Abstract
Analysis of the promoters of the bovine and human nuclear-encoded mitochondrial F0F1 ATP synthase α-subunit genes (ATPA) has identified several positive cis-acting regulatory regions that are important for basal promoter activity in human HeLa cells. We have previously determined that the binding of a protein factor, termed ATPF1, to an E-box sequence (CANNTG) located within one of these cis-acting regions is critical for transcriptional activation of the ATPA gene. In this article, we describe a second positive cis-acting regulatory element of the ATPA gene that is important for expression of the ATPA gene. We show that this cis-acting element also contains a binding site for a protein present in HeLa cells. On the basis of electrophoretic mobility shift patterns, oligonucleotide competition assays, and immunological cross-reactivity, we conclude that this protein factor is Yin-Yang 1 (YY1). Experiments carried out to examine the functional role of YY1 within the context of the A TP A promoter demonstrated that YY 1 acts as a positive regulator of the ATPA gene. For example, when the YY1 binding site of the ATPA promoter was placed upstream of a reporter gene it was found to activate transcription in transient transfection assays. In addition, disruption of the YY1 binding site in the ATPA gene resulted in a loss of transcriptional activity. Furthermore, in cotransfection experiments overexpression of YY1 in trans was found to activate transcription of ATPA promoter-CAT constructs. Thus, at least two positive trans-acting regulatory factors, ATPF1 and YY1, are important for expression of the bovine and human F0F1 ATP synthase α-subunit genes.
Keywords: Transcription factor, YY1, F0F1 ATP synthase, Mitochondria
MITOCHONDRIA provide most of the ATP for eukaryotic cells through the process of oxidative phosphorylation. The F0F1 ATP synthase complex is the central enzyme of the oxidative phosphorylation system synthesizing ATP from ADP and Pi utilizing energy stored by the electron transport chain. Biosynthesis of the mitochondrial ATP synthase complex requires the expression of genes encoded by two distinct genetic systems. For example, in mammalian cells two of the subunits of the ATP synthase complex are encoded by the mitochondrial DNA, whereas the remaining subunits are encoded by nuclear genes and imported posttranslationally into mitochondria [for reviews, see 3,41)].
The activities of the enzymes of the mitochondrial oxidative phosphorylation system vary greatly in response to a number of physiological conditions, including cell proliferation, development, differentiation, and hormonal stimulation [for reviews, see 3,20,28,41)]. A regulatory system must exist to coordinate the expression of the genes that encode proteins of the oxidative phosphorylation system to meet the varied energy needs of cells. The levels of the enzymes of the mitochondrial oxidative phosphorylation system are regulated, to a large extent, through transcriptional control, although regulation at a posttranscriptional level also plays an important role in the overall coordination of mitochondrial biogenesis (3,20,28).
Our laboratory has been analyzing the regulation of the expression of the nuclear gene (ATPA) that encodes the α-subunit of the bovine and human mitochondrial F0F1 ATP synthase complexes (7,19,31,43). By using a deletion and mutational analysis, we have identified several positive cis-acting regions of this gene that are important for expression in human HeLa cells (43). We have previously determined that an E-box element (CANNTG) located within one of these cis-acting regions plays a critical role in controlling the level of basal transcription. We have also demonstrated that a trans-acting factor present in HeLa cells, termed ATPF1, binds to this E-box element (43).
In this article, we report on a second positive cis-acting regulatory element that is required for basal expression of the ATPA gene. We demonstrate that a protein(s) present in HeLa nuclei binds to this cis-acting element. Based on electrophoretic mobility shift patterns, competition assays and reaction with antisera, we conclude that this protein is identical or closely related to the regulatory factor, Yin-Yang 1 (YY1) [for review, see (16)]. YY1 (also termed NF-E1, δ, CF1, FACT1, and UCRBP) is a zinc finger containing DNA binding protein belonging to the GLI-Kruppel family. YY1 has been found to play an important role in the expression of many cellular and viral genes. Interestingly, depending on the promoter context and the intracellular environment, YY1 can function as either an activator, a repressor, or an initiator of transcription (16). We demonstrate that YY1 acts as a positive regulator of the bovine and human ATPA genes.
The ATP synthase α-subunit gene is now the second nuclear gene encoding a protein of the mitochondrial oxidative phosphorylation system that has been shown to be regulated by YY1. Previously, Avadhani and coworkers had demonstrated that the mouse cytochrome c oxidase subunit Vb gene is positively regulated by YY1 (5). One mechanism of coordinating the expression of a large number of genes encoding proteins of the oxidative phosphorylation system is through common cis- and trans-acting regulatory factors. YY1 might represent such a factor.
MATERIALS AND METHODS
Transfection and Enzyme Assays
HeLa cells were grown on 10-cm dishes in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 5% calf serum. Transfections were carried out as described previously using the calcium phosphate coprecipitation method (43). Cells were typically transfected with 10–20 μg/dish of the CAT reporter plasmid DNA together with 2 μg/dish of the β-galactosidase expression plasmid, pCMV-β-gal. Approximately 48 h after transfection, cells were harvested and extracts were prepared. Chloramphenicol acetyltransferase (CAT) and β-galactosidase assays were carried out as described previously (43). The CAT activities of the extracts were normalized relative to the β-galactosidase activities. Promoter activity values represent the average of at least three separate transfections of three plates each. In some experiments, RNA was isolated from transfected cells as described (4) and the sites of transcription initiation were determined by primer extension analysis (4,31) using a radiolabeled CAT primer corresponding to nucleotides +2329 to +2348 of the pCAT-Basic vector (Promega). Primer extension products were separated on 8% polyacrylamide-urea sequencing gels and analyzed using a Phophorimager.
