Abstract
Expression of tissue-specific genes can be altered upon fusion of mammalian cells of different types. To resolve the genetic basis of this phenomenon and to identify components of the regulatory circuits that are involved, we have established a series of somatic cell hybrids between mouse T cells and L cells. These hybrids have an unusual and interesting phenotype. Unlike many hybrid cells studied, in which the expression of an entire set of tissue-specific genes was coordinately extinguished, in our T × L-cell hybrids only two out of seven T-cell-restricted genes were completely extinguished, whereas the other genes were repressed to various degrees. These hybrids extinguish the production of TCRβ and Thy-1 mRNA, repress the expression of TCRα, GATA-3, TCF-1, and LEF-1 genes to different extents, exhibit small changes in the level of CD3-ϵ mRNA, and continue to express the fibroblast-specific fibronectin gene, and the ets-1 gene. In this study we have evaluated for the first time the molecular mechanisms that underlie the repression of TCRα and TCRβ chain genes in T × L-cell hybrids. We have shown that multiple repression mechanisms, both direct and indirect, contribute to TCRα and TCRβ suppression. Repression of the expression of these genes correlated not only with the downregulation of GATA-3, TCF-1, and LEF-1 transcription factor expression, and with a change in the chromatin structure, but more importantly, with the activation of the silencer activity. Our study provides evidence for the existence of at least two negatively regulating elements, located at the TCRα enhancer-containing fragment and at the silencer region, which are active in our hybrid cells. We have shown that there was no correlation between the levels of GATA-3, TCF-1, and LEF-1 expression versus the level of TCRα mRNA in the independent hybrids. In contrast, both the silencer activity and the ability of the TCRα enhancer to downregulate thymidine kinase (TK) promoter activity were found to be in an inverse correlation with the ability of the different hybrid cells to express TCRα mRNA.
Keywords: Hybrid cells, Negative regulation, Silencer, TCRα, TCRβ
THE existence of a generalized system of gene repression has been proposed to account for the fact that only a small fraction of a given cell’s potential repertoire of genes is ever expressed. One of the remarkable features of tissue-specific gene expression is the magnitude of differential activity among various cell types. Both positive and negative modes of control are required to establish and maintain these functional differences. Positive regulation is exerted by the promoter and enhancer elements [reviewed in (33,46)], whereas negative regulation may be mediated through silencers, which are likely to be as important as enhancers in establishing lineage-specific gene expression [reviewed in (34)].
One approach to further elucidate the regulatory circuits that operate in repression of gene expression is that of somatic cell hybrids. Hybrid cells formed by fusing two distinctly different cell types generally fail to express the tissue-specific products of the parent cells, a phenomenon termed extinction. Extinction has been observed in both intra- and interspecies crosses and occurs in both heterokaryons and proliferating hybrid cells [reviewed in 6,25)].
One of the best studied biological system of cellular differentiation is T-cell development (12,39,40). This system provides several experimental advantages. First, T-cell differentiation involves the timely expression of different and well-characterized T-cell markers, such as the T-cell receptor (TCR)/CD3 complex of proteins. Second, the genes coding for these T-cell markers have been cloned and characterized. Third, expression of these genes is controlled by different regulatory elements (i.e., enhancers and silencers), of which some have been studied in detail. Identification of the T-cell-specific genes that undergo extinction in somatic cell hybrids and the subsequent identification of the target sequences that are involved in this suppression are crucial steps toward understanding the mechanism of negative regulation.
In the present study we have established somatic cell hybrids between a mouse T-cell line (BW5147) expressing the TCRβ, TCRα, Thy-1, and the CD3-ϵ T-cell-specific markers, and a mouse connective tissue-derived cell line (L cells), which is negative for these markers. Unlike many other somatic cell hybrids (11,43), these hybrids do not extinguish a whole set of differentiation specific traits, thus revealing an unusual phenotype. These T × L-cell hybrids express the ets-1 and fibronectin genes, extinguish the production of TCRβ and Thy-1 mRNAs, inhibit TCRα, GATA-3, LEF-1, and TCF-1 expression to various degrees, and exhibit a small apparent effect on the level of CD3-ϵ mRNA. In addition, we show that the TCRβ and TCRα enhancers linked to a heterologous reporter gene are targets for downregulation in the parental L cells and hybrid cells but not in the parent T-cell line. Moreover, we find that the TCRα enhancer downregulates the basal activity of the TK promoter. We have shown that changes in the chromatin structure of the TCRβ enhancer region and in expression of cellular trans-acting factors such as the GATA-3, LEF-1, and TCF-1 activators, participate in suppression of TCRα/β expression. Interestingly, the induction of the TCRα silencer activity also contributes to the inability of the hybrid cells to generate high levels of TCRα transcripts. Thus, like transcriptional activation, transcriptional repression of a gene is achieved through multiple, nonoverlapping molecular mechanisms.
MATERIALS AND METHODS
Cell Fusion
A 6-thioguanine-resistant subclone of the T-cell line BW5147 was fused with BUDR resistant (TK−) L cells (a connective tissue-derived cell line), by using 50% polyethylene glycol (15).
Cytogenetic Analysis
The cells were arrested by incubation for 30 min in the presence of 0.1 μg/ml colcemide (Gibco) and metaphase arrested cells were prepared (16).
DNA and RNA Blot Analysis
Genomic DNA was prepared, digested with restriction enzymes, electrophoresed on 1% agarose gels, and transferred to Nytran nylon membranes (Schleicher and Schull). Hybridization was performed with a random primer-labeled TCR-Jβ probe [1.8 kb BamHI/EcoRI fragment from puc-8Jbβ (38)].
Total RNA and poly(A)+ RNA were prepared, subjected to electrophoresis through a formaldehyde-containing 1% agarose gel, and transferred to Nytran filters. Hybridizations were performed with the following cDNA sequences, which were labeled with [α-32P]dCTP using a random priming kit (Amersham): fibronectin [a 0.5-kb EcoRI fragment (55)]; TCRα [a 1.0-kb EcoRI fragment from pTT11 (10)]; CD3-ϵ [a 1.4-kb EcoRI fragment from pDL1 (21)]; TCR-Cβ [a 434-bp HindIII/EcoRI fragment from pSPT672-CβT (29)]; Thy-1 [a 1.3-kb BamHI cDNA fragment from pCD-Thy-1 (56)]; GATA-3 [a 1.5-kb EcoRI fragment from pGEM7 (36)]; LEF-1 [a 1.4-kb EcoRI fragment (60)]; TCF-1 [a 0.7-kb NciI/EcoRI fragment from pM2A (50)]; and Ets-1 [a 435-bp PstI fragment (27)]. The filters were stripped in boiling water and rehybridized with 32P-labeled β-actin probe to control for the amount and integrity of mRNA loaded.
Plasmid Constructions and Transfections
To construct pBLCAT1.7βE, a 1.7-kb BglII restriction fragment containing the TCRβ enhancer was inserted into the BamHI site 5′ to the TK promoter regulating the chloramphenicol acetyl transferase (CAT) transcription unit in pBLCAT2 (kindly obtained from H. Clevers). To construct pBLCATαE515, a 515-bp PvuII-PvuII fragment encompassing the entire previously defined TCRα enhancer was cloned 5′ to the TK promoter driving CAT reporter gene in pBLCAT2. The plasmids pJ21, pJ21MoEnCα, and pJ21MoEnSilC were kindly obtained from A. Winoto and have previously been described (68).
The plasmids were tested by transient transfections into T cells using the DEAE-dextrane procedure (57) and into L and hybrid cells by the calcium phosphate precipitation method (26). To correct for differences in transfection efficiency we cotransfected 2 μg of the pCMV-βgal plasmid expressing β-galactosidase. At 44 h after transfection the cells were processed for β-galactosidase (22) and CAT activity (23). CAT products were quantitated on a Phospholmager by using ImageQuant software. Activities were expressed as percent conversion of CAM to the acetylated forms, and as relative CAT activities as specified in the legend to figures.
Isolation of Nuclei and DNase-I Treatments
Nuclei were prepared (8) and suspended in a solution of 10 mM Tris (pH 7.4), 10 mM NaCl, and 3 mM MgCl2 in a glass tube on ice. Nuclear concentrations were adjusted to a DNA concentration of 0.5 mg/ml. The nuclei were warmed to 37°C while shaking gently for 1 min, adjusted to 0.1 mM CaCl2, and digested for 2 min at 37°C with different concentrations of DNase-I ranging from 0 to 10 U/ml. Digestion was terminated by adding an equal volume of a solution of 20 mM Tris (pH 7.4), 200 mM NaCl, 2 mM EDTA, 1% SDS, and 200 μg/ml proteinase K. After overnight incubation at 37°C (or 1 h at 55°C) 1 volume of a 10 mM Tris (pH 7.4), 1 mM EDTA (TE) solution was added and the DNA was extracted once with phenol and three times with chloroform and precipitated with Na acetate and ethanol. DNA precipitates were spooled out, dissolved in TE before electrophoresis, and hybridized with the appropriate probes.
