Synthesis-dependent strand annealing is a primary mechanism in the repair of Cpf1-induced DSBs on homology repair templates; such templates are available and enable targeted gene replacement in rice.
Keywords: Cpf1, gene replacement, homology-directed DNA repair (HDR), ribozyme, rice (Oryza sativa L), synthesis-dependent strand annealing (SDSA)
Abstract
The recently developed CRISPR (clustered regularly interspaced short palindromic repeats)/Cpf1 system expands the range of genome editing and is emerging as an alternative powerful tool for both plant functional genomics and crop improvement. Cpf1-CRISPR RNA (crRNA) produces double strand DNA breaks (DSBs) with long 5'-protruding ends, which may facilitate the pairing and insertion of repair templates through homology-directed repair (HDR) for targeted gene replacement and introduction of the desired DNA elements at specific gene loci for crop improvement. However, the potential mechanism underlying HDR of DSBs generated by Cpf1-crRNA remains to be investigated, and the inherent low efficiency of HDR and poor availability of exogenous donor DNA as repair templates strongly impede the use of HDR for precise genome editing in crop plants. Here, we provide evidence of synthesis-dependent repair of Cpf1-induced DSBs, which enables us precisely to replace the wild-type ALS gene with the intended mutant version that carries two discrete point mutations conferring herbicide resistance to rice plants. Our observation that the donor repair template (DRT) with only the left homologous arm is sufficient for precise targeted allele replacement offers a better understanding of the mechanism underlying HDR in plants, and greatly simplifies the design of DRTs for precision genome editing in crop improvement.
Introduction
Double strand DNA breaks (DSBs) in target genes generated by Cas endonucleases are repaired by the error-prone non-homologous end joining (NHEJ) pathway or the precise homology-directed repair (HDR), or both NHEJ and HDR (Jinek et al., 2012; Cong et al., 2013; Zetsche et al., 2015). NHEJ often leads to a small insertion/deletion and is widely used in generating loss-of-function mutants. HDR utilizes a DNA donor repair template (DRT) that is flanked with sequences homologous to those adjacent to the DSBs to guide the DNA repair (Puchta, 1998). HDR can be used for targeted allele replacement or introduction of markers to target loci, providing a valuable tool for crop improvement. In principle, three potential mechanisms have been proposed for HDR of DSBs: single-strand annealing (SSA), synthesis-dependent strand annealing (SDSA), and the so-called double strand break repair (DSBR) model (Puchta, 1998). During SSA, both ends of the DSB carry complementary sequences. These molecules can then anneal to one another to form a chimeric DNA molecule with the 3' overhangs. Consequently, the sequences between the complementary sequences will be lost. Therefore, SSA is also classified as a non-conservative HDR-mediated DSB repair mechanism (Puchta and Fauser, 2013). In the cases of DSBR and SDSA, the homologous repair template can be supplied in cis or in trans. Following the DSB induction, 3' end invasion of a single strand into a homologous double strand occurs, resulting in a D-loop. Reparative synthesis is initiated using the newly paired strand as a template. In DSBR, DNA synthesis occurs at both broken ends so that genetic information is copied from both strands of the homologous sequences, which may lead to a crossover event (Puchta and Fauser, 2013). DSBR is a prominent mechanism for meiotic recombination (Osman et al., 2011). In SDSA, the DSB is at first resected and processed to generate 3' overhangs on both sides of the DSB. The 3' overhangs are then paired with the homologous arms of the DRT and are extended by DNA synthesis using the DRT as a template. Then, the newly synthesized strands withdraw from the DRT and anneal back to the locus (Puchta and Fauser, 2014; Paix et al., 2017). SDSA has been proposed as a repair mechanism when the single-stranded oligo DNA nucleotides (ssODNs) are used as the DRT (Paquet et al., 2016; Richardson et al., 2016; Kan et al., 2017). When dsDNAs are used as repair templates, different DSBs generated by Cas9 variants engage in different repair pathways and the polarity of the overhang structure is a critical determinant of DSB repair pathway choice in human cells (Bothmer et al., 2017). However, analyses of the sequence requirements for efficient repair of DSBs generated by Cas9 in human cells indicated that the repair process is more consistent with SDSA when either ssODNs or dsDNAs are used as repair templates (Paix et al., 2017). Thus, the mechanism underlying HDR of DSBs when a dsDNA is used as the DRT still remains inconclusive, especially in plants.
