Abstract
The 17-18S, 5.8S, and 25-28S rRNA species of eukaryotic cells are transcribed by RNA polymerase I into a single precursor molecule, from which external and internal spacer sequences are subsequently removed in an order series of nucleolytic reactions. Whereas the order of the cleavage reactions has long been established, only recently has significant progress been made in detailing the cis-acting elements and the trans-acting factors involved in this process. The use of recently developed systems for in vivo mutational analysis of yeast rDNA has greatly enhanced our knowledge of cis-acting structural features within the pre-rRNA, which are critical for correct and efficient removal of the spacer sequences. The same systems also allow a link to be forged between trans-acting processing factors and these cis-acting elements. In this review the newly obtained information will be summarized, focused predominantly on pre-rRNA processing in the yeast Saccharomyces cerevisiae.
Keywords: Ribosome, Processing, Precursor rRNA, Yeast
THE 17S–18S, 5.8S, and 26S–28S rRNAs of eukaryotic cells are transcribed by RNA polymerase I into a single precursor molecule, from which the external and internal spacer sequences are subsequently removed in an ordered series of steps (Fig. 1). Concomitantly with processing some 80 different ribosomal proteins associate with the primary transcript and the various precursor intermediates in a stepwise fashion.
FIG. 1.
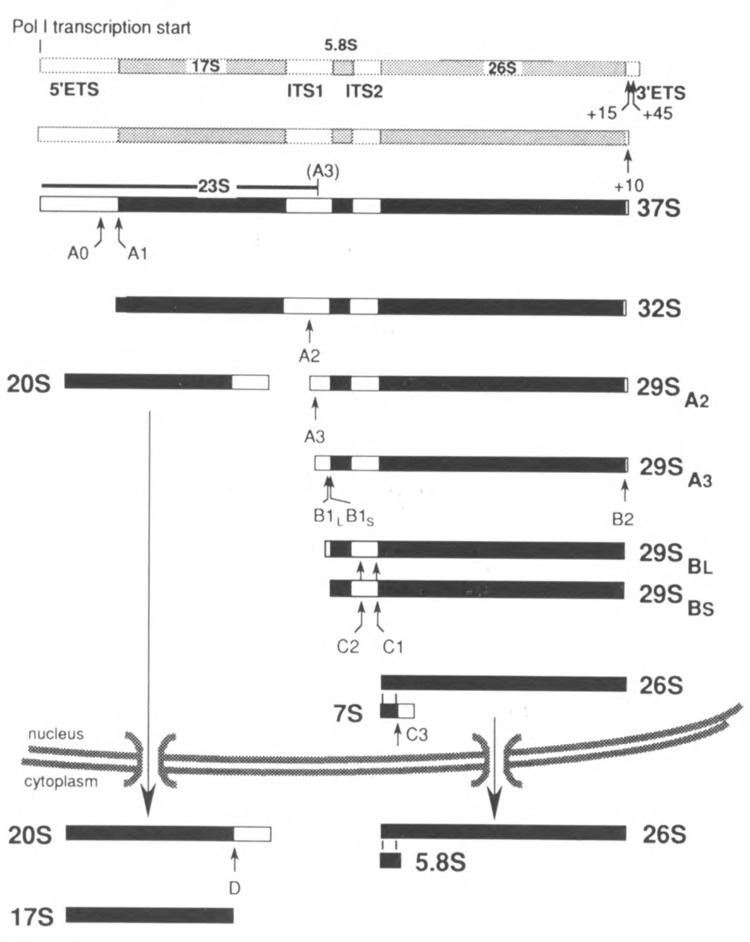
The major processing pathway of yeast pre-rRNA. Mature sequences are shown in black, spacer regions in white. Shaded species are not detectable as such in vivo but their existence can be inferred (see text). Note that all precursors are present in the form of RNP particles containing ribosomal proteins as well as nonribosomal components.
Although the sequence of events by which eukaryotic pre-rRNA matures has been known for quite some time, only recently has significant progress been made in detailing the molecular mechanisms of this process. In this review attention will be focused on pre-rRNA processing in Saccharomyces cerevisiae, as the progress to a large extent stems from studies on this unicellular eukaryote. For further information the reader is referred to several recent reviews (Raue and Planta, 1991; Woolford and Warner, 1991; Fournier and Maxwell, 1993; Mattaj et al., 1993; Eichler and Craig, 1994).
