Abstract
A binding site for the glucocorticoid receptor in the serum-inducible proliferin gene promoter has been reported to function as a composite glucocorticoid response element when fused to a minimal promoter. We now show that this element can also act as a glucocorticoid-independent negative regulator of transcription, both as an isolated element fused to a minimal promoter and within the context of the proliferin gene promoter. Furthermore, this element is recognized by a factor in mouse fibroblast cell extracts that is distinct from the glucocorticoid receptor and from AP-1, both of which have previously been shown to be able to bind to this site. The ability of this element to repress serum-inducible proliferin promoter activity is dependent on the position of this element with respect to the adjacent serum response region, and on the activity of a positive regulatory element located further upstream in the proliferin promoter.
Keywords: Proliferin gene expression, Glucocorticoid receptor, Transcription regulation
PROLIFERIN (PLF), also known as mitogen-regulated protein, is a member of the prolactin/growth hormone family in the mouse (12). In vivo, PLF synthesis occurs specifically in placental trophoblast giant cells (9,10,14), and the secreted protein has been found to regulate both angiogenesis (7) and uterine cell proliferation (21). In addition to its expression in placental trophoblasts, PLF synthesis has also been detected in a number of cell lines. In these cell cultures, PLF gene transcription is activated in response to the addition of serum, platelet-derived growth factor, or phorbol esters (3,6,11,12,20,22). In contrast, PLF transcription is repressed during the transition from an actively growing to a quiescent fibroblast (11), during differentiation of a multipotential fibroblast into a determined myoblast (25), and by addition of glucocorticoids (20).
Serum stimulation of PLF gene transcription in mouse fibroblast cell cultures is mediated by two adjacent elements, an AP-1 site at –231 to –224 and a second element from –223 to –204 that resembles the Sph I repeats in the simian virus 40 enhancer (6,20). Glucocorticoid inhibition of PLF gene expression was found to depend upon the glucocorticoid receptor, which binds to the PLF promoter sequences from –254 to –230, a site overlapping and upstream of the AP-1 element (20). Although this binding site is not similar in sequence to the consensus glucocorticoid response element (GRE), it can confer glucocorticoid regulation on a heterologous promoter (2). Surprisingly, the effect of this element in response to glucocorticoids varied depending upon the form of the AP-1 transcription factor present in the cell, such that the binding site functioned as a positive GRE in the presence of c-jun/c-jun homodimers, but a negative GRE with c-jun/c-fos heterodimers (2). This dual activity led to the designation of this element as a “composite” GRE, or cGRE (2).
The discovery of the PLF gene cGRE has provided an intriguing example of a complex regulatory element, and has led to the identification of similar elements in the human papilloma virus regulatory region (18) and the human cytosolic phospholipase A2 gene (19). Although the glucocorticoid receptor binding site in the PLF gene is the best characterized cGRE, to date it has only been analyzed in constructs in which the isolated element is fused to a heterologous promoter. We now report on our analysis of the action of this element within the context of the PLF promoter.
MATERIALS AND METHODS
Plasmid Constructions
Deletion mutations in the PLF promoter were generated by Bal 31 digestion of a genomic DNA fragment with boundaries at +61 (in the 5′ untranslated region) and –670, relative to the start site of transcription of PLF mRNA (13). End points of the deletions were determined by DNA sequence analysis. The – 251/– 232 mutant replaced 20 bp with a 10 bp linker. Promoter constructs were also prepared by transferring specific restriction fragments or synthetic double-stranded oligonucleotides to position – 33 in the Drosophila alcohol dehydrogenase gene promoter as described previously (2) or to position – 37 in the herpes simplex virus thymidine kinase gene promoter (17). All promoter constructs included the CAT gene as the reporter for transfection assays. Plasmids were propagated in E. coli DH5α cells and purified by CsCl/ethidium bromide equilibrium density gradient centrifugation.