Genome editing technologies enable precise modifications of DNA sequences in vivo and offer great promise for crop improvement. CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated Cas9) has revolutionized genome editing because of its simplicity and versatility (Cong et al., 2013; Miao et al., 2013; Shan et al., 2013; Ma et al., 2015; Sun et al., 2016; J. Li et al., 2018). CRISPR/Cpf1 (CRISPR from Prevoltella and Francisella 1), a new class 2 CRISPR/Cas system, was recently exploited as an alternative to the widely used SpCas9 in genome editing in many organisms including plants (Zetsche et al., 2015, 2017; Fonfara et al., 2016; Kim et al., 2016; Gao et al., 2017; Hu et al., 2017; Kim et al., 2017; Tang et al., 2017; Wang et al., 2017; Xu et al., 2017; S. Li et al., 2018; Zhong et al., 2018). Cpf1 utilizes a thymidine-rich protospacer adjacent motif (PAM) site ‘TTTN’ (Zetsche et al., 2015) or non-canonical ‘TYCV’ (S. Li et al., 2018), and is guided by a single CRISPR RNA (crRNA) (Zetsche et al., 2015). The Cpf1 crRNA (~43 nt) is shorter than that of SpCas9 single guide RNA (sgRNA) by 60 nucleotides and no trans-acting crRNA is needed (Fonfara et al., 2016). Moreover, Cpf1 produces long 5'-protruding ends, which may facilitate the pairing and insertion of repair templates (Zetsche et al., 2015). Thus, the CRISPR/Cpf1 system is emerging as an attractive tool for editing AT-rich regions (Begemann et al., 2017; Tang et al., 2017; Wang et al., 2017; S. Li et al., 2018). Herein, we coupled a plant codon-optimized LbCpf1 from Lachnospiraceae bacterium ND 2006 (Wang et al., 2017) with a simple ribozyme crRNA array driven by the OsU3 promoter (Gao and Zhao, 2014). We demonstrated that DRTs with either only the left homologous arm or two homologous arms function efficiently in achieving precise targeted replacement of the Acetolactate synthase (ALS) gene, which encodes a key enzyme for the biosynthesis of the branched chain amino acids leucine, isoleucine, and valine, and is a major target for ALS-inhibiting herbicides such as chlorsulfuron and bispyribac sodium (BS) in rice (Mazur et al., 1987). Our findings support the hypothesis that SDSA is involved in HDR of DSBs generated by Cpf1-crRNA in plant cells when the dsDNA is used as a DRT. Our observation that the left homologous arm alone is sufficient for HDR not only offers a better understanding of the mechanism underlying HDR in plants, but also greatly simplifies the design of DRTs for targeted allele replacement in crop plants.
Materials and methods
Construction of the CRISPR/LbCpf1-related vectors
The vectors used in this study were based on the vector pCXUN-LbCpf1 that replaced the ubiquitin-Cas9 in plasmid pCXUN-Cas9 (Sun et al., 2016) with ubiquitin-LbCpf1 in plasmid LbCpf1-OsU6 (Wang et al., 2017). The backbone of pCXUN-LbCpf1 contains a hygromycin-resistant gene (hpt) for callus selection. The SacI and PmeI sites in pCXUN-LbCpf1 were used for introducing the OsU3–RCR1–RCR2 expression cassette and the DRT, respectively (Supplementary Fig. S1 at JXB online).
The RCR1 unit was assembled through two rounds of overlapping PCRs. The first PCR was performed using primer set RCR1F2/RCR-common-R with the plasmid pRS316-RCR-GFP (Zhang et al., 2017) as a template, and the second one used primer set RCRF1/RCR-common-R using the PCR1 product as a template (Supplementary Table S1). The same procedure was used to obtain the RCR2 unit by utilizing the primers RCR2-F2/RCR-common-R and RCR-F1/RCR-common-R, respectively (Supplementary Table S1). The OsU3 promoter sequence was amplified with primer set OsU3F/OsU3R using plasmid pCXUN-Cas9-OsU3 as template (Supplementary Table S1). Because OsU3 promoter sequences were used in this experiment, we also placed an A before the first nucleotide of the target sequences. The full-length OsU3–RCR1–RCR2 cassette was obtained through five rounds of overlapping PCRs. The first PCR was performed with primer set OsU3F/OsU3-RCR1R using the OsU3 promoter sequence as template (Supplementary Table S1). The second PCR was performed with primer set RCR-Common-F/RCR1-10 random-R using the RCR1 unit as the template (Supplementary Table S1). Products of PCRs 1 and 2 were used as templates for the third PCR with the primer set OsU3-F/RCR1-10 random-R to generate the OsU3–RCR1 cassette (Supplementary Table S1). The fourth PCR was performed with primer set RCR2-10 random-F/SacI-RCR2-R using the RCR2 unit as the template (Supplementary Table S1). The products of PCRs 3 and 4 were used as templates for the fifth PCR with the primer set SacI-OsU3-F/SacI-RCR2-R to generate the OsU3–RCR1–RCR2 cassette. At the 5' end of the primer pair SacI-OsU3-F/SacI-RCR2-R, the sequences are homologous to the sequences outsides of the SacI site in pCXUN-LbCpf1. Then, the OsU3–RCR1–RCR2 fragment was cloned to the linearized pCXUN-LbCpf1 with SacI, by using the pEASY-Uni Seamless Cloning and Assembly Kit (TransGen Biotech, Beijing, China). The vector harboring OsU3–RCR1–RCR2 was named pCXUN-LbCpf1-OsU3-RCR1-RCR2.
The DRT with a mutated ALS sequence was synthesized by BGI (Beijing Genomics Institute-Shenzhen, Chinese Academy of Sciences, Shenzhen, China) (named as armed-DRT) (Supplementary Fig. S2B). The vector pCXUN-LbCpf1-OsU3-RCR1-RCR2-left-armed-DRT was obtained by overlapping PCRs. The first PCR was performed with primer set donor-armLF/donor-armLR1 using the synthesized armed-donor sequence as template. The second PCR was performed with primer set donor-armLF/donor-armLR2 using the product of PCR1 as the template. Products of PCR 2 were used as templates for the third PCR with the primer set pme-donor-armLF/pme-donor-armLR to generate the donor with only the left homology arm (Supplementary Fig. S2A). This fragment was cloned into the PmeI site of pCXUN-LbCpf1-OsU3-RCR1-RCR2 by using the Assembly Kit (TransGen Biotech). The final plasmid was named pCXUN-LbCpf1-OsU3-RCR1-RCR2-left-armed-DRT (Fig. 1A). PCR primers for vector construction are listed in Supplementary Table S1.