THE PROCESSING PATHWAY
As shown in Fig. 1, yeast pre-rRNA processing starts with removal of the 3′ETS (∼200 nt) by endonucleolytic cleavages at positions 45–50 and 15 nt downstream from the 3′ end of 26S rRNA (Kempers-Veenstra et al., 1986). The resulting intermediates are undetectable because they are rapidly converted into 37S pre-rRNA, which still carries 10 nt of the 3′ETS.
Next, the 5′ETS (699 nt) is removed by endonucleolytic cleavages at site A0, located between nt 609 and 610, immediately followed by a cut at site A1 corresponding to the 5′ end of the 17S rRNA sequence (Hughes and Ares, 1991; Beltrame et al., 1994). Recently, a strong primer extension stop has been reported 140 nt upstream from A0 (Beltrame et al., 1994), suggesting an additional processing site in the 5′ETS.
Processing at site Al is followed by cleavage at site A2, between nt 213 and 214 within the 363 nt long ITS1. In fact, the cleavages at sites AO, Al, and A2 are probably linked mechanistically (see below). The resulting 20S precursor to 17S rRNA is exported to the cytoplasm as a 43S preribosomal RNP particle where the remaining portion of ITS1 is removed by endonucleolytic cleavage at site D (Udem and Warner, 1973; Trapman and Planta, 1976). However, the 3′ terminus of the 20S intermediate-as determined by fingerprinting (De Jonge et al., 1977)-lies 4 nt upstream from the 5′ end of the 29SA2 precursor located by reverse transcription analysis (Henry et al., 1994; Van Nues, 1995), indicating that processing at site A2 may be a complex event.
The 29SA2 pre-rRNA is first cleaved at site A3 (also called A4; Lindahl et al., 1994) some 70 nt downstream from its 5′ end by the MRP endonuclease producing 29SA3 pre-rRNA (Henry et al., 1994). Processing at A3 can still occur even when cleavage at A2 is blocked by mutations either in cis or in trans (Beltrame et al., 1994; Henry et al., 1994), providing an alternative way to separate the two portions of the precursor. Whether the extended 20S intermediate having its 3′ end at site A3 can be converted into 17S rRNA as efficiently as the normal 20A pre-rRNA is somewhat controversial, however (Lindahl et al., 1994; Van Nues et al., 1994).
The majority (∼90%) of the 29A3 precursor is converted into 29SBS pre-rRNA exonucleolytically by the 5′ → 3′ exonucleases XRN1p and RAT1p. The remaining 10% is processed via a second, independent pathway of which the nature is still unclear, producing a set of 29SBL species that carry 6–8 extra nt at the 5′ end (Henry et at., 1994; Van Nues, 1995). Deletion of site A3 (Henry et al., 1994; Van Nues, 1995) or inactivation of RNase MRP (Shuai and Warner, 1991; Lindahl et al., 1992; Schmitt and Clayton, 1993) causes the majority of 29SA2 processing to be transferred to the B1L pathway, thus reversing the normal 1:8 ratio between the “long” and “short” forms of 5.8S rRNA. Furthermore, it appears that under these conditions ITS2 is removed first from the majority of the 29SA2 molecules before B1L cleavage takes place (Henry et al., 1994, and references therein). Although deletion of site A3 has very little, if any, effect on the levels of the three mature rRNA species, it causes a significant retardation in the growth rate of yeast cells dependent upon such mutant rDNA units (Van Nues, 1995). This is unlikely to be due to the abnormal ratio between long and short 5.8S rRNA-containing 60S sub-units as originally proposed to explain the lethality of mutations in RNase MRP (Schmitt and Clayton, 1993). However, the alternative pathway involving “premature” removal of ITS2 from the 29SA2 precursor might result in functionally defective 60S subunits due to mis-assembly. We have indeed found indications for a role of ITS2 in 60S subunit assembly (Musters et al., 1990; Van Nues et al., 1995). A scaffolding function for spacer sequences in ribosome assembly is further supported by the fact that point mutations in the 5′ETS of E. coli pre-rRNA cause a functional defect in the 30S subunits (Theissen et al., 1993).
Removal of ITS2 from the two 29SB precursor species proceeds via endonucleolytic cleavage at C2, which produces the 7S precursor to mature 5.8S rRNA. The immediate downstream product of cleavage at C2 is not detectable, indicating that generation of the 5′ end of mature 26S rRNA probably occurs by (virtually) simultaneous endonucleolytic cutting at C2 and C1, although very rapid exonucleolytic trimming of the intermediate presently cannot be excluded. Finally, the mature 3′ end of 5.8S rRNA is produced by cleavage at C3.