Electrophoretic Mobility Shift Assays
HeLa nuclear extracts were prepared using the procedure described by Dignam et al. (10). Electrophoretic mobility shift DNA-protein binding assays were performed as described previously (4,43), with modifications. Briefly, a 20-μl reaction mixture containing approximately 5 μg of nuclear extract protein and 0.25 μg of double-stranded poly(dI-dC) (Pharmacia) was incubated for 15 min at room temperature in binding buffer containing 10 niM Tris, pH 7.5, 60 mM KCl, 1 mM DTT, 1 mM EDTA, and 8% glycerol. After the addition of 32P-labeled probe, reaction mixtures were incubated for another 15 min. DNA-protein complexes were resolved on 4% polyacrylamide gels using 0.5 × TRIS-borate-EDTA as the running buffer. For competition experiments, a 100-fold molar excess of unlabeled competitor DNA was pre-incubated with nuclear extracts for 15 min prior to probe addition. To test the effect of various anti-sera, preimmune or immune serum was preincubated with nuclear extracts for 20 min at room temperature prior to the addition of probe.
Oligonucleotides and Antibodies
The following oligonucleotides and their complements (listed 5′-3′) were used in electrophoretic mobility shift assays:
NF-E1 consensus oligonucleotide: CGCTCCGCGGCCATCTTGGCGGCTGGT [(29); Santa Cruz Biotechnology Inc.);
NF-E1 mutant oligonucleotide: CGCTCCGCGATTATCTTGGCGGCTGGT (Santa Cruz Biotechnology Inc.);
adeno-associated virus P5 + 1 YY1 binding sequence: AGGGTCTCCATTTTGAAGCGGG (37,39);
rpL30 δ-binding sequence: CTCGCTCCCCGGCCATCTTGGCGGCTGGTGTTGG (2,17);
wild-type ATPA (+ 68 to +86 bp) oligonucleotide: CTCGGCCATTTTGTCCCAG (31);
ATPA mutant 1 (+ 68 to +86 bp) oligonucleotide: CTCAATTATTTTGTCCCAG;
ATPA mutant 2 (+ 68 to +86 bp) oligonucleotide: CTCGGCCAGGAAGTCCCAG.
Rabbit polyclonal antibodies against human YY1 were obtained from Santa Cruz Biotechnology Inc. (Santa Cruz, CA).
RESULTS
Nuclear Factor YY1 Binds to the ATPA Promoter
Our previous analysis of the bovine ATPA gene promoter revealed that the +25 to +136 bp region contains sequences essential for basal promoter activity in human HeLa cells (43). By using a 5′ deletion analysis, we have identified several positive cis-acting regions of the ATPA promoter that are important for basal promoter function. For example, deletion of the ATPA gene from +25 to +49 bp reduced basal promoter activity to levels approximately 4% of wild-type levels, whereas deletion to +72 bp further reduced promoter activity to levels approximately 0.1% of wild-type (43). We have also previously determined that the binding of a trans-acting regulatory factor, termed ATPF1, to an E-box element (CANNTG) located within the +25 to +48 bp region of the ATPA gene is critical for expression of this gene (43) (Fig. 1).
FIG. 1.
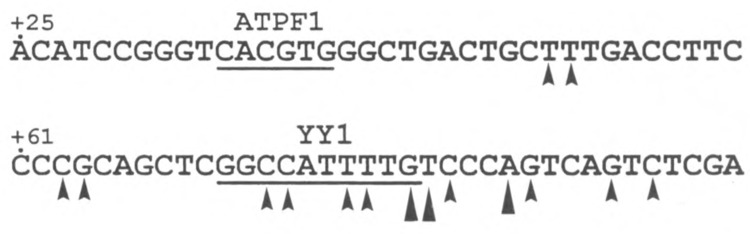
Sequence of a region of the bovine ATPA promoter. The nucleotide sequence of the +25 to +96 bp region of the bovine ATPA gene (31) is shown. The putative binding sites for the regulatory factors, ATPF1 (43) and YY1 (this article) are underlined. The sites of transcription initiation that map within this region of the bovine ATPA gene are indicated by arrowheads [(31); D. J. Pierce, unpublished observations). The sites of initiation that are located at identical positions in the bovine and human (1) ATPA genes are indicated by large arrowheads.
We examined the cis-acting sequences of the ATPA promoter that are located downstream of the +25 to +48 bp region. Experiments were first carried out to identify DNA-protein interactions using electrophoretic mobility shift assays. Initial studies were carried out using the +49 to +96 bp fragment of the ATPA gene as a probe. The results of these experiments revealed that HeLa nuclear protein(s) can bind to this region of the ATPA promoter resulting in a major shifted DNA-protein complex and several minor faster migrating species (Fig. 2, lane 2). Essentially all of the ATPA–HeLa complexes were competed by an excess of unlabeled ATPA fragment (Fig. 2, lane 3) but not by a nonspecific competitor DNA (lane 5), indicating the specificity of the HeLa-ATPA complexes. Similar DNA-protein complexes were formed when a smaller fragment (+ 68 to + 86 bp) of the ATPA promoter was used as a probe (Fig. 2, lane 7). The +68 to +86 bp region of the ATPA gene was also found to effectively compete for essentially all of the HeLa-ATPA complexes formed using the larger ATPA fragment (Fig. 2, lane 4).
FIG. 2.