RESULTS
Expression of T-Cell-Specific Genes in T × L-Cell Hybrids
Fusion of the T-lymphoma cell line (BW5147) with thymidine kinase-deficient (TK−) L cells gave rise to a set of hybrid cell lines. Ten selected clones were analyzed for the expression of T-cell differentiation traits. In some experiments all clones are shown. However, in others only two to four representative hybrids are shown, although more were analyzed. The selected clones differed in their morphology from the parental cells. The parental T cells were spherical and grew in suspension. The L cells grew as adherent fibroblast-like elongated cells. The hybrids were adherent and usually larger than either of the parental cells. In contrast to the morphology of the parental L cells, the hybrid cells had numerous long cellular extensions around their entire circumference.
Mouse–mouse hybrid cells have been shown to lose a few chromosomes in the first generations following fusion. After this, the chromosomal makeup of the hybrids becomes stable (19). The hybrid cells examined in the present study were either complete hybrids or at the most contained 10% fewer chromosomes than the sum number of their parental cells (on average, an L cell contains 46 ± 3 chromosomes and a BW5147 cell contains 34 ± 9 chromosomes).
BW5147 cells were found to be positive for the following T-cell-specific transcripts: TCRα, TCRβ, CD3-ϵ, and Thy-1. The hybrids were examined for the presence of these transcripts by RNA blot analysis. Figure 1 shows that the parental BW5147 T cells express a high level of TCRα transcripts, whereas L cells lack detectable TCRα mRNA. Only one of our hybrid cell lines express a high level of TCRα transcripts (but still lower than BW5147 TCRα mRNA), five hybrids display very low levels of these transcripts, and three show undetectable levels. The integrity of the RNAs was monitored in this and all following experiments with a β-actin probe (Fig. 1A). Low levels of TCRα mRNA in the hybrid cells could not be due to the loss of the chromosomes encoding these genes because Southern blot analysis of the DNA from parental and hybrid cells shows that all the hybrids possess the productively rearranged TCRα chain genes (data not shown).
FIG. 1.

Expression of TCRα and CD3-ϵ genes in T × L-cell hybrids. (A) Total RNA (15 μg) isolated from parental (BW5147 and L) and hybrid cells were electrophoresed on 1.0% agarose-formaldehyde gels, transferred to nitrocellulose, and hybridized with a 1.0-kb EcoRI DNA fragment containing the TCRα cDNA sequence. (B) RNA was hybridized with a 1.4-kb EcoRI DNA fragment containing the CD3-ϵ cDNA sequence. Blots were stripped and rehybridized with a β-actin probe. (A) Lanes 1–11: BW5147, L, Hy1A, Hy2A, Hy3A, Hy1B, Hy3B, Hy4B, Hy1C, Hy2C, and Hy4C, respectively. (B) Lanes 1–12: BW5147, L,Hy1A, Hy2A, Hy3A, Hy1B, Hy3B, Hy4B, Hy1C, Hy2C, Hy3C, and Hy4C, respectively.
The gene encoding the CD3-ϵ chain of the TCR/CD3 complex is uniquely transcribed in all T-lymphocyte lineage cells. Hybridization with a CD3-ϵ radioactive probe revealed that all the hybrids tested express the CD3-ϵ transcripts (Fig. 1B). The amount of the CD3-ϵ mRNA in our hybrids is between 5- and 20-fold lower than that in BW5147 cells. Thus, T × L-cell hybrids express low to intermediate levels of CD3-ϵ T-cell-specific mRNA, compared to the parental T-lymphoma cells.
Expression of Thy-1 and TCRβ Genes Is Repressed in T × L-Cell Hybrids
The same 10 independent hybrids were examined for the presence of Thy-1 transcripts. The Thy-1 gene is expressed in several tissues during development and the pattern varies considerably between species (13). The mouse protein is expressed in the nervous system, thymocytes, peripheral T cells, and, to a lesser extent, in several other tissues. We analyzed total cellular RNA from parental and hybrid cells on Northern blot using Thy-1 cDNA as a radioactive probe. Figure 2A shows that no hybridization could be detected to RNA extracted from the parental L cells or from the hybrids (Fig. 2A, lanes 2–12), whereas RNA from BW5147 T cells contains a high level of Thy-1 mRNA (Fig. 2A, lane 1). Furthermore, no hybridization signals were detected using 10 μg of Hy1A poly(A)+ mRNA, whereas a prominent band was observed with 1 μg of BW5147 total RNA (Fig. 2B, compare lane 3 to lane 4). Thus, Thy-1 transcripts are at least 100-fold more abundant in BW5147 cells than in the hybrid cells. The BW5147 Thy-1 genetic information is present in our hybrids. The absence of Thy-1 expression is not due to the loss of the genes encoding these molecules, because using restriction fragment length polymorphism analysis, we showed that the hybrid DNA contains the sum of the polymorphic Thy-1 hybridization bands. The lack of Thy-1 gene expression in the same type of hybrids (T × L) has been reported in the past (32,42), but in these early experiments there were no data indicating whether the Thy-1 mRNA is present or absent. Our results indicate that the extinction of Thy-1 expression in the hybrid cells occurs at the level of transcript accumulation.
FIG. 2.

Repression of Thy-1 gene expression in T × L-cell hybrids. (A) Total RNA (15 μg) from parental and hybrid cells were electrophoresed through formaldehyde/agarose gel, transferred to Nytran membranes, and hybridized with labeled Thy-1 cDNA probe. Lanes 1–12: BW5147, L, Hy1A, Hy2A, Hy3A, Hy1B, Hy3B, Hy4B, Hy1C, Hy2C, Hy3C, and Hy4C, respectively. (B) Different amounts of total BW5147 RNA (10, 5, and 1 μg; lanes 1–3, respectively) and poly(A)+ RNA of Hy1A (10, 5, and 1 μg; lanes 4–6, respectively) were hybridized with Thy-1 cDNA radioactive probe. The blots were stripped, rehybridized with a β-actin cDNA probe, and shown in the lower parts of this figure.
Hybridization of a similar RNA blot to a TCRβ probe revealed the expected 1.3-kb mRNA in the parental BW5147 T cells (Fig. 3A, B). In contrast, no hybridization could be detected with RNA extracted from parental L cells or from eight (out of 10 tested) independent hybrid cell lines (Fig. 3A, lanes 2–10). We prepared poly(A)+ mRNA from two hybrids, Hy1A and Hy3C, and analyzed them as described above. We estimate that in these hybrid cells the level of TCRβ mRNA is at least 100-fold lower than that in BW5147 cells. The lack of TCRβ expression cannot be explained by loss of the genes encoding these molecules because Southern blot analysis indicated that all of our 10 hybrids retained the TCR0 chain gene (rearranged and germ line) from both parents (Fig. 3C, lanes 1–5 show 5 out of 10 hybrids tested).
FIG. 3.

Repression of TCRβ chain mRNA in T × L-cell hybrids. (A) Total RNA (15 μg) from parental and hybrid cells were electrophoresed through formaldehyde/agarose gel, transferred to Nytran membranes, and hybridized with a labeled TCR-Cβ radioactive probe. Lanes 1–10: BW5147, L, Hy1A, Hy2A, Hy3A, Hy1B, Hy3B, Hy4B, Hy1C, and Hy3C, repectively. (B) Total BW5147 RNA (10, 5, 2.5, and 1 μg; lanes 1–4, respectively), poly(A)+ mRNA of Hy1A (2, 5, and 10 μg; lanes 5–7, respectively), and Hy3C poly(A)+ mRNA (1, 2, and 3 μg; lanes 8–10, respectively) were hybridized with a labeled TCR-Cβ chain probe. The blots were stripped and rehybridized with a β-actin cDNA probe. The blots containing the β-actin transcripts are shown in the lower parts of this figure. (C) Analysis of germ line and rearranged TCRβ chain genes in T × L cell hybrids. DNA of parental [L and T (BW5147)] and hybrid cells (15 μg) was digested with EcoRI, electrophoresed on 1% agarose gel, and transfered to Nytran paper. The blot was hybridized to a TCR-Cβ probe and autoradiographed. Hybrids 1–5 are Hy2A, Hy3A, Hy1B, Hy3B, and Hy2C respectively. Bands 4.4 kb and 2.3 kb represent the germ line TCR-Cβ gene and the 2.0-kb band represent the productively rearranged TCR-Cβ in T(BW5147) cells. Note that all the hybrids contain the 2.0-kb band.