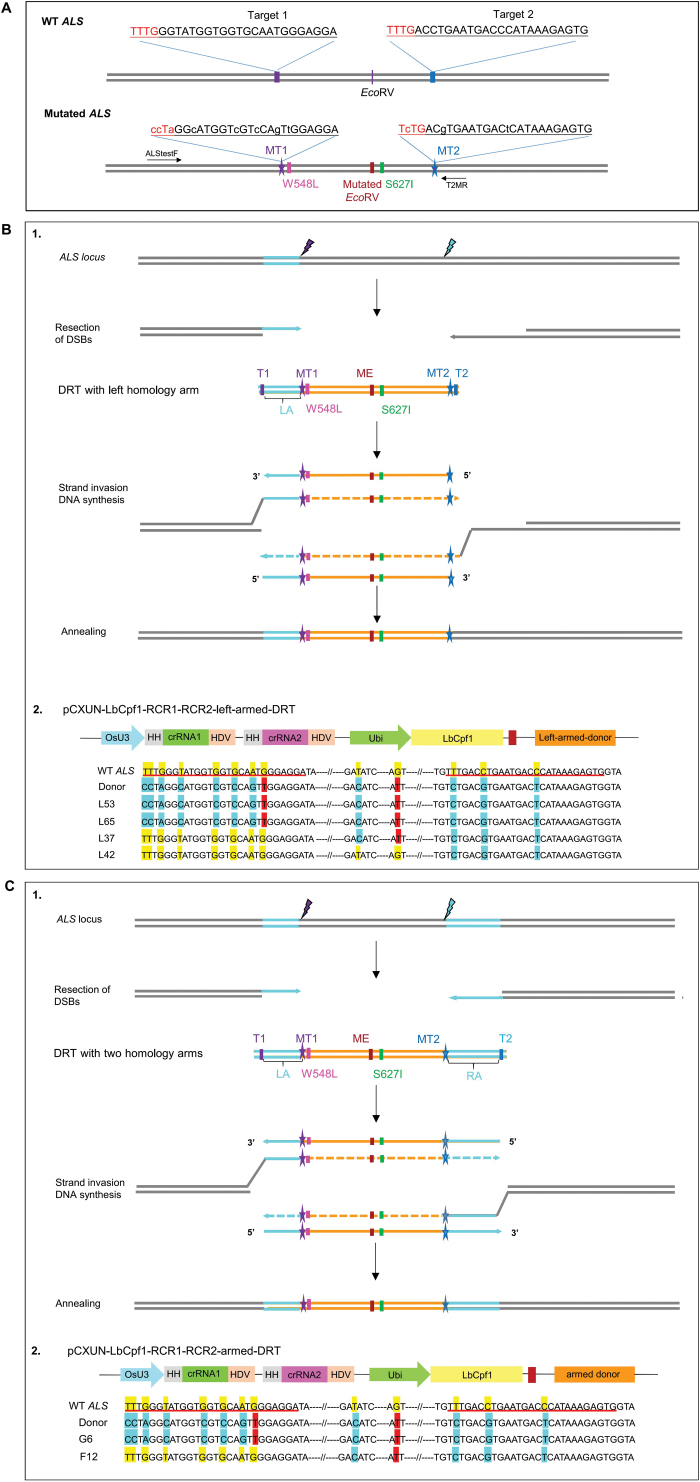
Fig. 1.
Homology-directed repair (HDR) of double strand breaks (DSBs) generated by the LbCpf1 endonuclease in rice. (A) A schematic description of HDR-mediated precise gene replacement in rice. A wild-type (WT) ALS gene fragment is removed by LbCpf1 and a couple of crRNAs. The LbCpf1 target sequences are underlined, and the PAM sites and the mutated PAM sites are underlined and highlighted in red. The WT ALS fragment is replaced by a mutant version that introduces changes of two discrete amino acid residues. The PAM sites and an EcoRV site are modified to prevent the replacement from further cleavage by LbCpf1/crRNAs and to facilitate detection of gene replacement events, respectively. (B) CRISPR/Cpf1-mediated HDR in rice calli using vector pCXUN-LbCpf1-OsU3-RCR1-RCR2-left-armed-DRT co-introduced with free left armed-DRT through CRISPR/Cpf1-mediated HDR. (B-1) A schematic description of the synthesis-dependent strand annealing (SDSA) pathway of HDR with the donor containing only the left homology arm. Each line corresponds to a DNA strand. Chromosomal DNA is in gray, DRT in orange, homology arms of DRT are in blue, and arrows refer to the 3' ends. T1, target 1; T2, target 2; MT1, mutated PAM site and mutated target 1; MT2, mutated PAM site and mutated target 2; LA, left arm; ME, mutated EcoRV. Resecting DSB creates 3' overhangs on each side of the DSB. The overhangs in the 3' end pair with complementary strands in the DRT and are extended. The newly synthesized strands withdraw from the donor and anneal back at the locus. (B-2) Sequence analyses of the representative HDR events. PCR products were amplified by allele-specific primer set ALStestF/T2MR as described in (A). The calli L53 and L65 had undergone precise HDR, whereas the calli L37 and L42 had undergone partial HDR. The sequences shadowed in yellow and blue represent the same bases as those of the wild type and the designed DRT, respectively. Specifically, the sequences shadowed in red indicate the expected targeted substitution. (C) CRISPR/Cpf1-mediated HDR in rice calli using vector pCXUN-LbCpf1-OsU3-RCR1-RCR2-armed-DRT co-introduced with free armed-DRT through CRISPR/Cpf1-mediated HDR. (C-1) A schematic description of the synthesis-dependent strand annealing (SDSA) pathway of HDR with a DRT containing two homologous arms. In this, and all other schematics, each line corresponds to a DNA strand. Chromosomal DNA is indicated in gray, DRT is in orange, the homology arms are in blue, and arrows indicate the 3' ends. T1, target 1; T2, target 2; MT1, mutated PAM site and mutated target 1; MT2, mutated PAM site and mutated target 2; LA, left arm; RA, right arm; ME, mutated EcoRV. (C-2) Sequence analyses of the representative HDR events. PCR products were amplified by allele-specific primer set ALStestF/T2MR as described in (A). The callus G6 had a precise HDR, whereas the callus F5 had undergone partial HDR. The sequences shaded in yellow and blue represent the wild-type and the designed donor repair template, respectively. Specifically, the sequences shaded in red indicate the expected targeted substitutions.
