Abstract
Aim:
The development of whole-genome screening methodologies for the detection of copy number variations (CNVs), such as array-based comparative genomic hybridization (aCHG), provides a much higher resolution than karyotyping leading to the identification of novel microdeletion and microduplication syndromes often associated with an autism spectrum disease (ASD) phenotype. The aim of the study was to determine CNVs of patients with ASD by using array-based comparative genomic hybridization.
Methods:
Fifty-three patients diagnosed with ASD between 20.01.2014 and 14.01.2015 were included in the study. Chromosome analysis of the patients was performed from peripheral blood cultures and analysed as normal. All patients were evaluated with P064C1 and P096A2 MLPA probes in terms of 16 mental retardation related syndromes. For aCGH method, SurePrint G3 Human microarrays 8x60K were used with genomic DNA isolated from peripheral blood.
Results:
According to results of 53 patients who were included in and performed with arrayCGH, 8 (15%) patients had CNVs classified as pathogenic or variant of unknown significance (VOUS) in the study. We detected a pathogenic NRXN1 gene partial CNV deletion (2p16.3) in two patients. Also we identified a 900 kb duplication of 4p15.31 including SLIT2 gene, and a 245 kb duplication of 15q11.2 including PWRN1 gene in one patient. Our other findings are considered to be a variant of unknown significance (VOUS).
Conclusion:
The results of the study support the literature knowledge, where the copy number variations that cannot be detected with conventional cytogenetics methods in terms of size may happen in patients with ASD.
Keywords: Autism, copy number variations, genomic hybridization, microarrays, karyotype
INTRODUCTION
Autism Spectrum Disorder (ASD) is a neurodevelopmental disorder characterized by persistent deficits in social communication and social interaction across multiple contexts, including deficits in social reciprocity, nonverbal communicative behaviors used for social interaction, and skills in developing, maintaining and understanding relationships. In addition to the social communication deficits, ASD requires the presence of restricted, repetitive patterns of behavior, interests and activities. These symptoms are present from early childhood and impair everyday functioning. Many individuals with ASD also have intellectual impairment and/or language impairment. In the etiology, heritability estimates for ASD have ranged from 37% to higher than 90%, based on twin concordance rates. Currently, as many as 15% of cases of ASD appear to be associated with a known genetic mutation, with different de novo copy number variants or de novo mutations in specific genes associated with the disorder in different families (1). Until recently, karyotyping has been the standard method for the detection of cytogenetic aberrations in patients with developmental disorders. However, there are some limitations to define microdeletions and microduplications by conventional karyotyping. The development of array based comparative genomic hybridization methods provide convenience for detection of copy number variations (CNVs) of some chromosomal regions including 15q13.2q13.3, 16p11.2, and 17p11.2 in ASD (2). The influence of CNVs that are rarely found in the pathogenesis of ASD have been accepted recently (3). The arrayCGH is now proposed as a first-tier test for intellectual disabilities, autism, and/or congenital anomalies to detect gains and losses (4). In this study, we used genome-wide high resolution arrayCGH in order to identify copy number aberrations in a cohort of 53 patients diagnosed with ASD. All patients were classified into different subpopulations regarding gender, phenotypic features and inheritance, which revealed specific patterns of CNVs within ASD.
METHODS
53 patients living in the Trakya region of Turkey with pre-diagnosis of ASD who were referred to Trakya University Child and Adolescent Psychiatry Department between the dates of 20 Jan 2015 and 08 Dec 2015 were included in our study. All patients were diagnosed as ASD by making evaluations and examinations according to DSM-V (1). Fourteen patients had a comorbidity with intellectual disability (ID) and sixteen patients had attention deficit hyperactivity disorder (ADHD). Four patients had two comorbidities with ID and ADHD. We used to assess their autistic findings by The Childhood Autism Rating Scale (CARS). Patients had CARS scores above 30 (cutoff for diagnosis of childhood autism), CARS is a 15-item behavior-rating scale designed to detect and quantify symptoms of autism as well as to distinguish them from other developmental disabilities. Each item on the CARS is scored on a Likert scale, from 1 (no signs of autism) to 4 (severe symptoms). The maximum CARS score is 60, and the cut-off for a diagnosis of autism is 30 (5). To assess their ADHD findings Conners Parent Rating Scale-Revised Short (CPRS-RS) was used. It consists of 27 items with three subscales (oppositional defiant, cognitive problems-inattentive, hyperactivity) and an auxiliary scale (ADHD index). Score value for each material ranging from 0-3. There are 4 answer choices. Not right (too rare), 0 points; somewhat true (sometimes), 1 point; quite correct (often, quite a lot), 2 points; very true (too often), 3 points. High score identified as having a lot of problems (6).