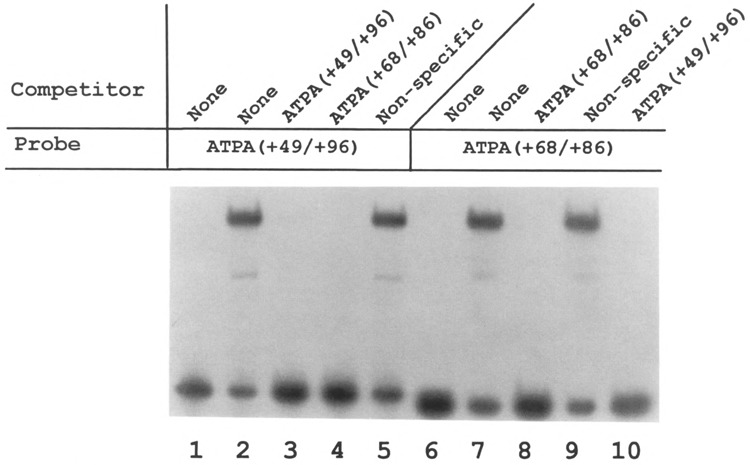
Binding of a HeLa nuclear factor(s) to the ATPA promoter. Electrophoretic mobility shift DNA-protein binding assays were carried out using 32P-labeled probes containing fragments of the bovine ATPA promoter (31,43) from +49 to +96 bp (lanes 1–5) or from +68 to +86 bp (lanes 6–10), together with HeLa cell nuclear extracts. Where indicated, a 100-fold molar excess of unlabeled DNA was added as a competitor. The nonspecific competitor DNA used in this experiment was sheared E. coli DNA. Lanes 1 and 6 are control reactions to which no HeLa nuclear extract was added.
This region of the ATPA promoter contains a sequence that resembles a sequence found in the promoter or enhancer regions of a number of cellular and viral genes, including the adeno-associated virus P5 (37), the immunoglobulin heavy and kappa light chains (29), ribosomal proteins, L30 and L32 (17), α-actin (32), c-fos (14), c-myc (32,33), ϵ- and γ-globins (15,30), cytochrome c oxidase subunit Vb (5), β-casein (26), serum amyloid A (24), murine leukemia virus (12), human papilloma virus-18 (6), murine intracisternal A-particles (35), human cytomegalovirus (23), and the human immunodeficiency virus (25) (Table 1). The sequence present in each of these genes has been shown to bind a nuclear factor termed YY1 [also called NF-E1, δ, CF1, F-ACT1, and UCRBP; for review, see (16)].
TABLE 1.
SEQUENCE COMPARISON OF A REGION OF THE ATPA PROMOTER WITH KNOWN YYI BINDING SITES
| Organism | Promoter/Enhancer | Binding Sequence |
|---|---|---|
| Bovine/human | ATPA | GGCCATTTTG |
| AAV | P5 (+l) | CTCCATTTTG |
| AAV | P5 (−60) | CGACATTTTG |
| Mouse | IgH (μE1) | GGCCATCTTG |
| Human | Igκ (E3′) | CTCCATCTTG |
| Mouse | rpL30 | GGCCATCTTG |
| Mouse | rpL32 | TGCCATCTGT |
| Mouse/chicken | α-actin | CGCCATATTT |
| Mouse | c-myc | GACCATTTTC |
| Human | c-fos | GTCCATATTA |
| Human | ε-globin | TATCATTTTG |
| Mouse | COXVb | GCCCATCTTG |
| Mouse | β-casein | AACCATTTTC |
| Rat | SAA1 | CACCATGTCA |
| MuLV | UCR | CGCCATTTTG |
| IAP | IUE | CGCCATCTTG |
| CMV | IE | GGCCATTTTA |
| HPV-18 | URR | TTTCATTAAT |
| HIV-1 | LTR | ACCCAGTACA |
Abbreviations used: AAV, adeno-associated virus; IgH (μE1), immunoglobulin heavy chain μE1 site; Igκ (E3′), immunoglobulin kappa 3′-enhancer; MuLV, murine leukemia virus; IAP, murine intracisternal A-particle; CMV, human cytomegalovirus; HPV-18, human papilloma virus type 18; HIV-1, human immunodeficiency virus type 1.
The relationship between the factor(s) binding to the ATPA promoter element and YY1 was assessed using several approaches. First, competition binding assays were carried out using oligonucleotides containing known YY1 binding sites. For these experiments, a labeled fragment containing the ATPA promoter sequence was used as a probe in electrophoretic mobility shift assays, together with an excess of oligonucleotides containing authentic YY1 binding sites as competitors. The results of these experiments indicate that oligonucleotides containing known YY1 binding sites (including P5 + 1, rpL30, and NF-E1) effectively compete for the binding of HeLa protein(s) to the ATPA promoter fragment (Fig. 3, lanes 4–6). In contrast, an oligonucleotide containing a mutant YYI (NF-E1) binding site did not compete for binding (Fig. 3, lane 7).
FIG. 3.
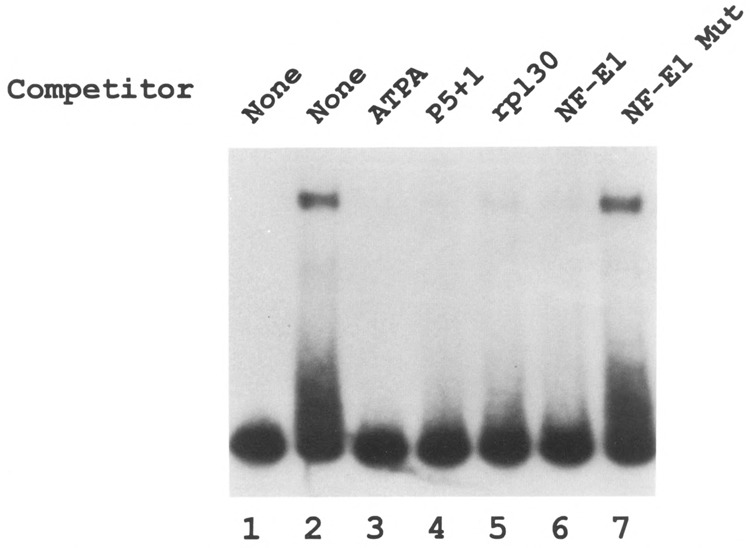
Oligonucleotides containing YY1 binding sites effectively compete for ATPA-HeLa. complexes. Electrophoretic mobility shift assays were carried out using a 32P-labeled fragment (+49 to +96 bp) of the ATPA gene as a probe, together with HeLa nuclear extracts. For competition assays, a 100-fold molar excess of various oligonucleotides was added as indicated: ATPA (+49 to +96 bp) (lane 3), P5 +1 (lane 4), rpL30 (lane 5), NF-E1 (lane 6), and mutant NF-E1 (lane 7). The sequences of the competitor oligonucleotides are listed in the Materials and Methods section. Lane 1 indicates labeled probe to which no HeLa cell extract was added.