For the vector pCXUN-LbCpf1-OsU3-RCR1-RCR2-armed-DRT, the donor fragment (Supplementary Fig. S2B) was amplified by PCR using primer set Pme-donorF/Pme-donorR with synthesized armed-DRT as template and cloned into the PmeI site of pCXUN-LbCpf1-OsU3-RCR1-RCR2 by using the Assembly Kit (TransGen Biotech). The final plasmid was named pCXUN-LbCpf1-OsU3-RCR1-RCR2-armed-DRT (Fig. 1B). PCR primers for vector construction are listed in Supplementary Table S1.
In vivo CRISPR/Cpf1-mediated HDR in rice calli
In order to provide enough donor fragments for HDR, both vectors and the free 549 bp left homology armed-DRT and the 670 bp armed-DRT with two homologous arms were co-introduced into rice (japonica cv. Zhonghua 11) calli with a molar ratio of 1:20 by particle bombardment Two days after bombardment, DNA from the calli was extracted using a DNA Quick Plant System (Tiangen, Beijing, China). PCR amplification was performed using EASY Taq polymerase (TransGen Biotech) employing 200 ng of genomic DNA as template. Each callus was tested individually by PCR and sequencing. The PCR products were generated using the allele-specific primer set ALSTestF/T2MR (Supplementary Table S1) with the upstream primer located in the genome sequence of the ALS gene outside of the left homology arm, whereas the downstream primer was an allele-specific primer (Fig. 1A). The obtained amplicons were cloned into the cloning vector pEasy-Blunt Zero (TransGen Biotech). At least 10 positive colonies for each sample were sequenced.
Rice transformation
The plasmids pCXUN-LbCpf1-OsU3-RCR1-RCR2-left-armed-DRT and pCXUN-LbCpf1-OsU3-RCR1-RCR2-armed-DRT were linearized by SacII and then mixed with the free DRTs at a molar ratio of 1:20, which were co-transformed into calli of a japonica rice (cv. Zhonghua 11) by particle bombardment following the protocol described previously (Li et al., 1993). Particle bombardments were performed using a PDS1000/He particle bombardment system (Bio-Rad, Hercules, CA, USA).
Rice calli induced from mature seeds were used for bombardment. After bombardment, the calli were selected on medium containing 50 mg l–1 hygromycin for 2 weeks to allow the growth of calli with the construct, either transiently expressed or stably integrated. Then the well-grown calli were transferred to induction medium with 0.4 μM BS. After 2 weeks, the vigorously grown calli were transferred to regeneration media to generate green plants.
Molecular characterization of the edited plants
Rice genomic DNA from ~0.2 g of leaf tissue was extracted using a DNA Quick Plant System (Tiangen). PCR amplification was performed using EASY Taq polymerase (TransGen Biotech) and 100 ng of genomic DNA as a template. All plants were tested individually with PCR-RE (PCR and restriction enzyme digestion assay) and sequencing. The PCR products amplified by the primer pair ALSTestF/ALSTestR (Supplementary Table S1) were digested with EcoRV and then directly sequenced to screen for the plants with a modified ALS gene. The sequence chromatograms were analyzed by a web-based tool (http://skl.scau.edu.cn/dsdecode/) to confirm the genotype and zygosity of the tested plants (Liu et al., 2015). Some PCR products were also cloned into the cloning vector P-easy (TransGen Biotech), and at least 10 positive colonies for each sample were sequenced. Primers for detection of the presence of LbCpf1, crRNA, and hptII are listed in Supplementary Table S1.
To investigate off-target effects, we selected three and two potential off-target sites based on the predictions of the Cas9-OFFinder tool (http://rgenome.net/cas-offinder/), for the target 1 and target 2, respectively (Supplementary Table S3). Site-specific genomic PCR and Sanger sequencing was used to determine the off-target effects. The primer sets are as listed in Supplementary Table S1.