CIS-ACTING ELEMENTS
Structural features of yeast pre-rRNA required for correct and efficient processing have been identified mainly by means of two different systems for in vivo mutational analysis in which the mutant rDNA units constitute either a minor (Musters et al., 1989) or the only (Henry et al., 1994; Lindahl et al., 1994; Venema et al., 1995) source of rRNA for the cells.
The first thing to be noted about the ds-acting processing elements is a partition into two groups. The portion of the pre-rRNA up to and including site A2 contains all elements necessary and sufficient for formation of 17S rRNA, whereas all elements essential for production of 5.8S/26S rRNA are located downstream from A2 (Van Nues et al., 1993, and references therein). Detailed identification of exacting elements has so far been limited predominantly to the spacer regions. However, specific portions of the mature sequences also are important (Kempers-Veenstra et al., 1986; Van Nues, 1995; R. E. Jeeninga & H. A. Raué, unpublished data).
3′ ETS
The cis-acting elements necessary and sufficient for removal of this spacer are located between positions −35 and +74 relative to the 3′ end of the 26S rRNA sequence as evident from a deletion analysis using minigenes. No detailed identification has as yet been carried out but comparison of different yeast species revealed conserved primary and secondary structural features that are feasible candidates (Kempers-Veenstra et al., 1986).
5′ ETS
Mutational analysis of the 5′ETS has been focused predominantly on a region containing a perfect 10 nt long sequence (positions 470–479) complementarity to the U3 snoRNA essential for the early processing steps. Precise deletion of this sequence blocks the cleavages at AO, Al, and A2, causing accumulation of the aberrant 23S pre-rRNA (Fig. 1). No mature 17S rRNA is produced from the mutant rDNA units but production of 26S rRNA is normal (Beltrame and Tollervey, 1992; Beltrame et al., 1994). These observations, and the fact that U3 snoRNA can be cross-linked to pre-rRNA in vivo at positions within and directly adjacent to the complementary sequence (Beltrame and Tollervey, 1992), identify the complementary region as the recognition site for U3 snoRNP. Additional structural features essential for 17S rRNA formation appear to be present in the 5′ETS (J. Venema, D. Tollervey, & H. A. Raué, unpublished data) but these remain to be characterized in detail.
ITS1
Chimeric S. cerevisiae pre-rRNAs containing T. delbrueckii, K. lactis, K. marxianus, or H. wingei ITS1 instead of the homologous spacer are correctly and efficiently converted into the mature species by the S. cerevisiae processing machinery (Van Nues et al., 1994). Each of these spacers conforms to the secondary structure model proposed by Yeh et al. (1990), even though significant sequence conservation is very limited (Fig. 2). Deletion analysis showed that the conserved sequence in domain III immediately downstream from site A2 is very important, though not absolutely required, for 17S rRNA formation. Production of 26S rRNA was not affected by deletion of this element (Lindahl et al., 1994; Van Nues et al., 1994). The remainder of domain III plays, at best, a minor role in processing. Likewise, most of domain II, including its conserved sequence element, is dispensable (Henry et al., 1994; Van Nues et al., 1994). However, almost complete removal of domain II did cause a strong growth defect, suggesting a role for this domain in assembly and/or nucleocytoplasmic transport of the small subunit (Van Nues, 1995).
FIG. 2.
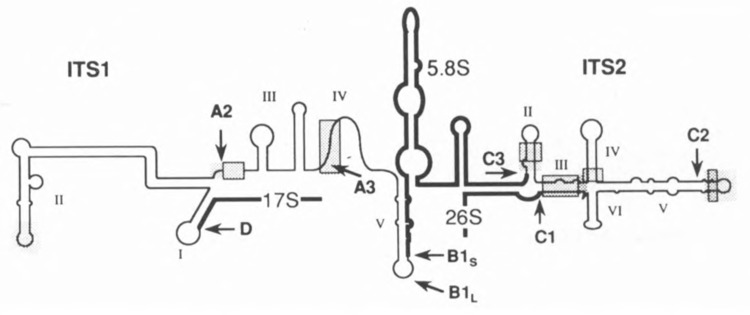
Schematic representation of the structure of the ITS1-5.8S-ITS2 region in the yeast 37S pre-rRNA (Yeh and Lee, 1991; Van Nues et al., 1993). Mature and spacer sequences are represented by bold and thin lines, respectively. Processing sites (cf. Fig. 1) are indicated by arrows. Highly conserved regions are indicated by shading and essential exacting processing elements so far identified are blocked.