To further confirm that YY1 binds to this region of the ATPA gene, electrophoretic mobility shift assays were carried out using purified YY1. The results of these experiments revealed that purified YY1 protein forms a complex with the ATPA promoter fragment that is similar to that formed with HeLa nuclear extracts (Fig. 4, lane 2). Next, specific antibodies against YY1 were used in electrophoretic mobility shift assays to confirm the binding of YY1 to the ATPA promoter. The results shown in Fig. 4 indicate that whereas preimmune serum had no effect on DNA-protein complex formation (lane 3), anti-YY1 antibodies specifically recognized the ATPA-HeLa complexes resulting in a super-shifted complex (lane 4). Furthermore, this super-shifted complex was effectively competed by an oligonucleotide containing an authentic YY1 binding site (Fig. 4, lane 5). These data reveal that a protein present in the ATPA-HeLa complexes represents YY1.
FIG. 4.
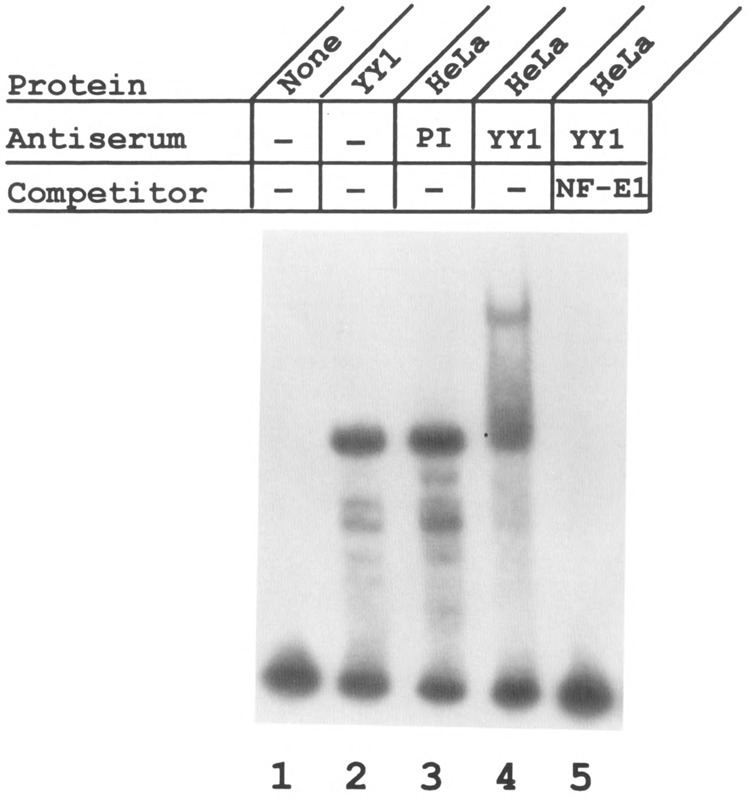
YY1 is present in HeLa-ATPA complexes. A fragment containing the +49 to +96 bp region of the ATPA gene was end-labeled and used as a probe in electrophoretic mobility shift assays. The binding reactions contained either no extract (lane 1), purified YY1 (Upstate Biotechnology) (lane 2), or HeLa nuclear extracts (lanes 3–5). In some reactions, preimmune serum (PI; 2 μl) (lane 3) or anti-YY1 antiserum (2 μl) (lanes 4 and 5) was added. In lane 5, a 100-fold molar excess of the NF-E1 oligonucleotide was added as a competitor.
YY1 Activates the ATPA Promoter
Experiments carried out in a number of laboratories have demonstrated that YY1 can act as either an activator or a repressor protein depending on promoter context and intracellular environment [reviewed by Hahn (16)]. To examine the functional role of YY1 within the context of the ATPA promoter, we cloned a 19-bp fragment of the ATPA gene (+68 to +86 bp) containing the YY1 binding site upstream of a reporter CAT gene. This construct was transfected into HeLa cells and the levels of CAT enzyme were determined. The results of these experiments demonstrated that this 19-bp fragment can activate transcription (Fig. 5). The levels of activity obtained with this fragment were approximately 11% of the levels achieved when using the basal ATPA (+25 to +136 bp) promoter (Fig. 5). As a control in these experiments, we cloned a fragment of the adeno-associated viral P5 promoter (P5 +1) (37) containing a YY1 element upstream of the reporter CAT gene and transfected this construct into HeLa cells. The YY1 element of the P5 promoter was found to repress transcription when tested in transient transfection assays in HeLa cells (Fig. 5), in agreement with previously published results (37). Thus, in our experimental system, activation is specific for the ATPA promoter. To verify that transcription initiates at authentic sites within the ATPA gene when the ATPA (+68 to +86 bp) fragment is transfected, primer extension experiments were performed using a primer complementary to the CAT gene. The results of these experiments indicated that initiation occurs at sites within the 19-bp fragment that correspond to start sites previously identified in the intact ATPA gene (1,31) (Fig. 6).
FIG. 5.
