Spatial patterning of gene expression in the bundle sheath of Arabidopsis is mediated by a conserved repetitive region in the SULTR2;2 promoter.
Keywords: Arabidopsis thaliana, bundle sheath, C3 photosynthesis, C4 photosynthesis, gene expression
Abstract
The bundle sheath provides a conduit linking veins and mesophyll cells. In the C3 plant Arabidopsis thaliana, it also plays important roles in oxidative stress and sulphur metabolism. However, the mechanisms responsible for the patterns of gene expression that underpin these metabolic specializations are poorly understood. Here, we used the Arabidopsis SULTR2;2 gene as a model to better understand mechanisms that restrict expression to the bundle sheath. Deletion analysis indicated that the SULTR2;2 promoter contains a short region necessary for expression in the bundle sheath and veins. This sequence acts as a positive regulator and is tolerant to multiple consecutive deletions indicating considerable redundancy in the cis-elements involved. It is highly conserved in SULTR2;2 genes of the Brassicaceae and is functional in the distantly related C4 species Flaveria bidentis that belongs to the Asteraceae. We conclude that expression of SULTR2;2 in the bundle sheath and veins is underpinned by a highly redundant sequence that likely represents an ancient and conserved mechanism found in families as diverse as the Asteraceae and Brassicaceae.
Introduction
The evolution of multicellularity is associated with individual cell types being able to undertake specialized roles within a tissue. In leaves, bundle sheath (BS) cells form a wreath-like structure around the vasculature, which appears to be analogous to the endodermis of roots (Esau, 1965). The role of BS cells is best characterized in C4 species, which partition photosynthesis between the BS and mesophyll cells. In most C4 plants, after HCO3− is initially fixed into C4 acids by phosphoenolpyruvate carboxylase in mesophyll cells, these C4 acids diffuse to the BS where CO2 is released and refixed by ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO). Decarboxylation of C4 acids in the BS generates a high concentration of CO2 around RuBisCO that suppresses the oxygenase activity of the enzyme and in so doing reduces photorespiration (Hatch, 1987). Thus, in C4 species, the BS is specialized to allow efficient fixation of CO2 in the Calvin–Benson–Bassham cycle. In some C4 plants, the BS is also modified in terms of light capture. For example, in maize and sorghum photosystem II does not fully assemble in the BS (Kubicki et al., 1994), but components of cyclic electron transport are more abundant in the BS compared with mesophyll cells (Takabayashi et al., 2005). In addition to these changes associated with photosynthesis, the C4 BS is also modified to preferentially undertake starch synthesis and degradation, as well as the initial steps of sulphur assimilation (Friso et al., 2004).
In C3 plants, the role of the BS is less clearly defined. It is thought to help maintain hydraulic integrity of the xylem (Sade et al., 2014), regulate flux of metabolites in and out of the leaf (Leegood, 2008) and act as a starch store (Miyake and Maeda, 1976). The C3 BS is less important for photosynthesis than that of C4 species. However, although only around 15% of all chloroplasts of the C3 leaf are found in BS cells (Kinsman and Pyke, 1998), reducing photosynthesis in these cells compromises growth and seed production (Janacek et al., 2009). Thus, although less photosynthetic than the C4 BS, this physiological analysis indicates that the BS of C3 plants is also specialized. This notion is consistent with analysis of gene expression in this cell type. For example quantification of transcripts available for translation indicate that the Arabidopsis BS is likely important in sulphur metabolism, glucosinolate biosynthesis, trehalose metabolism and detoxification of reactive oxygen species (Aubry et al., 2014). In summary, in both C3 and C4 plants mechanisms must operate to restrict the expression of some genes to BS cells.
To date, most studies of the mechanisms responsible for preferential gene expression in the BS have used C4 species (Hibberd and Covshoff, 2010). In the C4 dicotyledon Flaveria trinervia the glycine decarboxylase P-subunit (GLDPA) gene contains two promoters, one proximal to the coding region and the other more distal. Activity of the distal promoter is high but not cell-type specific. However, in the presence of the proximal promoter, transcripts derived from the distal promoter are degraded in mesophyll cells by nonsense-mediated RNA decay of incompletely spliced transcripts (Engelmann et al., 2008; Wiludda et al., 2012). Despite the phylogenetic distance between the Brassicaceae and the Asteraceae the GLDPA promoter from F. trinervia is able to generate BS-specific activity in C3 Arabidopsis (Engelmann et al., 2008; Wiludda et al., 2012). In Amaranthus hybridus 5′ and 3′ untranslated regions (UTRs) of the RbcS1 gene act to restrict accumulation of the glucoronidase (GUS) reporter to the BS of C4Flaveria bidentis and appear to function as enhancers of translation (Patel et al., 2006). Lastly, in C4Gynandropsis gynandra preferential expression of NAD-ME1 and -2 genes in the BS is associated with coding sequence rather than UTRs or promoter elements (Brown et al., 2011). The motifs underpinning this regulation are a pair of duons that play a dual role in coding for amino acids as well as the spatial patterning of gene expression associated with the C4 leaf. Although these duons are present in C3 Arabidopsis and many other land plants they do not act to generate cell-specific expression in the ancestral C3 state (Brown et al., 2011; Reyna-Llorens et al., 2018). In summary, current evidence indicates that gene expression in the BS of C4 species is controlled by a variety of mechanisms, some of which involve regulatory codes that are derived from the ancestral C3 state.
However, our understanding of how gene expression is restricted to the BS in C3 species is poor. A small number of promoters including SHORT-ROOT (Dhondt et al., 2010), SCARECROW (Wysocka-Diller et al., 2000), and SULTR2;2 (Takahashi et al., 2000) have been reported to drive BS-specific expression in Arabidopsis, but neither the molecular nature of cis-regulatory elements controlling their expression, nor transcription factors that control expression in the BS have been identified. An increased understanding of these processes would not only advance our understanding of mechanisms underpinning cell-specific gene expression in multicellular leaves, but also provide insight into whether C4 gene expression is built on pre-existing mechanisms found in C3 species.