The Fibroblast-Specific Gene, Fibronectin, Is Expressed in T × L-Cell Hybrids
To study whether the fibronectin gene is expressed in the hybrid cells, we hybridized parental and hybrid mRNAs with a fibronectin radioactive probe. This hybridization revealed a prominent transcript of about 8.5 kb in L cells and in our T × L-cell hybrids (4 out of 10 hybrids are shown), but not in BW5147 T cells (Fig. 4). Thus, the fibronectin gene is expressed in a dominant manner in the hybrid cells.
FIG. 4.
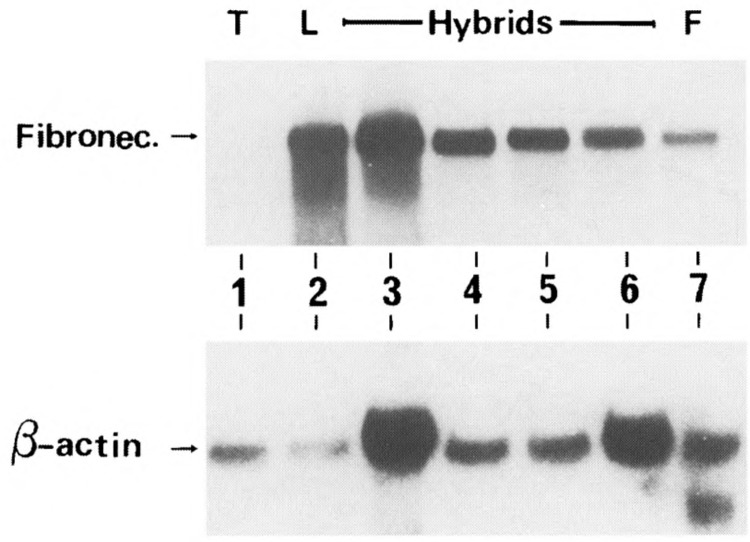
Expression of fibronectin gene in T × L-cell hybrids. Total RNA (15 μg/lane) of the parental (T, L) and hybrid cells were electrophoresed, blotted, and hybridized with a labeled fibronectin radioactive probe. Lanes 1–7: BW5417, L, Hy1B, Hy2A, Hy2C, Hy3A, and normal embryonic fibroblast from C3H mouse (F), respectively. The blot was stripped, rehybridized with a β-actin cDNA probe, and is shown in the lower part of this figure.
The TCRβ Chain Enhancer Activity and Chromatin Structure Are Altered in T × L-Cell Hybrids
The apparent inability of the hybrids to transcribe the TCRβ chain gene may be due to different causes, one of which is altered chromatin structure. Many transcribed genes contain sites in chromatin that are hypersensitive to DNase-I (49,58,69); such sites often encompass DNA sequences that are nucleosome free in vivo and important for gene control [reviewed in (18)]. Previously published results indicated that the TCRβ chain gene contains two DNase-I hypersensitive sites, DHD1 and DHD2, located downstream of the Cβ2 region (28). Hypersensitivity in these two regions was expressed in a tissue-specific manner. The DHD1 site was found to be located in the TCRβ enhancer. This enhancer element has been identified in a 550-bp fragment located 5 kb downstream of the Cβ2 region (37,48). We compared the chromatin structure of the TCRβ gene in the parental cells and in two representative hybrid clones. Nuclei from these cells were incubated with a range of DNase-I concentrations. Subsequently, DNA was prepared, digested with BamHI and Bg1I, and probed with a 1.0-kb fragment derived from the most 5′ region of the BamHI-Bg1I fragment (Fig. 5A). The BamHI-Bg1I digested BW5147 DNA gave rise to a band of 6.6 kb (Fig. 5B), and with increasing amounts of DNase-I, a second band of 3.5 kb appeared. The location of this hypersensitivity site (indicated in Fig. 5B by an arrow) coincided with the previously published DHD1 site. Interestingly, unlike the BW5147 cell line, the 3.5-kb subband was not observed in L and hybrid cells at any DNase-I concentrations used. An additional subband of 6.0 kb was strongly apparent in L cells but also visible in T and hybrid cells. The significance of this band is not clear. Thus, the hybrid cells lack the T-cell-specific DNase-I hypersensitive site, which is located in the TCRβ enhancer.
FIG. 5.
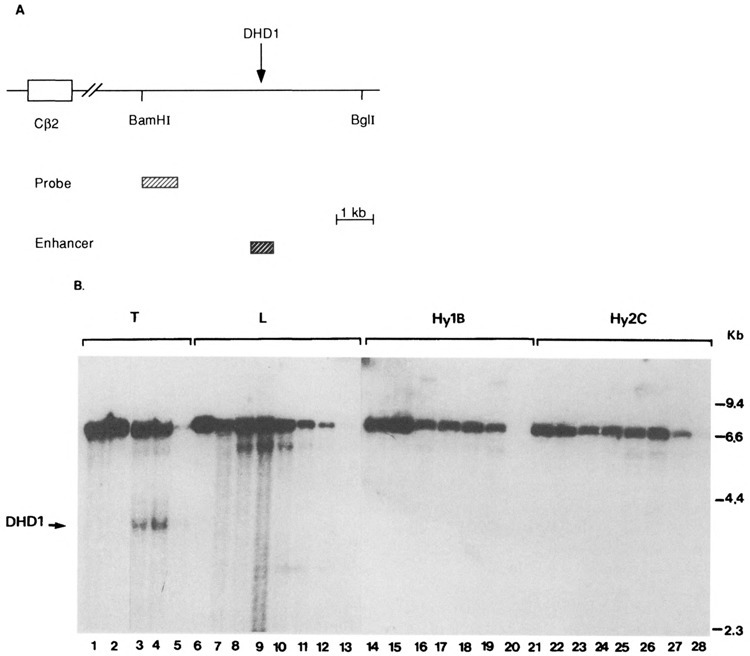
Mapping of DNase-I hypersensitive regions in parental and hybrid cell lines. (A) A map of the mouse TCRβ 3′ region. The region examined for DNase-I hypersensitivity is downstream of the Cβ2 region between the two indicated BamHI and BglI restriction sites. The probe used for Southern blots and the region encompassing the TCRβ enhancer are indicated. (B) DNase-I hypersensitivity of the TCRβ BamHI-BglI region in T cells (lanes 1–5), L cells (lanes 6–13), Hy1B (lanes 14–20), and Hy2C (lanes 21–28). DNase-I-treated DNA was double digested with BamHI and BglI and analyzed by Southern blotting. Lanes 1, 6, 14, and 21 are from nuclear samples lysed without prewarming to 37°C. The absence of subbands in these samples indicates the absence of endogenous nuclease activity during preparation of nuclei. We treated the nuclei at 37°C for 2 min. The T-cell nuclei (lanes 2–5) were treated with 0, 2, 3, and 5 units of DNase-I, respectively. The L-cell nuclei (lanes 7–13) and Hy2C nuclei (lanes 22–28) were treated with 0, 1, 2, 3, 4, 5, and 10 units of DNase-I, respectively. The Hy1B nuclei (lanes 15–20) were treated with 0, 2, 3, 4, 5, and 10 units of DNase-I, respectively. The nuclear incubations in the absence of added DNase-I reveal the extent of endogenos nuclease cleavage within the given time period. The arrow indicates the position of a T-cell-specific subband generated from the hypersensitive DNase-I site defined in the DHD1 region (28). This band is absent in the hybrids. There is an additional subband of 6 kb that is generated by DNase-I, and appears in all cell lines and thus is not a T-cell-specific band.
