Abstract
The genome of a wild-type solventogenic Clostridium sp. strain LJ4 that could directly convert undetoxified lignocellulosic hydrolysate to butanol and tolerate high concentration of furan and phenolic derivates occurring in the lignocellulosic hydrolysate is described. 16S rDNA gene sequencing and analysis indicated that it is closely related to Clostridium acetobutylicum. The genome size of strain LJ4 is 3.90 Mp, which has a G + C content of 30.72% and encodes 2711 proteins. It also has one 0.19 Mp plasmid with 181 predicted encoding proteins. Alcohol dehydrogenases (ADs) and a nicotinamide adenine dinucleotide phosphate (NADPH)-dependent flavin mononucleotide (FMN) reductase were identified, which may play key roles in inhibitors’ resistance in lignocellulosic hydrolysate.
Keywords: Clostridium, Draft genome sequencing, Furan and phenolic derivates, Alcohol dehydrogenases, NADPH-dependent FMN reductase
Butanol has been considered as a potential biofuel, which could be directly used without modification of current engines. In addition, butanol contains higher energy content for a given volume than ethanol. Solventogenic Clostridium species are able to produce butanol through acetone–butanol–ethanol (ABE) fermentation process (Lee et al. 2016; Shanmugam et al. 2018). However, ABE fermentation shows several major drawbacks, including the high cost of substrates, low productivity and high recovery costs (Baral and Shah 2014; Gu et al. 2011). To overcome these obstacles, one option is to utilize renewable lignocellulosic materials as substrates, such as corn fiber, wheat straw and bagasse fiber. However, during the pretreatment process of lignocellulosic materials, various lignocellulose-derived microbial inhibitory compounds (LDMICs) are generated along with reducing sugars, which significantly inhibit the cell growth and metabolism (Ezeji et al. 2007). Although series of inhibitor removal technologies have been developed including physical and chemical methods; however, it may not be economically viable due to the high operation cost and potential loss of fermentable sugars (Dong et al. 2018; Sharma et al. 2017). To make bioconversion of lignocellulosic biomass to butanol more economically feasible, development of an inhibitor-tolerant strain is still crucial.
In the present study, Clostridium sp. strain LJ4, a strictly anaerobic and gram-positive solventogenic bacterium was isolated from the compost containing soil sample in Pearl Spring, Nanjing, China. The culture has been preserved in China Center for Type Culture Collection (Wuhan, China) under the deposit number of CCTCC M2017754. Strain LJ4 could directly produce butanol from the undetoxified lignocellulosic hydrolysate. When 60 g/L of glucose was supplemented in the undetoxified lignocellulolytic hydrolysate, it could produce 13.9 g/L of butanol, 5.6 g/L of acetone, 1.17 g/L of ethanol, 9.3 g/L of acetic acid and 3.6 g/L of butyric acid. Furthermore, it could tolerate up to 5 g/L of furfural, 5 g/L of 5-hydroxymethyl-furfural (HMF), 1.9 g/L of ferulic, 1 g/L of p-coumaric and 4 g/L of vanillin. Compared with reported solventogenic Clostridium strains, strain LJ4 could resist higher concentrations of inhibitors, indicating the potential application for butanol production from undetoxified lignocellulosic materials (Table 1) (Ezeji et al. 2007; Guo et al. 2012; Jurgens et al. 2012). To determine the reason for the increased resistance to inhibitors, the draft genome of strain LJ4 was accordingly sequenced and putative genes for enzymes likely to be involved in the process were identified (Chen et al. 2018).
Table 1.
