PubMed] [Google Scholar]
- Gothandam KM, Kim ES, Cho H, Chung YY. 2005. OsPPR1, a pentatricopeptide repeat protein of rice is essential for the chloroplast biogenesis. Plant Molecular Biology 58, 421–433. [DOI] [PubMed] [Google Scholar]
- Hajirezaei MR, Börnke F, Peisker M, Takahata Y, Lerchl J, Kirakosyan A, Sonnewald U. 2003. Decreased sucrose content triggers starch breakdown and respiration in stored potato tubers (Solanum tuberosum). Journal of Experimental Botany 54, 477–488. [DOI] [PubMed] [Google Scholar]
- Hung WF, Chen LJ, Boldt R, Sun CW, Li HM. 2004. Characterization of Arabidopsis glutamine phosphoribosyl pyrophosphate amidotransferase-deficient mutants. Plant Physiology 135, 1314–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Li M, Liang N, et al. 2007. Molecular cloning and function analysis of the stay green gene in rice. The Plant Journal 52, 197–209. [DOI] [PubMed] [Google Scholar]
- Jung KH, Hur J, Ryu CH, Choi Y, Chung YY, Miyao A, Hirochika H, An G. 2003. Characterization of a rice chlorophyll-deficient mutant using the T-DNA gene-trap system. Plant & Cell Physiology 44, 463–472. [DOI] [PubMed] [Google Scholar]
- Kim JE, Myong K, Shim WB, Yun SH, Lee YW. 2007. Functional characterization of acetylglutamate synthase and phosphoribosylamine-glycine ligase genes in Gibberella zeae. Current Genetics 51, 99–108. [DOI] [PubMed] [Google Scholar]
- Kusaba M, Ito H, Morita R, et al. 2007. Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. The Plant Cell 19, 1362–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi K, Sakata C, Nakamura T, Kawasaki S, Yoshimura A, Iba K. 2011. A plastid protein NUS1 is essential for build-up of the genetic system for early chloroplast development under cold stress conditions. The Plant Journal 68, 1039–1050. [DOI] [PubMed] [Google Scholar]
- Lee S, Kim JH, Yoo ES, Lee CH, Hirochika H, An G. 2005. Differential regulation of chlorophyll a oxygenase genes in rice. Plant Molecular Biology 57, 805–818. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. 1987. Chlorophylls and carotenoids – pigments of photosynthetic biomembranes. Methods in Enzymology 148, 350–382. [Google Scholar]
- Liu W, Fu Y, Hu G, Si H, Zhu L, Wu C, Sun Z. 2007. Identification and fine mapping of a thermo-sensitive chlorophyll deficient mutant in rice (Oryza sativa L.). Planta 226, 785–795. [DOI] [PubMed] [Google Scholar]
- Ma J Ren D, Wu G Zhu X, Ma L, Ling Y Sang X, He G. 2014. Genetic analysis and gene mapping of a marginal albino leaf mutant mal in rice. The Crop Journal 40, 591–599. [Google Scholar]
- Ma L, Sang X, Zhang T, et al. 2017. ABNORMAL VASCULAR BUNDLES regulates cell proliferation and procambium cell establishment during aerial organ development in rice. New Phytologist 213, 275–286. [DOI] [PubMed] [Google Scholar]
- Park YJ, Song ES, Kim YT, Noh TH, Kang HW, Lee BM. 2007. Analysis of virulence and growth of a purine auxotrophic mutant of Xanthomonas oryzae pathovar oryzae. FEMS Microbiology Letters 276, 55–59. [DOI] [PubMed] [Google Scholar]
- Ren D, Yu H, Rao Y, et al. 2018. ‘Two-floret spikelet’ as a novel resource has the potential to increase rice yield. Plant Biotechnology Journal 16, 351–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnorr KM, Nygaard P, Laloue M. 1994. Molecular characterization of Arabidopsis thaliana cDNAs encoding three purine biosynthetic enzymes. The Plant Journal 6, 113–121. [DOI] [PubMed] [Google Scholar]
- Smith PM, Atkins CA. 2002. Purine biosynthesis. Big in cell division, even bigger in nitrogen assimilation. Plant Physiology 128, 793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su N, Hu ML, Wu DX, et al. 2012. Disruption of a rice pentatricopeptide repeat protein causes a seedling-specific albino phenotype and its utilization to enhance seed purity in hybrid rice production. Plant Physiology 159, 227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences, USA 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto H, Kusumi K, Noguchi K, Yano M, Yoshimura A, Iba K. 2007. The rice nuclear gene, VIRESCENT 2, is essential for chloroplast development and encodes a novel type of guanylate kinase targeted to plastids and mitochondria. The Plant Journal 52, 512–527. [DOI] [PubMed] [Google Scholar]
- Sugimoto H, Kusumi K, Tozawa Y, Yazaki J, Kishimoto N, Kikuchi S, Iba K. 2004. The virescent-2 mutation inhibits translation of plastid transcripts for the plastid genetic system at an early stage of chloroplast differentiation. Plant & Cell Physiology 45, 985–996. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong QL, Cho Y, Barate AK, Kim S, Watarai M, Hahn TW. 2015. Mutation of purD and purF genes further attenuates Brucella abortus strain RB51. Microbial Pathogenesis 79, 1–7. [DOI] [PubMed] [Google Scholar]
- Wang P, Gao J, Wan C, Zhang F, Xu Z, Huang X, Sun X, Deng X. 2010. Divinyl chlorophyll(ide) a can be converted to monovinyl chlorophyll(ide) a by a divinyl reductase in rice. Plant Physiology 153, 994–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z-W, Lv J, Xie S-Z, et al. 2017. OsSLA4 encodes a pentatricopeptide repeat protein essential for early chloroplast development and seedling growth in rice. Plant Growth Regulation 84, 249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Zhang X, He B, et al. 2007. A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiology 145, 29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Xu J, Huang L, et al. 2016. PGL, encoding chlorophyllide a oxygenase 1, impacts leaf senescence and indirectly affects grain yield and quality in rice. Journal of Experimental Botany 67, 1297–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SC, Cho SH, Sugimoto H, Li J, Kusumi K, Koh HJ, Iba K, Paek NC. 2009. Rice virescent3 and stripe1 encoding the large and small subunits of ribonucleotide reductase are required for chloroplast biogenesis during early leaf development. Plant Physiology 150, 388–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liu LL, Cai MH, et al. 2015. A point mutation of magnesium chelatase OsCHLI gene dampens the interaction between CHLI and CHLD subunits in rice. Plant Molecular Biology Reporter 33, 1975–1987. [Google Scholar]
- Zhang T, Li Y, Ma L, et al. 2017. LATERAL FLORET 1 induced the three-florets spikelet in rice. Proceedings of the National Academy of Sciences, USA 114, 9984–9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zrenner R, Stitt M, Sonnewald U, Boldt R. 2006. Pyrimidine and purine biosynthesis and degradation in plants. Annual Review of Plant Biology 57, 805–836. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.