Simultaneous deletion of domains IV and V leads to a severe reduction in the levels of mature 5.8S and 26S rRNA while leaving 17S rRNA formation unaffected. Surprisingly, removal of either domain individually has very little effect (Van Nues et al., 1994). However, deletion of domain IV shifts the major processing pathway from B1S to B1L as a result of the absence of the conserved sequence element containing processing site A3 (Henry et al., 1994; Van Nues, 1995). Removal of only domain V has no significant consequences for the ratio of B1S to B1L processing. However, this deletion mutant can form an alternative domain V-like structure in which the 5′ terminal sequence of 5.8S rRNA pairs with the 3′ terminal portion of domain IV even though the latter has little sequence identity to domain V (Van Nues, 1995).
ITS2
In contrast to ITS1, functional conservation of ITS2 extends only as far as the closely related T. delbrueckii yeast species (Van der Sande et al., 1992; Van Nues et al., 1993; Van Nues et al., 1995). Nevertheless, the c/s-acting elements critical for ITS2 processing proved to be confined to the regions displaying strong sequence conservation within the secondary structure model proposed by Lee and coworkers (Yeh and Lee, 1990) (Fig. 2), which is phylogenetically well supported (Van der Sande et al., 1992; Van Nues et al., 1995). The outer region of domain IV, all of domain VI, and a large internal portion of domain V can be individually deleted without noticeable effect on the formation of 26S rRNA. Remarkably, combining the neutral domain V deletion with either of the neutral domain IV or VI deletions had a dramatic negative effect on 26S rRNA production, probably reflecting a requirement for higher-order structure within ITS2 (Van Nues et al., 1995). Furthermore, even mutations that appear to be completely neutral with respect to processing significantly retard cellular growth, providing further support for a scaffolding role of ITS2 in 60S sub-unit assembly (Musters et al., 1990; Van Nues et al., 1995).
TRANS-ACTING FACTORS
Ribosomal Proteins
As rRNA processing and assembly into ribosomal subunits are overlapping processes, ribosomal proteins are obvious candidates for trans-acting factors. Indeed, disturbance of either 17S or 5.8S/26S rRNA maturation has been observed upon depletion or mutation of a number of different r-proteins belonging to the small or large sub-unit, respectively (reviewed in Van Nues et al., 1993). So far, no examples of r-proteins that are absolutely required for any of the processing steps have been found.
snoRNPs
These are so far the best-studied examples of nonribosomal trans-acting factors involved in pre-rRNA processing. At least 14 different snoRNP species have been characterized in yeast cells, of which only three are essential (see Fournier and Maxwell, 1993; Mattaj et al., 1993, for reviews). One of these is U3 discussed above, which is crucial for processing at sites AO, Al, and A2 (Hughes and Ares, 1991; Beltrame et al., 1994). Cleavage at the latter two sites is also absolutely dependent upon the U14 and snR30 snoRNPs (Li et al., 1990; Morrissey and Tollervey, 1993; Beltrame et al., 1994). A fourth species, snR10, is very important, but not absolutely essential, for cleavage at Al and A2 (Tollervey, 1987; Beltrame et al., 1994). Because depletion of any one of these snoRNPs causes essentially the same phenotype, it has been suggested that they are part of a large processing complex or “processome” assembled on the 5′ terminal portion of 37S pre-rRNA (Fournier and Maxwell, 1993). U3 and U14 snoRNPs associate with the pre-rRNA via RNA-RNA interactions in the 5'ETS and the mature 17S rRNA sequences, respectively (Jarmolowski et al., 1990; Beltrame et al., 1994). The molecular basis for the assembly of snR30 and snR10 is unknown, but the conserved sequence at site A2 could be involved. RNase MRP may also be part of the processome because either mutations in this RNP or deletion of the sequence around site A3 cause deviations in the order of the early processing events (Shuai and Warner, 1991; Lindahl et al., 1992; Schmitt and Clayton, 1993; Van Nues, 1995).