Cell Culture, DNA Transfections, and CAT Assays
Mouse L cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% calf serum, 2 mM glutamine, and 10 units/ml each penicillin and streptomycin. Approximately 2 × 106 L cells were transferred to each 10-cm dish 24 h before DNA transfection (15 μg/dish) with DEAE-dextran (15). After 4 h, medium was removed and cells were exposed to a DMSO shock; cultures were then fed fresh medium containing either 0.5% or 15% calf serum. Cultures were harvested 48 h posttransfection. Protein extracts from transfected L cell cultures were prepared by repeated freeze-thaw lysis in 0.25 M Tris-HCl, pH 7.8. Extract volumes with equal amounts of protein, as determined by a dye binding assay (1), were incubated with [14C]chloramphenicol (New England Nuclear), and reaction products were re-solved by thin-layer chromatography (5). CAT activity was quantified by excising regions of the thin-layer chromatography plate for liquid scintillation counting or by analysis on a Molecular Dynamics phosphorimager. Construct activities were analyzed by at least four independent transfections.
Electrophoretic Mobility Shift Assays
Double-stranded oligonucleotides for electrophoretic mobility shift assays (shown below) were labeled with [α-32P]dATP by filling in the recessed ends with the Klenow fragment of DNA polymerase I. Competitor DNAs were blunt-end, double-stranded oligonucleotides or fragments excised from a plasmid vector. DNAs were added to whole-cell extracts prepared from mouse L fibroblasts as described (16). Binding reactions were carried out in 25 μl of 10 mM Tris-HCl, pH 7.8, 50 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 5% (v/v) glycerol, 0.2 mM phenylmethylsulfonyl fluoride, 10 ng (105 cpm) DNA probe, 25 μg of protein extract, and 1 μg of poly(dl-dC). In some reactions, 1–2 μl of normal rabbit serum, an anti-serum against c-fos (which completely blocks AP-1 binding activity in mouse fibroblast extracts) (6), or a monoclonal antibody that recognizes the mouse glucocorticoid receptor (4) were also added. Reactions were incubated for 20 min at 25°C, and protein-DNA complexes were separated from free DNA by 6% polyacrylamide gel electrophoresis in 44.5 mM Tris base, 1.25 mM EDTA, and 44.5 mM boric acid.
Oligonucleotide Sequences
The oligonucleotides utilized for the electrophoretic mobility shift experiments were the wild-type PLF gene cGRE from – 254 to – 230: sense 5′ - TCGACGGCTACTCACAGTATGATTTGT TTTG, and antisense 5′-GATCCAAAACAAAT CATACTGTGAGTAGCCG; mutant cGRE (mutations in lower case): sense 5′-TCGACGGC TACTCACAagcTtcTagaTTTTG, and antisense 5′ - GATCCAAAAtctAgaAgctTGTGAGTAGCCG); upstream PLF gene positive element (– 275 to – 253): sense 5′-CGGGATCCTTATGAGGAAGACATAGTT GTGG, and antisense: 5′-AATTCCACAACT ATGTCTTCCTCATAAGGATCCCG); and PLF AP-1 element and flanking sequence from –234 to –219: sense 5′-AATTCGTTTTAGTCAGAG CAT, and antisense 5′-CGATGCTCTGACT AAAACG. The tyrosine aminotransferase GRE (5′-CTGTACAGGATGTTCTAGCTAC) was excised from a plasmid clone (provided by Keith Yamamoto) as a 5-mer of the GRE.
RESULTS
Function of the cGRE in the PLF Promoter
The activity of the isolated PLF gene cGRE has been characterized in transfected HeLa cells (2), but endogenous PLF gene expression occurs in immortalized mouse fibroblasts (11,22). Mouse L cell fibroblasts have proven to be a useful background for PLF promoter transfection studies, because in these cells a fragment of the proliferin promoter to –670 displays serum, phorbol ester, and glucocorticoid regulation (13,20). Therefore, we first tested the activity of the isolated cGRE constructs from Diamond et al. (2) in transfected mouse L fibroblasts to determine if this element has detectable activity in this cell system.