As a step towards uncovering mechanisms that underpin expression in BS cells, we sought to identify mechanisms in cis associated with the SULTR2;2 gene of C3 Arabidopsis, which encodes a low-affinity sulphur transporter (Takahashi et al., 2000). Elements in the promoter sequence that regulate the spatial patterning and the strength of gene expression were identified. Specifically, preferential expression in the BS is mediated by a repetitive region that is highly conserved within orthologous genes from species of the Brassicaceae. This region acts to enhance expression in the BS and veins rather than repressing expression in mesophyll cells. Furthermore, the SULTR2;2 promoter from Arabidopsis generates expression in BS and vein cells in the C4 species F. bidentis that belongs to the Asteraceae. The most parsimonious explanation for this finding is that a common transcription factor is shared by these phylogenetically dispersed species, and that it functions in both the veins and BS cells of both C3 and C4 species.
Materials and methods
Cloning of promoter–reporter gene constructs and 5′ rapid amplification of cDNA ends
All DNA fragments created by PCR were confirmed by DNA sequencing. A full-length promoter construct was generated via PCR with Arabidopsis Columbia-0 (Col-0) genomic DNA. Subsequent constructs were generated using this as a template. Restriction sites were added to the respective fragments by PCR and fragments were inserted into pBI121 or a partially modified pBI121. Region 2 with internal deletions was synthesized by GenScript and swapped with the full-length region 2 of 2::5.2::GUS to generate the internal deletion constructs. Total RNA from leaves of wild type Arabidopsis Col-0 plants was extracted, DNase I treated and purified with the RNeasy® Plant Mini Kit (Qiagen, Hilden, Germany). cDNA was generated from 1 µg RNA and then 5′ rapid amplification of cDNA ends (RACE)-PCR performed using the Advantage® 2 DNA Polymerase Mix (Clontech Laboratories, Mountain View, CA, USA) or Phusion® HF DNA Polymerase (Thermo Fisher Scientific, Waltham, MA, USA). Two nested 3′ oligonucleotides were used, AtSultr2;2–11 and AtSultr2;2–13, both binding in the cDNA of AtSULTR2;2. PCR products were cloned and confirmed via colony PCR. Correct clones were the subjects of plasmid preparation and sequencing.
Transformation and plant growth
Arabidopsis ecotype Col-0 was transformed using floral dipping (Clough and Bent, 1998; Logemann et al., 2006) using Agrobacterium tumefaciens strain AGL1. Flaveria bidentis was transformed as described previously (Chitty et al., 1999). Successful transformations of Arabidopsis or F. bidentis were tested by PCR. Before transplanting to soil, positive transformants of Arabidopsis were selected on kanamycin. Seeds were sterilized by washing twice for 5 min with 20% (v/v) DanKlorix (Colgate-Palmolive, New York, USA) and 0.02% (v.v) Triton X-100 and four times with sterile water. Stratification of seeds was performed at 4 °C for 48 h prior to spreading them on half-strength Murashige–Skoog medium pH 5.7 containing 10 g l−1 (w/v) sucrose (Sigma-Aldrich, St Louis, MO, USA), 0.5 g l−1 MES (Biomol, Hamburg, Germany), 2.13 g l−1 (w/v) Murashige–Skoog basal salts (Duchefa Biochemie, Haarlem, Netherlands), 0.75% agar (SERVA Electrophoresis, Heidelberg, Germany), 50 µg ml−1 kanamycin (Sigma-Aldrich) and 100 mg l−1 Cefotaxim (Fresenius Kabi Deutschland, Bad Homburg, Germany). Plants were transferred to 14 h light/10 h dark with temperatures of 23 °C day and 20 °C night and a light intensity of 90 µmol m−2 s−1.
Visual and quantitative analysis of reporter genes
To take account of effects caused by transgene insertion into different genomic locations, between nine and 41 independent T0 plants were analysed for each construct. Histochemical analysis was performed as described previously (Engelmann et al., 2008). For Arabidopsis 3- to 4-week-old rosette leaves or 10- to 14-day-old seedlings and for F. bidentis the sixth leaves of 40–50 cm-tall plants were used. Transverse sections were prepared manually using a razor blade. Stained leaves were imaged using light microscopy. Quantification of GUS activity was performed via a fluorimetric assay (Jefferson et al., 1987) using two to four leaves of 3- to 4-week-old T0 Arabidopsis plants or the fifth leaf of 40–50 cm-tall T0F. bidentis plants. For F. bidentis, an entire leaf was wrapped in aluminium foil, frozen in liquid nitrogen, pulverized using a hammer, and approximately 300 mg of powdered tissue transferred to 1.5 ml reaction tubes. Arabidopsis leaves were directly harvested into 1.5 ml reaction tubes and frozen in liquid nitrogen. One volume of extraction buffer was added (100 mM Na2HPO4 pH 7.5, 1 mM EDTA, 0.1% N-laurylsarcosine, 0.1% Triton X-100, 20% methanol) and the material homogenized. The homogenate was then centrifuged at 15000 g and 4 °C for 5 min, and the clear supernatant used for measurements of protein content and GUS activity. The incubation buffer used to measure GUS activity was identical to the extraction buffer except that 4-methylumbelliferyl-β-D-glucuronide (MUG) was added to a final concentration of 1 mM. The Mann–Whitney U-test was used to determine statistical differences between datasets. Imaging of H2B::YFP was performed on a Zeiss LSM 780 confocal laser-scanning microscope, and yellow fluorescent protein (YFP) fluorescence excited at 514 nm with emission detected between 517 and 569 nm.
Results
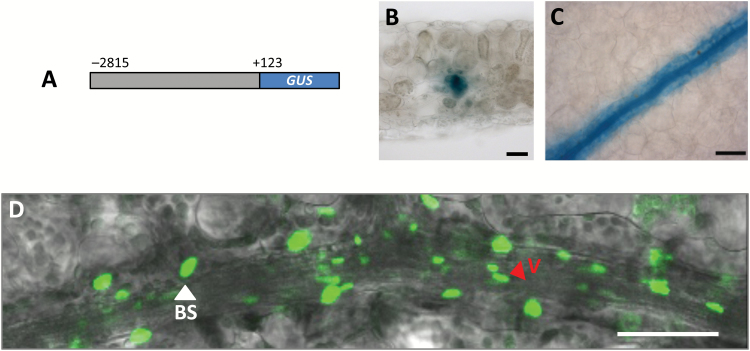
Nucleotides −2815 to +123 relative to the predicted SULTR2;2 translational start site have previously been reported to generate expression in the BS of Arabidopsis (Takahashi et al., 2000). We confirmed this finding (Fig. 1A–C). Staining was evident in vascular tissue as well as the BS but there was no evidence that GUS accumulated in mesophyll cells (Fig. 1A–C). A translational fusion between the YFP and the nuclear localized histone 2B protein under control of the SULTR2;2 promoter labelled nuclei of BS cells and vascular tissue (Fig. 1D) and indicated that the presence of GUS in the BS and veins was due to gene expression and not diffusion of the dye outwards from to vascular tissue. Consistent with previous reports (Chytilova et al., 1999), it was noticeable that vascular nuclei were elongated and rod-like whilst the BS contained larger, more spherical nuclei (Fig. 1D). We conclude that nucleotides between −2815 and +123 of SULTR2;2 are sufficient to drive gene expression in vascular and BS cells of Arabidopsis, and thus sought to understand the sequences responsible.