Results
Left armed-DRT successfully mediated HDR in rice calli
To investigate whether dsDNA with only a left homologous arm can effectively mediate HDR, we first tested this strategy in a transient assay in rice calli to replace the wild-type ALS gene with the mutated version (Fig. 1A). We designed a plasmid named pCXUN-LbCpf1-OsU3-RCR1-RCR2-left-armed-DRT, which harbors cassettes expressing LbCpf1, two crRNAs, and a designed DRT with only a 97 bp left homologous arm (left armed-DRT) (Fig. 1B-1; Supplementary Figs S1A, 2A). The two crRNAs could cut out a fragment from the wild-type ALS gene in the presence of LbCpf1. The 549 bp DRT (Supplementary Fig. S2A) contained two desired mutations (W548L and S627I) that rendered rice plants resistant to multiple ALS-inhibiting herbicides (Mazur et al., 1987). We also introduced synonymous mutations into the two crRNA target sequences and a restriction site inside the DRT. We then flanked the DRT with the same two crRNA target sequences to enable the release of DRT from the vector in vivo (Supplementary Fig. S2A). Both the vector and the free 549 bp left armed-DRT were co-introduced into rice (japonica cv. Zhonghua 11) calli with a molar ratio of 1:20 by particle bombardment in order to enrich the availability of DRTs. After 2 d, genomic DNA was extracted from the bombarded calli to determine whether the intended gene replacement through SDSA was achieved following SDSA-mediated HDR with only left armed-DRT (Fig. 1B-1). By using the allele-specific primer set ALSTestF/T2MR (Table S1), we detected the precise replacement of the wild-type ALS gene with the mutant version (Fig. 1B-2). We also observed partial HDR events, by which only one end was replaced (Fig. 1B-2), suggesting that there might be template switching during the repair of DSBs (Smith et al., 2007).
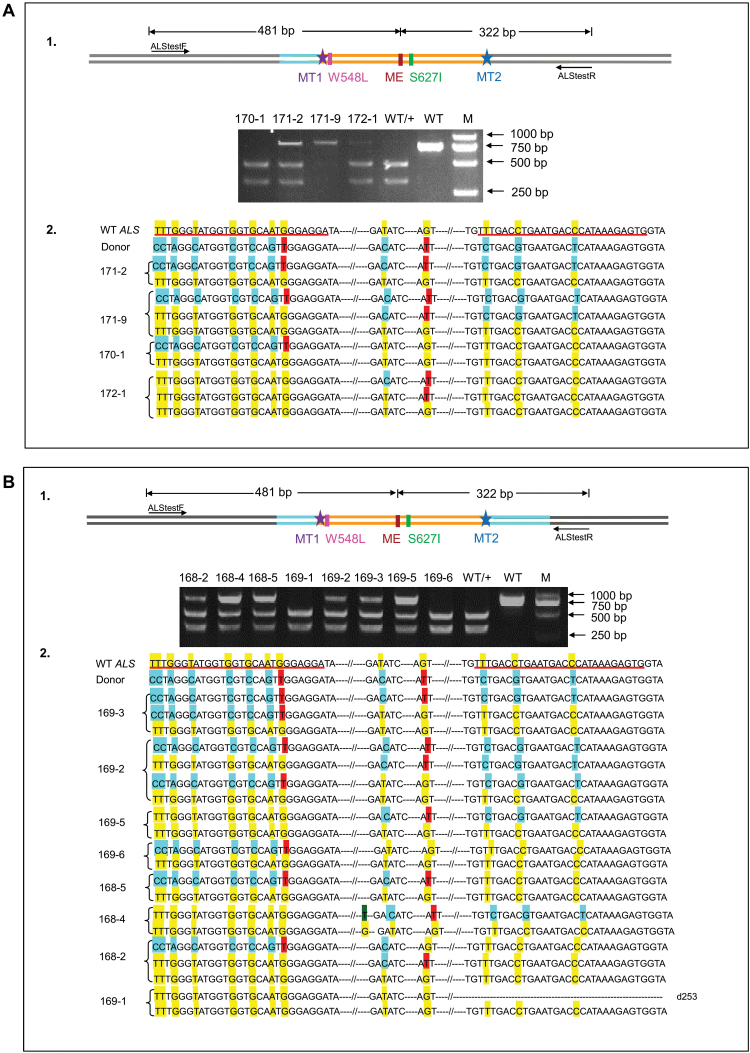
Fig. 2.
Generation of stable precisely edited rice lines through SDSA-mediated HDR. (A) Only the left homology arm strategy for CRISPR/LbCpf1-mediated HDR in rice. (A-1) PCR-RE analyses of the different genotypes. PCR products amplified by primers ALStestF/R were digested with EcoRV (GACATC). M, DL2000; WT, wild-type; T1, target 1; T2, target 2; MT1, mutated PAM site and mutated target 1; MT2, mutated PAM site and mutated target 2; ME, mutated EcoRV; WT/+, EcoRV cuts the PCR products of the wild type, resulting in 481 bp and 322 bp fragments. Successful HDR leads to EcoRV-resistant bands. (A-2) Sequence analyses of the HDR events. Line 171-2 has one allele with precise HDR, while the other is wild type. Line 171-9 is a chimeric line with three alleles: a precise HDR, a partial HDR, and a wild type. Line 170-1 has a heterozygous partial HDR. Line 172-1 is a chimeric line with three alleles: one base pair substitution, a partial HDR, and a wild type. The sequences shaded in yellow and blue represent wild-type and the designed donor repair template, respectively. Specifically, the sequences shaded in red indicate the expected targeted substitution. (B) Generation of stable precisely edited rice lines using pCXUN-LbCpf1-OsU3-RCR1-RCR2-armed donor vector through CRISPR/Cpf1-mediated HDR. (B-1) PCR-RE analyses of the different genotypes. PCR products amplified by primers ALStestF/R were digested with EcoRV (GACATC). M, DL2000; WT, wild-type; T1, target 1; T2, target 2; MT1, mutated PAM site and mutated target 1; MT2, mutated PAM site and mutated target 2; ME, mutated EcoRV; WT/+, EcoRV cuts the PCR products of the wild type, resulting in 481 bp and 322 bp fragments. EcoRV failed to digest the PCR products if HDR was successful. (B-2) Representative sequences of the different genotypes. Line 169-3 is a chimeric line with three alleles: one precise HDR, one partial HDR, and a wild type. Line 169-2 is a chimeric line with four alleles: a precise HDR, two partial HDR events, and a wild type. Lines 168-4, 168-5, 169-5, and 169-6 have one partial HDR, except for a synonymous G to T base substitution at position 1911 bp in Line 168-4. The 168-2 is a chimeric line with three alleles. The first and second allele have partial HDR, while the third allele is wild type. Line 169-1 has a 253 bp deletion. The sequences shaded in yellow and blue represent wild-type and the designed donor repair template, respectively. Specifically, the sequences shaded in red indicate the expected targeted substitutions. In d#, the # refers to the number of base pairs deleted from the target sites. Different numbers of lines indicate independent lines developed from resistant calli.