A construct containing one copy of this element (plfG, with PLF promoter sequences from – 254 to – 230) had no discernable effect on transcription from the alcohol dehydrogenase gene minimal promoter, but inclusion of three copies of the element (plfG3) resulted in a marked decrease in CAT activity, even in the absence of glucocorticoids (Fig. 1). The ability of the isolated PLF gene cGRE to act as a glucocorticoid-independent negative element in L cells represented an unexpected activity for this element. In comparison, one or three copies of a previously uncharacterized segmzent of the PLF gene promoter (from – 274 to – 252) placed upstream of the alcohol dehydrogenase minimal promoter conferred a higher level of reporter expression (Fig. 1). Thus, the region from – 274 to – 252 appears to encompass a positive transcriptional regulatory element.
FIG. 1.
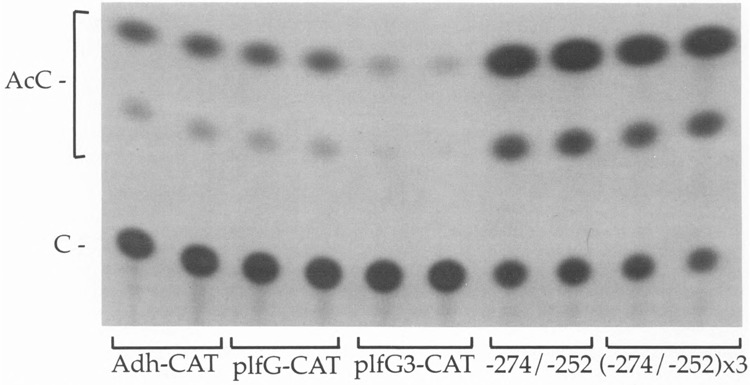
Activity of the cGRE in mouse fibroblasts. Duplicate transfections of mouse L fibroblasts (maintained in 15% serum) were carried out with the alcohol dehydrogenase minimal promoter linked to CAT (Adh-CAT), and this same vector in which one copy (plfG-CAT) or three copies (plfG3-CAT) of the PLF cGRE, or one or three copies of the PLF promoter sequence from – 274 to – 252, were inserted. CAT reporter activity, measured as the average conversion of chloramphenicol (C) to acetylated forms (AcC) for the duplicate samples, was: Adh-CAT, 12.3%; plfG-CAT, 10.9%; plfG3-CAT, 2.2%; – 274/– 252-CAT, 69.9%; and (– 272/ – 252) × 3-CAT, 78.2%.
To determine if the cGRE can also act as a negative element within the natural PLF promoter, a series of 5′ deletion mutant promoters was assayed by transfection into mouse L cells. Deletion to position – 268 upstream of the transcription start site resulted in a promoter equivalent to the –670 bp promoter in basal (0.5% serum) and serum-stimulated activity (Fig. 2). Further deletion to – 256 caused a significant decrease in both basal and serum-stimulated activity, as well as in the magnitude of the serum induction. That this construct is serum inducible is consistent with the presence of an intact serum response region (– 231 to – 204), but the decreased activity of the – 256 deletion indicates that sequences between – 268 and – 256 are critical for the activity of the positive element that was identified in Fig. 1. This analysis also revealed that the cGRE (between –254 and –230) does act as a glucocorticoid-independent negative element in the PLF promoter, because a deletion to –232 (removing the cGRE) resulted in a promoter with much stronger activity and serum induction than the deletion to – 256. Based on the results of four independent transfections, the – 232 promoter was 4.3 ± 0.4-fold more active than the – 256 promoter in the presence of serum, and the serum inducibility of the – 232 promoter was 2.7 ± 0.4-fold greater than the – 256 promoter. An intermediate deletion to – 244 generated a promoter with reduced activity relative to the – 670, – 268, and – 232 bp promoters, indicating that the sequence between – 244 and – 232 (5′-AGTATGA TTTGTT-3′) is sufficient for the function of the negative element.
FIG. 2.