Fig. 1.
The SULTR2;2 promoter drives expression in veins and bundle sheath cells of Arabidopsis. (A–C) Nucleotides −2185 to +123 of SULTR2;2 are sufficient to generate preferential accumulation of the GUS reporter in bundle sheath and vein cells. (D) H2B::YFP fusion marks the larger bundle sheath nuclei (white arrowhead) as well as the smaller, more elongated nuclei of the vasculature (red arrowhead). Histological GUS assays were allowed to proceed for 23 h (B, C). Scale bars: 500 µm (B, C) or 50 µm (D).
A short region that is sufficient to activate gene expression in bundle sheath and veins
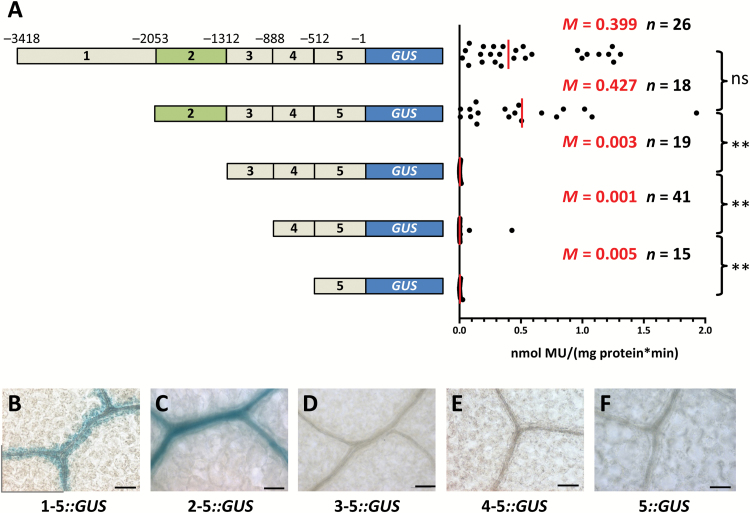
To better understand sequences that generate expression in BS and veinal cells, a 5′ deletion series was generated (Fig. 2A). The nucleotides downstream of the predicted translational start site of SULTR2;2 were not required to direct expression to BS and vasculature (Fig. 2A; Supplementary Fig S1 at JXB online). Subsequent deletions to the promoter were made and are hereafter referred to as regions 1–5. Removal of region 1 had no significant effect on either activity or spatial accumulation of GUS (Fig. 2A, C) indicating that no essential cis-regulatory elements are located within this section. Deletion of region 2 resulted in total loss of GUS activity and staining (Fig. 2A, D), and removal of regions 3 and 4 had no further effect (Fig. 2A, E, F). These data therefore indicate that nucleotides in region 1 do not impact on promoter activity, but that region 2 is necessary for BS and veinal expression in mature leaves. A separate deletion series involving slightly smaller regions indicated that the construct containing nucleotides from −2815 to +123 led to stronger GUS activity than those from −3418 to −1 (Supplementary Figs S1, S2). This indicates that either there is a repressor within nucleotides −3418 to −2815, and/or that there is an enhancer within nucleotides −1 to +123. However, these quantitative regulators did not abolish expression in the BS and vasculature and so we did not dissect them further. Rather, subsequent analysis was focussed on region 2, which was necessary for expression in BS and veinal cells.
Fig. 2.
A 715-nucleotide region in the SULTR2;2 promoter that is necessary for bundle sheath and veinal expression. (A) Schematic representation of the 5′ deletion series (left) with GUS activities (right). (B–F) GUS accumulation from each deletion. Deletion of region 2 abolished accumulation of GUS. Data from GUS activity assays include the median (M) indicated by red lines and the number (n) of independent lines. Statistical significance for each pairwise comparison is marked by a bracket to the right (ns, non-significant; **P<0.01). Histological GUS assays were allowed to proceed for 23 h (B), 4 h (C) and 6 days (D–F). Scale bars: 50 µm.
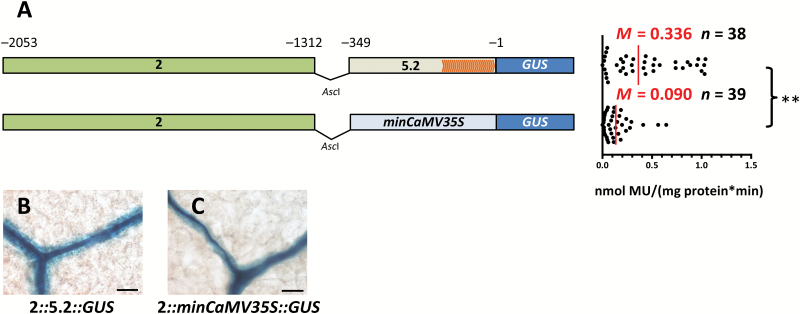
The lack of GUS accumulation in the BS and veins, and loss of promoter activity once region 2 is removed could be because this region contains cis-elements that generate expression specifically in these cells or because it drives ubiquitous expression but regions 3 to 5 contain elements that restrict activity to the BS and veins. To investigate this possibility, 5′ rapid amplification of cDNA ends was first used to define the transcriptional start site of SULTR2;2 (Supplementary Fig. S3). No single strong transcription start site was detected but rather multiple transcripts were initiated from position -125 onwards (Supplementary Fig. S2). Nucleotides spanning −349 to −1 were therefore considered likely to be sufficient for transcriptional initiation and are hereafter referred to as the core promoter. Fusion of region 2 to this core promoter led to MUG conversion that was comparable to that from the full-length promoter (Figs 2A, 3A) and was also sufficient to direct accumulation of GUS to veins and the BS (Fig. 3B). To exclude the possibility that the core promoter includes cis-elements necessary for BS- and vein-specific expression, region 2 was also fused to the minimal CaMV35S promoter, which does not drive significant expression. Although this led to 4-fold lower GUS activity than the full-length promoter (Fig. 3A), GUS accumulation was still restricted to veins and the BS (Fig. 3C). We conclude that although nucleotides −349 to −1 are not able to generate expression in the BS and veins (Fig. 2F), they likely represent the core promoter of SULTR2;2. In contrast, nucleotides −2053 to −1312 are sufficient to restrict expression to the BS and vascular tissue of Arabidopsis.