Inhibitors’ tolerance comparison of solventogenic Clostridium strains
| Furfural (g/L) | HMF (g/L) | Ferulic (g/L) | p-Coumaric (g/L) | Vanillin (g/L) | References | |
|---|---|---|---|---|---|---|
| C. beijerinckii BA101 | 3 | 3 | 0.3 | 0.3 | – | Ezeji et al. (2007) |
| C. acetobutylicum CICC8012 | 1 | 1 | 0.5 | 0.5 | 2 | Jurgens et al. (2012) |
| C. beijerinckii IB4 | 1 | 1 | 1 | 0.5 | 0.5 | Guo et al. (2012) |
| C. beijerinckii NCIMB8052 | – | – | 0.3 | 0.5 | 0.3 | Guo et al. (2012) |
| Clostridium sp. strain LJ4 | 4 | 5 | 1.9 | 1 | 4 | This study |
“–” means the inhibitor was not investigated in the study
Strain LJ4 was cultivated in mineral salts medium added with glucose as the sole carbon source for 5 days and its genomic DNA was extracted using G+ bacteria genomic DNA kit (ZOMANBIO, China). 16S rDNA gene of strain LJ4 was first amplified from its genome. Based on the 16S rDNA gene sequence comparison in NCBI website, strain LJ4 belongs to the genus of Clostridium. Sequences were then aligned using Clustal W interface in MEGA 5.01 based on the neighbor joining method, and bootstrap values were based on 1000 replicates (Fig. 1). The nearest neighbor of strain LJ4 is C. acetobutylicum strain ATCC824 (Accession number NR_074511.2), which shares 99.78% similarity with strain LJ4 calculated by ANI. Furthermore, the genomic DNA of strain LJ4 was broken into small fragments with an average of 500 bp using a nebulization technique, and then DNA libraries were accordingly constructed. The draft genome was sequenced using Illumina HiSeq 2500 system (Table 2). The result contained 19,725,674 raw reads of chromosome and 192,126 raw reads of plasmid. However, in high-throughput sequencing, some sequencing errors often occur and the sequence quality is relatively low. To obtain higher quality and more accurate biological information analysis results, the original sequencing data needs to be optimized by Cutadapt (version 1.9.1) (Martin 2011). The standard included removed DNA spike-in, adaptors and ambiguous or low-quality reads (the reads containing more than 10% N or the length less than 75 bp after trimming and the quality score of bases are less than 20). Paired reads have 92.47% of bases with quality score Q30 higher than the raw 85.48% (Q30 is the quality score specified by Illumina, indicating the probability of errors in basecalling lower than 0.1%). Furthermore, based on the optimized data, the filtered reads (16,473,780 of chromosome, 192,126 of plasmid) were assembled using de novo through the method of de Bruijn graphs by software of Velvet (version 1.2.10) (Zerbino and Birney 2008).
Fig. 1.
Phylogenetic tree of Clostridium sp. strain LJ4 using the neighbor joining method (MEGA5.01) based on its 16S rDNA gene sequences. The taxon named after strain and appended accession number in brackets and the strain with a T for Type Culture in superscript. The scale represents the genetic distance between sequences. Numbers displayed on the branches are the bootstrap support obtained through 1000. The full names of the abbreviations for the culture collections: ATCC American Type Culture Collection, NCIMB National Collections of Industrial, Food and Marine Bacterial, DSM Deutsche Sammlung von Mikroorganismen und Zellkulturen, JCM Japan Collection of Microorganisms Replications
Table 2.