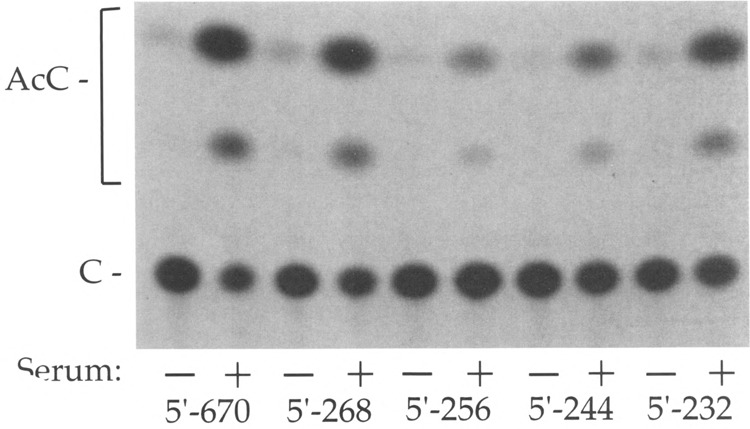
Activity of PLF promoter mutants. A series of PLF promoter-CAT reporter constructs with progressive 5′ deletions was transfected into mouse L cells. Conversion of chloramphenicol (C) to acetylated forms (AcC) for cells maintained in 0.5% (-) or 20% ( + ) serum, respectively, was: 5′ – 670, 3.2% and 73.1%; 5′ – 268, 3.6% and 59.2%; 5′ – 256, 1.6% and 9.3%; 5′ –2 44, 1.3% and 15.2%; 5′ – 232, 2.3% and 47.2%.
The ability of sequences within the cGRE to act as a glucocorticoid-independent repression element was also tested by mutation of this region (from – 251 to – 232) within the – 670 bp promoter. Compared to the wild-type promoter, the mutant promoter lacking the cGRE/negative element displayed a greater activity in transfected mouse L cells maintained in low serum (Fig. 3). From four independent transfections, the promoter mutated at the cGRE was 2.5 ± 0.7-fold stronger than the wild-type promoter in low serum. A significant difference between the wild-type and mutant promoters under high serum conditions was not detected, probably because of the inclusion of the upstream positive element in these constructs.
FIG. 3.
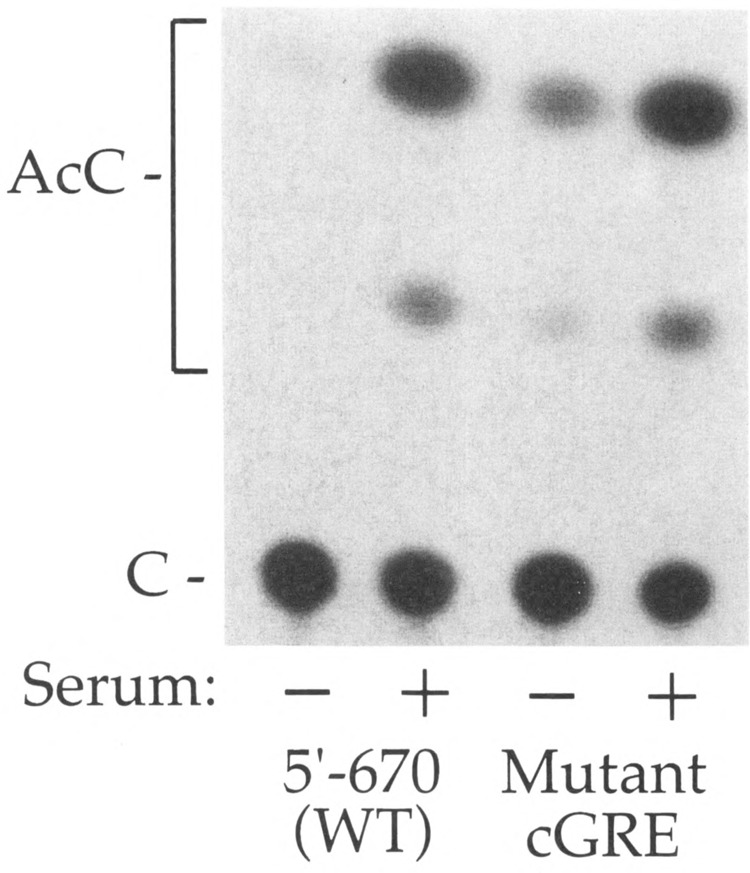
Activity of the cGRE in the PLF promoter. The wild-type PLF promoter 5′ (– 670) and a linker-substitution mutant – 251 / – 232 in this – 670 promoter background were linked to the CAT gene at position + 61 in the PLF 5′ untranslated region. These DNA constructs were transfected into mouse L cells. After transfection, cultures were maintained for 2 days in 0.5% or 15% serum. Cell extracts were prepared and assayed for CAT enzymatic activity. Conversion of chloramphenicol (C) to acetylated forms (AcC) in 0.5% serum (−) and 15% serum (+), respectively, was 3.3% and 41.5% for wild-type, and 14.0% and 55.3% for the cGRE mutant.