So far RNase MRP is the only snoRNP in yeast known to be involved in processing of the 29SA2 precursor into 5.8S/26S rRNA. Vertebrate cells, however, contain a U8 snoRNA, associated with fibrillarin, which is essential for removal of both the 3′ETS and ITS2 in Xenopus oocytes (Peculis and Steitz, 1993, 1994). The linkage between these two processing events, which has also been observed in the fission yeast S. pombe (Melekhovets et al., 1994), suggests that the 3′ terminal portion of the pre-rRNA is assembled into its own processing complex. As specific mutations in Noplp, the yeast homolog of mammalian fibrillarin, impair maturation of 26S rRNA (Tollervey et al., 1993), yeast cells may contain a hitherto undetected homolog of U8 snoRNP.
An interesting recent development is the discovery of additional fibrillarin-containing vetrebrate snoRNPs, of which the RNA components show complementarity to evolutionarily conserved sequences in one of the mature rRNAs (Pellizoni et al., 1994, and references therein). Their role is still unclear, but it has been proposed that these snoRNPs control the coordinate assembly of the r-proteins with the pre-rRNA, a suggestion supported by the fact that some mutations in yeast Noplp (fibrillarin) disturb 60S subunit assembly (Tollervey et al., 1993).
Nucleases
Our knowledge of the nucleolytic enzymes involved in yeast pre-rRNA processing at present is still very limited. As discussed above, the RNase MRP cleaves at site A3 and the XRN1p and RAT1p exonucleases are responsible for the trimming of the 29SA3 precursor to 29Sbs as well as for the degradation of the ITS1 fragment liberated by the 20S → 17S conversion (Stevens et al., 1991). It is noteworthy that RNase MRP is structurally related to RNase P. The two enzymes share the Poplp protein (Lygerou et al., 1994), and their RNA components have similar secondary structures (Schmitt et al., 1993). Finally, the RNA82p endonuclease has been implicated in processing of the 3′ ETS, which is impaired in a yeast strain carrying a mutation in the RNA82 gene that also interferes with 3′ processing of 5S rRNA and maturation of a dimeric tRNA precursor (Piper et al., 1983; Kempers-Veenstra et al., 1986; Piper and Stråby, 1989).
Other Trans-Acting Proteins
In addition to the protein components of the snoRNPs discussed above, more than 10 yeast proteins have been implicated in rRNA processing on the basis of genetic evidence. Although the actual role of most of these proteins is still very poorly understood, in general their mutation or depletion predominantly affects maturation of either 17S or 5.8S/26S rRNA, thus lending further support to the existence of two relatively independent processing complexes.
At present we can distinguish two families among these proteins. The GAR family, encompassing Ssb1p, Nsr1p, Nop3p, Gar1p, and Nop1p (fibrillarin), is characterized by a common structural motif rich in glycine and arginine residues that a specifically binds RNA (Russell and Tollervey, 1992, and references therein). Because (except for Gar1p) these proteins also contain one or more copies of the RNA binding RRM motif, it has been suggested that they play a role in the folding of pre-rRNA. Ssb1p is present in snR10 RNP (Clark et al., 1990), whereas Gar1p is found in both snR10 and snR30 RNPs (Girard et al., 1992). Nop3p is found both in the nucleolus and the nucleoplasma and, therefore, may play a role in the delivery of components involved in pre-rRNA processing and assembly, including ribosomal proteins, to the nucleolus (Russell and Tollervey, 1992). Nsr1p was originally identified as a protein that recognizes nuclear localization signals (NLS) of karyophilic proteins. Thus, it could promote ribosomal assembly by interacting with the NLSs of the r-proteins on the one hand and the pre-rRNA-through its RRM motifs-on the other (Xue and Mélèse, 1994). Nop4p/Nop77p does contain multiple RRM domains but lacks a GAR motif (Sun and Woolford, 1994). It interacts genetically with Noplp (Bergèss et al., 1994).
The second family consists of the proteins Sbp4p, Dsr1p, and CA9p that contain the conserved sequence motifs characteristic of ATP-dependent RNA helicases (Sachs and Davis, 1990; Ripmaster et al., 1992; O’Day and Abelson cited in Eichler and Craig, 1994). These proteins, therefore, could be involved in the structural rearrangements of the pre-RNA that occur during processing and assembly.