HDR in rice calli mediated by DRT with two homologous arms
Similarly, we designed a plasmid named pCXUN-LbCpf1-OsU3-RCR1-RCR2-armed-DRT that harbors cassettes expressing LbCpf1, two crRNAs, and a designed DRT with two homologous arms (armed-DRT) (Figs 1C-1; Supplementary Fig. S1B). This 670 bp armed-DRT was the same as the 549 bp left armed-DRT shown above, except that it also has a 121 bp right homologous arm (Supplementary Fig. S2B). Following SDSA (Fig. 1C-1), we observed the intended precise replacement of the wild-type ALS gene with the mutant version and partial homologous recombination events when this armed-donor was used (Fig. 1C-2).
Generation of stable edited rice plants by HDR using either left armed-DRT or DRT with two arms
We then investigate whether we can generate stable precisely edited rice lines using the two strategies described above. For vectors with left armed-DRT and those with armed-DRT, 152 and 164 calli were bombarded, respectively. The calli that survived one round of selection pressure of hygromycin were subsequently transferred onto induction medium and regeneration medium with 0.4 µM BS. Then, the regenerated plants were used for PCR-RE. PCR primer set ALStestF/R, which is located at the genome site beyond the two homologous arms in DRT, was designed to amplify an ALS fragment from both the wild-type ALS locus and the edited ALS, but not from the plasmid (Fig. 2; Supplementary Table S1). All plantlets developed from one callus were treated as a pool. The PCR products were then digested with EcoRV. The plantlets in a pool that gave a PCR-RE result different from that of the wild type were then transferred into soil individually and were tested with PCR-RE and sequenced. No obvious phenotypic variations were observed between these lines and the non-transformed wild-type control. In total, 71 and 94 plants developed from 15 and 20 resistant calli for the two different vectors, respectively, were selected for further analyses (Supplementary Table S2).
For vector pCXUN-LbCpf1-OsU3-RCR1-RCR2-left-armed-DRT, among the 71 plants recovered, PCR-RE and sequencing analyses identified one heterozygous line (Line 171-2) and one chimeric line (Line 171-9) with one allele containing the precise replacement as designed, and two lines (Lines 170-1 and 172-1) with partial HDR at either the W548L or the S627I locus (Fig. 2A; Supplementary Table S2). For vector pCXUN-LbCpf1-OsU3-RCR1-RCR2-armed-DRT, among 94 plants recovered, PCR-RE and sequencing analyses identified two chimeric genotypes with one allele that had the expected precise gene replacement (Line 169-2 and 169-3), and five genotypes with partial replacements (Lines 168-2, 168-4, 168-5, 169-5, and 169-6) (Fig. 2B; Supplementary Table S2). Different genotypes observed in these lines indicated that they were independent lines. No off-target effects were observed at predicted potential off-target sites in these tested lines (Table S3).
Discussion
Cpf1-induced DSBs enable targeted gene replacement in rice
We precisely replaced the wild-type ALS gene with the intended mutant version that carries two discrete point mutations conferring herbicide resistance to rice plants through HDR of LbCpf1-induced DSBs by using DRT either with only a left homologous arm or with both homologous arms (Fig. 2). The achievement of precise gene replacement events using only left armed-DRT not only further our understanding of the potential mechanism underlying HDR of DSBs, but also greatly simplifies the design of DRTs for precision genome editing in crop improvement. However, we observed a relatively lower efficiency of CRISPR/Cpf1-mediated ALS gene replacement in this study, compared with that of CRISPR/Cas9-mediated gene replacement in our previous studies (Sun et al., 2016; J. Li et al., 2018). The difference in efficiency probably results from the fact that the editing activity of Cpf1 is not as high as that of Cas9 as reported by others and our previous study (Ma et al., 2015; Wang et al., 2017; S. Li et al., 2018). Despite the lower efficiency, CRISPR/LbCpf1-mediated gene replacement targets sequences that cannot be edited by Cas9 due to the differences in PAM requirement (Jinek et al., 2012; Cong et al., 2013). LbCpf1 utilizes a thymidine-rich PAM ‘TTTV’ (Zetsche et al., 2015), which enables targeting AT-rich regions of a genome such as 5'- and 3'-untranslated regions and the promoter of a gene of interest. It is usually more difficult to find suitable target sequences in the above-mentioned regions for SpCas9, which has a PAM requirement of NGG. Therefore, CRISPR/Cpf1-mediated gene editing is particularly useful for precisely knocking-in a gene of interest or replacing a gene/promotor at a targeted specific locus, or inserting a small DNA fragment that encodes an epitope tag or fluorescence protein at the 5' or 3' end of a gene.
HDR of Cpf1-induced DSBs in rice is consistent with the proposed SDSA model
When a dsDNA DRT is used as the repair template, the Cas9-(D10A) induced 5' overhang DNA structure and DRTs with an asymmetric left homology arm are particularly amenable to gene conversion/replacement (Bothmer et al., 2017). Cpf1 cleavage produces 5'-protruding sticky ends, which may facilitate the pairing and insertion of the DRT through microhomology-mediated end joining (MMEJ) (Zetsche et al., 2015). In this study, we added two crRNA targets at each end of the DRTs. We achieved either precise gene replacement or partial HDR events both in transient assays and in stable edited rice plants (Figs 1B, C, 2). However, we did not detect any events harbouring the integration of the entire DRT with the target sequences at each end, indicating that the DSBs are indeed resected through exo- and/or endonucleolytic processing of the 5' end to yield the 3' overhangs on both sides of the DSBs (Bothmer et al., 2017; Paix et al., 2017). The invasion of 3' end single strands may lead to the formation of a Holliday junction (HJ) and paired with the homologous arms of the DRT, and then extended by DNA synthesis, thus resulting in conservative precise gene replacement (Puchta, 1998). Our observations of the successful achievement of HDR events when a DRT with only the left homologous arm was used as repair template are consistent with a replicative repair process that requires pairing between a 3'-homology arm on the donor and homologous sequences in the genome on at least one side of the DSBs, as demonstrated recently in human cells (Paix et al., 2017). Besides, we also observed the template switching between DRT and wild-type sequences as shown in some partial HDR events (Figs 1B, C, 2), indicating the occurrence of template switching due to unstable replication forks during multiple rounds of dissociation and re-invasion (Smith et al., 2007). This phenomenon, again, is similar to the recently reported precision gene editing using Cas9 endonuclease in human cells (Paix et al., 2017).
Taken together, we here demonstrate synthesis-dependent repair of Cpf1-induced DSBs, which enables us to replace the wild-type ALS gene precisely with the intended mutant version that carries two discrete point mutations conferring herbicide resistance to rice plants. Our finding that donor with only the left homology arm is sufficient to achieve targeted gene replacement enables us to gain a better mechanistic understanding of HDR of DSBs for targeted gene replacement in various organisms and simplifies the design of DRTs to improve agriculturally important traits in crops through CRISPR/Cpf1-mediated genome editing.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. The map of plasmids used in this study.
Fig. S2. Sequences of the DRTs.
Table S1. The primer sets used in this study.
Table S2. Characterization of regenerated rice plants in the T0 generation.
Table S3. Analysis of potential off-target effects.
Author contributions
LQX and YDZ conceived the project. SYL, JYL, JHZ, WMD, JDF and SS performed the experiments. LQX and YDZ wrote the manuscript.
Supplementary Material
Acknowledgements
We would like to thank Professor Jian-Kang Zhu who generously provided us with the LbCpf1 plasmid. This work is partly funded by the Ministry of Science and Technology of China (2016YFD0100500), the Chinese Ministry of Agriculture and Rural Affairs (2018ZX0801016B), and the Central Non-Profit Fundamental Research Funding supported by the Institute of Crop Sciences, Chinese Academy of Agricultural Sciences (CAAS) (S2018QY05). YDZ is supported by a startup project from Hua-Zhong Agricultural University, China. The authors have filed a patent application based on the system developed in this paper.