The TCRβ enhancer contains several different motifs (24,59), is necessary for T-cell-specific expression of a TCRβ chain gene in transgenic mice, and directs transcriptional activation in transient transfections of T cells. To assess the involvement of this enhancer in TCRβ chain extinction in the hybrids, we transiently transfected the parental and hybrid cells with two different vectors (Fig. 6A): 1) pBLCAT2 (a plasmid carrying only the TK promoter and the CAT transcription unit), and 2) pBLCAT1.7βE (a similar plasmid carrying the T-cell-specific TCRβ enhancer). To overcome the bias of different transfection efficiencies into various cell lines, all transfections were repeated in at least three independent experiments and corrected to the levels of β-galactosidase produced by cotransfection of the pCMV-β-galactosidase plasmid. In all subsequent cotransfection experiments, CAT activity was normalized in the same fashion. As expected, CAT expression driven by the TK promoter and TCRβ enhancer was significantly higher in T cells than L cells. In L cells, transfection of pBLCAT1.7βE gave rise to a lower CAT activity than transfection of pBLCAT2. This could be due to negatively regulating transcription factors expressed in nonlymphoid cells that bind to the TCRβ enhancer, as was previously reported (30). Interestingly, transfection of pBLCAT1.7βE DNA into three representative hybrid cells, Hy3A, Hy1B, and Hy2C, yielded a very low level of CAT expression, similar to CAT activity driven by the pBLCAT2 vector alone (Fig. 6B). Similar results were obtained when we transfected the identical CAT reporter plasmid driven by a smaller fragment (0.8 kb) encompassing the TCRβ enhancer. This low level of CAT expression implies that the inability of the hybrid cells to generate TCRβ transcripts is at least in part due to inactivition of the TCRβ enhancer.
FIG. 6.
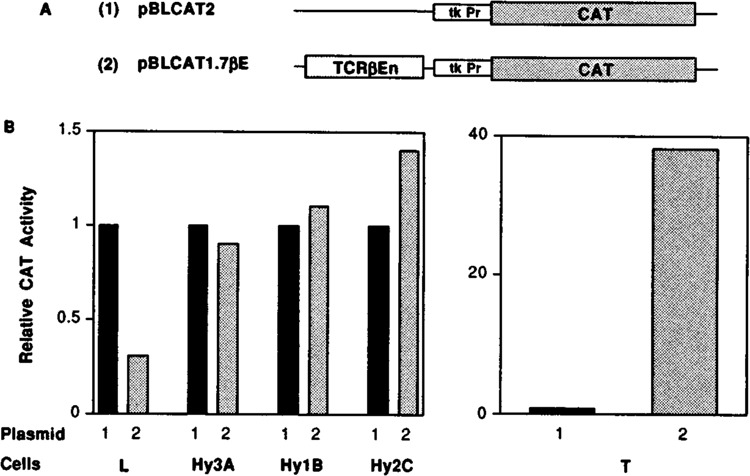
Expression of CAT gene linked to TCRβ chain enhancer is downregulated in T × L-cell hybrids. (A) A schematic representation of pBLCAT2 and pBLCAT1.7βE plasmids. The pBLCAT2 plasmid contains the TK promoter inserted in front of the CAT transcription unit, and the pBLCAT1.7βE contains the 1.7-kb BamHI fragment harboring the TCRβ enhancer cloned at the BamHI site located 5′ to the TK promoter. (B) Parental (BW5147, and L) and hybrid cells were cotransfected with 15 μg of either pBLCAT2 or pBLCAT1.7βE together with 2 μg of pCMV-β-gal DNA. After 48 h, cells were harvested, lysed, and CAT activities were determined and quantitated on a Phospholmager by using ImageQuant Software. The percent conversion of each separate transfection was normalized to the β-galactosidase activity. The values for percent conversion, presented as means ± SDs, corresponding to pBLCAT2 and pBLCAT1.7βE in L, BW5147, Hy3A, Hy1B, and Hy2C are 14.8 ± 1.5, 4.4 ± 0.5, 0.7 ± 0.1, 26.6 ± 3.1, 19.3 ± 2.2, 17.4 ± 2.4, 19.5 ± 2.3, 21.5 ± 2.5, 9 ± 1.1, and 12.6 ± 1.5, respectively. For calculation of relative CAT activity, the activity obtained from pBLCAT2 construct in each cell line was arbitrarily assigned a value of 1. The other values observed are relative to that of pBLCAT2 and are the mean values of three to four independent experiments.
The TCRα Enhancer Is a Target for Repression in T × L-Cell Hybrids
To determine whether suppression of TCRα mRNA in hybrid cells also correlates with down-regulation of the TCRα enhancer activity, we transiently transfected parental and hybrid cells with CAT reporter gene driven by either TK promoter alone (pBLCAT2) or together with a PvuII-PvuII 515-bp fragment encompassing the entire previously defined TCRα enhancer cloned 5′ to the promoter (pBLCATαE515). As shown in Fig. 7, in T cells the TCRα enhancer fragment induced CAT activity by at least 30-fold (Fig. 7B, right panel, lanes 1 and 2). To our surprise, upon transfection of pBLCATαE515 into parental L-cells and into three representative hybrid cells, Hy3A, Hy1B and Hy2C, the TCRα enhancer-containing fragment reduced CAT activity to about 50% of that seen with the TK promoter alone (transfections were repeated more than 10 times). Thus, in the hybrid cells, the 515-bp fragment not only did not activate the TK promoter but also repressed the promoter activity below its basal level. Hy3A expresses the highest level of TCRα mRNA, whereas Hy2C is the lowest TCRα-expressing hybrid cell line. However, the latter still expresses a higher level of TCRα mRNA than L cells. It is interesting to note that the TCRα enhancer repressing activity is higher in Hy2C than in Hy3A; thus, it inversely correlates with TCRα mRNA expression in these cells. Therefore, changes in the cellular activity of trans-acting factors, such as induction of repressors, acting on the TCRα enhancer, most probably contribute to the inability of the hybrid cells to generate high levels of TCRα transcripts.
FIG. 7.

The TCRα: enhancer represses the TK promoter activity. (A) A schematic representation of pBLCAT2 and pBLCATαE515 plasmids. The pBLCAT2 plasmid contains the TK promoter inserted in front of the CAT transcription unit and the pBLCATαE515 carries the TCRα PvuII-PvuII 515-bp enhancer fragment cloned 5′ to the TK promoter. (B) Parental (BW5147 and L) and hybrid cells (Hy3A, Hy1B, Hy2C) were cotransfected with 15 μg of either pBLCAT2 or pBLCATαE515 together with 2 μg of pCMV-βgal DNA. CAT activities were determined and quantitated as described in the legend to Fig. 6B. The values for percent conversion, presented as means ± SDs, corresponding to pBLCAT2 and pBLCATαE515 in L, BW5147, Hy3A, Hy1B, and Hy2C are 4.08 ± 0.5, 1.22 ± 0.1, 0.91 ± 0.14, 34.3 ± 7.85, 1.75 ± 0.09, 1.06 ± 0.05, 3.16 ± 0.85, 1.7 ± 0.2, 2.61 ± 0.25, and 0.98 ± 0.13, respectively. For calculation of relative CAT activity, the activity obtained from pBLCAT2 construct in each cell line was arbitrarily assigned a value of 1. The other values observed are relative to that of pBLCAT and are the mean values of 10 independent experiments.
T × L-Cell Hybrids Express Ets-1 and Repress LEF-1, TCF-1, and GATA-3 Expression
A large number of studies have shown that in different cell types there is a coordinate induction of sets of tissue-specific genes. The use of common sets of tissue-specific transcription factors to regulate multiple lineage-specific genes represents an economical mechanism for coordinate gene expression during development. Thus, it is not surprising that the TCRα and TCRβ enhancers share overlapping sets of nuclear binding sites. They include, among others, the GATA-3, Ets-1, TCF-1, and LEF-1/TCF-1α (denoted LEF-1) transcription factors (40). Thus, we were interested in correlating the downregulation of the TCRβ and TCRα enhancer activities in T × L-cell hybrids with changes in the mRNAs encoding proteins interacting with the enhancer motifs. Therefore, mRNA from parental and hybrid cells was analyzed for TCF-1, LEF-1, GATA-3, and Ets-1 expression. BW5147 T cells contain mRNA for TCF-1, LEF-1, and GATA-3 transcription factors, whereas the L cells do not express detectable levels of these T cell-restricted transcription factors. Interestingly, all T × L-cell hybrids tested (only two are shown) show low but detectable levels of TCF-1, LEF-1, and GATA-3 mRNAs (Fig. 8A). We have examined the presence of GATA-3 and LEF-1 proteins in the nuclear extracts of the parental and hybrid cells. As expected from the Northern analysis, extracts derived from T × L-cell hybrids express low but detectable GATA-3 and LEF-1 binding activities (data not shown). Our results suggest that the low levels of T-cell-specific transcription factors present in our hybrids most probably contribute to the low level of TCRα expression and to the extinction of TCRβ expression.
FIG. 8.