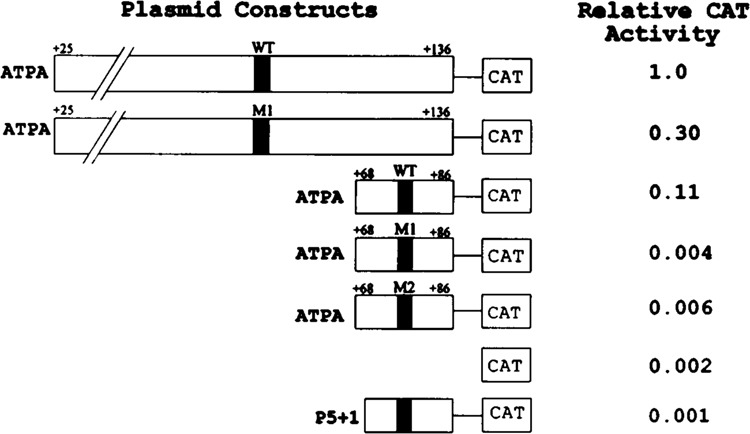
The YY1 binding site in the ATPA gene acts as an activator of transcription. The +25 to +136 bp or the +68 to +86 bp region of the ATPA gene was cloned into the vector, pCAT-Basic (Promega). Similarly, the same fragments of the ATPA gene containing mutations (M1, M2) in the YY1 element were cloned into the pCAT-Basic vector. In addition, the P5 (+1) region of the adeno-associated virus P5 (37) was cloned into pCAT-Basic. Wild-type (WT) YY1 elements are indicated by closed boxes and mutated YY1 elements by hatched boxes. HeLa cells were transfected with each of the indicated CAT reporter plasmids, together with pCMV-β-gal plasmid DNA. Cells were harvested after 48 h and assayed for CAT and β-galactosidase activities as described in the Materials and Methods section. The CAT activities of the transfected cells were normalized relative to the β-galactosidase activities.
FIG. 6.
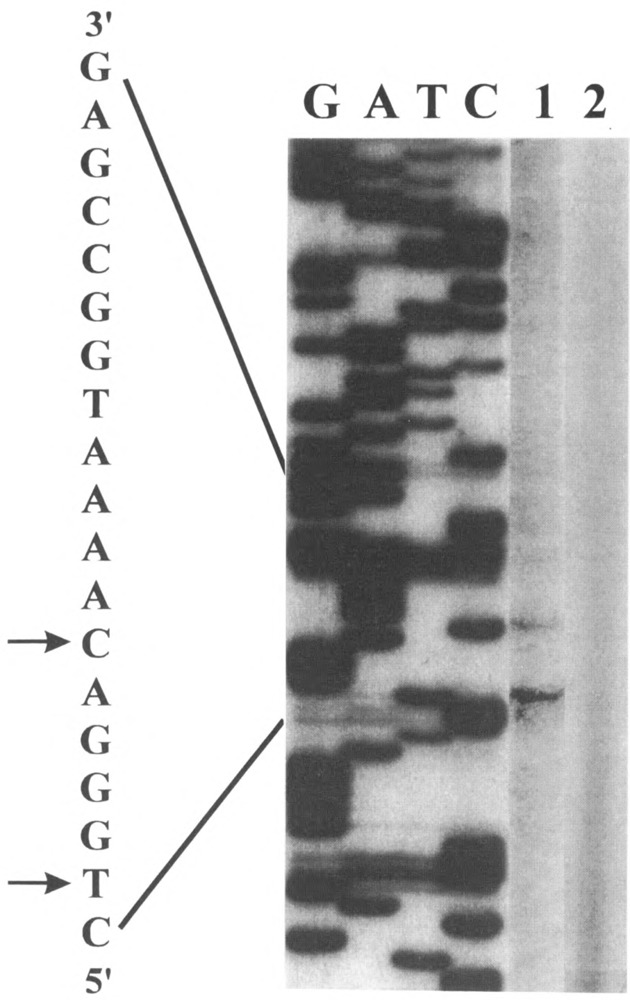
Sites of transcription initiation of DNA constructs transfected into HeLa cells. HeLa cells were transfected with either the ATPA (+68 to +86 bp) fragment cloned into the pCAT-Basic vector or the pCAT-Basic vector DNA alone. Total RNA was isolated from cells approximately 48 h after transfection. A 5′ end-labeled CAT primer DNA was annealed to approximately 50 μg of RNA from the transfected cells and extended with Moloney murine leukemia virus reverse transcriptase. Lane 1 represents primer extension products from cells transfected with the ATPA (+68/+86 bp)–CAT DNA and lane 2 with the pCAT-Basic DNA. The sequencing ladder (labeled G, A, T, C) was generated by using the same CAT primer and the ATPA (+68/+86 bp)-CAT plasmid DNA as the template. The transcription start sites are indicated by arrowheads on the antisense sequence.
Next, ATPA promoter constructs containing site-specific mutations at the YY1 binding site were examined. First, the effect of these mutations on YY1 binding was analyzed using the wild-type and mutant ATPA fragments as probes in electrophoretic mobility shift assays. The results of these experiments demonstrated that both of the mutant ATPA fragments had completely lost YY1 binding activity when compared with the wild-type fragment (Fig. 7, lanes 7 and 9). In addition, neither of the mutant fragments could compete for binding to the wild-type ATPA fragment (Fig. 7, lanes 4 and 5). Next, the effect of these mutations on ATPA promoter activity was tested in transient transfection assays. The results of these experiments revealed that both mutant 1 and mutant 2 ATPA–CAT constructs showed marked reduction in promoter activity whether examined within the context of the basal ATPA promoter or the truncated 19-bp fragment (Fig. 5). Thus, mutations that prevent YY1 binding to the ATPA promoter result in a loss of transcriptional activation. The combined results of these experiments indicate that YY1 is a positive regulator of ATPA transcription.
FIG. 7.