The remaining proteins show no structural features that are immediately indicative for their role in pre-rRNA processing. However, both Prp20p (Kadowaki et al., 1993) and Srp1p (Yano et al., 1994; Görlich et al., 1994) appear to be part of the nuclear import machinery. It should be kept in mind that the role of genetically defined trans-acting proteins can be even more indirect (e.g., they may affect the intracellular level of crucial components of the processing machinery) (Hermann-Le Denmat et al., 1994; Hess et al., 1994).
ACKNOWLEDGEMENTS
Work in the authors’ laboratory was supported in part by the Netherlands Foundation for Chemical Research with financial aid from the Netherlands Organization for Scientific Research. We thank all colleagues who sent us reprints and preprints of their work.
REFERENCES
- Beltrame M., Henry Y., and Tollervey D. (1994), Nucleic Acids Res 22, 4057–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrame M. and Tollervey D. (1992), EMBO J, 11, 1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergès T., Petfalski E., Tollervey D., and Hurt E. C. (1994), EMBO J 13, 3136–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M. W., Yip M. L. R., Campbell J., and Abelson J. (1990), J Cell Biol 11, 1741–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jonge P., Klootwijk J., and Planta R. J. (1977), Eur J Biochem 72, 361–369. [DOI] [PubMed] [Google Scholar]
- Eichler D. C. and Craig N. (1994), Prog Nucleic Acids Res Mol Biol 49, 197–239. [DOI] [PubMed] [Google Scholar]
- Fournier M. J. and Maxwell E. S. (1993), Trends Biochem Sci 18, 131–135. [DOI] [PubMed] [Google Scholar]
- Girard J. P., Lehtonen H., Caizergues-Ferrer M., Amalric F., Tollervey D., and Lapeyre B. (1992), EMBO J 11, 673–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D., Prehn S., Laskey R. A., and Hartmann E. (1994), Cell 79, 767–778. [DOI] [PubMed] [Google Scholar]
- Henry Y., Wood H., Morrissey J. P., Petfalski E., Kearsey S., and Tollervey D. (1994), EMBO J 13, 2452–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann-Le Denmat S., Werner M., Sentenac A., and Thuriaux P. (1994), Mol Cell Biol 14, 2905–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess S. M., Stanford D. R., and Hopper A. K. (1994), Nucleic Acids Res 22, 1265–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J. and Ares M. (1991) EMBO J 10, 4231–4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmolowski A., Zagorski J., Li H. V., and Fournier M. J. (1990), EMBO J 9, 4503–4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T., Goldfarb D., Spitz L. M., Tartakoff A. M., and Ohno M. (1993), EMBO J 12, 2929–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempers-Veenstra A. E., Oliemans J., Offenberg H., Dekker A. F., Piper P. W., Planta R. J., and Klootwijk J. (1986), EMBO J 5, 2703–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. V., Zagorski J., and Fournier M. J. (1990), Mol Cell Biol 10, 1145–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl L., Archer R. H., and Zengel J. M. (1992), Nucleic Acids Res 20, 295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl L., Archer R., and Zengel J. M. (1994), Nucleic Acids Res 22, 5399–5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lygerou Z., Mitchell P., Petfalski E., Seraphin B., and Tollervey D. (1994) Genes Dev 8, 1423–1433. [DOI] [PubMed] [Google Scholar]
- Mattaj I. W., Tollervey D., and Seraphin B. (1993) FASEB J 7, 47–53. [DOI] [PubMed] [Google Scholar]
- Melekhovets Y. F., Good L., Elela A., and Nazar R. N. (1994), J Mol Biol 239, 170–180. [DOI] [PubMed] [Google Scholar]
- Morrissey J. P. and Tollervey D. (1993), Mol Cell Biol 13, 2469–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musters W., Planta R. J., Van Heerikhuizen H., and Raué H. A. (1990), in The Ribosome: Structure, Function and Evolution (Hill W. E., Dahlberg A. E., Garrett R. A., Moore P. B., Schlessinger D., and Warner J. R., eds.), American Society of Microbiology, Washington, DC, pp. 435–442. [Google Scholar]