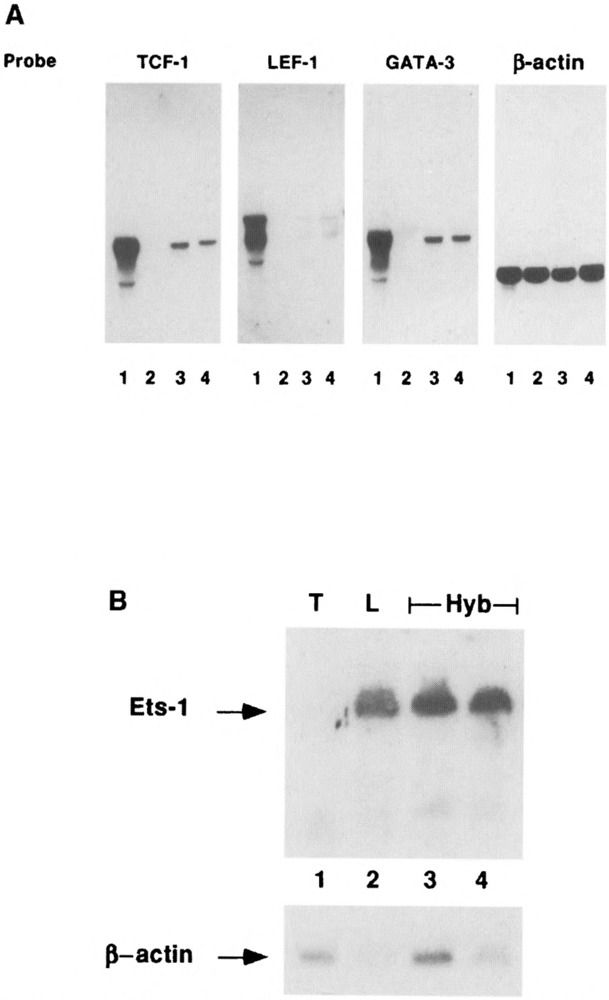
T × L-cell hybrids express Ets-1 and low levels of GATA-3, LEF-1/TCF-1α, and TCF-1 mRNAs. (A) RNA (15 μg/lane) isolated from BW5147 (lane 1), L cells (lane 2), Hy3A (lane 3), and Hy1B (lane 4) cell lines were electrophoresed on 1% agarose-formaldehyde gel, transferred to Nitran filter, and hybridized sequentially with GATA-3, LEF-1, TCF-1, and β-actin radioactive probes. (B) RNA (15 μg/lane) isolated from parental BW5147 (lane 1), L cells (lane 2), Hy3A (lane 3), and Hy1B (lane 4) cell lines were electrophoresed, blotted, and hybridized with a labeled ets-1 probe.
To our surprise, L cells express ets-1 mRNA, whereas no ets-1 mRNA was detected in BW5147 T cells. Interestingly, our hybrids (only 2 out of 10 are shown) express ets-1 mRNA at similar levels to that found in parental L cells, emphasizing that the fibroblastic-like phenotype is dominant in these hybrid cells.
The TCRα Silencer Is Active in T × L-Cell Hybrids
We have studied the possibility that repression of TCRα expression occurs through a direct mechanism in which the non-T cells contribute a negative factor(s) that directly repress regulatory sequences within the TCRα gene. Previously, several lineage-specific transcriptional silencers, located 5′ of the murine TCRα enhancer, have been identified (68). These silencers are required to restrict the activity of the TCRα enhancer to the α/β T-cell lineage. It was shown that these silencers were capable of repressing the activity of ubiquitously active enhancer, such as the Moloney enhancer, in non-α/β T cells. We analyzed the presence of the silencer activity in three representative hybrid cells. Parental and hybrid cells were transfected with pJ21, pJ21MoEnCα, and pJ21MoEnSilC plasmids (Fig. 9A). Transfections of these plasmids into parental cells showed that the TCRα silencer was active in L cells and inactive in T cells (data not shown). Interestingly, the presence of the TCRα silencer in the pJ21MoEnSilC construct repressed the CAT activity driven by the Moloney enhancer in all hybrids tested (Fig. 9B). This α silencer represses CAT activity to a lower level in Hy1B and Hy2C than in Hy3A. As discussed above, hybrids 1B and 2C express lower levels of TCRα mRNA compared with Hy3A cells, which express the highest level of TCRα mRNA among 10 independent hybrid clones (Fig. 1A, lane 5), and the lowest level of silencer activity (Fig. 9C). Thus, we conclude that the T × L-cell hybrids harbor the silencer activity and, furthermore, there is an apparent inverse correlation between the level of TCRα mRNA versus the levels of the silencer activity in the hybrid cell clones analyzed (Fig. 9C).
FIG, 9.

The T × L-cell hybrids harbor the previously identified TCRα silencer activity. (A) A schematic representation of pJ21 (1), pJ21MoEnCα (2), and pJ21MoEnSilC (3) plasmids, kindly obtained from A. Winoto. The pJ21 (1) contains the c-fos promoter inserted in front of the CAT transcription unit; the pJ21MoEnCα (2) contains the Moloney enhancer and a fragment of the Cα exon (0.5 kb), the pJ21MoEnSilC encompasses silI (0.4 kb) and SilII (0.33 kb), both cloned 3′ to the Moloney enhancer fragment. (B) Hybrid cells were cotransfected with 15 μg of pJ21, pJ21MoEnCα, and pJ21MoEnSilC together with 2 μg of pCMV-βgal DNA. CAT and β-galactosidase activities were assayed as described in the legend to Fig. 6B. The values for percent conversion, presented as means ± SDs, corresponding to pJ21, pJ21MoEnCα, and pJ21MoEnSilC in Hy3A, Hy1B, and Hy2C are 1.5 ± 0.3, 43.5 ± 5.1, 30.45 ± 2.9, 1.4 ± 0.2, 31 ± 5.1, 8.26 ± 0.9, 0.8 ± 0.1, 14.5 ± 1.2, and 2.36 ± 0.36, respectively. For calculation of relative CAT activity, the activity obtained from pJ21 in each cell line was arbitrarily assigned a value of 1. The other values observed are relative to that of pJ21 and are the mean values of four independent experiments. Silencer activity was calculated as the ratio between plasmids 2 and 3 CAT activities. (C) The relative level of TCRα mRNA (calculated as the ratio between the level of TCRα mRNA in Hy3A, Hy1B, and Hy2C, and the level of TCRα mRNA in BW5147 cells, normalized to β-actin mRNA, and averaged for four independent experiments) was plotted against the silencer activity calculated as described in (B). The values for the relative TCRα mRNA corresponding to Hy3A, Hy1B, and Hy2C are 4.7%, 1.5%, and 0.8%, respectively.
DISCUSSION
The aim of this work was to study the molecular mechanisms that play a role in downregulation of T-cell-specific genes. Somatic cell hybrids demonstrated different phenotypes, of which extinction of specific cellular functions is most frequently observed (14). The underlying mechanisms regulating the extinction phenomenon are thought to mirror, at least in part, the repertoire of regulatory mechanisms controlling mammalian cell differentiation [review in (6)]. Extinction of a whole set of differentiated traits has been observed in several combinations of hybrid systems. The generality of extinction was suggested based on investigations assaying hepatoma × fibroblast hybrid cells (11), and compatible results were obtained in myeloma × fibroblast and myeloma × T somatic cell hybrids (3,43,54). In this study we have shown that fusion of T cells to L cells yields a set of hybrids with an unusual and interesting phenotype. These hybrids, as expected, continuously express the fibroblast marker, fibronectin, in a dominant manner, and extinguish the production of TCRβ and Thy-1 mRNAs. However, the expression of TCRα, CD3-ϵ, GATA-3, LEF-1, and TCF-1 was not extinguished but was inhibited to different degrees. We do not know the reasons for the unusual phenotype of our hybrids, but these cell lines provide an attractive biological system to decipher the different molecular mechanisms that take part in conferring negative regulation upon the expression of these genes. We have studied the molecular mechanisms that contribute to the suppression of two T-cell-specific genes (i.e., TCRα and TCRβ), and have shown for the first time that both direct and indirect repression mechanisms operate in T × L-cell hybrids. Our data indicate that no single mechanism represses the entire trait of T-cell-specific genes in the hybrid cells. It is likely that these genes are shut off by several tissue-specific extinguishers (TSE) and only the combined effects of multiple regulatory genes result in suppression of T-cell-specific gene expression, as was shown for liver-specific gene extinction in hepatoma × fibroblast hybrids (7).