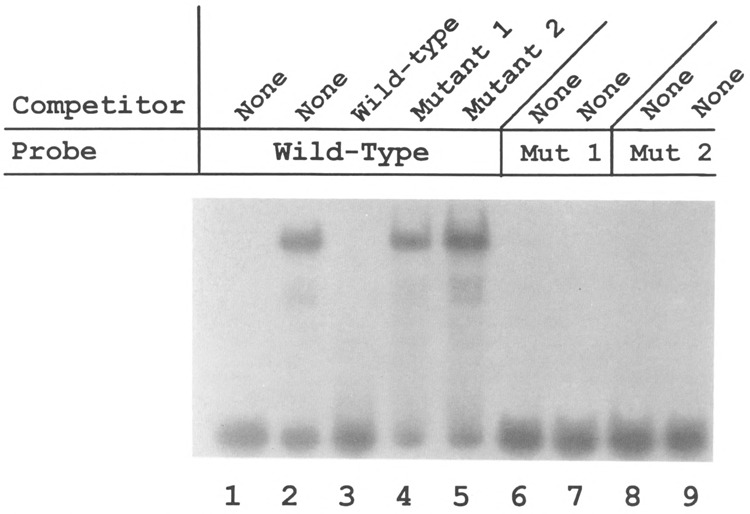
Mutant forms of the ATPA promoter do not bind YY1. Electrophoretic mobility shift DNA-protein binding assays were carried out using labeled oligonucleotides containing either the wild-type ATPA (+68 to +86 bp) sequence (lanes 1–5), the mutant 1 ATPA (+68 to +86 bp) sequence (lanes 6 and 7), or the mutant 2 ATPA (+68 to +86 bp) sequence (lanes 8 and 9). Where indicated, a 100-fold molar excess of either wild-type, mutant 1, or mutant 2 ATPA oligonucleotide was added as a competitor. Lanes 1, 6, and 8 represent control reactions to which no HeLa nuclear extract was added. Wild-type ATPA (+68 to +86 bp) sequence: CTCGGCCATTTTGTCCCAG; mutant 1 ATPA sequence: CTCAATTATTTTGTCCCAG; mutant 2 ATPA sequence: CTCGGCCAGGAAGTCCAG.
YY1 Is a Trans-Activator of the ATPA Promoter
To further confirm that YY1 exerts a positive effect on ATPA gene expression, we tested whether overexpression of YY1 in trans could activate ATPA–reporter constructs in cotransfection assays. For these experiments, a plasmid expressing YY1 under the control of the cytomegalovirus (CMV) promoter (pCMV-YY1) was cotransfected into HeLa cells, together with a reporter plasmid containing the CAT gene driven by the ATPA promoter. The CMV vector alone was used as a control in parallel transfections. The results of these experiments revealed that when pCMV-YY1 was cotransfected together with the ATPA (+25/+136 bp)–CAT construct, expression of the CAT gene from the ATPA promoter was increased significantly (Fig. 8). In contrast, no trans-activation was observed when the pCMV vector alone was transfected. As an additional control, we determined that expression of YY1 in trans did not affect transcription of a CAT reporter containing the SV40 promoter and enhancer driving expression of the CAT gene (pSVCAT) (Fig. 8).
FIG. 8.
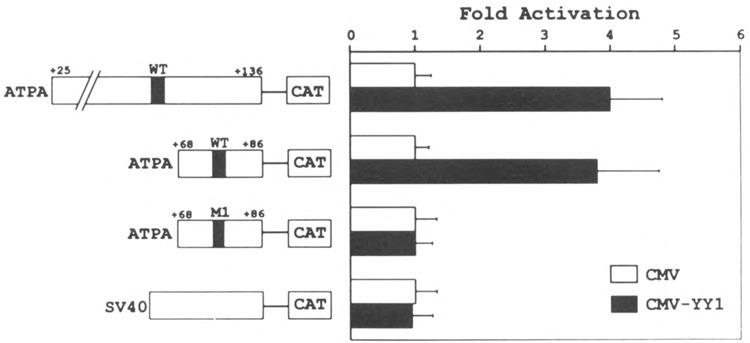
YY1 transactivates ATPA–CAT reporters. HeLa cells were cotransfected with either 10 μg of pATPA wild-type (+25/ +136 bp)/CAT, pATPA wild-type (+68/+86 bp)/CAT, pATPA mutant 1 (+68/+86 bp)/CAT, or pSVCAT plasmid DNAs, together with 10 μg of either pCMV (open bars) or pCMV-YY1 (closed bars), and 2 μg of pCMV-β-gal DNA. Cells were harvested after 48 h and assayed for β-galactosidase and CAT activities as described in the Materials and Methods section. The CAT activities of the cells that were transfected with pCMV-YY1 DNA were compared to those with pCMV DNA (fold activation).
To determine whether the cotransfected YY1 activated the ATPA promoter construct through its binding site, mutated and truncated ATPA–CAT constructs were also tested in cotransfection assays. The truncated 19-bp ATPA (+68 to +86 bp) construct contains only the YY1 binding site whereas the mutated ATPA construct disrupts the binding of YY1 (Fig. 7). The results of these experiments revealed that the truncated ATPA (+68/+86 bp)–CAT construct was also trans-activated by coexpression of the YY1 plasmid (Fig. 8). In contrast, there was little or no trans-activation of the mutant 1 ATPA (+68/+86 bp)–CAT construct by YY1 (Fig. 8). The results of these cotransfection experiments demonstrate that recombinant YY1 can trans-activate the ATPA gene and that this trans-activation is dependent on the YY1 binding site in the ATPA promoter.
DISCUSSION
In this study, we demonstrate that the nuclear factor YY1 [also termed NF-E1, δ, CF1, UCRBP, and F-ACT1 (16)] plays an important activator role in the transcriptional regulation of the bovine and human F0F1, ATP synthase α-subunit genes. Evidence for this includes: 1) competition with oligonucleotides containing known YY1 binding sites and the use of purified YY1 and polyclonal antibodies revealed that YY1 was a component of complexes formed with HeLa extracts and the ATPA gene promoter; 2) a synthetic oligomer containing the YY1 sequence of the ATPA promoter was found to activate expression of a heterologous CAT gene; 3) site-directed mutagenesis of the YY1 binding region in the ATPA gene led to a decrease in the transcriptional activity of reporter constructs; and 4) overexpression of YY1 in trans-activated ATPA–CAT constructs in transient cotransfection assays.