Factor Binding to the PLF Gene cGRE/Negative Element
The glucocorticoid-independent activity of the cGRE in the native PLF promoter suggested that this element can be recognized by a factor in addition to the glucocorticoid receptor. To determine if this element is bound specifically by a factor present in mouse fibroblasts, whole-cell extracts from mouse L cells were incubated with the radio-labeled cGRE and subjected to polyacrylamide gel electrophoresis (Fig. 4). A protein-DNA complex was detected with the wild-type cGRE probe (lane 1) but not with a cGRE mutated at residues – 243 through – 234 (lane 2). The complex was specific because it was competed by excess wild-type (lane 3), but not mutant (lane 4), cGRE DNA.
FIG. 4.
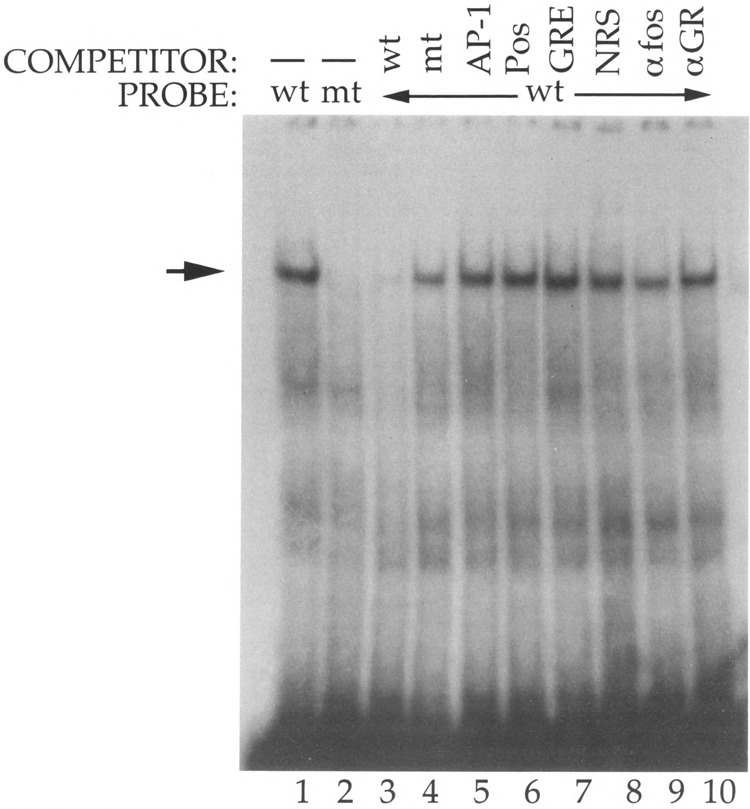
Protein binding to the cGRE/negative element. Double-stranded oligonucleotides corresponding to the wild type (lane 1) or mutant (lane 2) PLF negative element (from – 254 to – 230) were radiolabeled and incubated with mouse L cell whole-cell extracts. The mutated cGRE was altered at residues – 243 through – 234. Specific binding (arrow) was determined by competition with a 50-fold molar excess of unlabeled double-stranded wild-type (lane 3) or mutant (lane 4) negative element. Additional competition binding reactions included a 50-fold molar excess of the PLF gene AP-1 element (lane 5), the PLF positive element from – 274 to – 253 (lane 6), or five copies of the tyrosine aminotransferase GRE (lane 7). Parallel reactions were also incubated with normal rabbit serum (lane 8), an antiserum against c-fos (lane 9), or a monoclonal antibody against the glucocorticoid receptor (lane 10). Bound and free DNA were separated by polyacrylamide gel electrophoresis.