Genome features of Clostridium sp. strain LJ4
| Features | Value of chromosome | Value of plasmid |
|---|---|---|
| Reads | 19,725,674 (raw reads) 16,473,780 (filtered reads) |
192,126 |
| Scaffolds | 21 | 1 |
| GC content (%) | 30.72 | 30.91 |
| Total number of genes | 3886 | 181 |
| Total number of proteins | 3711 | 181 |
The draft genome was predicted to include 3892 protein-coding sequences including 181 sequences located in the plasmid by the software of prodigal (version 3.02) (Hyatt et al. 2010). Non-coding RNAs including 38 rRNAs and 66 tRNAs were obtained by comparing with the Rfam (version 12.0) database (Nawrocki et al. 2014). For better understanding, these genes were annotated in non-redundant protein (NR), gene ontology (GO), kyoto encyclopedia of genes and genomes (KEGG) and cluster of orthologous groups of proteins (COG) databases. It is worth mentioning that genes were classified into 23 kinds of protein functions in COG database, including defense systems, energy production and replication, recombination and repair, etc., which contribute to inhibitors’ tolerance in lignocellulosic hydrolysate. Generally, toxic inhibitors would be completely or partially metabolized to less toxic compounds by inhibitor tolerating strains through limitation of the uptake or enhancement of the extrusion, or regulation of expression levels of transcription factors associated with toxicity tolerance and some heat shock proteins (Liu et al. 2018; Zhang et al. 2015). Within the chromosome of strain LJ4, gene 3-352 [alcohol dehydrogenases (ADs)] and gene 3-409 [a nicotinamide adenine dinucleotide phosphate (NADPH)-dependent flavin mononucleotide (FMN) reductase] were found, which may be responsible for converting toxic inhibitors into less toxic compounds. The amino acid sequences of predicted genes were searched for similarity against protein data bank archive (pdb) database in the NCBI website, and top 10 hits were chosen to study the phylogeny of respective enzymes. Sequences were aligned using ClustalW interface in MEGA 5.01 based on the neighbor joining method, and bootstrap values were based on 1000 replicates (Fig. 2). Ads (gene 3-352) exhibited 57% similarity with the annotated bacterial sequence Yqhd from E. coli (Accession number: 4QGS_A). And gene 3-409 (NADPH-dependent FMN reductase) showed 40% with the annotated bacterial sequence Ylr011wp from Saccharomyces cerevisiae (Accession number 1T0I_A). ADs have been reported to convert furfural and HMF into their relevant alcohols with the consumption of intracellular energy and reducing power (NADH/NADPH), which has been described in ethanologenic microorganisms including S. cerevisiae and Zymomonas mobilis (Agrawal and Chen 2011; Liu et al. 2008). For instance, S. cerevisiae could convert 2.88 g/L of furfural into 2.94 g/L furfuryl alcohol with the catalysis of ADs (Liu et al. 2005). Similarly, C. acetobutylicum ATCC824 could also convert 1.63 g/L of furfural into 1.66 g/L furfuryl alcohol (Zhang et al. 2012) with the catalysis of ADs, suggesting that strain LJ4 may also adopt the similar mechanism to reduce the toxicity of furfurals. Phenolic compounds, such as ferulic acid can be metabolized by phenolic acid decarboxylase and reductase in Lactobacillus sp. and S. cerevisiae (Filannino et al. 2015; Mukai et al. 2014). Phenolic acids reduction involves a hydrogen donor and the re-oxidation of the reduced co-factor NADH, which may provide metabolic advantage through NAD+ regeneration (Filannino et al. 2015; Liu et al. 2017). Within solventogenic Clostridium sp., NADPH-dependent FMN reductase has been reported to be responsible for ferulic acid tolerance. For instance, 0.50 g/L of ferulic acid was completely converted into the less toxic phenolic compound of hydroferulic acid by C. beijerinckii 4693: int after 12 h (Liu et al. 2017). Though the detailed molecular mechanisms remained unclear, the existence of NADPH-dependent FMN reductase within strain LJ4 may play key roles for ferulic acid tolerance. Taken together, due to the presence of ADs and NADPH-dependent FMN reductase, strain LJ4 could tolerate high concentration of inhibitors in lignocellulosic hydrolysate. However, future studies are still needed to characterize these enzymes and elaborate the tolerance mechanism clearly.
Fig. 2.