Transcription of TCRα and TCRβ genes is regulated by multiple cis-acting elements such as promoters, enhancers, silencers, and a locus control region (LCR) (17,40). The TCRα and TCRβ enhancer regions enhance lymphoid cell-specific transcription (37,67). Our transient transfection experiments employing the CAT reporter gene driven by a ubiquitous promoter and either TCRα or TCRβ enhancer show that suppression is exerted upon a newly transfected reporter gene driven by these enhancers.
Similar to other enhancers, the TCRα and TCRβ enhancers harbor overlapping sets of binding sites for ubiquitous and lineage-restricted transcription factors such as LEF-1, TCF-1, GATA-3, and Ets-1 (24,40). These proteins appear to play important roles in regulating the expression of TCRα/β genes during T-cell development and activation (60,61,64). LEF-1 is expressed during most stages of T-cell development, and in murine pre-B cells (51,60). Unlike LEF-1, TCF-1 is not expressed in cells of the B lineage but is rather limited to T cells (51). LEF-1 gene destruction does not affect the T lineage (62), whereas in mice carrying a homozygous germ-line mutation in the TCF-1 gene, thymocyte development is blocked at the transition from the CD8+ immature single-positive to the CD4+/CD8+ double-positive stage (63). Thus, TCF-1 controls an essential step in thymocyte development. Functional and biochemical characterization of LEF-1 and TCF-1 indicate that these proteins participate in regulation of the TCRα enhancer (50,60,65). We have found that our T × L-cell hybrid cells contain low levels of TCF-1 and LEF-1 mRNAs and proteins. However, previously published data show, in a different T × L-cell hybrid combination (EL4 × B82), that TCR × expression is extinguished and is accompanied by a complete loss of LEF-1 expression (70). We do not know the reason for this discrepancy; however, it is probably due to the different T- and fibroblast-like cell lines used for fusion, just as different results were obtained in analyzing Oct-2 expression in different T × myeloma hybrid combinations (53,71). Like TCF-1 and LEF-1, GATA-3 is a lineage-restricted transcription factor that is expressed very early in T-cell development, suggesting its involvement in T-lymphocyte differentiation (20,36). Our results show that expression of GATA-3 was considerably lower in our hybrids compared to the parental T cells. Thus, our finding that repression of TCRα and TCRβ expression is accompanied by repression of GATA-3, LEF-1, and TCF-1 transcription factors is in agreement with the notion that an indirect mechanism plays a role in suppression in of these genes in our hybrids.
Loss of an essential activator(s) has been demonstrated in several hybrid systems, notably in pituitary × fibroblast (47), myeloma × fibroblast (3,35), insulinoma × fibroblast (41), and hepatoma × fibroblast hybrids (7,9). In most cases, it was found that extinction of growth hormone, immunoglobulin, α1-antitrypsin, insulin, and albumin genes was accompanied by extinction of the transactivators Pit-1/GHF1, Oct-2, LF-B1, IEF1, and HNF-1, respectively, which bind to crucial sequences in the promoter/enhancer regulatory elements. More recently, we have shown that expression of the B-cell-specific coactivator BOB.1/OBF.1 is switched off in myeloma × fibroblast hybrids (54). Whereas loss of a tissue-specific activator has often been correlated with gene extinction, it has rarely followed that provision of this factor to the hybrid cells reactivates the relevant genes (7,53). An exception was the finding that uninterrupted expression of Oct-2 in myeloma × T hybrids was enough to maintain the expression of B-cell-specific genes. This unique effect may reflect the position of Oct-2 within the hierarchy of genes that control the B-cell-specific expression (53). Due to the observed redundancy in transcriptional activator binding sites in many enhancers, the lack of a single transcription factor would have had only a modest effect on enhancer activity. Thus, redundancy in TCRα and TCRβ enhancers can most probably compensate for the loss of a single transcription factor. Therefore, lack of either GATA-3 or TCF-1 or LEF-1 activity alone most likely will not lead to significant loss of TCRα and -β transcriptional function. Probably, the effect of a combined loss of several transcriptional activators on enhancer activity will be more dramatic. This suggestion is strengthened by our findings that the expression of three different prominent transcription factors was considerably repressed in T × L-cell hybrids.
It is likely that additional mechanisms contribute to the downregulation of TCRα/β gene expression in our hybrid cells. One possible mechanism involves a repressor(s) that directly regulates the genes themselves (44). Our study provides evidence of the possible existence of at least two negatively regulating mechanisms in our hybrids. One mechanism involves the previously identified TCRα silencer activity (68). We have shown that in our hybrids there is an apparent inverse correlation between the level of TCRα mRNA versus the level of the silencer activity. Hy3A, which expresses the highest level of TCRα mRNA, shows the lowest level of the silencer activity, whereas Hy2C exhibits the lowest level of TCRα mRNA and the highest level of silencer activity. The second negatively regulating mechanism involves the TCRα enhancer-containing fragment, which down-regulates the basal activity of the TK promoter. This suggests for the first time that this fragment contains a silencer activity, the identity of which we have not yet evaluated. Interestingly, the ability of this new silencer to downregulate the TK promoter activity also inversely correlates with the ability of the hybrid clones to express TCRα mRNA.
Studying the expression of Ets-1 in our hybrids was especially intriguing for two reasons: 1) Ets binding sites have been identified within several T-cell-specific regulatory sequences such as the TCRα, TCRβ enhancers [(31,39); reviewed in (45)], and 2) Ets-1 is known to be preferentially expressed in thymocytes and T cells, and its expression is developmentally regulated during thymic ontogeny (4,5). To our surprise, Ets-1 mRNA was detected in the parental L cells and was absent in the parental T-cell line. Our findings of Ets-1 expression in L cells are consistent with previously published data showing Ets-1 expression in endothelial cells and astrocytes (1,66). Interestingly, it was previosly shown that Ets-1 specifically represses activity of the complete TCRβ enhancer in human T Jurkat cells (52). Our preliminary co-transfection experiments introducing an Ets-1 expression vector with a CAT reporter gene driven by either the TCRα or TCRβ enhancer led to a modest repression of these enhancers’ activity, suggesting a repressive role for Ets-1 in our hybrid cells (data not shown).
We have studied an additional aspect of the molecular mechanism that underlies extinction of TCRβ mRNA. We have shown that the shutdown of TCRβ transcription is accompanied by a change in the chromatin structure of the TCRβ enhancer region. This provides a general mechanism for prohibiting TCRβ gene activation in inappropriate cell type. We have previously also shown that in embryonal × fibroblast hybrid cells, extinction of Oct-3/4 gene expression is correlated with changes in the chromatin structure of the promoter/enhancer regulatory upstream region (2). Thus, extinguished genes are in a less accessible chromatin configuration than their expressed counterparts. Our results provide a strong basis for the concept that analogously to cell-type transcriptional activation, the degree of suppression of a single tissue-specific gene is determined by the combined action of multiple repression mechanisms.
ACKNOWLEDGEMENTS
We wish to thank Dr. U. Nudel for the β-actin probe, Dr. L. Hood for the TCRβ and TCRα plasmids, Dr. C. Terhorst for the CD3-ϵ probe, Dr. H. Clevers for the TCF-1 probe, Dr. D. Engel for the GATA-3 probe, Dr. R. Grosschedl for the LEF-1 probe, Dr. B. Graves for the ets-1 gene, and Dr. A. Winoto for the pJ21, pJ21MoEnCα, and pJ21MoEnSilc plasmids. We also acknowledge the assistance of Ms. Gillian Hirst in preparing this manuscript. This work was supported by the Paul Ehrlich Center for the study of normal and leukemic cells (to R.L. and Y.B.) and by a project grant from the Israel Cancer Research Fund (to Y.B.).