Addition of an excess amount of DNA containing a cluster of five GREs did not reduce binding to the cGRE (lane 7). Thus, the factor binding to the cGRE appears to be distinct from the glucocorticoid receptor. Because the cGRE has also been reported to have weak AP-1 binding sites (2,8), we also examined the ability of the PLF gene AP-1 element to compete for protein binding to the cGRE; no competition was observed (lane 5), suggesting that the bound factor does not correspond to a jun-fos heterodimer or a jun-jun homodimer. Addition of an antiserum against c-fos (which is effective at blocking binding to the PLF gene AP-1 site and to other AP-1 sites) (4,6) or against other AP-1 components (data not shown), or an antibody against the glucocorticoid receptor did not disrupt binding (lanes 9-10), consistent with these factors not being present in the complex. Finally, addition of the unlabeled positive regulatory element (from – 275 to – 253) also failed to compete with the cGRE for factor binding (lane 6), indicating that the proteins recognizing these two PLF promoter elements are distinct.
Interaction of the cGRE/Negative Element and the Serum Response Region
The ability of the cGRE/negative element to repress serum-inducible transcription suggests that this element is able to interfere with the activity of the adjacent serum response region. To test this directly, the serum response region (– 231 to – 204) alone or the serum response region along with the cGRE/negative element (– 256 to – 204) were transferred upstream of the herpes virus thy-midine kinase gene minimal promoter. As seen in Fig. 5, the – 231 to – 204 region alone was able to confer serum responsiveness to the minimal promoter, as demonstrated previously (20). Addition of the GRE/negative element inhibited transcription in response to serum, demonstrating that this element can block the activity of the serum response region.
FIG. 5.
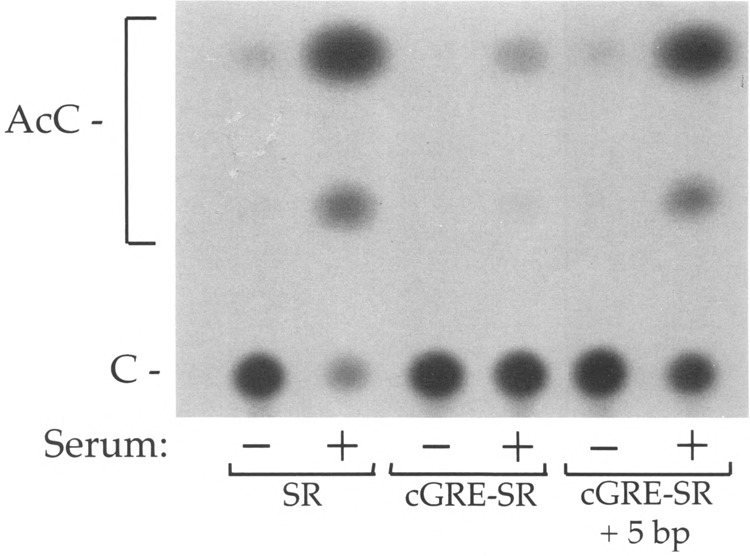
Interaction of the cGRE/negative element and the serum response region. The PLF serum response region from – 234 to – 204 alone (SR), the serum response region with the cGRE/negative element (– 256 to – 204, designated cGRE-SR), and the serum response region with the cGRE/negative element separated by an additional 5 bp were fused to the herpes virus thymidine kinase minimal promoter. These DNAs were transfected into mouse L cells, which were then maintained for 2 days in 0.5% or 15% serum. Conversion of chloramphenicol to acetylated forms in 0.5% serum (–) and 15% serum ( + ), respectively, was 4.1% and 91.3% for – 234/– 204; 0.5% and 6.6% for – 256/– 204; and 3.0% and 61.9% for – 256/– 204 ( + 5 bp).