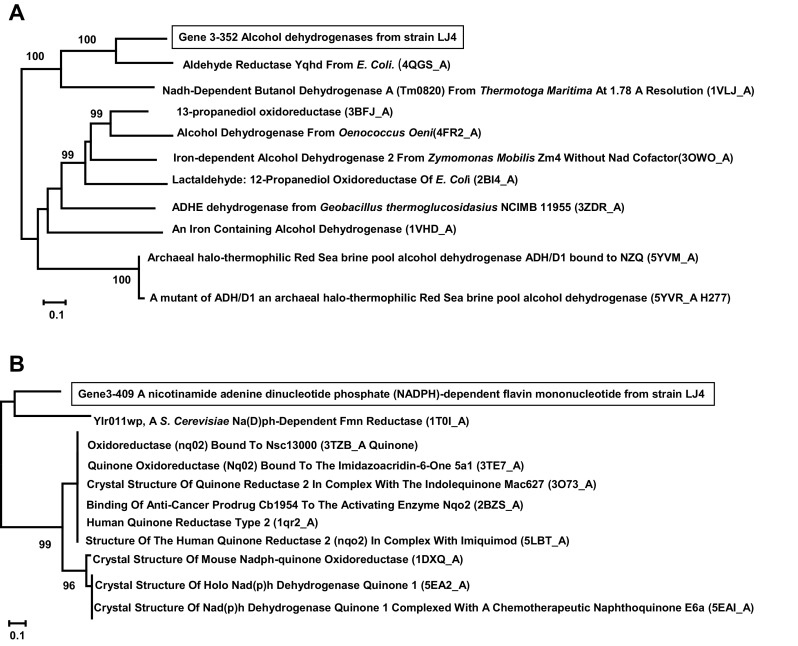
Neighbor joining tree calculated by ClustalW for a gene 3-352 Alcohol dehydrogenases, b gene 3-409 A nicotinamide adenine dinucleotide phosphate (NADPH)-dependent flavin mononucleotide from Clostridium sp. LJ4 constructed by MEGA 5.01. The taxon named after enzyme and appended accession number in brackets. The scale represents the genetic distance between sequences. Numbers displayed on the branches are the bootstrap support obtained through 1000 replications
Acknowledgements
This work was supported by the Jiangsu Province Natural Science Foundation for Youths (BK20170993, BK20170997), the Project of State Key Laboratory of Materials-Oriented Chemical Engineering (KL16-08), the Jiangsu Synergetic Innovation Center for Advanced Bio-Manufacture (XTE1840), the Key Science and Technology Project of Jiangsu Province (BE2016389), and the National Natural Science Foundation of China (no. 21706125, no. 21727818, no. 21706124, no. 31700092).
Compliance with ethical standards
Conflict of interest
The authors have declared there was no conflict of interest.
Footnotes
Contributor Information
Min Jiang, Email: bioengine@njtech.edu.cn.
Fengxue Xin, Email: xinfengxue@njtech.edu.cn.
References
- Agrawal M, Chen RR. Discovery and characterization of a xylose reductase from Zymomonas mobilis ZM4. Biotechnol Lett. 2011;33:2127–2133. doi: 10.1007/s10529-011-0677-6. [DOI] [PubMed] [Google Scholar]
- Baral NR, Shah A. Microbial inhibitors: formation and effects on acetone-butanol-ethanol fermentation of lignocellulosic biomass. Appl Microbiol Biotechnol. 2014;98:9151–9172. doi: 10.1007/s00253-014-6106-8. [DOI] [PubMed] [Google Scholar]
- Chen C, Sun C, Wu YR. The draft genome sequence of a novel high-efficient butanol-producing bacterium Clostridium diolis strain WST. Curr Microbiol. 2018;75:1011–1015. doi: 10.1007/s00284-018-1481-5. [DOI] [PubMed] [Google Scholar]
- Dong JJ, Han RZ, Xu GC, Gong L, Xing WR, Ni Y. Detoxification of furfural residues hydrolysate for butanol fermentation by Clostridium saccharobutylicum DSM 13864. Bioresour Technol. 2018;259:40–45. doi: 10.1016/j.biortech.2018.02.098. [DOI] [PubMed] [Google Scholar]
- Ezeji T, Qureshi N, Blaschek HP. Butanol production from agricultural residues: impact of degradation products on Clostridium beijerinckii growth and butanol fermentation. Biotechnol Bioeng. 2007;97:1460–1469. doi: 10.1002/bit.21373. [DOI] [PubMed] [Google Scholar]
- Filannino P, Bai Y, Di CR, Gobbetti M, Gänzle MG. Metabolism of phenolic compounds by Lactobacillus spp. during fermentation of cherry juice and broccoli puree. Food Microbiol. 2015;46:272–279. doi: 10.1016/j.fm.2014.08.018. [DOI] [PubMed] [Google Scholar]
- Gu Y, et al. Economical challenges to microbial producers of butanol: feedstock, butanol ratio and titer. Biotechnol J. 2011;6:1348–1357. doi: 10.1002/biot.201100046. [DOI] [PubMed] [Google Scholar]
- Guo T, Tang Y, Zhang QY, Du TF, Liang DF, Jiang M, Ouyang PK. Clostridium beijerinckii mutant with high inhibitor tolerance obtained by low-energy ion implantation. J Ind Microbiol Biotechnol. 2012;39:401–407. doi: 10.1007/s10295-011-1017-5. [DOI] [PubMed] [Google Scholar]
- Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgens G, Survase S, Berezina O, et al. Butanol production from lignocellulosics. Biotechnol Lett. 2012;34:1415–1434. doi: 10.1007/s10529-012-0926-3. [DOI] [PubMed] [Google Scholar]
- Lee SH, Yun EJ, Kim J, Sang JL, Um Y, Kim KH. Biomass, strain engineering, and fermentation processes for butanol production by solventogenic Clostridia. Appl Microbiol Biotechnol. 2016;100:1–17. doi: 10.1007/s00253-015-7019-x. [DOI] [PubMed] [Google Scholar]
- Liu ZL, Slininger PJ, Gorsich SW. Enhanced biotransformation of furfural and hydroxymethylfurfural by newly developed ethanologenic yeast strains. Appl Biochem Biotechnol. 2005;121(1–3):451–460. doi: 10.1385/ABAB:121:1-3:0451. [DOI] [PubMed] [Google Scholar]
- Liu ZL, Moon J, Andersh BJ, Slininger PJ, Weber S. Multiple gene-mediated NAD(P)H-dependent aldehyde reduction is a mechanism of in situ detoxification of furfural and 5-hydroxymethylfurfural by Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2008;81:743–753. doi: 10.1007/s00253-008-1702-0. [DOI] [PubMed] [Google Scholar]
- Liu J, Guo T, Yang T, Xu J, Tang C, Liu D, Ying H. Transcriptome analysis of Clostridium beijerinckii adaptation mechanisms in response to ferulic acid. Int J Biochem Cell B. 2017;86:14–21. doi: 10.1016/j.biocel.2017.02.009. [DOI] [PubMed] [Google Scholar]
- Liu J, Lin Q, Chai X, Luo Y, Guo T. Enhanced phenolic compounds tolerance response of Clostridium beijerinckii NCIMB 8052 by inactivation of Cbei_3304. Microb Cell Fact. 2018;17:35. doi: 10.1186/s12934-018-0884-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17
- Mukai N, Masaki K, Fujii T, Iefuji H. Single nucleotide polymorphisms of PAD1 and FDC1 show a positive relationship with ferulic acid decarboxylation ability among industrial yeasts used in alcoholic beverage production. J Biosci Bioeng. 2014;118:50. doi: 10.1016/j.jbiosc.2013.12.017. [DOI] [PubMed] [Google Scholar]
- Nawrocki EP, Burge SW, Bateman A, et al. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 2014;43(D1):D130–D137. doi: 10.1093/nar/gku1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugam S, Sun C, Zeng X, Wu YR. High-efficient production of biobutanol by a novel Clostridium sp. strain WST with uncontrolled pH strategy. Bioresour Technol. 2018;256:543–547. doi: 10.1016/j.biortech.2018.02.077. [DOI] [PubMed] [Google Scholar]
- Sharma HK, Xu C, Qin W (2017) Biological pretreatment of lignocellulosic biomass for biofuels and bioproducts: an overview waste and biomass valorization. 10.1007/s12649-017-0059-y
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Han B, Ezeji TC. Biotransformation of furfural and 5-hydroxymethyl furfural (HMF) by Clostridium acetobutylicum ATCC 824 during butanol fermentation. New Biotechnol. 2012;29:345–351. doi: 10.1016/j.nbt.2011.09.001. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ujor V, Wick M, Ezeji TC. Identification, purification and characterization of furfural transforming enzymes from Clostridium beijerinckii NCIMB 8052. Anaerobe. 2015;33:124–131. doi: 10.1016/j.anaerobe.2015.03.005. [DOI] [PubMed] [Google Scholar]