Ethical board approval of the study was taken from Trakya University Scientific Research Ethical Board with the number of 2014-29. Informed consent forms were signed by the parents of all patients included in the study. Before chromosome analysis, we examined all patients physically to exclude any of the major congenital dysmorphic abnormality which may be associated with a syndrome. Chromosome analysis of the patients was performed with GTL banding (450-550 band level) from peripheral blood cultures. All patients were evaluated in terms of 16 mental retardation related syndromes with P064.C1 and P096.A2 Multiplex Ligation Dependent Probe Amplification (MLPA) probe kits (MRC-Holland, Amsterdam, Netherlands). Genomic DNA (gDNA) was isolated from peripheral blood samples of patients according to QIAamp Blood kit protocol (Qiagen, Hilden, Germany). gDNA samples were measured with NanoDrop (ThermoScientific, USA). gDNA concentration of samples was optimized as 50 ng/µl for arrayCGH study. For arrayCGH method, SurePrint G3 Human microarrays 8x60 K (Agilent Technologies, Palo Alto, CA, USA) were used according to manufacturer’s instructions. Microarray slides were scanned with 3 µm resolution in Agilent Microarray Scanner System. Feature Extraction 12.0.1.1 and Cytogenomics 2.9.2.4 (Agilent Technologies, Palo Alto, CA, USA) software’s were used for the analysis of the samples. Aberrant signals including 4 or more adjacent probes were considered as genomic CNVs. CNVs smaller than 10 kb were evaluated as false positive and excluded from results. The detected CNVs were analysed and classified with well-known benign and disease database such as DGV (http://projects.tcag.ca), ClinGen-International Standarts for Cytogenomic Arrays Consortium Database (Clinical Genome Resource- https://www.clinicalgenome.org/), Decipher (Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources-http://decipher.sanger.ac.uk/), Simons Foundation Autism Research Initiative Gene (SFARI Gene), reports in OMIM (Online Mendelian Inheritance in Man) and related literature. CNVs are classified as pathogenic, variant of uncertain clinical significance(VOUS) and benign according to American College of Medical Genetics guideline (4). This study has received financial support from Trakya University Scientific Research Project Committee with the number TÜBAP-2014/81.
RESULTS
In our study, 14 female (26%) and 39 (74%) male, total of 53 subjects who were diagnosed as ASD were included. Mean age of the patients was ±9.2 years. All patients had normal karyotype structure in terms of numerical and structural chromosomal abnormalities that can be detected with conventional cytogenetic methods. However, deletion in 22q13.33 chromosomal locus was detected in one patient after analysis among 16 syndromes associated with mental retardation by using P064.C1 and P096.A2 MLPA probe mix. In the literature, 22q13.33 deletion was reported as Phelan-McDermid syndrome. Since clinical and physical manifestations of the patient matched with Phelan-McDermid syndrome, the patient was not included arrayCGH experiments. According to results of 52 patients who were included in and performed with arrayCGH, 8 (15%) patients had CNVs classified as pathogenic or variant of unknown significance (VOUS). The demographic information, CNV regions, CNV size, aberration type, classification of CNVs and clinical diagnosis data of the patients are shown in Table 1. We detected a NRXN1 gene partial deletion (2p16.3) in two patients. Also we found a 900 kb duplication of 4p15.31 including SLIT2 gene and a 245 kb duplication of 15q11.2 including PWRN1 gene in one patient. Our other findings considered to be a variant of unknown significance (VOUS) are OPTN, MCM10 and CCDC3 genes CNV duplication at 10p13, GRIP1 gene partial deletion at 12q14.3, CHRNA7 gene partial duplication at 15q13.3, miR128-1, ZRANB3, RHDM3 CNV deletions at 2q21.3 and MACROD2 gene partial deletion at 20p12.1.
Table 1.
Summary of abnormal copy number variations detected in cases with Autism Spectrum Disorders (N:8 of 53 patient)ID (intellectual disability), ASD (Autism Spectrum Disorder), ADHD (attention deficit hyperactivity disorder), VOUS (variation of unknown significance), Chromosome coordinates from human reference sequence Feb. 2009 (GRCh37/hg19).