The close proximity of the cGRE/negative element to the serum response region further suggests that direct interactions between factors bound at these sites might be important for the observed inhibition. Alternatively, these factors might act independently by contacting the basal transcription machinery bound at the TATA box. To distinguish between these possibilities, the cGRE/negative element was moved 5 bp further upstream of the serum response region. If the first model is correct, then moving the cGRE/negative element 5 bp further upstream from the serum response region, thereby rotating the relative positions of the elements by 180°, should disrupt an interaction between bound factors and restore serum inducibility. As shown in Fig. 5, the effect of the 5 bp insertion between the cGRE/negative element and the serum response region was to restore full serum-inducible activity. Thus, the effect of the cGRE/negative element in the PLF promoter is dependent on its position relative to the adjacent serum response region.
DISCUSSION
Characterization of the PLF gene promoter cGRE in this study has revealed a previously undetected function for this element, namely the glucocorticoid-independent repression of the adjacent serum response region. This activity represents a third function of this element, in addition to the glucocorticoid- and jun/jun homodimer-dependent stimulation of transcription from a minimal promoter, and the glucocorticoid- and jun/fos-dependent repression of transcription from a minimal promoter (2). The glucocorticoid-independent activity of the cGRE is detected both within the context of the PLF promoter and in constructs in which the cGRE and the PLF serum response region are transferred upstream of a minimal promoter. We therefore now refer to this element as a cGRE/negative element.
The mechanism of action of the cGRE in response to glucocorticoids appears to involve binding of both the glucocorticoid receptor and AP-1 (2,8). The ability of the cGRE/negative element to behave as a glucocorticoid-independent repression element suggests that it can also be recognized by a factor in addition to the glucocorticoid receptor. Indeed, binding studies reveal that a protein present in mouse L fibroblasts specifically recognizes this element, and that this factor is neither the glucocorticoid receptor nor a form of AP-1. Other studies that have suggested that the function of the PLF cGRE may depend on factors in addition to AP-1 and the glucocorticoid receptor (23,26).
Interaction of the cGRE/negative element with the factor detected in fibroblast extracts may obscure the adjacent serum response region for protein binding, or might form nonproductive protein-protein interactions with the factors bound to the serum response region. Both of these models are consistent with the finding that the negative element acts in a position-dependent manner. The loss of activity that results from moving this element 5 bp further away from the serum response region suggests that negative regulation is more likely to result from the action of this regulatory factor on the adjacent serum response region than on the more distant basal transcription complex at the TATA box and transcription start site.
This study has also revealed the presence of another positive regulatory element located between residues – 268 and – 252 in the PLF gene promoter. The region of the PLF promoter between – 268 and – 204 thus contains at least four elements (this positive element, the cGRE/negative element, and the AP-1 and Sph I components of the serum response region) that contribute to the response of the promoter to serum (Fig. 6). In addition to its ability to act as a positive element when linked to a minimal promoter, the sequence from – 268 to – 252 can interfere with repression by the cGRE/negative element under high serum conditions. One possible mechanism for this action of the – 268/– 252 positive element is that factors bound to this site and to the cGRE/negative element might interact, perhaps providing a “fine-tuning” mechanism for regulating the activity of the adjacent serum response region. Thus, growth-regulated PLF gene transcription may be controlled by modulating the relative activities of at least four distinct factors.
FIG. 6.
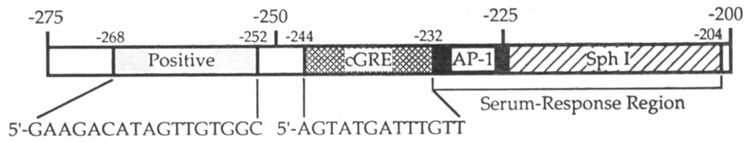
Schematic diagram of the PLF gene transcriptional regulatory region. Four elements have been defined in the region spanning from – 204 to – 268, including two elements (AP-I and Sph I-like) that constitute the serum response region (6,20), the cGRE/negative element (shown with the boundaries and corresponding sequence sufficient for the glucocorticoid-independent negative regulatory activity determined in this report), and the upstream positive element.
ACKNOWLEDGEMENTS
We thank Keith Yamamoto for tyrosine amino-transferase GRE and plfG plasmid DNAs, Bruce Spiegelman for antiserum against c-fos, Robert Harrison for an antibody against the glucocorticoid receptor, and Jack Groskopf and Barbara Wu for many helpful discussions. This work was supported by grant HD29962 from the National Institutes of Health. J.C.M. was supported by a predoctoral training grant from the National Institutes of Health.