References
- Begemann MB, Gray BN, January E, Gordon GC, He Y, Liu H, Wu X, Brutnell TP, Mockler TC, Oufattole M. 2017. Precise insertion and guided editing of higher plant genomes using Cpf1 CRISPR nucleases. Scientific Reports 7, 11606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothmer A, Phadke T, Barrera LA, et al. 2017. Characterization of the interplay between DNA repair and CRISPR/Cas9-induced DNA lesions at an endogenous locus. Nature Communications 8, 13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfara I, Richter H, Bratovič M, Le Rhun A, Charpentier E. 2016. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532, 517–521. [DOI] [PubMed] [Google Scholar]
- Gao L, Cox DBT, Yan WX, Manteiga JC, Schneider MW, Yamano T, Nishimasu H, Nureki O, Crosetto N, Zhang F. 2017. Engineered Cpf1 variants with altered PAM specificities. Nature Biotechnology 35, 789–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Zhao Y. 2014. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. Journal of Integrative Plant Biology 56, 343–349. [DOI] [PubMed] [Google Scholar]
- Hu X, Wang C, Liu Q, Fu Y, Wang K. 2017. Targeted mutagenesis in rice using CRISPR–Cpf1 system. Journal of Genetics and Genomics 44, 71–73. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan Y, Ruis B, Takasugi T, Hendrickson EA. 2017. Mechanisms of precise genome editing using oligonucleotide donors. Genome Research 27, 1099–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Kim J, Hur JK, Been KW, Yoon SH, Kim JS. 2016. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nature Biotechnology 34, 863–868. [DOI] [PubMed] [Google Scholar]
- Kim H, Kim ST, Ryu J, Kang BC, Kim JS, Kim SG. 2017. CRISPR/Cpf1-mediated DNA-free plant genome editing. Nature Communications 8, 14406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhang X, Sun Y, Zhang J, Du W, Guo X, Li S, Zhao Y, Xia L. 2018. Efficient allelic replacement in rice by gene editing: a case study of the NRT1.1B gene. Journal of Integrative Plant Biology (in press). [DOI] [PubMed] [Google Scholar]
- Li L, Qu R, de Kochko A, Fauquet C, Beachy RN. 1993. An improved rice transformation system using the biolistic method. Plant Cell Reports 12, 250–255. [DOI] [PubMed] [Google Scholar]
- Li S, Zhang X, Wang W, Guo X, Wu Z, Du W, Zhao Y, Xia L. 2018. Expanding the scope of CRISPR/Cpf1-mediated genome editing in rice. Molecular Plant (in press). [DOI] [PubMed] [Google Scholar]
- Liu W, Xie X, Ma X, Li J, Chen J, Liu YG. 2015. DSDecode: a web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations. Molecular Plant 8, 1431–1433. [DOI] [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, et al. 2015. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Molecular Plant 8, 1274–1284. [DOI] [PubMed] [Google Scholar]
- Mazur BJ, Chui CF, Smith JK. 1987. Isolation and characterization of plant genes coding for acetolactate synthase, the target enzyme for two classes of herbicides. Plant Physiology 85, 1110–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J, Guo D, Zhang J, Huang Q, Qin G, Zhang X, Wan J, Gu H, Qu LJ. 2013. Targeted mutagenesis in rice using CRISPR–Cas system. Cell Research 23, 1233–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman K, Higgins JD, Sanchez-Moran E, Armstrong SJ, Franklin FC. 2011. Pathways to meiotic recombination in Arabidopsis thaliana. New Phytologist 190, 523–544. [DOI] [PubMed] [Google Scholar]
- Paix A, Folkmann A, Goldman DH, Kulaga H, Grzelak MJ, Rasoloson D, Paidemarry S, Green R, Reed RR, Seydoux G. 2017. Precision genome editing using synthesis-dependent repair of Cas9-induced DNA breaks. Proceedings of the National Academy of Sciences, USA 114, E10745–E10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquet D, Kwart D, Chen A, Sproul A, Jacob S, Teo S, Olsen KM, Gregg A, Noggle S, Tessier-Lavigne M. 2016. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533, 125–129. [DOI] [PubMed] [Google Scholar]
- Puchta H. 1998. Repair of genomic double-strand breaks in somatic plant cells by one-sided invasion of homologous sequences. The Plant Journal 13, 331–339. [Google Scholar]
- Puchta H, Fauser F. 2013. Gene targeting in plants: 25 years later. International Journal of Developmental Biology 57, 629–637. [DOI] [PubMed] [Google Scholar]
- Puchta H, Fauser F. 2014. Synthetic nucleases for genome engineering in plants: prospects for a bright future. The Plant Journal 78, 727–741. [DOI] [PubMed] [Google Scholar]
- Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE. 2016. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR–Cas9 using asymmetric donor DNA. Nature Biotechnology 34, 339–344. [DOI] [PubMed] [Google Scholar]
- Shan Q, Wang Y, Li J, et al. 2013. Targeted genome modification of crop plants using a CRISPR–Cas system. Nature Biotechnology 31, 686–688. [DOI] [PubMed] [Google Scholar]
- Smith CE, Llorente B, Symington LS. 2007. Template switching during break-induced replication. Nature 447, 102–105. [DOI] [PubMed] [Google Scholar]
- Sun Y, Zhang X, Wu C, He Y, Ma Y, Hou H, Guo X, Du W, Zhao Y, Xia L. 2016. Engineering herbicide-resistant rice plants through CRISPR/Cas9-mediated homologous recombination of acetolactate synthase. Molecular Plant 9, 628–631. [DOI] [PubMed] [Google Scholar]
- Tang X, Lowder LG, Zhang T, et al. 2017. A CRISPR–Cpf1 system for efficient genome editing and transcriptional repression in plants. Nature Plants 3, 17018. [DOI] [PubMed] [Google Scholar]
- Wang M, Mao Y, Lu Y, Tao X, Zhu JK. 2017. Multiplex gene editing in rice using the CRISPR–Cpf1 system. Molecular Plant 10, 1011–1013. [DOI] [PubMed] [Google Scholar]
- Xu R, Qin R, Li H, Li D, Li L, Wei P, Yang J. 2017. Generation of targeted mutant rice using a CRISPR–Cpf1 system. Plant Biotechnology Journal 15, 713–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, et al. 2015. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR–Cas system. Cell 163, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Heidenreich M, Mohanraju P, et al. 2017. Multiplex gene editing by CRISPR–Cpf1 using a single crRNA array. Nature Biotechnology 35, 178. [DOI] [PubMed] [Google Scholar]
- Zhang T, Gao Y, Wang R, Zhao Y. 2017. Production of guide RNAs in vitro and in vivo for CRISPR using ribozymes and RNA polymerase II promoters. Bio-Protocol 7, e2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z, Zhang Y, You Q, et al. 2018. Plant genome editing using FnCpf1 and LbCpf1 uucleases at redefined and altered PAM sites. Molecular Plant (in press). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.