REFERENCES
- 1. Amouyel P.; Gegonne A.; Delacourte A.; Defossez A.; Stehelin D. Expression of ETS proto-oncogenes in astrocytes in human cortex. Brain Res. 447:149–153; 1988. [DOI] [PubMed] [Google Scholar]
- 2. Ben-Shushan E.; Pikarsky E.; Klar A.; Bergman Y. Extinction of Oct-3/4 gene expression in embryonal carcinoma × fibroblast somatic cell hybrids is accompanied by changes in the methylation status, chromatin structure, and transcriptional activity of the Oct-3/4 upstream region. Mol. Cell. Biol. 13:891–901; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bergman Y.; Strich B.; Sharir H.; Ber R.; Laskov R. Extinction of Ig genes expression in myeloma × fibroblast somatic cell hybrids is accompanied by repression of the oct-2 gene encoding a B-cell specific transcription factor. EMBO J. 9:849–855; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bhat N. K.; Fisher R. J.; Fujiwara S.; Ascione R.; Papas T. S. Temporal and tissue-specific expression of mouse ets genes. Proc. Natl. Acad. Sci. USA 84:3161–3165; 1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bhat N. K.; Komschlies K. L.; Fujiwara S.; Fisher R. J.; Mathieson B. J.; Gregorio T. A.; Young H. A.; Kasik J. W.; Ozato K.; Papas T. S. Expression of ets genes in mouse thymocyte subsets and T cells. J. Immunol. 142:672–678; 1989. [PubMed] [Google Scholar]
- 6. Boshart M.; Nitsch D.; Schutz G. Extinction of gene expression in somatic cell hybrids — a reflection of important regulatory mechanisms. Trends Genet. 9:240–245; 1993. [DOI] [PubMed] [Google Scholar]
- 7. Bulla G. A.; DeSimone V.; Cortese R.; Fournier R. E. K. Extinction of α1-antitrypsin gene expression in somatic cell hybrids: Evidence for multiple controls. Genes Dev. 6:316–327; 1992. [DOI] [PubMed] [Google Scholar]
- 8. Burch J. B. E.; Weintraub H. Temporal order of chromatin structural changes associated with activation of the major chicken vitellogenin gene. Cell 33:65–76; 1983. [DOI] [PubMed] [Google Scholar]
- 9. Cerrghini S.; Yaniv M.; Cortese R. Hepatocyte dedifferentiation and extinction is accompanied by a block in the synthesis of mRNA coding for the transcription factor HNF1/LFB1. EMBO J. 9:2257–2263; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chien Y.-H.; Becker D. M.; Lindsten T.; Okamura M.; Cohen D.; Davis M. M. A third type of murine T-cell receptor gene. Nature 312:31–35; 1984. [DOI] [PubMed] [Google Scholar]
- 11. Chin A. C.; Fournier R. E. K. A genetic analysis of extinction: Trans-regulation of 16 liver-specific genes in hepatoma-fibroblast hybrid cells. Proc. Natl. Acad. Sci. USA 84:1614–1618; 1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Clevers H. C.; Owen M. J. Towards a molecular understanding of T-cell differentiation. Immunol. Today 12:86–92; 1991. [DOI] [PubMed] [Google Scholar]
- 13. Cooper E. L.; Mansour M. H. Cell surface antigen Thy-1. New York: Marcel Dekker; 1989:197–219. [Google Scholar]
- 14. Davidson R. L. Gene expression in somatic cell hybrids. Annu. Rev. Genet. 8:195–218; 1974. [DOI] [PubMed] [Google Scholar]
- 15. Davidson R. L.; Gerald P. S. Improved techniques for the induction of mammalian cell hybridization by polyethylene glycol. Somat. Cell Genet. 2:165–176; 1976. [DOI] [PubMed] [Google Scholar]
- 16. Dev V. G.; Tantravahi R. In: Techniques in somatic cell genetics. New York: Plenum Publishing; 1982:493–511. [Google Scholar]
- 17. Diaz P.; Cado D.; Winoto A. A locus control region in the T cell receptor α/δ locus. Immunity 1:207–217; 1994. [DOI] [PubMed] [Google Scholar]
- 18. Eissenberg J. C.; Cartwright I. L.; Thomas G. H.; Elgin S. C. R. Selected topics in chromatin structure. Annu. Rev. Genet. 91:485–536; 1985. [DOI] [PubMed] [Google Scholar]
- 19. Ephrussi B. Hybridization of somatic cells. Princeton, NJ: Princeton University Press; 1972. [Google Scholar]
- 20. George K. M.; Leonard M. W.; Roth M. E.; Lieuw K. H.; Kioussis D.; Grosveld F.; Engel J. D. Embryonic expression and cloning of the murine GATA-3 gene. Development 120:2673–2686; 1994. [DOI] [PubMed] [Google Scholar]
- 21. Gold D. P.; van Dongen J. J. M.; Morton C. C.; Bruns G. A. P.; van der Elsen P.; Geurts van Kessel A. H. M.; Terhorst C. The gene encoding the ϵ subunit of the T3/T-cell receptor complex maps to chromosome 11 in humans and to chromosome 9 in mice. Proc. Natl. Acad. Sci. USA 84:1664–1668; 1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gorman C. M.; Merlino G. T.; Willingham M. C.; Pastan I.; Howard B. H. The rouse sarcoma virus long terminal repeat is a strong promoter when introduced into a variety of eukaryotic cells by DNA-mediated transfection. Proc. Natl. Acad. Sci. USA 79:6777–6781; 1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gorman C. M.; Moffat L. E.; Howard B. H. Recombinant genomes which express chloramphenicol acetyl transferase in mammalian cells. Mol. Cell. Biol. 2:1044–1051; 1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gottschalk L. R.; Leiden J. M. Identification and functional characterization of human T-cell receptor β gene transcriptional enhancer: Common nuclear proteins interact with the transcriptional regulatory elements of the T-cell receptor α and β genes. Mol. Cell. Biol. 10:5486–5495; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gourdeau H.; Fournier R. E. K. Genetic analysis of mammalian cell differentiation. Annu. Rev. Cell Biol. 6:69–94; 1990. [DOI] [PubMed] [Google Scholar]
- 26. Graham F.; Van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456–457; 1973. [DOI] [PubMed] [Google Scholar]
- 27. Gunther C. V.; Nye J. A.; Bryner R. S.; Graves B. J. Sequence-specific DNA binding of the protooncoprotein ets-1 defines a transcriptional activator sequence within the long terminal repeat of the Moloney murine sarcoma virus. Genes Dev. 4:667–679; 1990. [DOI] [PubMed] [Google Scholar]
- 28. Hashimoto Y. T cell receptor β gene has two downstream DNase I hypersensitive regions. Possible mechanisms of tissue- and stage-specific gene regulation. J. Exp. Med. 169:2097–2107; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hedrick S. M.; Cohen D. I.; Nielsen E. A.; Davis M. M. Isolation of cDNA clones encoding T-cell-specific membrane-associated proteins. Nature 308:149–153; 1984. [DOI] [PubMed] [Google Scholar]
- 30. Henderson A. J.; McDougall S.; Leiden J.; Calame K. L. GATA elements are necessary for the activity and tissue specificity of the T-cell receptor beta-chain transcriptional enhancer. Mol. Cell. Biol. 14:4286–4294; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ho, I.-C; Bhat N. K.; Gottschalk L. R.; Lindsten T.; Thompson C. B.; Papas T. S.; Leiden J. M. Sequence specific binding of human Ets-1 to the T-cell receptor α gene enhancer. Science 250:814–818; 1990. [DOI] [PubMed] [Google Scholar]
- 32. Hyman R.; Kelleher R. Absence of Thy-1 antigen in L-cell × mouse lymphoma hybrids. Somatic Cell Genet. 1:335–343; 1975. [DOI] [PubMed] [Google Scholar]
- 33. Johnson P. F.; McKnight S. L. Eukaryotic transcriptional regulatory proteins. Annu. Rev. Biochem. 58:799–839; 1989. [DOI] [PubMed] [Google Scholar]
- 34. Johnson A. D. The price of repression. Cell 81:655–658; 1995. [DOI] [PubMed] [Google Scholar]
- 35. Junker S.; Pedersen S.; Schreiber E.; Matthias P. Extinction of an immunoglobulln κ promoter in cell hybrids is mediated by the octamer motif and correlates with suppression of Oct-2 expression. Cell 61:467–474; 1990. [DOI] [PubMed] [Google Scholar]
- 36. Ko L. J.; Yamamoto M.; Leonard M. W.; George K. M.; Ting P.; Engel J. D. Murine and human T-lymphocyte GATA-3 factors mediate transcription through a cis-regulatory element within the human T-cell receptor δ gene enhancer. Mol. Cell. Biol. 11:2778–2784; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krimpenfort P.; de Jong R.; Uematsu Y.; Dembic Z.; Ryser S.; von Bohmer H.; Steinmetz M.; Berns A. Transcription of T cell receptor β-chain genes is controlled by a downstream regulatory element. EMBO J. 7:745–750; 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kronenberg M.; Goverman J.; Haars R.; Malissen M.; Kraig E.; Phillips L.; Delovitch T.; Suciu-Foca N.; Hood L. Rearrangement and transcription of the β-chain genes of the T-cell antigen receptor in different types of murine lymphocytes. Nature 313:647–653; 1985. [DOI] [PubMed] [Google Scholar]