Fig. 3.
The SULTR2;2 promoter contains a region that is sufficient to activate expression in bundle sheath and veinal cells. (A) Schematic representation of constructs containing nucleotides −2053 to −1312 combined with either the core promoter of SULTR2;2 or the minimal CaMV35S promoter (left) and quantitative analysis of expression from each construct based on the GUS activity assay (right). Orange arrowheads within the core promoter indicate transcription start sites obtained by 5′-RACE. (B, C) Both constructs are sufficient to generate GUS accumulation in the bundle sheath. Data from GUS activity assays include the median (M) indicated by red lines and the number (n) of independent lines. Statistical significance is indicated to the right (**P<0.01). Histological GUS assays were allowed to proceed for 5 h (B) and 22 h (C). Scale bars: 50 µm.
AtSULTR2;2 contains multiple redundant sequences mediating expression in the bundle sheath and vein
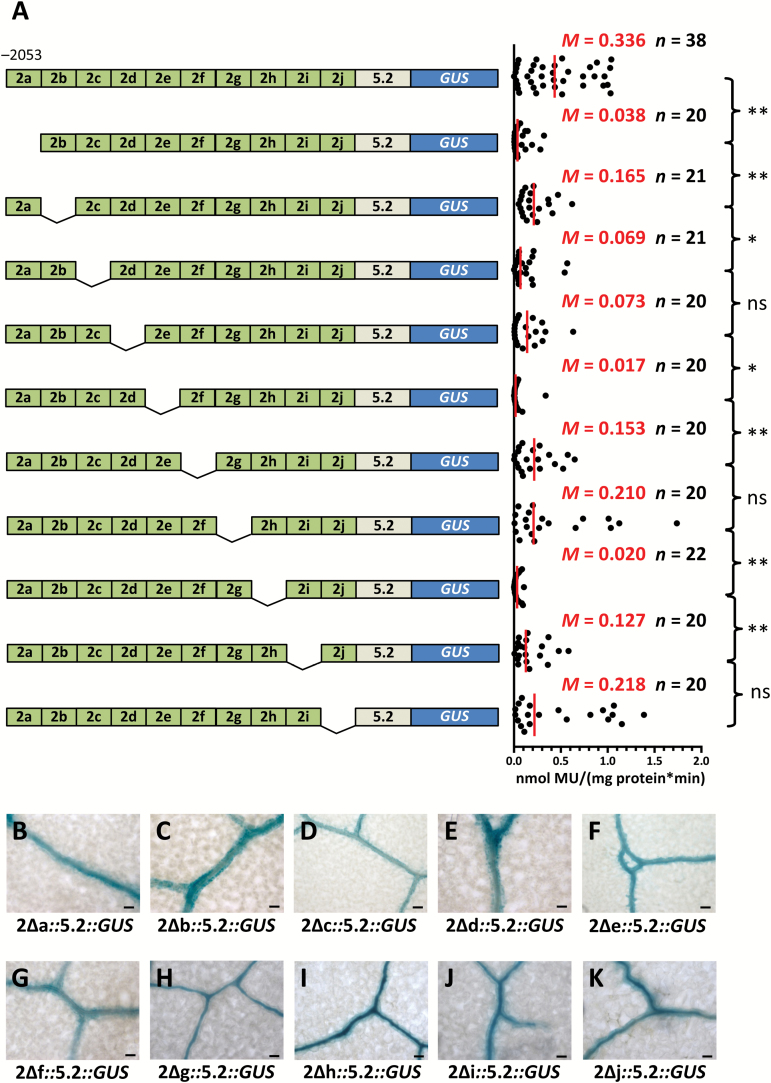
Having established that nucleotides −2053 to −1312 relative to the predicted translational start site are sufficient for BS and vein expression, an unbiased approach to further dissect this region was adopted. Ten consecutive deletions were made and each was fused to the core promoter of SULTR2;2 (Fig. 4A). Strikingly, none of these deletions resulted in total loss of GUS activity (Fig. 4A), nor was GUS staining lost from BS and veins (Fig. 4B–K), indicating that despite an absence of repeated cis-elements in this region, significant functional redundancy in regulatory elements must mediate this patterning of gene expression. However, it was notable that GUS activity declined to varying degrees compared with the intact region (Fig. 4A), suggesting that either these redundant cis-elements act additively, or that this region contains quantitative elements regulating gene expression.
Fig. 4.
SULTR2;2 contains a region with multiple redundant regions capable of directing bundle sheath and veinal expression. (A) Schematic representation of internal deletion series constructs (left) and quantitative analysis of expression from each construct based on the GUS activity assay (right). Except for the last deletion representing 66 bp, each construct lacks consecutive 75-bp sequences. These internal deletions modify the GUS activity (A), but none abolishes accumulation of GUS in the bundle sheath (B–K). Data from GUS activity assays include the median (M) indicated by red lines and the number (n) of independent lines. Statistical significance for each pairwise comparison is marked by a bracket to the right (ns, non-significant; *P<0.05; **P<0.01). Histological GUS assays were allowed to proceed for 23 h (B), 16 h (C), 48 h (D), 23 h (E), 48 h (F), 8 h (G), 3 h (H), 7 h (I), 3 h (J), and 19 h (K). Scale bars: 50 µm.