- Musters W., Venema J., Van der Linden G., Van Heerikhuizen H., Klootwijk J., and Planta R. J. (1989), Mol Cell Biol 9, 551–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peculis B. A. and Steitz J. A. (1993), Cell 73, 1233–1245. [DOI] [PubMed] [Google Scholar]
- Peculis B. A. and Steitz J. A. (1994), Genes Dev 18, 2241–2255. [DOI] [PubMed] [Google Scholar]
- Pellizoni L., Crosio C., Campioni N., Loreni F., and Pierandrei-Amaldi P. (1994), Nucleic Acids Res 22, 4607–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper P. W., Bellatin J. A., and Lockheart A. (1983), EMBO J 2, 353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper P. W. and Stråby K. B. (1989), FEBS Lett 250, 311–316. [DOI] [PubMed] [Google Scholar]
- Raué H. A. and Planta R. J. (1991), Prog Nucleic Acids Res Mol Biol 41, 91–129. [DOI] [PubMed] [Google Scholar]
- Ripmaster T. L., Vaughn G. P., and Woolford J. L. (1992), Proc Natl Acad Sci USA 89, 11131–11135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell I. D. and Tollervey D. (1992), J Cell Biol 119, 737–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A. B. and Davis R. W. (1990), Science 247, 1077–1079. [DOI] [PubMed] [Google Scholar]
- Schmitt M. E., Bennett J. L., Dairaghi D. J., and Clayton D. A. (1993), FASEB J 7, 208–213. [DOI] [PubMed] [Google Scholar]
- Schmitt M. E. and Clayton D. A. (1993), Mol Cell Biol 13, 7935–7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai K. and Warner J. R. (1991), Nucleic Acids Res 19, 5059–5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens A., Hsu C. L., Isham K. R., and Larimer R. W. (1991) J Bacteriol 173, 7024–7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C. and Woolford J. L. (1994) EMBO J 13, 3127–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G., Thelen L., and Wagner R. (1993), J Mol Biol 233, 203–218. [DOI] [PubMed] [Google Scholar]
- Tollervey D. (1987), EMBO J 6, 4169–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D., Lehtonen H., Jansen R., Kern H., and Hurt E. C. (1993), Cell 72, 443–457. [DOI] [PubMed] [Google Scholar]
- Trapman J. and Planta R. J. (1976), Biochim Biophys Acta 442, 265–274. [DOI] [PubMed] [Google Scholar]
- Udem S. A. and Warner J. R. (1973), J Mol Biol 65, 227–242. [DOI] [PubMed] [Google Scholar]
- Van der Sande C. A. F. M., Kwa M., Van Nues R. W., Van Heerikhuizen H., Raué H. A., and Planta R. J. (1992), J Mol Biol 223, 899–910. [DOI] [PubMed] [Google Scholar]
- Van Nues R. W. (1995), Ph.D. thesis, Vrije Universiteit, Amsterdam. [Google Scholar]
- Van Nues R. W., Venema J., Planta R. J., and Raué H. A. (1993), in The Translational Apparatus (Nierhaus K. H., Subramanian A. R., Erdmann V. A., Franceschi F., and Wittmann-Liebold B., eds.), Plenum Press, New York, pp. 151–162. [Google Scholar]
- Van Nues R. W., Rientjes J. M. J., Vander Sande C. A. F. M., Zerp S. F., Sluiter C., Venema J., Planta R. J., and Raué H. A. (1994), Nucleic Acids Res 22, 912–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nues R. W., Rientjes J. M. J., Morré S. A., Mollee E., Planta R. J., Venema J., and Raué H. A. (1995), J Mol Biol 250, in press. [DOI] [PubMed] [Google Scholar]
- Venema J., Dirks-Mulder A., Faber A. W., and Raué H. A. (1995), Yeast 11, 145–156. [DOI] [PubMed] [Google Scholar]
- Woolford J. L. and Warner J. R. (1991), in The Molecular and Cellular Biology of the Yeast Saccharomyces cerevisiae (Broach J. R., Pringle J. R., and Jones E. W., eds.), CSH Lab. Press, New York, pp. 587–626. [Google Scholar]
- Xue Z. and Mélèse T. (1994), Trends Cell Biol, 4, 414–417. [DOI] [PubMed] [Google Scholar]
- Yano R., Oakes M. L., Tabb M. M., and Nomura M. (1994), Proc Natl Acad Sci USA 91, 6880–6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh L.-C., Thweatt R., and Lee J. C. (1990), Biochemistry 29, 5911–5918. [DOI] [PubMed] [Google Scholar]
- Yeh L.-C. C. and Lee J. C. (1990), J Mol Biol 211, 699–712. [DOI] [PubMed] [Google Scholar]
- Yeh L.-C. C. and Lee J. C. (1991), J Mol Biol 228, 827–839. [DOI] [PubMed] [Google Scholar]