| Patient nr. | Age | Gender | Chromosomal location | Chromosome Coordinates | Array aberration | Size (bp) | Gene | Clinical diagnosis | Classification |
|---|---|---|---|---|---|---|---|---|---|
| 7 | 6 | M | 12q14.3 | 67,062,346-67,189,836 | Deletion | 127.490 | GRIP1 | ID-ASD-ADHD | VOUS |
| 20 | 5 | M | 15q13.3 | 32,098,670-32,426,869 | Duplication | 328.199 | CHRNA7 | ID-ASD-ADHD | VOUS |
| 23 | 10 | F | 2p16.3 | 51,137,071-51,314,430 | Deletion | 177.359 | NRXN1 | ASD | Pathogenic |
| 29 | 8 | M | 20p12.1 | 14,700,040-14,875,770 | Deletion | 175.730 | MACROD2 | ASD-ADHD | VOUS |
| 31 | 7 | M | 4p15.31 | 19,316,566-20,299,465 | Duplication | 982.899 | SLIT2 | ID-ASD-ADHD | VOUS |
| 15q11.2 | 24,587,026-24,833,382 | Duplication | 246.356 | PWRN1 | VOUS | ||||
| 45 | 3 | F | 2p16.3 | 51,083,410-51,122,150 | Deletion | 38.740 | NRXN1 | ASD | Pathogenic |
| 52 | 3 | M | 10p13 | 13,020,390-13,251,208 | Duplication | 230.818 | OPTN, MCM10, CCDC3 | ASD | VOUS |
| 54 | 5 | M | 2q21.3 | 136,019,030-136,447,036 | Deletion | 428.006 | miR128-1,ZRANB3, R3HDM1 | ASD | VOUS |
DISCUSSION
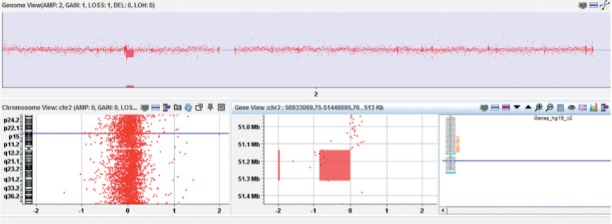
CNVs have repeatedly been found to cause or predispose to ASD (7). Roberts et al. determined the rate of CNVs in ASD patient group as 20% (8). Schaefer et al. found significant CNV in 22% (14 of 68) and in another study Shen et al. reported 18.2% copy number abnormalities in patients (9, 10). The overall diagnostic yield in our study was 15% for patients with ASD and similar to literature.177 kb long partial NRXN1 deletion has been identified in patient number 23 who has typical autism findings (Figure 1). Also 39 kb long partial NRXN1 deletion was identified in patient number 45. NRXN1 gene is mapped at 2p16.3 chromosomal locus, and encoded a protein function in synaptic transmission (11). Neurexins are a protein family functioning as cell adhesion molecules and receptors in nervous system. Cell adhesion is significant for providing coordination of synaptic activity in brain. Moreover, cell adhesion is the ultimate process in learning, memory and cognition by synapse formation, durability and plasticity. Copy number variations and point mutations of NRXN1 gene have been anticipated to be related with autism (11–13). Harrison et al. stated that autism diagnosis and heterozygous NRXN1 deletion in female twin is associated (12). Xie et al. in their recent study determined a microduplication in CHRNA7 gene, mapped at 15q13.3 chromosomal region, in a case clinically diagnosed as autism spectrum disorder (14). Partial NRXN1 deletion found in our two independent patients supports these studies. There has been association between 15q13.3 chromosomal region microdeletions and autism, behavioral deficits and other neuropsychiatric disorders, but the pathogenity of microduplications have not been clarified yet. Bachielli et al. had reported a copy number variation in a patient with ASD and moderate cognitive inefficiency previously (15). Leblond et al. also in their study indicated duplications at SHANK3 gene along with CHRNA7 gene in two cases and proposed a multi effect model for autism-genetic basis (16). We detected a partial CHRNA7 gene duplication encompassing 328.199 kb at 15q13.3 chromosomal locus in patient number 20. GRIP1 is a protein structure that plays a significant role in dendritic cell development. Dendritic development abnormalities is a process manifested in neurologic and neurodevelopmental disorder such as schizophrenia, Down Syndrome, Fragile X syndrome, Angelman Syndome, Rett Syndrome, and autism (17). In this study, a GRIP1 gene partial deletion at 12q14.3 chromosomal region was detected in patient number 7. GRIP1 gene variations have been reported to be efficient in social behaviors and autism phenotype (18). Furthermore, Han et al. reported that GRIP1/2 mediated signaling pathways are important in behavioural deficits related to ASD in mice (19). Although our findings are compatible with the literature, GRIP1 gene function in autism patients should be investigated in further studies. SLIT2 and RHOA genes are components of synaptogenesis/axon pathway which is important in ASD. SLIT2 has been thought to be related with nerve system development and function (20). Martin et al. reported a duplication in a case of ADHD (21). We identified in patient number 31 a 982 kb long duplication spanning SLIT2 gene at 4p15.31 region. In the same patient, we detected a PWRN1 gene partial duplication, mapped at 15q11.2 where genes related with Prader-Willi syndrome are located (22). Bolton et al. in their study remarked that 15q11-q13 duplications in Prader-Willi/Angelman critical region are related to mental disorders in different levels and motor coordination problems. However, association between ASD and 15q11-q13 duplications has not been enlightened yet (23). CNVs in SLIT2 and PWRN1 genes located at different locus may promote multifactorial genetic model concept in ASD. A 175 kb long deletion spanning MACROD2 gene translational region at chromosome 20p12.1 locus was detected in one patient in this study. According to recent genome studies, a significant correlation between MACROD2 CNVs and schizophrenia, brain infarction and encephalization has been reported (24). Also, MACRO-domain region has been thought to play an important role in post-translation mechanism and sirtuin biology process like DNA repair, heterochromatin formation and long term memory formation (25). Jones et al. reported that MACROD2 gene is a candidate risk factor for ASD-like traits in the general population (26). We suggest that further studies in patients and ASD-ADHD diagnosis in different population can contribute to the literature. