Abstract
Nuclear receptors are model transcription factors. This highly conserved super family of ligand binding transcription factors includes estrogen, progesterone, retinoic acid, thyroid hormone, vitamin D receptors, and several orphan receptors. Nuclear receptors function as homodimers, heterodimers, or monomers. Human thyroid hormone, retinoic acid, vitamin D, and several orphan receptors prefer to work as heterodimers with retinoic X receptor (RXR). RXR function is regulated by its cognate ligand 9-cis-retinoic acid. In some cases heterodimers of RXR are subject to regulation by two different ligands. Mammalian cells are not entirely suited to study pure heterodimeric functions because they contain a repertoire of endogenous receptors and their ligands. Yeast does not contain nuclear receptors or its ligands. Ligand-dependent function of several human nuclear receptors has been reconstructed in yeast. Yeast can be used as a model system to dissect interaction between various heterodimeric partners. The molecular genetics and the speed of doing the experiments in yeast allows us to rapidly clone mammalian cofactors that prefer to work with different heterodimeric partners. Once the human geneome sequence is complete, we predict that the total number of human nuclear receptors will increase from 150 to 500. Novel and efficient cell-based systems will be needed to understand the function of orphan receptors. Yeast is an ideal system to identify pure heterodimeric partners and discover novel ligands for orphan receptors. The advantages and disadvantages of yeast and mammalian system to study nuclear receptor function are discussed.
Keywords: Human nuclear receptors, Targeting transcriptional regulation, Nuclear receptor function in yeast
THE CHALLENGE
Signal transduction has several control points that are attractive drug targets (30,65). Among myriad of targets, regulation of transcription remains an unexploited area for therapeutic intervention. Transcriptional regulation is a complex process that involves interaction of regulatory transcription factors with the basal transcription machinery, to activate or repress target genes [for reviews on transcription see (10,22,24,29,68)]. Complete sequence analysis of H. influenza genome suggests that about 5% of the total genes encode for transcription apparatus (14). If we are to extend these estimates to the human genome, then it appears that about 5000 genes are responsible for selective regulation of 100,000 human genes, during growth and development. The number of regulatory transcription factors (TFs) is likely to be a small proportion of the 5000 genes involved in gene transcription. It is clear from these estimates that a limited repertoire of transcription factors governs the tissue- and cell-specific anatomy and physiology. How do cells make use of the limited number of transcription factors to differentially regulate gene expression? The answer lies in combinatorial interaction between various transcription factors to generate multiplicity that in turn endows the cell with diversity in TFs. Thus, NFκB, a regulatory TF, may interact with c-Jun and ATF-2 to specifically regulate INFβ gene in one case whereas it can interact with c-Jun:cFos and NF-AT to regulate IL-2 gene in T cells (53,70). Thus, oligomeric and combinatorial interaction is a common theme among all the TFs. The critical question is how to decipher the oligomeric nature of the superamolecular complex of TFs that occupy the target gene. Nuclear receptors are model transcription factors. The issue of multiplicity and diversity and how to exploit these features for selective drug targeting is the main subject of this review. We have borrowed heavily from the retinoid, thyroid, and vitamin D receptor field to illustrate several examples of heterodimeric interplay. The advantages of yeast to study interaction between heterodimers, basal transcription machinery, and other mammalian coregulatory proteins, in the presence or absence of cognate ligands, is also discussed.
THE MAIN QUESTION
Currently about 150 nuclear receptors have been discovered (49). Ligands for most of these nuclear receptors have not been identified as yet; hence they are known as “orphan receptors.” We predict that most (if not all) of the human cDNAs will be made public by the end of 1996, via expression sequence tag (EST) approaches (1). It is reasonable to assume that with the completion of human genome sequences, this number is likely to increase up to 500. With such a dramatic increase in the number of nuclear receptors, elucidation of their biological role will clearly lag behind. Because many of the receptors heterodimerize with retinoic X receptors (RXR) as well as other receptors, it is essential for us to know the heterodimeric partner that targets a promoter of interest. In some cases both the heterodimeric partners are responsive to their cognate ligands. Some of the key questions addressed in this article are: How do nuclear receptors select heterodimeric partners and what is the role of DNA response elements in this selection? Can we develop yeast cell-based systems that will allow studies on pure heterodimeric interactions, in order to discover novel ligands for orphan nuclear receptors?
NUCLEAR RECEPTORS AND THEIR MODE OF ACTION
Nuclear receptors control diverse aspects of growth, development, and homeostasis by regulating the transcription of complex networks of genes. Members of this superfamily (Fig. 1) are transcription factors that modulate the activity of promoters in target genes through direct association with specific DNA sequences called hormone response elements (HREs). Typically these receptors bind to direct repeats (DRs) whereas some also bind as symmetrical repeats that are either inverted (IR) or everted (ER) (3,6,19,51,71,75,76). A graphic representation of the DNA binding properties of these receptors on DR, IR, and ER response elements is shown in Fig. 2. These receptors exert their regulatory effects on target gene expression by binding as monomers, homodimers, or heterodimers to hexameric AGGTCA core motifs in HREs that have subtle variations in sequences, spacing, and orientation. The nuclear receptor superfamily can be broadly subdivided into four classes based upon their dimerization and DNA binding properties [see (69)]. The first two subclasses are ligand-inducible receptors. Class I includes the known steroid receptors that function as ligand-induced homodimers and bind to DNA half-sites organized as inverted repeats (see Fig. 2). Class II includes all other known ligand-dependent receptors exclusive of steroid hormones and consists predominantly of the thyroid, retinoid, and vitamin D receptors, which heterodimerize with RXR (35,36,44,50,80,87). The members of this class of receptors share significant homology at primary structure level, as illustrated in Fig. 1.
FIG. 1.
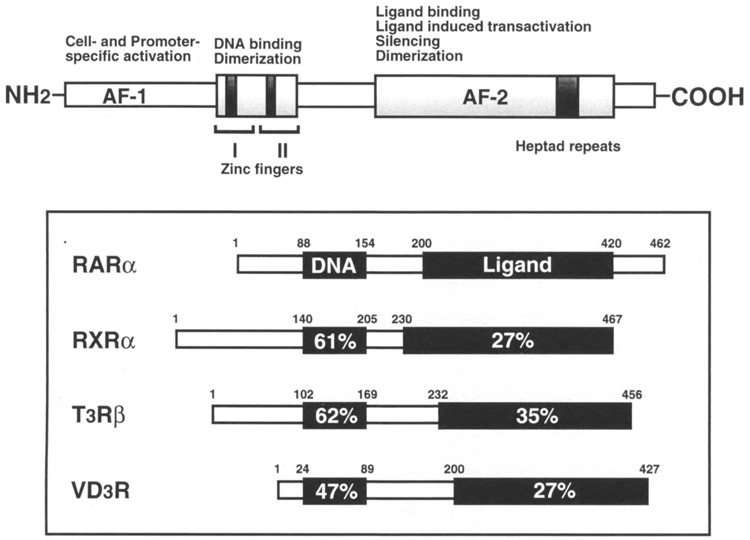
Comparison of nuclear receptor family of related proteins. Nuclear receptors contain highly conserved modular structures composed of two zinc finger DNA binding domain and the carboxy-terminal ligand binding domain. The detailed properties of different domains are summarized in the diagram. The percent identity of amino acids between representative members of the nuclear receptor superfamily is also shown. The amino-terminal of the receptor proteins is highly variable and may be involved in promoter or cell-specific transactivation. The two transcription activation functions (AF), AF-1 and AF-2, of the receptor proteins are distributed between amino- and carboxy-terminals.
FIG. 2.
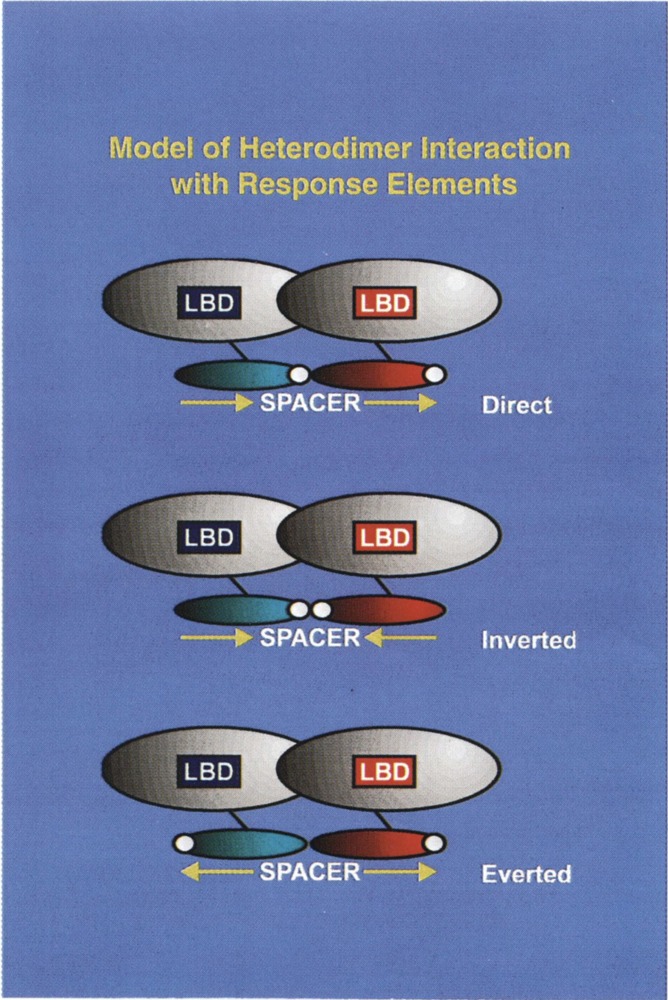
Model for the binding of heterodimer receptor interaction with direct repeat (DR), inverted repeat (IR), and everted repeat (ER) elements. The important features of this model are: the ligand binding domain (LBD) present at the carboxy-terminus mediates dimerization. The ability of these heterodimers to bind to DR, IR, or ER elements implies that the DNA binding domains are rotationally flexible with respect to the carboxy-terminal dimerization interface of the receptors. In one DR element RXR has been demonstrated to be located on the upstream half-site, with heterodimeric partner, RAR, TR, or VDR binding to the downstream half-site of the recognition motif. This polarity of RXR and the heterodimeric partner also determines the optimum spacing between the half-sites of the recognition motif (for further discussion see the New Concepts in Regulation of Heterodimer Function section).
Of particular interest, the specific ligand (or hormone activator) of responsive nuclear receptors can be generated from one of three established sources to date: i) the ligand may be synthesized at a site other than the target cell in the classic endocrine fashion (e.g., thyroid hormone); ii) the ligand may be generated within the target cell from a precursor (apohormone) such as the isomerizaton of all trans-retonic acid to 9-cis-retinoic acid (9c-RA) (e.g., the specific ligand and RXR); iii) the ligand is synthesized intracellularly where it is only active locally and is not secreted (e.g., prostaglandin). The class III and class IV subgroups contain primarily receptors whose cognate ligands have as yet to be identified and are therefore designated to be orphan receptors. Class III receptors bind primarily to direct repeat HREs as homodimers, whereas class IV typically binds to extended core sites as monomers (see Fig. 2).
Structural Features
As shown in Fig. 1, the highly conserved DNA binding domain is characteristic of the nuclear receptor superfamily and mediates the recognition of specific base pairs within the core binding motif through two interdependent zinc finger structures that form major groove contacts with base pairs that define the core recognition motif [(46) and references therein]. Moreover, amino acids immediately carboxy-terminal to the zinc finger, termed the T box, are involved in defining the part of the dimerization interface between RXR homodimers bound to an RXR-specific response element, and an adjacent amino acid cluster, termed the A box, is necessary for the recognition of specific base pairs at the 5′ end of the core recognition motif. Although differences occur in A and T boxes for the nuclear receptor within each subgroup, such as RXR vs. RAR vs. TR, sequences are highly conserved within a specific receptor subtype (isotype) (e.g., RXRα vs. RXRβ vs. RXRγ). Additionally, it has been established that the high-affinity binding of heterodimers is dependent not only upon these DNA binding regions but also upon carboxy-terminal dimerization function present within the ligand binding domain of each receptor (12,20,21,44,74,80).
Class I and II nuclear receptors achieve transcriptional regulation through autonomous transcription activation functions (AF) wherein AF-1 domain is constitutive and located in the N-terminal part of the receptor and the AF-2 or τ (Tau-4) domain is ligand dependent and is located in the C-terminal domain (see Fig. 1 for detailed structural comparison) (9,15,23,31,52,64,72,83). Numerous studies of carboxyl-terminal domain of the nuclear receptors have established that it contains many overlapping functions that mediate ligand-dependent activation and repression, receptor homo- and heterodimerization, as well as ligand binding (20,49). The binding of ligand to the receptor is believed to induce a conformational change in receptors that leads to activation of transcription (55,61,69,81).
Mechanistic Aspects
The mechanisms whereby ligand-activated DNA-bound receptors transmit their transactivating “signal” to the basal transcription machinery and the factors underlying hormonal activation are not precisely known. Transcriptional activators are thought to modulate transcription by promoting or stabilizing the assembly of preinitiation complexes, which may involve direct or indirect actions on components of the basal transcription machinery [see reviews (62,85)]. However, experimental evidence supports the existence of bridging molecules, which are termed coactivators, that function as transcription intermediary factors (TIFs) or adaptors and are thought to mediate the interaction of transactivators with the basal transcriptional machinery (24). Moreover, transcriptional interference can be demonstrated by the presence of a strong activator that suppresses the activity of other transactivational factors by sequestering putative bridging factors [see reviews (45,59)] or by occupying a surface required to mediate or receive trans-acting signals through a phenomenon of surface saturation (66). Recent detailed studies in the most C-terminal part of the nuclear receptor AF-2 (τ) domain has revealed a negatively charged amphipathic α-helical region, which has a high degree of conservation between class 1 and class II receptors and which regulates transactivation as well as transcriptional interference (i.e., squelching between class I and class II receptors). These observations indicate that this region is probably part of a surface of interaction for either a general putative coactivator or basal transcription factor. Alternatively, it could assist a putative bridging factor utilized to initiate transcription by both class I and class II receptors in the presence of cognate ligands.
HETERODIMERIZATION – A KEY PROPERTY OF NUCLEAR RECEPTORS
Key Role of RXR
The formation of heterodimers by RXRs with the other members of class II nuclear receptors facilitates their binding to DNA response elements containing degenerate Xn-AGGTCA core motif half-sites in natural genes, which are configured as DRs (76), IRs (75,84), ERs (3,11,60,71). Because the ultraspiracle receptor (which is the RXR homologue in Drosophila) is a partner for the ecdysone receptor mediating morphogenesis, hetero-dimerization is a biological phenomenon that must have developed prior to the divergence of vertebrates and invertebrates and has been evolutionary preserved as a critical component of endocrine signaling pathways. The common heterodimerization partner RXR also has an overlapping role in retinoid signaling, and three mammalian RXR isoforms subserve these heterodimerization functions (18). These findings strongly link vitamin A signaling to many nuclear receptor transcriptional pathways.
The formation of heterodimers by class II nuclear receptors leads to several models of receptor–receptor interaction with response elements containing differently configured conserved domains that function in an interdependent manner to mediate protein–DNA and protein–protein interactions [see review (49)]. Protein–DNA interactions are mediated by the highly conserved DNA binding domain within each receptor that defines the nuclear receptor superfamily, whereas the protein–protein interactions necessary for the formation of heterodimers are mediated by extensive C-terminal dimerization interfaces contained within the ligand binding domain [see review (20)]. Thus, for DRs (which promote head to tail arrangements of nuclear receptor dimers on target DNA), the base pair sequences that separate two core binding sites can regulate the DNA binding and transcriptional effects of RXR for spacers of 1, 2, 3, 4, and 5 nucleotides for heterodimerization with either PPAR and RXR, RAR, VDR, T3R, and RAR respectively (33,36,51,76). The optimal gap length for each heterodimer is determined by protein–protein contacts that appropriately position the DBDs of RXR and its partner (37,56,57,73,86). The formation of RXR heterodimers with other class II receptors such as RAR, TR, and VDR increases not only the efficiency of binding to DNA but also results in specific response element repertoires. However, in contrast to DRs, which promote head to tail orientations, homodimers have very weak DNA binding and transactivation on DRs and instead bind with higher affinity to IR and ER symmetrical (palindromic) HREs. Additionally, TR has a much higher preference than RXR for binding to PuGGTCA (G motif) over a PuGTTCA (T motif) and is also more efficiently bound to DR4 when the PuGGTCA is the 3′ end motif than rather than the 5′ end motif (DR4G/T). Such observations suggest that the binding of RXR/TR heterodimers occurs with a designated polarity (to result in anisotropic complexes) (48,86). As revealed by cross-linking experiments using full-length receptors, RXR is almost invariably located at the 5′ end to either TR or RAR on DR + 4 and DR + 5 elements, respectively [see review (20)]. The ability of homo- and heterodimeric receptors to bind to palindromic (ER or IR) and DR orientations of the core recognition motif implies that the DBDs are rotationally flexible with respect to the C-terminal dimerization interface. Because a change of spacing by one nucleotide for a DR-DNA target requires the RXR partner to rotate approximately 36° around the double helix and be translated 3.4 Å, the DNA spacing between core motifs is critical in determining heterodimer DBD complex formation on direct repeat HREs. When heterodimers bind to DRs, RXR is located on the upstream core recognition motif with its heterodimeric partner bound to the downstream motif (see Fig. 3). Hence, through the formation of an asymmetric interface between DBD of the RXR and its heterodimeric partner, the optimal spacing between core recognition motifs and the preferential binding affinity to specific heterodimers is favored. Although ER or IR HREs also promote transactivation from homodimers, they may also transactivate heterodimers but to a lesser degree of specificity (51). Thus, an IR + O of the AGGTCA motif (TRE-PAL) can function as a common response element not only for TRs but also RARs and RXRs. Similarly, the everted palindromes present in the chicken lysozyme and medium-chain fatty acyl coenzyme A dehydrogenase genes can support nonspecific transcriptional responses to both RARs and TRs. Such observations suggest that studies of transcriptional responses of class II receptors by promoters containing DR elements initially proposed to respond according to a relatively simple spacing “the 1 to 5 rule” for determining receptor-ligand specificity (76) may not apply to all biological conditions. In fact, the subsequent demonstration that such receptors can bind to DNA as either homodimers or heterodimers and the identification of additional hormonal response elements in natural genes supports a more complex picture of transcriptional regulation. From studies of many genes, under a variety of experimental conditions, it can be concluded that whereas the spacing between DRs as well as superficially for ERs and IRs may limit the degree of cooperatively between dimerization interfaces, it does not impose an absolute restriction on the hormonal responses of nuclear receptor heterodimers, because they can also be influenced by heterodimer subtypes, cellular and promoter contexts, as well as accessory basal transcriptional machinery coregulators (see below).
FIG. 3.
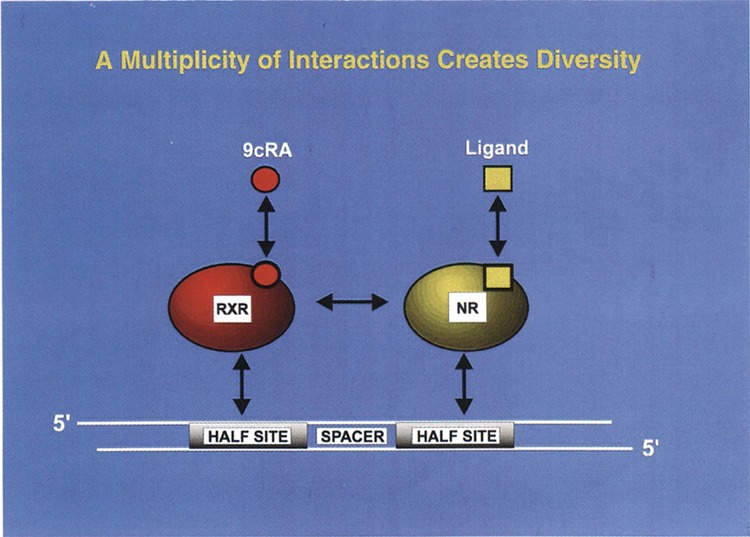
A multiplicity of interaction creates diversity. Heterodimerization is a key property of nuclear receptors and other transcription factors. In this diagram the retinoic X receptor (RXR) has been shown to heterodimerize with a variety of nuclear receptors (NR). The functional interaction between the receptor heterodimers is governed by DNA sequence of the half-site recognition motif, the spacer sequence, protein–protein interaction, protein–DNA interaction, and the cognate ligands. The model is primarily driven by reconstruction of ligand-dependent function of human nuclear receptors in yeast. Dual ligand function for nuclear receptor heterodimers is also a very important feature of this model. [Adapted from Kephart et al. (34).]
Since multiple subtypes derived from different chromosomes have been detected for TRα and β (40), RXRα, β, and γ, as well as RARα, β, and γ (18), and these receptors are differentially expressed in mammalian tissues, the potential diversity of different nuclear receptor class II heterodimer combinations is enormous. A simple calculation to estimate the number of different combinatorial interactions between RXRα and other heterodimeric partners has suggested about 400,000 possible combinations (5). Not only are there subtle differences in molecular size as well as structure for each of these receptor subtypes (isotypes), but also variant isoforms, which arise through alternate splicing and promoter usage, have been discovered for each receptor subtypes (i.e., RARα 1, 2, and 3), which can differ in their expression and function. The formation of heterodimers with differences in subtype and isoform and their functions on a variety of differently configured HREs undoubtedly result in conformational changes that could control the accessibility of the LBD domain to cognate ligands and its contacts with the basal transcription machinery.
The interaction of cognate ligands with TR/RXR heterodimers can lead to four states of receptor occupancy: 1) both TR and RXR receptors unoccupied, 2) RXR occupied and TR unoccupied, 3) RXR unoccupied and TR occupied, and 4) both TR and RXR receptors occupied. Thus, it is possible that effects on dimerization and/or transcriptional activation could have different consequences for each state of receptor occupancy. Theoretically, the effects of a single cognate ligand of a heterodimeric–DNA complex could be dependent not only upon the subtle con-formational differences resulting from the formation of diverse isotype and isoform combinations of heterodimers but also their consequent interactions with DNA sequences in target genes with differences in half-site sequence orientation and spacing between half-sites, as well as the heterogeneities in promoter context and tissue-specific transcriptional coregulatory factors. Such biological conditions will also determine both the action of each cognate ligand on the heterodimers and whether there will be further dual ligand-induced transcriptional enhancement or suppression (see Fig. 3). Despite the complexities of transcriptional regulation that can arise by the formation of the heterodimers–DNA complexes, such diversity can be exploited nevertheless to discover specific combinatorial targets of transcriptional regulation (see the section on Yeast Facilitates Dual Ligand Responsivity of Heterodimers). The elucidation of new pathways of heterodimer interactions with the coregulators of transcription can provide unique opportunities for selectively regulating transcription by controlling the contact of cognate ligands with the basal transcription machinery.
NEW CONCEPTS IN REGULATION OF HETERODIMER FUNCTION
Polarity of Heterodimer Interaction
As discussed above and reviewed in detail elsewhere (20), studies of thyroid hormone and retinoic acid receptor interactions with RXRs have raised the possibility that specific orientations in both the DNA binding domain and C-terminal dimerization domain interfaces may contribute to the binding site preferences of homodimeric and heterodimeric receptor complexes (37). Because the DBD of the TR is proposed to preferentially rotate by approximately 180° with respect to the C-terminal dimerization interface, such an alignment with RXR would result in a head to tail configuration that would position the RXR over the upstream binding site (5′ end) of a DR + 4 HRE and facilitate the formation of the asymmetric interface between DBDs that determines optimal spacing of core recognition motifs. Similarly, DR + 5 spacing would favor RXR occupying the upstream (5′ end) binding site and RAR the downstream (3′ end) site and implies that the cooperative interface formed between DBDs also contributes to a polarity in the arrangement of heterodimer binding sites. Using mammalian cells, several groups have established that the polarity of DNA binding by RXR/TR or RXR/RAR het-erodimers can modulate ligand responsivity such that only the 3′ end downstream site is responsive to ligand whereas the RXR heterodimer partner, which is in a 5′ end upstream location, is tran-scriptionally suppressed (i.e., a silent partner) (17). Interestingly, RXR can be responsive to 9c-RA when it heterodimerizes with specific orphan receptors (17,41,49). The responsivity of RAR to its cognate ligand [i.e., all-transretinoic acid (at-RA)] on a DR + 1 compared to DR + 5 HRE when bound as a heterodimer with RXR [which results in opposite polarities of RXR/RAR heterodimeric binding to the asymmetrically orientated half-sites (38)] induces transactivation when RAR is on the downstream (3′ end) half-site on DR + 5 but not when RAR is bound to the upstream half-site on a DR + 1. However, mutations in RAR and RXR that reverse binding have been observed to alter the pattern of transcriptional response to RAR-specific ligands.
Regulation of Heterodimeric Function by Cell and Promoter Context
Studies assessing identical TR/RXR-TREs complexes in CV1 and HeLa cell lines have established that the ligand binding and transactivation effects of 9c-RA are suppressed in the absence of T3 and that combined 9c-RA and T3 could not enhance ligand-dependent transactivation to levels greater than those observed for T3 alone (16,17). However, studies in JEG3 mammalian cells using identical nuclear receptor heterodimers detected enhanced dual ligand transactivation in the presence of a rat growth hormone promoter or palindromic sequences but not when DR + 4 response elements were present (63). In agreement, the expression of VDR/RXRα heterodimers in ML breast cancer cells or ultraspiracle-deficient Drosophila SL3 cells could also achieve dual (vitamin D + 9c-RA) ligand-dependent transactivational enhancement on a mouse osteopontin natural gene promoter but not a palindromic vitamin D response element (6). In contrast, studies of VDR action in monkey kidney, Cos7 cells, and the rat osteosarcoma cell line ROS17/2.8 (which have endogenous RXRα and β but not γ) have shown that 1,25 (OH)2 D3 action could not be enhanced by co-added 9c-RA (13).
Regulation of Transcription by Cell-Specific Coregulators
Studies of nuclear receptor function in different eukaryotic cell lines using identical heterodimer–DNA sequences have demonstrated that the action of each cognate ligand for the heterodimer partner is differentially regulated by variations in cellular context. Such differences between cell types demonstrate the important regulatory role of local environmental accessory cellular factors in determining whether a putative transcription initiation complex will be either assembled or inhibited. These observations indicate that cell line-related differences in compressor or coactivator proteins that link these activated heterodimeric DNA complexes and their ligands to the RNA polymerase II holoenzyme complex play important roles in transcriptional regulation. Unliganded nuclear receptor heterodimers can inhibit transactivation by preventing formation of the preinitiation complex. Indeed, because regions in the hinge and N-terminal part of ligand binding domain of the thyroid hormone receptor are also required for silencing, the existence of additional interacting factors that are essential for ligand-independent repression has been postulated. In accordance with these speculations, 270 kDa (N-CoR) and 168 kDa (SMRT) proteins have been cloned from mammalian cell libraries as TR and RAR associated compressors (TRACs), which dominate over ligand-dependent transcriptional activation events (8,32). Such proteins bind with two unliganded receptors and are released upon ligand binding. The carboxyl-termini of both proteins contain a receptor interaction domain whereas the amino-terminal portion contains two novel repressor motifs. In the absence of ligand, the repressor remains associated with the receptor and this results in a strong inhibition of basal transcriptional activity of the associated promoter. However, in the presence of their cognate ligand, the TRACs repressor can be dissociated and the suppression relieved to allow net activation. Because an intact C-terminus transactivation (AF-2) domain is essential for the dissociation of the co-factor and thereby linkage between transcriptional suppression and activation through this mechanism can be postulated (8). Several positive cofactors have been additionally identified to include Tripl (42,79), TIF-1 (42), RIP140, and RIP160 (also known as ERAP160) (7,25). The Trip1 is homologous to the yeast SUG1 protein and interacts with AF-2/τ c domain of TR/RAR and VDR as well as estogen receptor and minimally with RXR (43,78) in a hormone-dependent fashion whereas the ERAP160 and RIP140 proteins binds to the AF-2/τ region in a hormone-dependent fashion to estrogen receptor, RAR, and TR. Moreover, the amino-terminus of TR interacts specifically with TFIIB (4) and the AF-2/τ c of RXR directly interacts with TATA binding protein, both in vivo and in vitro (67), thereby suggesting that an individual receptor may have multiple pathways of activation. Although TRACs have been observed to be ubiquitous, subtle differences have been observed between different mammalian cells (32). Additionally, the ligand-induced reversibility of TRACs was reported to be dependent upon the polarity of heterodimer binding unless the TR or RAR hinge region, which binds these corepressors, is inactivated by structural mutations (39). These observations suggest that mammalian TRACs are dominant over coactivators with respect to ligand-independent transactivation of heterodimers in either a positive or negative transactivational mechanism and that polarity of heterodimeric binding to the DNA sequences results in allosteric mechanisms that can be influenced by the action of cognate ligands.
ROLE OF YEAST IN DECIPHERING SPECIFIC HUMAN HETERODIMERIC FUNCTIONS
Yeast Facilitates Dual Ligand Responsivity of Heterodimers
Although TR and RAR receptors preferentially bind to specific HREs in target gene as heterodimeric complexes with RXRs, the action of each cognate ligand and the role of accessory cellular factors that differentially regulate the transcription responses to heterodimeric-DNA complex are not well understood. Studies in most mammalian cell lines have demonstrated that unliganded TR and RAR function as transcriptional silencers and that the formation of heterodimers with RXRs inhibits 9c-RA binding and the activation of transcription (16,17). However, from parallel studies in yeast, we and others have demonstrated that TR functions as an activator on Ir palindromes and TRE palindromes (54,58) and that TR/RXR heterodimers not only enhance transactivation when complexed with a DR + 4 HRE but also that the RXR partner of the heterodimer is also responsive to 9c-RA (26,82). These observations confirm the important regulatory role of cell-specific accessory transcriptional cofactors in controlling the eukaryotic gene expression. In the cellular context of yeast, our studies have discovered that RXR is shifted from its function as a silent heterodimer partner to an active receptor that is not only responsive to 9c-RA but can also promote dual ligand (9c-RA + T3) enhanced transactivation. This dual ligand-dependent increase in transcription was only observed with TRβ:RXRγ, but not with TRβ:RXRα heterodimers (82). Such discordant observations in yeast compared to mammalian cells also suggest that inhibition of 9c-RA responsiveness in mammalian cells may be mediated by TRAC-like repressor and represents a tissue-specific phenomenon. It is therefore likely that the primitive eukaryote yeast is devoid of comparable TRAC-like corepressor proteins recently discovered in mammalian cells (8,32) These repressors interact with the hinge region of (D subdomain of the ligand binding region) of TR and RAR, but not RXR, vitamin D, or other known members of the type I steroid nuclear receptor family. Receptor-specific (T3 or at-RA) ligands are required to relieve basal repression and promote activation of transcription. In the absence of specific corepressor TRAC proteins and/or the presumptive presence of dominant coactivator or adaptor proteins in yeast, RXR can escape from the inhibitory effects of TR heterodimerization in the mammalian cells to permit the C-terminus LBD of RXR to contact directly with 9c-RA and the specific basal transcription apparatus involved in RNA polymerase II activation. Similarly, from studies on the interactions between human vitamin D receptor (VDR) and human RXRα, mouse RXRβ-2 and mouse RXRγ, we have observed that RXR/VDR heterodimers showed ligand-dependent transactivation from natural VDREs present in rat renal 24-hydroxylase gene by RXR subtype-specific VDR heterodimers but not from consensus VDREs, which bound with high affinity as VDR-RXR complexes (34). Enhanced transactivation of VDR was observed only with RXRα or RXRγ and not with RXRβ, even though the latter heterodimer bound tightly to DNA. Hence, the reconstruction of renal 24-hydroxylase promoter in yeast is very similar with respect to the transactivation potential of specific VDREs and the fold activation is very similar to that previously observed in osteosarcoma cells (34). Thus, the absence of endogenous nuclear receptors and ligands as well as the likely unique cellular context of transcriptional cofactors present in yeast can facilitate the detection of novel regulatory pathways that are unique to mammalian cells. These features can also be applicable to the discovery of specific co-regulators of heterodimer function. Also the action of cognate ligands and their anologues on novel protein–protein and protein–DNA interaction with the basal transcription machinery can be systematically evaluated.
PROS AND CONS OF USING THE YEAST SYSTEM TO STUDY HETERODIMERIC INTERACTIONS
It should be clear to the reader that we use the yeast system as a tool to address mammalian nuclear receptor properties that are not easily studied in mammalian cells. Although the yeast system by no means replaces the mammalian system, both systems can complement each other in studies of eukaryotic transcription regulation, depending upon the nature of the question. Table 1 summarizes advantages and disadvantages of the yeast and mammalian systems with respect to study of nuclear receptors.
TABLE 1.
PROS AND CONS OF USING YEAST TO STUDY NUCLEAR RECEPTORS
| Yeast | Tissue Culture Cells |
|---|---|
| Ligand-dependent transactivation of nuclear receptors possible. Compounds that act as antagonists in tissue culture cells behave as partial agonists in yeast. | Cell and promoter selective agonist and antagonist function can be reconstructed. |
| Pure heterodimeric and multimeric interactions possible with nuclear receptors. | Not possible due to endogenous nuclear receptors. |
| Ligands are not metabolized. | Ligands are metabolized in mammalian cells. |
| Unique (simple) basal transcription context. | Physiological relevance of the unique context, depending upon the promoter and cell line. |
| Can produce large amounts of a receptor protein. | Incompatible with cell physiology. |
| Complementation cloning to discover novel orphans or partners (i.e., RIPs and TRIPs). | Not possible or very time consuming. |
| Stable transformants. HTS engine, parallel screens possible using robotics. | Transfection of DNA required. Data from stable cell lines may not be reliable. |
Ligand-Dependent Responses
As discussed previously, because of its simpler (or primitive) RNA polymerase II context, hetero-dimers expressed in yeast with RXR retain a potential to respond to both the ligands depending upon the specific DNA configuration and half-site spacing. This is an advantage, as we can explore the ligand binding sites of both the monomers that make up the functional transcription factor. So far RXR has been identified as one of the promiscuous heterodimeric partners; with the advent of more heterodimeric partners to be discovered from about 80% of the human genome yet to be made public, the yeast system is likely to play an important role in discovery of new ligands. As outlined in Table 1, it is important to recognize that although yeast is quite faithful in profiling compounds that have shown agonist responses in mammalian cells, it does not faithfully profile molecules that act as antagonists in mammalian cells. For example, human estrogen and progesterone receptor antagonists behave as partial agonists in the yeast system [(34,47,77), Graumann et al., unpublished data]. We therefore suggest that the yeast system not be used for detail profiling of antagonist activities. However, it is relevant to note that the agonist activities of various class I and class II receptors in yeast have provided a very sensitive cell-based system to monitor the majority of ligand-dependent responses (2,26-28,34,47,54, 58,77).
Pure Heterodimeric Interactions
One of the advantages of the yeast system is that almost all of its genome has been sequenced (or will be made public by the end of 1996). Thus far, to our knowledge, no endogenous RAR, RXR, TR, or VDR or orphan receptor-like sequences have been found in yeast. Although it is remarkable that most human nuclear receptor ligands are very responsive in yeast, the main advantage of yeast lies with its primitve environment that is devoid of nuclear receptors. Any transcriptional responses, constitutive or ligand dependent, are purely the result of the receptor genes that have been transformed into yeast. In this respect one is studying pure heterodimeric or homodimeric receptor interaction that is not possible in mammalian cells. How the yeast system can be exploited to discover receptor-targeted ligands can be illustrated from the example of COUP, a family of orphan receptors that are considered to be homodimeric repressors. If we wish to identify a ligand that will block the suppresser function of the receptors, yeast can be easily screened to discover COUP-specific ligands. Nuclear receptors including COUP do render some constitutive transactivation response in yeast (Tsai et al., personal communication). These constitutive properties in yeast can in turn be used as signals to identify agonists/antagonists. Such ligands can be further screened in appropriate mammalian cell systems to confirm and validate the profile of the ligands. By analogy, the yeast system is being used as an engine to discover ligands.
Another important feature of using the yeast system is that cells are grown in synthetic minimal medium, and unlike tissue culture cells that need fetal calf serum, the transcriptional responses observed in yeast are not subject to endogenous ligands present in tissue culture medium. Some of the ligands that have been tested by our laboratory (i.e., 9c-RA, vitamin D3, T3, estrogen, and progesterone) do not undergo any significant chemical modification in yeast as has been the case with some of the ligands in tissue culture cells. Because the ligand-dependent responses observed in yeast are pure and entirely driven by the added ligands, the yeast system is therefore quite amenable to high throughput screening to discover novel ligands.
Complementation Cloning
Development of several molecular biological tools (i.e., variety of plasmids, regulated promoters, genetic markers) and the power of yeast molecular genetics have greatly enhanced our ability to clone mammalian genes that cooperate or complement a function on yeast-expressed human nuclear receptors. The yeast system has been used by a number of laboratories to clone ligand-dependent or independent corepressors and coactivators of the nuclear receptors (8,32,43). The value of using yeast to study nuclear receptors is twofold. First, yeast genetics can be used to identify yeast coregulators that are essential for ligand-dependent transactivation of nuclear receptors. The yeast genes SWI1, SWI2, and SWI3, which are required for transcription of several yeast genes, were also found to be essential for glucocorticoid receptor function in yeast (79). The SWI homologs in higher eukaryotes have been discovered and their function can be reconstrucetd in yeast for further studies (24). Second, the partners of human nuclear receptors can be cloned by transforming mammalian cDNA expression libraries to complement a desired function in yeast. This approach has been employed by number of investigators, as described above. Using this approach, Lee et al. (43) have identified a human protein that binds to TRβ and is a homolog of yeast SUG1 gene. Thus, evolutionary conservation of RNA pol II apparatus between yeast and mammals can be used in a number of ways to study yeast or mammalian coregulator interaction with nuclear receptors. This is the single most useful feature of the yeast molecular genetics that has allowed us to identify functional and perhaps physiologically relevant partners. In this respect, studies on human nuclear heterodimers in yeast are of special significance as we set out to reconstruct human nuclear receptor supermolecular complexes in yeast.
FUTURE PERSPECTIVES
Numerous findings of nuclear receptor heterodimeric interaction can be summarized in Fig. 3. Driven by the studies performed in yeast, this model proposes that both cognate ligands of the heterodimeric partners can be functional on a target gene. Although RXR has been largely presumed to work as a silent partner in mammalian cells, exceptions have been increasingly documented (41). We predict that several opportunities will arise when dual ligand action may be activated. This feature has not been fully exploited in mammalian cells because of the limited number of laboratories that have focused nuclear receptor studies on more or less the same cell lines. Our findings also suggest that 1, 2, 3, 4, and 5 base pair spacer rule could also be more flexible than previously surmised. As described in Fig. 3, these multiple features create a significant amount of diversity that is presumably exploited by cells to modulate their physiology. The heterodimeric properties of human nuclear receptors observed in yeast should certainly be exploited to discover novel ligands for therapeutic benefit. We have previously calculated the number of different heterodimers that are possible with a variety of response elements, monomeric partners, and their cognate ligands (5). Conservative calculations suggest that combinations of about half a million different structures are possible. The critical question is how to decipher which of these structures are operative on a target gene. We believe that reconstruction of an appropriate heterodimer function in yeast in the presence of a consensus or a natural response element is one of the several promising approaches that can be followed by the discovery of a novel ligand. A new ligand targeting a desired heterodimer can then be tested in physiological settings or in a homologous cell line to profile the agonist or antagonist activities. We suggest that the yeast system could become a major player in the discovery of novel ligands.
There are about 150 nuclear receptor genes known thus far (49). This number has evolved when only about 20% of the genome has been sequenced (or made public). Once the whole genome is sequenced, our task to identify physiologically relevant partners will become very daunting. It is time consuming and tedious to identify nuclear receptor heterodimeric partners by biochemical means. A molecular genetic approach that is amenable to high throughput screens is highly desirable. Another critical element of this strategy is the ease of identifying ligands by using the yeast system. As the nuclear receptor field enters the next decade and the excitement of several new orphans yet to be discovered increases, novel methods to study their structure and function will have to be developed. The systems and approaches described above is one such approach.
ACKNOWLEDGEMENTS
Some of the work discussed in this article was performed during T.R.B.’s tenure at Smith Kline Beecham Pharmaceuticals, King of Prussia, PA. T.R.B. is grateful to Dr. Daniel Kephart of Smith Kline Beecham, Dr. Sotirios Karathanasis of Wy-eth Ayerst, Dr. James Wittliff of University of Louisville, Dr. Alois Jungbauer of IAM, Vienna, Austria, and Dr. Sohaib Khan of University of Cincinnati Medical School for their critique, support, and encouragement. This work was partly supported by VLI Research grant to T.R.B. The valuable assistance of Carolyn Walfish in preparation of the manuscript is gratefully acknowledged. This work was also supported in part by grants to P.G.W. from The Medical Research Council of Canada #MT-12604, The W. Garfield Weston Foundation, The Brian Davidson Memorial Fund of The Ontario Grocery Industry Foundation, The Saul A. Silverman Family Foundation and Temmy Latner/Dyncare, The Meadowcroft Group, and The Mount Sinai Hospital Department of Medicine Research Fund.
REFERENCES
- 1. Adams M. D.; Kerlavage A. R.; Fleichmann R. D.; Fuldner R. A.; et al. Initial assessment of the human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequences. Nature 377(Suppl. 28):3–20; 1995. [PubMed] [Google Scholar]
- 2. Allegretto E. A.; McClurg M. R.; Lazarchik S. B.; Clemm D. L.; Kerner S. A.; Elgort M. G.; Boehm M. G.; White S. K.; Pike J. W.; Heyman R. A. Transactivation properties of retinoic acid and retinoid X receptors in mammalian cells and yeast: Correlation with hormone binding and effects of metabolism. J. Biol. Chem. 268:26625–26633; 1993. [PubMed] [Google Scholar]
- 3. Baniahmad A.; Steiner C.; Kohne A. C.; Renkawitz R. Modular structure of a chicken lysozyme silencer: Involvement of an unusual thyroid hormone receptor binding site. Cell 61:505–514; 1990. [DOI] [PubMed] [Google Scholar]
- 4. Baniahmad A.; Ha I.; Reinberg D.; Tsai S.; Tsai M. J.; O’Malley B. W. Interaction of human thyroid hormone receptor β with transcription factor TFIIB may mediate target gene derepression and activation by thyroid hormone. Proc. Natl. Acad. Sci. USA 90:8832–8836; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Butt T. R.; Karathanasis K. L. Transcription factors as drug targets: Opportunities for therapeutic selectivity. GeneExpr. 4:319–336; 1995. [PMC free article] [PubMed] [Google Scholar]
- 6. Carlberg C.; Bendik I.; Wyss A.; Meier E.; Sturzenbecker L. J.; Grippo J. F.; Hunziker W. Two nuclear signalling pathways for vitamin D. Nature 361:657–660; 1993. [DOI] [PubMed] [Google Scholar]
- 7. Cavailles V.; Cauvious S.; L’Horset F.; Lopez G.; Hoare S.; Kushner P. J.; Parker M. G. Nuclear factor RIP140 modulates transactivational activation for the estrogen receptor. EMBO J. 14:3741–3751; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen J. D.; Evans R. M. A transcriptional corepressor that interacts with nuclear hormone receptors. Nature 377:454–457; 1995. [DOI] [PubMed] [Google Scholar]
- 9. Danielian P. S.; White R.; Lees J. A.; Parker M. G. Identification of a conserved region required for hormone dependent transactivational activation by steroid hormone receptors. EMBO J. 11:1025–103; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Drapkin R.; Merino A.; Reinberg D. Regulation of RNA polymerase II transcription. Curr. Opin. Cell. Biol. 5:469–476; 1993. [DOI] [PubMed] [Google Scholar]
- 11. Farsetti A.; Desvergne B.; Hallenbeck P.; Robbins J.; Nikodem V. M. Characterization of myelin basic protein thyroid hormone response element and its function in the context of a native and heterologous promoter. J. Biol. Chem. 267:15784–15788; 1992. [PubMed] [Google Scholar]
- 12. Fawell S. E.; Lees J. A.; White R.; Parker M. G. Characterization and colocalization of steroid binding and dimerization activities in the mouse estrogen receptor. Cell 60:953–962; 1990. [DOI] [PubMed] [Google Scholar]
- 13. Ferrara J.; McCuaig K.; Hendy J.; Uskokovic M.; White J. H. High potency transcriptional activity by 16-ene derivators of 1,25 dihydroxy vitamin D3 . J. Biol. Chem. 269:2971–2981; 1994. [PubMed] [Google Scholar]
- 14. Fleischmann R. D.; Adams M. D.; White O.; et al. Whole-genome random sequencing and assembly of haemophilus influenzae Rd. Science 269:469–512; 1995. [DOI] [PubMed] [Google Scholar]
- 15. Folkers G. E.; van der Leade B. J.; van der Saag P. T. The retinoic acid receptor-beta 2 contains two separate cell-specific transactivation domains, at the N-terminus and in the ligand-binding domains. Mol. Endocrinol. 7:616–627; 1993. [DOI] [PubMed] [Google Scholar]
- 16. Force W. K.; Tillman J. B.; Sprung C. N.; Spindler S. R. Homodimer and heterodimer DNA binding and transcriptional responsiveness of triiodothyronine (T3) and 9-cis-retinoic acid are determined by the number and order of high affinity half-sites in a T3 response element. J. Biol. Chem. 269:8863–8871; 1994. [PubMed] [Google Scholar]
- 17. Forman B. M.; Umesono K.; Chen J.; Evans R. M. Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell 81:541–550; 1995. [DOI] [PubMed] [Google Scholar]
- 18. Giguere V. Retinoic acid receptor and cellular retinoid binding proteins: Complex interplay in retinoid signalling. Endocr. Rev. 15:61–79; 1994. [DOI] [PubMed] [Google Scholar]
- 19. Glass C. K.; Holloway J. M.; Devary O. B. V.; Rosenfeld M. G. The thyroid hormone receptor binds with opposite transcriptional effects to a common sequence motif in thyroid hormone and estrogen response elements. Cell 54:313–323; 1988. [DOI] [PubMed] [Google Scholar]
- 20. Glass C. K. Differential recognition of target genes by nuclear receptor monomers, dimers, and heterodimers. Endocr. Rev. 15:391–407; 1994. [DOI] [PubMed] [Google Scholar]
- 21. Glass C. K.; Lipkin S. M.; Devary O. B. V.; Rosenfeld M. G. Positive and negative regulation of gene transcription by a retinoic acid-thyroid hormone receptor heterodimer. Cell 59:697–708; 1989. [DOI] [PubMed] [Google Scholar]
- 22. Goodrich J. A.; Tjian R. TBP-TAF complexes: Selectivity factors for eukaryotic transcription. Curr. Opin. Cell. Biol. 6:403–409; 1994. [DOI] [PubMed] [Google Scholar]
- 23. Gronemeyer H. Control of transcription activation by steroid hormone receptors. FASEB J. 6:2524–2529; 1992. [DOI] [PubMed] [Google Scholar]
- 24. Guarente L. Transcriptional coactivators in yeast and beyond. Trends Biol. Sci. 20:517–521; 1995. [DOI] [PubMed] [Google Scholar]
- 25. Halachmi S.; Marden E.; Martin G.; MacKay H.; Abbondanza C.; Brown M. Estrogen receptor-associated proteins: Possible mediators of hormone-induced transcription. Science 264:1455–1458; 1994. [DOI] [PubMed] [Google Scholar]
- 26. Hall B. L.; Smit-McBride Z.; Privalsky M. L. Reconstitution of retinoid X receptor and combinatorial regulation of other nuclear receptors in the yeast Saccharomyces cerevisiae . Proc. Natl. Acad. Sci. USA 90:6929–6933; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Heery D. M.; Zacharewski T.; Pierrat B.; Gronemeyer H.; Chambon P.; Losson R. Efficient transactivation by retinoic acid receptors in yeast require retinoid X receptors. Proc. Natl. Acad. Sci. USA 90:4281–4285; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Heery D. M.; Pierrat B.; Gronemeyer H.; Chambon P.; Losson R. Homo- and heterodimers of retinoid X receptor (RXR) activate transcription in yeast. Nucleic Acids Res. 22:726–731; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hernandez N. TBP, a universal eukaryotic transcription factor? Genes Dev. 7:1291–1301; 1993. [DOI] [PubMed] [Google Scholar]
- 30. Hill C. S.; Treisman R. Transcriptional regulation by extracellular signals mechanisms and specificity. Cell 80:199–211; 1995. [DOI] [PubMed] [Google Scholar]
- 31. Hollenberg S. M.; Evans R. M. Multiple and cooperative transactivation domains of the human glucocorticoid receptor. Cell 55:899–906; 1988. [DOI] [PubMed] [Google Scholar]
- 32. Horlein H. J.; Naar A. M.; Heinzel T.; Torchia J.; Gloss B.; Kurokawa R.; Ryan A.; Kamei Y.; Soderstrom M.; Glass C. K.; Rosenfeld M. G. Ligand-independent repression by thyroid hormone receptor mediated by a nuclear receptor corepressor. Nature 377:397–403; 1995. [DOI] [PubMed] [Google Scholar]
- 33. Issemann I.; Prince R. A.; Tugwood J. D.; Green S. The retinoid X receptor enhances the function of the peroxisome proliferator activated receptor. Biochimie 75:251–256; 1993. [DOI] [PubMed] [Google Scholar]
- 34. Kephart D. D.; Walfish P. G.; DeLuca H.; Butt T. R. RXR isotype identity directs human vitamin D receptor heterodimer transactivation from the 24-hydroxylase vitamin D response elements in yeast. Mol. Endocrinol. 10(4):408–419; 1996. [DOI] [PubMed] [Google Scholar]
- 35. Kliewer S. A.; Umesono K.; Mangelsdorf D. J.; Evans R. M. Retinoid X receptor interacts with nuclear receptors in retinoic acid, thyroid hormone and vitamin D3 signalling. Nature 355:446–449; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kliewer S. A.; Umesono K.; Noonan D. J.; Heyman R. A.; Evans R. M. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature 358:771–774; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kurokawa R.; Yu V. C.; Naar A.; Kyakumoto S.; Han Z.; Silverman S.; Rosenfeld M. G.; Glass C. K. Differential orientation of DNA-binding domain and carboxy terminal dimerization and interface regulate binding site selection by nuclear receptor hetero-dimers. Genes Dev. 7:1423–1435; 1993. [DOI] [PubMed] [Google Scholar]
- 38. Kurokawa R.; Direnzo J.; Boehm M.; Sugarman J.; Gloss B.; Rosenfeld M. G.; Heyman R. A.; Glass C. K. Regulation of retinoid signalling by receptor polarity and allosteric control of ligand binding. Nature 371:528–531; 1994. [DOI] [PubMed] [Google Scholar]
- 39. Kurokawa R.; Soderstrom M.; Horlein A.; Halachmi S.; Brown M.; Rosenfeld M. G.; Glass C. K. Polarity-specific activites of retinoic acid receptors determined by a co-repressor. Nature 377:451–454; 1995. [DOI] [PubMed] [Google Scholar]
- 40. Lazar M. A. Thyroid hormone receptors: Multiple forms, multiple possibilities. Endocr. Rev. 14:184–193; 1993. [DOI] [PubMed] [Google Scholar]
- 41. Leblanc B. P.; Stunnenberg H. G. 9-cis retinoic acid signalling: Changing partners causes some excitment. Genes Dev. 9:1811–1816; 1995. [DOI] [PubMed] [Google Scholar]
- 42. Le Douarin B.; Zechel C.; Gamier J. M.; Lutz Y.; Tora L.; Pierrat B.; Heery D.; Gronemeyer H.; Chambon P.; Losson R. The N-terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors is fused to β-raf in the oncogenic protein T18. EMBO J. 14:2020–2033; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee J. L.; Ryan F.; Swafffield J. C.; Johnston S. A.; Moore D. D. Interaction of thyroid-hormone receptor with a conserved transcriptional mediator. Nature 374:91–94; 1995. [DOI] [PubMed] [Google Scholar]
- 44. Leid M.; Kastner P.; Lyons R.; Nakshatri H.; Saunders M.; Zacharewski T.; Chen J-Y.; Staub A.; Garnier J-M.; Mader S.; Chambon P. Purification, cloning, and RXR identity of the HeLa cell factor with which RARa or TR heterodimerizes to bind target sequences efficiently. Cell 68:377–395; 1992. [DOI] [PubMed] [Google Scholar]
- 45. Lewin B. Commitment and activation at pol II promoters: A tail of protein–protein interactions. Cell 61:1161–1164; 1990. [DOI] [PubMed] [Google Scholar]
- 46. Luisi B. F.; Xu W. X.; Otwinowski Z.; Freedman L. P.; Yamamoto K. R.; Sigler P. B. Crystallographic analysis of the interaction of glucocorticoid receptor with DNA. Nature 352:497–505; 1991. [DOI] [PubMed] [Google Scholar]
- 47. Lyttle C. R.; Damian-Matsumura P.; Juul H.; Butt T. R. Human estrogen receptor regulation in a yeast model system and studies on receptor agonists and antogonists. J. Steroid Biochem. Mol. Biol. 42:677–685; 1992. [DOI] [PubMed] [Google Scholar]
- 48. Mader S.; Chambon P.; White J. H. Defining a minimal estrogen receptor DNA binding domain. Nucleic Acids Res. 21:1125–1132; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mangelsdorf D. J.; Evans R. M. The RXR hetero-dimers and orphan receptors. Cell 83:841–50; 1995. [DOI] [PubMed] [Google Scholar]
- 50. Marks M. S.; Hallenbeck P. L.; Nagata T.; Segars J. H.; Apella E.; Nikodem V. M.; Ozato K. H-2RIIBP (RXRβ) heterodimerization provides a mechanism for combinatorial diversity in the regulation of retinoic acid and thyroid hormone responsive genes. EMBO J. 11:1419–1435; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Naar A. M.; Boutin J. M.; Lipkin S. M.; Yu V. C.; Holloway J. M.; Glass C. K.; Rosenfeld M. G. The orientation and spacing of core DNA-binding motifs dictates selective transcriptional responses to three nuclear receptors. Cell 65:1267–1279; 1991. [DOI] [PubMed] [Google Scholar]
- 52. Nagpal S.; Saunders M.; Kastner P.; Durand B.; Nakshatri H.; Chambon P. Promoter context- and response element-dependent specificity of transcriptional activation and modulating functions of retinoic acid receptors. Cell 70:1007–1019; 1992. [DOI] [PubMed] [Google Scholar]
- 53. Nolan G. P. NF-AT-AP-1 and Rel-bZIP: Hybrid vigor and binding under the influence. Cell 77:795–798; 1994. [DOI] [PubMed] [Google Scholar]
- 54. Ohashi H.; Yang Y.-F.; Walfish P. G. Rat liver c-erb A β1 thyroid hormone receptor is a constitutive activator in yeast (Saccharomyces cerevisiae): Essential role of domains D, E, and F in hormone-independent transcription. Biochem. Biophys. Res. Commun. 178:1167–1175; 1991. [DOI] [PubMed] [Google Scholar]
- 55. Parker M. G. Steroid and related receptors. Curr. Opin. Cell. Biol. 5:499–504; 1993. [DOI] [PubMed] [Google Scholar]
- 56. Perlmann T.; Rangarajan P. N.; Umesono K.; Evans R. M. Determinants for selective RAR and TR recognition of direct repeat HREs. Genes Dev. 7:1411–1422; 1993. [DOI] [PubMed] [Google Scholar]
- 57. Predki P. F.; Zambie D.; Sarkar B.; Giguere V. Ordered binding of retinoic acid and retinoid-X receptor to asymmetric response elements involves determinants adjacent to DNA-binding domains. Mol. Endocrinol. 8:31–39; 1994. [DOI] [PubMed] [Google Scholar]
- 58. Privalsky M. L.; Sharif M.; Yamamoto K. R. The viral erbA oncogene protein, a constitutive repressor in animal cells as a hormone regulated activator in yeast. Cell 63:1277–1286; 1990. [DOI] [PubMed] [Google Scholar]
- 59. Ptashne M.; Gann A. A. F. Activators and targets. Nature 346:329–331; 1990. [DOI] [PubMed] [Google Scholar]
- 60. Raisher B. D.; Gulick T.; Zhang Z.; Strauss A. W.; Moore D. D.; Kelly D. P. Identification of a novel retinoid-response element in the promoter region of the medium chain acyl-co-enzyme dehydrogenasegene. J. Biol. Chem. 267:20264–20269; 1992. [PubMed] [Google Scholar]
- 61. Renaud J.-P.; Rochel N.; Ruff M.; Vivat V.; Chambon P.; Gronemeyer H.; Moras D. Crystal structure of the RAR-γ ligand-binding domain bound to all-trans retinoic acid. Nature 378:681–689; 1995. [DOI] [PubMed] [Google Scholar]
- 62. Roeder R. G. The complexities of eukaryotic transcription initiation: Regulation of preinitiation complex assembly. Trends Biochem. Sci. 16:402–408; 1991. [DOI] [PubMed] [Google Scholar]
- 63. Rosen E. D.; O’Donnell A.; Koenig A. J. Ligand-dependent synergy of thyroid hormone-retinoid X receptors. J. Biol. Chem. 267:22010–22013; 1992. [PubMed] [Google Scholar]
- 64. Saatcioglou F.; Deng T.; Karin M. A novel cis element mediating ligand-independent activation by c-Erb-A: Implications for hormonal regulation. Cell 75:1095–1105; 1993. [DOI] [PubMed] [Google Scholar]
- 65. Saltiel A. R. Signal transduction pathways as drug targets. Sci. Am. Sci. Med. Nov/Dec:58–67; 1995. [Google Scholar]
- 66. Scholer H. R.; Ceisiolka T.; Gruss P. A nexus between Oct-4 and E1A: Implications for gene regulation in embryonic stem cells. Cell 66:291–304; 1991. [DOI] [PubMed] [Google Scholar]
- 67. Schulman A. G.; Chakvarti D.; Juguilon H.; Romo A.; Evans R. M. Interaction between the retinoid X receptor and a conserved region of the TATA-binding protein mediate hormone-dependent transactivation. Proc. Natl. Acad. Sci. USA 92:8288–8292; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Struhl K. Duality of TBP, the universal transcription factor. Science 263:1103–1104; 1995. [DOI] [PubMed] [Google Scholar]
- 69. Stunnenberg H. G. Mechanisms of transactivation by retinoic acid receptors. Bioessays 15:309–315; 1993. [DOI] [PubMed] [Google Scholar]
- 70. Thanos D.; Maniatis T. NF-KB: A lesson in family values. Cell 80:529–532; 1995. [DOI] [PubMed] [Google Scholar]
- 71. Tini M.; Otulakowski G.; Breitman M. L.; Tsui L.-C; Giguere V. An everted repeat mediates retinoic acid induction of gamma-F crystallin gene: Evidence of a direct role for retinoids and lens development. Genes Dev. 7:295–307; 1993. [DOI] [PubMed] [Google Scholar]
- 72. Tora L.; White J.; Brou C; Tasset D.; Webster N.; Scheer E.; Chambon P. The human estrogen receptor has two independent nonacidic transcriptional activation functions. Cell 59:477–487; 1989. [DOI] [PubMed] [Google Scholar]
- 73. Towers T. L.; Luisi B. F.; Asianov A.; Freedman L. P. DNA target selectivity for vitamin D3 receptor: Mechanisms of dimer binding of asymmetric repeat element. Proc. Natl. Acad. Sci. USA 90:6310–6314; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tsai S. Y.; Carlstedt-Duke J.; Weigel N. L.; Dahlman K.; Gustafsson J.-A.; Tsai M.-J.; O’Malley B. W. Molecular interactions of steroid hormone receptor with its enhancer element: Evidence for receptor dimer formation. Cell 55:361–369; 1989. [DOI] [PubMed] [Google Scholar]
- 75. Umesono K.; Giguere V.; Glass C. K.; Rosenfeld M. G.; Evans R. M. Retinoic acid and thyroid hormone induced gene expression through a common responsive element. Nature 336:262–265; 1988. [DOI] [PubMed] [Google Scholar]
- 76. Umesono K.; Murakami K. K.; Thompson C. C.; Evans R. M. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, vitamin D3 receptors. Cell 65:1255–1266; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Vegeto E.; Allan G. F.; Schrader W. T.; Tsai M-J.; McDonnell D. P.; O’Malley B. R. The mechanism of RU486 antagonism is dependent on the conformation of the carboxy-terminal tail of the human progestrerone receptor. Cell 69:703–713; 1992. [DOI] [PubMed] [Google Scholar]
- 78. vom Baur E.; Zechel C.; Heery D.; Heine M.; Gamier J. M.; Vivat V.; Le Douarin B.; Gronemeyer H.; Chambon P.; Losson R. Differential ligand-dependent interaction between AF-2 activation domain of nuclear receptor and putative transcriptional intermediary factors in mSUGl and TIF1. EMBO J. 15:110–124; 1996. [PMC free article] [PubMed] [Google Scholar]
- 79. Yoshinaga S. K.; Peterson C. L.; Herskowitz I.; Yamamoto K. R. Role of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science 258:1598–1604; 1992. [DOI] [PubMed] [Google Scholar]
- 80. Yu V. C.; Delsert C; Andersen B.; Holloway J. M.; Devary O. B. V.; Naar A. M.; Kim S. Y.; Boutin J. M.; Glass C. K.; Rosenfeld M. G. RXRβ: A co-regulator that enhances binding of retinoic acid, thyroid hormone, and vitamin D receptors to their cognate response elements. Cell 67: 1251–1266; 1991. [DOI] [PubMed] [Google Scholar]
- 81. Wagner R. L.; Apriletti J. W.; McGrath M. E.; West B. L.; Baxter J. D.; Fletterick R. J. A structural role for hormone in the thyroid hormone receptor. Nature 378:690–697; 1995. [DOI] [PubMed] [Google Scholar]
- 82. Walfish P. G.; Yang Y.-F.; Chang L. A.; Yoganathan T.; Butt T. R. Cross-talk between thyroid hormone and specific retinoid X receptor subtypes in yeast selectively regulates cognate ligand actions. (submitted). [PMC free article] [PubMed]
- 83. Webster N.; Green S.; Jin J. R.; Chambon P. The hormone-binding domains of the estrogen and glucocorticoid receptors contain an inducible transcription activation function. Cell 54:199–207; 1988. [DOI] [PubMed] [Google Scholar]
- 84. Williams G. R.; Harney J. W.; Forman B. M.; Samuels H. H.; Brent G. A. Oligomeric binding of T3 receptor is required for maximal T3 response. J. Biol. Chem. 266:19636–19644; 1991. [PubMed] [Google Scholar]
- 85. Zawel L.; Reinberg D. Advances in RNA polymerase II transcription. Curr. Opin. Cell. Biol 4:488–495; 1992. [DOI] [PubMed] [Google Scholar]
- 86. Zechel C.; Shen X.-Q.; Chambon P.; Gronemeyer H. Dimerization interfaces formed between the DNA binding domains determine the cooperative binding of RXR/RAR and RXR/TR hetero-dimers to DR5 and DR4 elements. EMBO J. 13: 1414–1424; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zhang X.-K.; Hoffmann B.; Tran P.B.-V.; Graupner G.; Pfahl M. Retinoid X receptor is an auxiliary protein for thyroid hormone and retinoic acid receptors. Nature 355:441–446; 1992. [DOI] [PubMed] [Google Scholar]