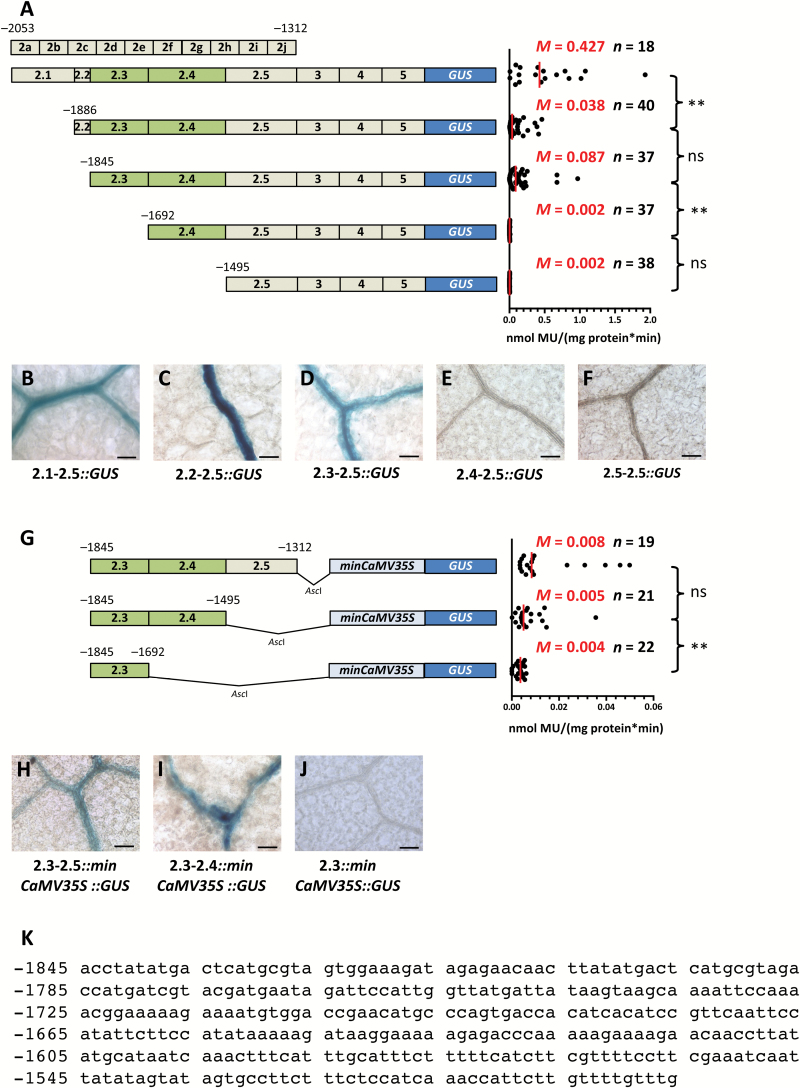
To better understand the extent to which cis-elements in this section of the promoter act redundantly, larger deletions were made from position −2053 (Fig. 5A). This generated five regions, hereafter referred to as subregions 2.1–2.5. Deletion of subregion 2.1 resulted in a strong reduction of GUS activity (Fig. 5A) implying that this region contains a quantitative enhancer element. However, accumulation of GUS was maintained in the BS and veins (Fig. 5C). Deleting subregion 2.2 had no clear additional impact on either this spatial patterning or activity (Fig. 5A, D). However, the subsequent deletion of subregion 2.3 caused loss of GUS activity and also loss of GUS staining in BS and vascular tissue (Fig. 5A, E). These data indicate that cis-regulatory elements mediating BS and vein expression are situated in subregion 2.3, or that quantitative elements in this region mask qualitative elements in distal subregions. To address these options, 3′ deletions of region 2 were also created (Fig. 5G). As subregions 2.1 and 2.2 had little impact on expression in the BS and vasculature, the last three subregions were fused to the minimal CaMV35S promoter. Expression from each of these three constructs was low (Fig. 5G); however, expression could be observed from the construct containing subregions 2.3, 2.4, and 2.5 (Fig. 5H). Removal of subregion 2.5 resulted in loss of GUS in rosette leaves although cotyledons still showed patchy staining restricted to the BS and vascular tissue (Fig. 5I). Once subregion 2.4 was removed, GUS activity was further reduced and GUS staining was no longer detectable even in seedlings (Fig. 5G, J). We conclude that subregion 2.3 contains cis-regulatory elements necessary (Fig. 5E) and regions 2.3 and 2.4 (Fig. 5I) are sufficient for expression of AtSULTR2;2 in the BS and veins.
Fig. 5.
A short region of the SULTR2;2 promoter that is necessary for expression in the bundle sheath and vein tissue. (A) Schematic representation of deletions made to region 2 of the SULTR2;2 promoter (left) and quantitative analysis of expression from each construct based on the GUS activity assay (right). Activity was no longer detectable when subregions 4 and 5 were removed. (B–F) Histochemical staining of leaves indicated that subregion 3 is required for expression in the bundle sheath. (G) Schematic representation of 3′ deletion constructs placed upstream of the minimal CaMV35S promoter (left) and quantitative analysis of expression from each construct based on the GUS activity assay (right). (H–I) Nucleotides from −1845 to −1495 are sufficient to drive expression in the bundle sheath of leaves (H) and cotyledons (I), respectively. (J) Deletion of nucleotides −1692 to −1312 abolishes accumulation of GUS in the BS. Data from GUS activity assays include the median (M) indicated by red lines and the number (n) of independent lines. Statistical significance for each pairwise comparison is marked by a bracket to the right (ns, non-significant; **P<0.01). (K) Sequence of the 350 bp region that is sufficient for expression in bundle sheath strands of Arabidopsis. Histological GUS assays were allowed to proceed for 4 h (B), 47 h (C), 4 h (D) 6 d (E, F), 5 d (H), 2 d (I), and 29 h (J). Scale bars: 50 µm.
The cis-regulatory elements necessary for expression of AtSULTR2;2 in BS and veins therefore appear to be located in a 350-nucleotide region of the promoter. As finer-scale deletions had failed to identify the exact cis-elements responsible for this phenotype (Fig. 4) a phylogenetic approach was undertaken. Orthologues of AtSULTR2;2 were identified from seven species of the Brassicaceae. Alignments of sequences 5 kb upstream of each orthologue indicated that with the exception of Arabidopsis lyrata, which contains a 446 nucleotide insertion, region 2 is highly conserved (Fig. 6). However, no short sequences or motifs within this sequence that may restrict expression to the BS specifically could be identified. Although the results of this alignment therefore do not identify a specific cis-element that could be bound by a transcription factor responsible for generating expression in the BS and vein, they do support the functional analysis and implicate subregions 2.3 and 2.4 as critical components of the SULTR2;2 promoter for this expression.
Fig. 6.