Transcription factor YY1, a zinc finger-containing protein, binds to cis-acting elements in a number of cellular and viral genes [for review, see (16)]. YY1 is a multifunctional regulatory factor. Depending on the promoter context and the intracellular environment, YY1 can act as either an activator, a repressor, or an initiator of transcription. For example, YY1 represses the adeno-associated virus P5 promoter (37,39), the immunoglobulin kappa 3′ enhancer (29), the human papilloma virus-18 promoter (6), the skeletal α-actin promoter (21,32), the c-fos promoter (14), the β-casein promoter (26), the serum amyloid A promoter (24), the γ- and ϵ-globin genes (30), the Moloney murine leukemia virus promoter (12), and the human immunodeficiency virus promoter (6). In contrast, YY1 is an activator for the c-myc promoter (33), the immunoglobulin heavy chain intronic enhancer (29), the murine intracisternal A-particle upstream enhancer (35), the promoters of the ribosomal proteins, L30 and L32 (2,9,17), the promoter of the mouse cytochrome c oxidase subunit Vb gene (5), the transcriptional regulatory region of LINE-1 (38), the B19 parvovirus P6 promoter (27), and the Surf-1/Surf-2 promoter (13). In this study, we demonstrate that YY1 acts as an activator of the bovine and human ATPA genes.
The mammalian ATPA gene is constitutively expressed in most (or all) tissues. Like many other housekeeping genes, the promoter of the ATPA gene lacks a canonical TATA box and has multiple sites of transcription initiation (1,31) (see Fig. 1). Another feature of many ubiquitously expressed genes is the presence of a pyrimidine-rich sequence beginning with a CA at or around the cap site or in the 5′-untranslated region, which appears to play a role in transcription initiation. The pyrimidine-rich sequence in several of these genes has been shown to bind the regulatory factor, YY1 (2,5,17). The YY1 element in the ATPA gene is located within a cluster of initiation sites and is just upstream of two major transcription start sites that map at identical positions in both the bovine and human genes (see Fig. 1). A direct role for YY1 in transcription initiation has been shown for the adeno-associated virus P5 (37,42) and the mouse cytochrome c oxidase subunit Vb genes (5).
Although YY1 is present in many different tissues and can generally be categorized as a ubiquitous transcription factor, there are a number of ways that the effect of YY1 can be regulated. For example, the activity of YY1 can be modulated through interaction with other proteins. Several laboratories have demonstrated that the adenovirus E1A protein (39), the oncoprotein, c-Myc (40), and the nucleolar protein, B23 (18) can all reverse the transcriptional repression exerted by YY1. In addition, the amount of YY1 changes in certain cell types in response to differentiation, such as when cultured embryonic chicken myoblasts differentiate into myotubes (22). Furthermore, YY1 has been found to compete with certain activators (including SRF, NF-κB, GATA, and mammary gland factor) for the binding to overlapping binding sites [see (14,15,22,24,26,30)]. Repression by YY1 is relieved when these activators displace YY1.
The ATP synthase α-subunit gene is now the second nuclear gene encoding a subunit of mammalian mitochondrial oxidative phosphorylation system that has been shown to regulated by the transcription factor, YY1. Previously, it had been demonstrated that the mouse cytochrome c oxidase subunit Vb gene contains a YY1 binding site that functions as an initiator sequence binding protein that directs transcriptional activation (5). Biogenesis of the mitochondrial oxidative phosphorylation system requires the coordinated expression of a large number of genes encoded by both the nuclear and mitochondrial genomes. One mechanism of coordinating the expression of the genes encoding proteins of the oxidative phosphorylation system is through common cis- and trans-acting regulatory factors. YY1 now joins NRF-1 (11,45) and GABP (8,44) as regulatory factors that may be involved in coordinating the expression of oxidative phosphorylation genes to meet cellular energy requirements.
In summary, in this study we demonstrate that the multifunctional regulatory factor, YY1, plays a critical role in activation of the bovine and human F0F1 ATP synthase α-subunit genes. YY1 is the second positive trans-acting regulatory factor that has been shown to be required for expression of these genes. We have previously determined that the binding of a protein factor, termed ATPF1, to an E-box element (CANNTG) is essential for transcriptional activation of the bovine and human ATPA genes (43). Preliminary data indicate that there are synergistic interactions between these two activation elements (E. M. Jordan and G. A. M. Breen, unpublished observations).
ACKNOWLEDGEMENTS
We thank Dr. Thomas Shenk (Princeton University), Dr. Edward Seto (The University of Texas Health Science Center at San Antonio), Dr. Robert Perry (Fox Chase Cancer Center), and Dr. Michael Atchison (The University of Pennsylvania) for the generous gifts of plasmid constructs, oligonucleotides, and/ or antibodies. This work was supported by a Public Health Service Grant GM41738 from the National Institutes of Health and a Grant-in-Aid from the American Heart Association, Texas Affiliate to G.B.