- 39. Leiden J. M. Transcriptional regulation during T-cell development: The α TCR gene as a molecular model. Immunol. Today 13:22–30; 1992. [DOI] [PubMed] [Google Scholar]
- 40. Leiden J. M. Transcriptional regulation of T cell receptor genes. Annu. Rev. Immunol. 11:539–570; 1993. [DOI] [PubMed] [Google Scholar]
- 41. Leshkowitz D.; Walker M. D. Extinction of insulin gene expression in hybrids between β cells and fibroblasts is accompanied by loss of the putative β-cell-specific transcription factor IEF1. Mol. Cell. Biol. 11:1547–1552; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liang W.; Cohen E. Somatic hybrid of thymus leukemia (+) and (−) cells forms thymus leukemia antigens but fails to undergo modulation. Proc. Natl. Acad. Sci. USA 72:1873–1877; 1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lieberman S. A.; Hines M. D.; Bergsagel P. L.; Kuehl W. M.; Eckhardt L. A. Coordinate silencing of myeloma-specific genes in myeloma × T lymphoma hybrids. J. Immunol. 151:2588–2600; 1993. [PubMed] [Google Scholar]
- 44. Macleod C. L.; Minning L.; Gold D. P.; Terhorst C. Wilkinson M. Negative trans-regulation of T-cell antigen receptor/T3 complex mRNA expression in murine T-lymphoma somatic cell hybrids. Proc. Natl. Acad. Sci. USA 83:6989–6993; 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Macleod K.; Leprince D.; Stehelin D. The ets gene family. Trends Biol. Sci. 17:251–256; 1992. [DOI] [PubMed] [Google Scholar]
- 46. Maniatis T.; Goodbourn S.; Fischer J. A. Regulation of inducible and tissue-specific gene expression. Science 236:1237–1245; 1987. [DOI] [PubMed] [Google Scholar]
- 47. McCormick A.; Wu D.; Castrillo J.-L.; Dana S.; Jennine S.; Thompson E. B.; Karin M. Extinction of growth hormone expression in somatic cell hybrids involves repression of the specific trans-activator GHF-1. Cell 55:379–389; 1988. [DOI] [PubMed] [Google Scholar]
- 48. McDougall S.; Peterson C.; Calame K. A transcriptional enhancer 3’ of Cβ2 in the T cell receptor β locus. Science 241:205-^08; 1988. [DOI] [PubMed] [Google Scholar]
- 49. McGhee J. D.; Wood W. I.; Dolan M.; Engel J. D.;Felsenfeld G. A 220 base pair region at the 5′ end of the chicken adult β-globin gene is accessible to nuclease digestion. Cell 27:45–55; 1981. [DOI] [PubMed] [Google Scholar]
- 50. Oosterwegel M.; van de Wetering M.; Dooijes D.; Klomp L.; Winoto A.; Georgopoulos K.; Meijlink F.; Clevers H. Cloning of murine TCF-1, a T cell specific transcription factor interacting with functional motifs in CD3-ϵ and T cell receptor enhancers. J. Exp. Med. 173:1133–1142; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Oosterwegel M.; van de Wetering M.; Timmerman J.; Kruisbeek A.; Destree O.; Meijlink F.; Clevers H. Differential expression of the HMG box factors TCF-1 and LEF-1 during murine embryogenesis. Development 118:439–448; 1993. [DOI] [PubMed] [Google Scholar]
- 52. Prosser H. M.; Wotton D.; Gegonne A.; Ghysdael J.; Wang S.; Speck N. A.; Owen M. J. A phorbol ester response element within the human T-cell receptor β-chain enhancer. Proc. Natl. Acad. Sci. USA 89:9934–9938; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Radomska H. S.; Shen C.-P.; Kadesch T.; Eckhardt L. A. Constitutively expressed Oct-2 prevents immunoglobulin gene silencing in myeloma × T cell hybrids. Immunity 1:623–634; 1994. [DOI] [PubMed] [Google Scholar]
- 54. Reich L.; Sharir H.; Ber R.; Wirth T.; Bergman Y.; Laskov R. Phenotypic dominance and extinction of gene expression in murine myeloma × fibroblast somatic cell hybrids. Somat. Cell Mol. Genet. 22:1–20, 1996. [DOI] [PubMed] [Google Scholar]
- 55. Schwarzbauer J. E.; Tamkun J. W.; Lemischka I. R.; Hyness R. O. Three different fibronectin mRNAs arise by alternative splicing within the coding region. Cell 35:421–431; 1983. [DOI] [PubMed] [Google Scholar]
- 56. Silver J. Structural organization of the Thy-1 gene. In: Cell surface antigen Thy-1. New York: Marcel Dekker; 1989:241–269. [PubMed] [Google Scholar]
- 57. Sompayrac L. M.; Danna K. J. Efficient injection of monkey cells with DNA of simian virus 40. Proc. Natl. Acad. Sci. USA 78:7575–7578; 1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Stalder J.; Larsen A.; Engel J. D.; Olan M.; Groudine M.; Weintraub H. Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNase-I. Cell 20:451–460; 1980. [DOI] [PubMed] [Google Scholar]
- 59. Takeda J.; Cheng A.; Mauxion F.; Nelson C. A.; Newberry R. D.; Sha W. C.; Sen R.; Loh D. Y. Functional analysis of the murine T-cell receptor β enhancer and characteristics of its DNA-binding proteins. Mol. Cell. Biol. 10:5027–5035; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Travis A.; Amsterdam A.; Belanger C.; Grosschedl R. LEF-1, a gene encoding a lymphoid-specific HMG domain protein, regulates T-cell receptor α enhancer function. Genes Dev. 5:880–894; 1991. [DOI] [PubMed] [Google Scholar]
- 61. van de Wetering M.; Oosterwegel M.; Dooijes D.; Clevers H. Identification and cloning of TCF-1, a T lymphocyte-specific transcription factor containing a sequence-specific HMG box. EMBO J. 10:123–132; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. van Genderen C.; Okamura R.M.; Farinas I.; Quo R.-G.; Parslow T. G.; Bruhn L.; Grosschedl R. Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1-deficient mice. Genes Dev. 8:2691–2703; 1994. [DOI] [PubMed] [Google Scholar]
- 63. Verbeek S.; Izon D.; Hofhuis F.; Robanus-Maandag E.; te Riele H.; van de Wetering M.; Oosterwegel M.; Wilson A.; Robson MacDonald H.; Clevers H. An HMG-box-containing T-cells factor required for thymocyte differentiation. Nature 374:70–74; 1995. [DOI] [PubMed] [Google Scholar]
- 64. Waterman M. L.; Jones K. A. Purification of TCF-1 alpha, a T-cell-specific transcription factor that activates the T-cell receptor C α gene enhancer in a context-dependent manner. New Biol. 2:621–636; 1990. [PubMed] [Google Scholar]
- 65. Waterman M. L.; Fischer W. H.; Jones K. A. A thymus-specific member of the HMG protein family regulates the human T cell receptor Cα enhancer. Genes Dev. 5:656–669; 1991. [DOI] [PubMed] [Google Scholar]
- 66. Wernert N.; Raes M.-B.; Lassalle M.-P.; Dehouck B.; Gosselin B.; Van-denbunder B.; Stehelin D. c-ets-1 proto-oncogene is a transcription factor expressed in endothelial cells during tumor vascularization and other forms of angiogenesis in humans. Am. J. Pathol. 140:119–127; 1992. [PMC free article] [PubMed] [Google Scholar]
- 67. Winoto A.; Baltimore D. A novel, inducible and T cell-specific enhancer located at the 3′ end of the T cell receptor α locus. EMBO J. 8:729–733; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Winoto A.; Baltimore D. αβ lineage-specific expression of the α T cell receptor gene by nearby silencers. Cell 59:649–655; 1989. [DOI] [PubMed] [Google Scholar]
- 69. Wu C. The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase-I. Nature 286:854–860; 1980. [DOI] [PubMed] [Google Scholar]
- 70. Yamada T.; Hitomi Y.; Shimizu K.; Ohki M.; Oikawa T. Extinction of T cell receptor alpha-chain gene expression accompanied by loss of the lymphoid enhancer-binding factor 1 (LEF-1) in murine somatic cell hybrids. Mol. Cell. Biol. 13:1943–1950; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yu H.; Porton B.; Shen L.; Eckhardt L. Role of the octamer motif in hybrid cell extinction of immunoglobulin gene expression: Extinction is dominant in a two enhancer system. Cell 58:441–448; 1989. [DOI] [PubMed] [Google Scholar]