The region necessary for bundle sheath and veinal expression of SULTR2;2 is highly conserved in the Brassicaceae. The promoter of AtSULTRT2;2 was aligned against sequences ~5 kb upstream of genes from seven additional species of the Brassicaceae. With the exception of Arabidopsis lyrata, which contains a 446 bp insertion, region 2 is highly conserved. Black boxes indicate strong similarity of sequences, grey boxes sequences not matching the consensus sequence, and black lines gaps. The level of similarity is also indicated above the alignment. High peaks in green mark strong similarity, low peaks in red, poor similarity.
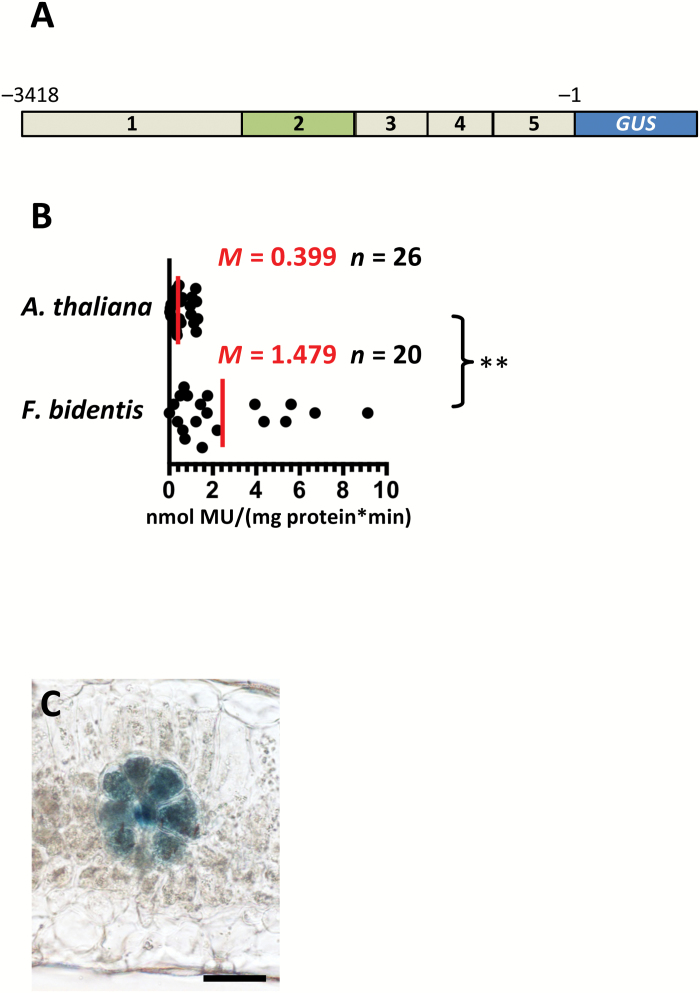
The AtSULTR2;2 promoter is capable of driving bundle sheath and vein expression in C4Flaveria bidentis
As the GLDPA promoter from the C4 species F. trinervia is able to confer BS and veinal expression in C3 Arabidopsis (Engelmann et al., 2008), we next tested whether the Arabidopsis SULTR2;2 promoter would lead to expression in the BS and veins of C4F. bidentis. GUS activity in transgenic F. bidentis plants was about fourfold higher than that in Arabidopsis (Fig. 7A, B). However, histochemical analysis of mature leaves revealed a very similar expression pattern to that in Arabidopsis with strong GUS accumulation in BS and vascular tissue but not in mesophyll cells (Fig. 7C). This indicates that transcription factors from F. bidentis recognize cis-regulatory elements from the Brassicaceae that generate expression in both the BS and veinal cells. The most parsimonious explanation for this finding is that these sequences represent part of an ancient and conserved mechanism that restricts gene expression to BS and vasculature of dicotyledenous leaves.
Fig. 7.
The Arabidopsis SULTR2;2 promoter generates strong bundle sheath and vein expression in the C3 species Flaveria bidentis. (A) Schematic representation of the sequence placed into Flaveria bidentis. (B) Quantitative analysis of expression from each construct based on the GUS activity assay. To facilitate comparison, GUS data from the same construct in C3 Arabidopsis are included. (C) Representative image of transverse section of Flaveria bidentis after histochemical staining for GUS. Data from GUS assays include the median (M) indicated by red lines and the number (n) of independent lines. Statistical significance is indicated to the right (**P<0.01). Histological GUS assays were allowed to proceed for 4 h. Scale bar: 50 µm.
Discussion
SULTR2;2 contains a short region that activates expression in the bundle sheath and veins
SULTR2;2 encodes a low affinity transporter that facilitates movement of sulphate from the vascular bundle to palisade cells in the leaf (Takahashi et al., 2000). Consistent with this function, analysis of both GUS and the histone2B::YFP fusion indicated that the SULTR2;2 promoter directs expression to veins as well as to BS cells. It was notable that none of the various deletions we made to this promoter led to expression being restricted to either veins or BS cells. The GLDPA promoter from F. trinervia also drives expression in both veins and BS cells of Arabidopsis (Engelmann et al., 2008). These findings therefore imply that the SULTR2;2 and GLDP genes may be controlled by gene regulatory networks that are shared by these cell types. It is possible that these cell types share some gene regulatory networks because they are derived from the same lineage (Dengler and Nelson, 1999; Soros and Dengler, 2001).
Within the SULTR2;2 promoter, one specific region that impacted on gene expression in the BS and veins was identified. SULTR2;2 is located directly upstream of a second sulphur transporter gene, SULTR1;2, which is also preferentially expressed in bundle sheath strands (Aubry et al., 2014). Because they are transcribed from opposite strands it is possible that the regulatory mechanisms described here control the expression of both genes. The sequence identified, consisting of 350 nucleotides, is both necessary and sufficient for generating expression in BS and veins. No specific cis-elements that are repeated across this entire region were identified. Moreover, it was also not possible to identify sequences that were shared between this region and the small number of promoters (Wysocka-Diller et al., 2000; Knappe et al., 2003; Dhondt et al., 2010; Wiludda et al., 2012) that have been reported to drive expression in BS cells of Arabidopsis (Supplementary Fig. S4). Small consecutive deletions within this region failed to abolish this spatial patterning implying that multiple contiguous elements act redundantly to generate strong and stable expression in the BS and vasculature. The only detectable impact of deleting any part of this region was for strength of expression to be reduced. We therefore propose that either multiple independent modules contained within this region act additively, or that distinct quantitative elements are co-located, and at least partially overlapping, with cis-elements that determine this expression in BS and veinal cells. Redundancy of this sort has previously been reported for the promoter of Phenylalanine Ammonia Lyase2, which drives xylem-specific expression in tobacco (Leyva et al., 1992; Hatton et al., 1995), DORNRÖSCHEN-LIKE of Arabidopsis, which contains three functionally redundant enhancers (Comelli et al., 2016), and EVEN-SKIPPED (EVE) from Drosophila melanogaster, where a minimal enhancer is sufficient to direct expression of EVE to the second stripe, but surrounding binding sites increase the robustness of this patterning during genetic and environmental perturbations (Ludwig et al., 2011). Thus, although the exact role of redundancy in the regulation of SULTR2;2 is unclear, it may also increase robustness in the control of gene expression during environmental perturbations, and/or increase patterning precision (Barolo, 2012; Payne and Wagner, 2015).