REFERENCES
- 1. Bradford M. M. Anal. Biochem. 72:248–254; 1976. [DOI] [PubMed] [Google Scholar]
- 2. Diamond M. I.; Miner J. N.; Yoshinaga S. K.; Yamamoto K. R. Science 249:1266–1272; 1990. [DOI] [PubMed] [Google Scholar]
- 3. Fienup V. K.; Jeng M. H.; Hamilton R. T.; Nilsen-Hamilton M. J. Cell. Physiol. 129:151–158; 1986. [DOI] [PubMed] [Google Scholar]
- 4. Gametchu B.; Harrison R. W. Endocrinology 114:274–279; 1984. [DOI] [PubMed] [Google Scholar]
- 5. Gorman C. M.; Moffat L. F.; Howard B. H. Mol. Cell. Biol. 2:1044–1051; 1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Groskopf J. C.; Linzer D. I. H. Mol. Cell. Biol. 14:6013–6020; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jackson D.; Volpert O.; Bouck N.; Linzer D. I. H. Science 266:1581–1584; 1994. [DOI] [PubMed] [Google Scholar]
- 8. Kerppola T. K.; Luk D.; Curran T. Mol. Cell. Biol. 13:3782–3791; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee S.-J.; Nathans D. Endocrinology 120:208–213; 1987. [DOI] [PubMed] [Google Scholar]
- 10. Lee S.-J.; Talamantes F.; Wilder E.; Linzer D. I. H.; Nathans D. Endocrinology 122:1761–1768; 1988. [DOI] [PubMed] [Google Scholar]
- 11. Linzer D. I. H.; Nathans D. Proc. Natl. Acad. Sci. USA 80:4271–4275; 1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Linzer D. I. H.; Nathans D. Proc. Natl. Acad. Sci. USA 81:4255–4259; 1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Linzer D. I. H.; Mordacq J. C. EMBO J. 6:2281–22881 1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Linzer D. I. H.; Lee S.-J.; Ogren L.; Talamantes F.; Nathans D. Proc. Natl. Acad. Sci. USA 82:4356–4359; 1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lopata M. A.; Cleveland D. W.; Sollner-Webb B. Nucleic Acids Res. 12:5707–5717; 1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marais R.; Wynne J.; Treisman R. Cell 73:381–393; 1993. [DOI] [PubMed] [Google Scholar]
- 17. McKnight S. L.; Kingsbury R. Science 217:316–324; 1982. [DOI] [PubMed] [Google Scholar]
- 18. Mittal R.; Kumar K. U.; Pater A.; Pater M. M. Mol. Endocrinol. 8:1701–1708; 1994. [DOI] [PubMed] [Google Scholar]
- 19. Miyashita A.; Crystal R. G.; Hay J. G. Nucleic Acids Res. 23:293–3011 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mordacq J. C.; Linzer D. I. H. Genes Dev. 3:760–769; 1989. [DOI] [PubMed] [Google Scholar]
- 21. Nelson J. T.; Rosenzweig N.; Nilsen-Hamilton M. Endocrinology 136:283–288; 1995. [DOI] [PubMed] [Google Scholar]
- 22. Nilsen-Hamilton M.; Shapiro J. M.; Massoglia S. L.; Hamilton R. T. Cell 20:19–28; 1980. [DOI] [PubMed] [Google Scholar]
- 23. Pearce D.; Yamamoto K. R. Science 259:1161–1165; 1993. [DOI] [PubMed] [Google Scholar]
- 24. Shida M. M.; Ng Y.-K.; Soares M. J.; Linzer D. I. H. Mol. Endocrinol. 7:181–188; 1993. [DOI] [PubMed] [Google Scholar]
- 25. Wilder E. L.; Linzer D. I. H. Mol. Cell. Biol. 9:430–441; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yoshinaga S. K.; Yamamoto K. R. Mol. Endocrinol. 5:844–853; 1991. [DOI] [PubMed] [Google Scholar]