REFERENCES
- 1. Akiyama S.; Endo H.; Inohara N.; Ohta S.; Kagawa Y. Biochim. Biophys. Acta 1219:129–140; 1994. [DOI] [PubMed] [Google Scholar]
- 2. Atchison M.; Meyuhas O.; Perry R. P. Mol. Cell Biol. 9:2067–2074; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Attardi G.; Schatz G. Annu. Rev. Cell Biol. 4:289–333; 1988. [DOI] [PubMed] [Google Scholar]
- 4. Ausubel F. M.; Brent R.; Kingston R. E.; Moore D. D.; Seidman J. G.; Smith J. A.; Struhl K., eds. Current protocols in molecular biology. New York: Wiley Interscience; 1994. [Google Scholar]
- 5. Basu A.; Park Y.; Atchison M. L.; Carter R. S.; Avadhani R. G. J. Biol. Chem. 268:4188–4196; 1993. [PubMed] [Google Scholar]
- 6. Bauknecht T.; Angel P.; Royer H.-D.; zur-Hausen H. EMBO J. 11:4607–4617; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Breen G. A. M. Biochem. Biophys. Res. Commun. 152:264–269; 1988. [DOI] [PubMed] [Google Scholar]
- 8. Carter R. S.; Bhat N. K.; Basu A.; Avadhani N. G. J. Biol. Chem. 267:23418–23426; 1992. [PubMed] [Google Scholar]
- 9. Chung S.; Perry R. P. (1993) Nucleic Acids Res. 21:3301–3308; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dignam J. D.; Lebovitz R. M.; Roeder R. G. Nucleic Acids Res. 11:1475–1489; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Evans M. J.; Scarpulla R. C. Genes Dev. 4:1023–1034; 1990. [DOI] [PubMed] [Google Scholar]
- 12. Flanagan J. R.; Becker K. G.; Ennist D. L.; Gleason S. L.; Driggers P. H.; Levi B.-Z.; Appela E.; Ozato K. Mol. Cell Biol. 12:38–44; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gaston K.; Fried M. FEBS Lett. 347:289–294; 1994. [DOI] [PubMed] [Google Scholar]
- 14. Gualberto A.; LePage D.; Pons G.; Mader S. L.; Park K.; Atchison M. L.; Walsh K. Mol. Cell Biol. 12:4209–4214; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gumucio D. L.; Heilstedt-Williamson H.; Gray T. A.; Tarle S. A.; Shelton D. A.; Tagle D. A.; Slightom J. L.; Goodman M.; Collins F. S. Mol Cell Biol. 12: 4919–4929; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hahn S. Curr. Biol. 2:152–155; 1992. [DOI] [PubMed] [Google Scholar]
- 17. Hariharan N.; Kelley D. E.; Perry R. P. Proc. Natl. Acad. Sci. 88:9799–9803; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Inouye C. J.; Seto E. J. Biol. Chem. 269:6506–6510; 1994. [PubMed] [Google Scholar]
- 19. Jordan E. M.; Breen G. A. M. Biochim. Biophys. Acta 1173:115–117; 1993. [DOI] [PubMed] [Google Scholar]
- 20. Kagawa Y.; Ohta S. Int. J. Biochem. 22:219–229; 1990. [DOI] [PubMed] [Google Scholar]
- 21. Lee J.-S.; Galvin K. M.; Shi Y. Proc. Natl. Acad. Sci. USA 90:6145–6149; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee T.-C; Shi Y.; Schwartz R. J. Proc. Natl. Acad. Sci. USA 89:9814–9818; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu R.; Baillie J.; Sissons J. G. P.; Sinclair J. H. Nucleic Acids Res. 22:2453–2459; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lu S.-Y.; Rodriguez M.; Liao W. S.-L. Mol. Cell Biol. 14:6254–6263; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Margolis D. M.; Somasundaran M.; Green M. R. J. Virol. 68:905–910; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Meier V. S.; Broner G. Mol. Cell Biol. 14:128–137; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Momoeda M.; Kawase M.; Jane S. M.; Muyamura K.; Young N. S.; Kajigaya S. J. Virology 68:7159–7168; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nagley P. Trends Genet. 7:1–4; 1991. [DOI] [PubMed] [Google Scholar]
- 29. Park K.; Atchison M. L. Proc. Natl. Acad. Sci. USA 88:9804–9808; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peters B.; Mereezhinskaya N.; Diffley J. F. X.; Noguchi C. T. J. Biol. Chem. 268:3430–3437; 1993. [PubMed] [Google Scholar]
- 31. Pierce D. J.; Jordan E. M.; Breen G. A. M. Biochim. Biophys. Acta 1132:265–275; 1992. [DOI] [PubMed] [Google Scholar]
- 32. Riggs K. J.; Merrell K. T.; Wilson G.; Calame K. Mol. Cell Biol. 11, 1765–1769; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Riggs K. J.; Salique S.; Wong K.-K.; Merrell K. T.; Lee J.-S.; Shi Y.; Calame K. Mol. Cell Biol. 13:7487–7495; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sambrook J.; Fritsch E.; Maniatis T. Molecular cloning. A Laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 35. Satyamoorthy K.; Park K.; Atchison M. L.; Howe C. C. Mol. Cell Biol. 13:6621–6628; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Seto E.; Lewis B.; Shenk T. Nature 365:462–464; 1993. [DOI] [PubMed] [Google Scholar]
- 37. Seto E.; Shi Y.; Shenk T. Nature 354:241–145; 1991. [DOI] [PubMed] [Google Scholar]
- 38. Singer M. F.; Krek V.; McMillan J. P.; Swergold G. D.; Thayer R. E. Gene 135:183–188; 1993. [DOI] [PubMed] [Google Scholar]
- 39. Shi Y.; Seto E.; Chang L. S.; Shenk T. Cell 67:377–388; 1991. [DOI] [PubMed] [Google Scholar]
- 40. Shrivastava A.; Saleque S.; Kalpana G. V.; Artandi S.; Goff S. P.; Calame K. Science 262:1889–1892; 1993. [DOI] [PubMed] [Google Scholar]
- 41. Tzagoloff A.; Myers A. M. Annu. Rev. Biochem. 55:249–285; 1986. [DOI] [PubMed] [Google Scholar]
- 42. Usheva A.; Shenk T. Cell 76:1115–1121; 1994. [DOI] [PubMed] [Google Scholar]
- 43. Vander Zee C. A.; Jordan E. M.; Breen G. A. M. J. Biol. Chem. 269:6972–6977; 1994. [PubMed] [Google Scholar]
- 44. Virbasius J. V.; Virbasius C.-m. A.; Scarpulla R. C. Genes Dev. 7:380–392; 1993. [DOI] [PubMed] [Google Scholar]
- 45. Virbasius C-m. A.; Virbasius J. V.; Scarpulla R. C. Genes Dev. 7:2431–2445; 1993. [DOI] [PubMed] [Google Scholar]