Compared with other examples of elements that restrict gene expression to BS and veinal cells of C3 species, this single block of sequence from SULTR2;2 that acts as a positive regulator of transcription appears to operate via relatively simple mechanisms. For example, the F. trinervia GLDPA generates BS and vein expression (Engelmann et al., 2008) because of a complex interplay between transcriptional and post-transcriptional processes. These are mediated by distal and proximal sequences relative to the translational start site, leading to repression of GLDP expression in mesophyll cells (Wiludda et al., 2012). We therefore propose that the positive regulator located upstream of SULTR2;2 could be used as a synthetic module to manipulate or engineer processes in BS and vein cells of Arabidopsis.
The bundle sheath and vein element of SULTR2;2 is conserved in the Brassicaceae and functional in the Asteraceae
Alignment of SULTR2;2 promoters from multiple species of the Brassicaceae did not reveal an individual shared cis-element but rather highlighted a sequence that was conserved across the whole of region 2. This sequence conservation argues for relatively strong purifying selection compared with the rest of the SULTR2;2 promoter and also implies that this region may function as part of a widely conserved positive regulator of gene expression in BS and veins of the Brassicaceae. Consistent with this proposal and indicating that these regulatory elements may be even more ancient, when the SULTR2;2 promoter was placed into the phylogenetically distant C4 species F. bidentis it was also recognized by trans-factors that restricted gene expression to the C4 BS and vasculature. This is analogous to the behaviour of the GLDPA promoter from C4F. trinervia, which is able to restrict expression to the BS and veins of Arabidopsis (Engelmann et al., 2008; Wiludda et al., 2012). Currently, the crown ages of the rosids and asterids are estimated to be 108–117 and 107–117 million years ago (Wikström et al., 2001; Sanderson et al., 2004) indicating that these clades diverged in the early Cretaceous. Whilst for both SULTR2;2 and GLDP it is possible that different mechanisms lead to BS and vein expression in species of the Brassicaceae and Asteraceae, it seems more likely that expression of each gene is determined by ancient and highly conserved cis-regulatory codes that have been maintained since these clades diverged from their last common ancestor. Although the plant vasculature is thought to have started to evolve from 450 to 430 million years ago (Ye, 2002; Furuta et al., 2014), to our knowledge there are no clear estimates of when the BS originated. It would be intriguing if regulatory networks operating in both the veins and BS cells are uncovered that can be associated with the evolution of the vasculature in early diverging lineages of land plants.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Nucleotides −3418 to −1 relative to the predicted translational start site generate preferential accumulation of the GUS reporter in bundle sheath cells.
Fig. S2. Additional deletion series generated for SULTRT2;2.
Fig. S3. Alignment of 5′ ends of cDNAs obtained via 5′ rapid amplification of cDNA ends.
Fig. S4. Dot plots indicating lack of conservation between the SULTR2;2 promoter and others reported to drive expression in the bundle sheath of Arabidopsis.
Supplementary Material
Acknowledgements
Work performed in PW’s laboratory (SK, KP, MK, UG) was supported by the Deutsche Forschungsgemeinschaft through the Cluster of Excellence in Plant Sciences Duesseldorf-Cologne (EXC 1028). HW was supported by a BBSRC PhD studentship. We thank the Center for Advanced imaging (HHU Düsseldorf) for provision and technical assistance of the Zeiss LSM 780 laser-scanning microscope.
References
- Aubry S, Smith-Unna RD, Boursnell CM, Kopriva S, Hibberd JM. 2014. Transcript residency on ribosomes reveals a key role for the Arabidopsis thaliana bundle sheath in sulfur and glucosinolate metabolism. The Plant Journal 78, 659–673. [DOI] [PubMed] [Google Scholar]
- Barolo S. 2012. Shadow enhancers: frequently asked questions about distributed cis-regulatory information and enhancer redundancy. BioEssays 34, 135–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NJ, Newell CA, Stanley S, Chen JE, Perrin AJ, Kajala K, Hibberd JM. 2011. Independent and parallel recruitment of preexisting mechanisms underlying C₄ photosynthesis. Science 331, 1436–1439. [DOI] [PubMed] [Google Scholar]
- Chitty JA, Furbank RT, Marshall JS, Chen Z, WC T. 1999. Genetic transformation of the C4 plant, Flaveria bidentis. The Plant Journal 6, 949–956. [Google Scholar]
- Chytilova EVA, Macas J, Galbraith DW. 1999. Green fluorescent protein targeted to the nucleus, a transgenic phenotype useful for studies in plant biology. Annals of Botany 83, 645–654. [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Comelli P, Glowa D, Chandler JW, Werr W. 2016. Founder-cell-specific transcription of the DORNRÖSCHEN-LIKE promoter and integration of the auxin response. Journal of Experimental Botany 67, 143–155. [DOI] [PubMed] [Google Scholar]
- Dengler N, Nelson T. 1999. Leaf structure and development in C4 plants. In: Sage RF, Monson RK, eds. C4 Plant Biology. San Diego: Academic Press. [Google Scholar]
- Dhondt S, Coppens F, De Winter F, Swarup K, Merks RM, Inzé D, Bennett MJ, Beemster GT. 2010. SHORT-ROOT and SCARECROW regulate leaf growth in Arabidopsis by stimulating S-phase progression of the cell cycle. Plant Physiology 154, 1183–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelmann S, Wiludda C, Burscheidt J, Gowik U, Schlue U, Koczor M, Streubel M, Cossu R, Bauwe H, Westhoff P. 2008. The gene for the P-subunit of glycine decarboxylase from the C4 species Flaveria trinervia: analysis of transcriptional control in transgenic Flaveria bidentis (C4) and Arabidopsis (C3). Plant Physiology 146, 1773–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau K. 1965. Plant anatomy. New York: John Wiley and Sons. [Google Scholar]
- Friso G, Giacomelli L, Ytterberg AJ, Peltier JB, Rudella A, Sun Q, Wijk KJ. 2004. In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. The Plant Cell 16, 478–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta KM, Hellmann E, Helariutta Y. 2014. Molecular control of cell specification and cell differentiation during procambial development. Annual Review of Plant Biology 65, 607–638. [DOI] [PubMed] [Google Scholar]
- Hatch MD. 1987. C4 photosynthesis: a unique blend of modified biochemistry, anatomy and ultrastructure. Biochimica Biophysica Acta 895, 81–106. [Google Scholar]
- Hatton D, Sablowski R, Yung MH, Smith C, Schuch W, Bevan M. 1995. Two classes of cis sequences contribute to tissue-specific expression of a PAL2 promoter in transgenic tobacco. The Plant Journal 7, 859–876. [DOI] [PubMed] [Google Scholar]
- Hibberd JM, Covshoff S. 2010. The regulation of gene expression required for C4 photosynthesis. Annual Review of Plant Biology 61, 181–207. [DOI] [PubMed] [Google Scholar]
- Janacek SH, Trenkamp S, Palmer B, et al. 2009. Photosynthesis in cells around veins of the C3 plant Arabidopsis thaliana is important for both the shikimate pathway and leaf senescence as well as contributing to plant fitness. The Plant Journal 59, 329–343. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO Journal 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsman EA, Pyke KA. 1998. Bundle sheath cells and cell-specific plastid development in Arabidopsis leaves. Development 125, 1815–1822. [DOI] [PubMed] [Google Scholar]
- Knappe S, Löttgert T, Schneider A, Voll L, Flügge UI, Fischer K. 2003. Characterization of two functional phosphoenolpyruvate/phosphate translocator (PPT) genes in Arabidopsis–AtPPT1 may be involved in the provision of signals for correct mesophyll development. The Plant Journal 36, 411–420. [DOI] [PubMed] [Google Scholar]
- Kubicki A, Steinmüller K, Westhoff P. 1994. Differential transcription of plastome-encoded genes in the mesophyll and bundle-sheath chloroplasts of the monocotyledonous NADP-malic enzyme-type C4 plants maize and Sorghum. Plant Molecular Biology 25, 669–679. [DOI] [PubMed] [Google Scholar]
- Leegood RC. 2008. Roles of the bundle sheath cells in leaves of C3 plants. Journal of Experimental Botany 59, 1663–1673. [DOI] [PubMed] [Google Scholar]
- Leyva A, Liang X, Pintor-Toro JA, Dixon RA, Lamb CJ. 1992. cis-Element combinations determine phenylalanine ammonia-lyase gene tissue-specific expression patterns. The Plant Cell 4, 263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann E, Birkenbihl RP, Ülker B, Somssich IE. 2006. An improved method for preparing Agrobacterium cells that simplifies the Arabidopsis transformation protocol. Plant Methods 2, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig MZ, Manu, Kittler R, White KP, Kreitman M. 2011. Consequences of eukaryotic enhancer architecture for gene expression dynamics, development, and fitness. PLoS Genetics 7, e1002364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake H, Maeda E. 1976. Development of bundle sheath chloroplasts in rice seedlings. Canadian Journal of Botany 54, 556–565. [Google Scholar]
- Patel M, Siegel AJ, Berry JO. 2006. Untranslated regions of FbRbcS1 mRNA mediate bundle sheath cell-specific gene expression in leaves of a C4 plant. The Journal of Biological Chemistry 281, 25485–25491. [DOI] [PubMed] [Google Scholar]
- Payne JL, Wagner A. 2015. Mechanisms of mutational robustness in transcriptional regulation. Frontiers in Genetics 6, 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyna-Llorens I, Burgess SJ, Reeves G, Singh P, Stevenson SR, Williams BP, Stanley S, Hibberd JM. 2018. Ancient duons may underpin spatial patterning of gene expression in C4 leaves. Proceedings of the National Academy of Sciences, USA 115, 1931–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sade N, Shatil-Cohen A, Attia Z, Maurel C, Boursiac Y, Kelly G, Granot D, Yaaran A, Lerner S, Moshelion M. 2014. The role of plasma membrane aquaporins in regulating the bundle sheath-mesophyll continuum and leaf hydraulics. Plant Physiology 166, 1609–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson MJ, Thorne JL, Wikström N, Bremer K. 2004. Molecular evidence on plant divergence times. American Journal of Botany 91, 1656–1665. [DOI] [PubMed] [Google Scholar]
- Soros CL, Dengler NG. 2001. Ontogenetic derivation and cell differentiation in photosynthetic tissues of C3 and C4 Cyperaceae. American Journal of Botany 88, 992–1005. [PubMed] [Google Scholar]
- Takabayashi A, Kishine M, Asada K, Endo T, Sato F. 2005. Differential use of two cyclic electron flows around photosystem I for driving CO2-concentration mechanism in C4 photosynthesis. Proceedings of the National Academy of Sciences, USA 102, 16898–16903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Watanabe-Takahashi A, Smith FW, Blake-Kalff M, Hawkesford MJ, Saito K. 2000. The roles of three functional sulphate transporters involved in uptake and translocation of sulphate in Arabidopsis thaliana. The Plant Journal 23, 171–182. [DOI] [PubMed] [Google Scholar]
- Wikström N, Savolainen V, Chase MW. 2001. Evolution of the angiosperms: calibrating the family tree. Proceedings of the Royal Society of London. Series B: Biological Sciences 268, 2211–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiludda C, Schulze S, Gowik U, Engelmann S, Koczor M, Streubel M, Bauwe H, Westhoff P. 2012. Regulation of the photorespiratory GLDPA gene in C4Flaveria: an intricate interplay of transcriptional and posttranscriptional processes. The Plant Cell 24, 137–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka-Diller JW, Helariutta Y, Fukaki H, Malamy JE, Benfey PN. 2000. Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development 127, 595–603. [DOI] [PubMed] [Google Scholar]
- Ye ZH. 2002. Vascular tissue differentiation and pattern formation in plants. Annual Review of Plant Biology 53, 183–202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.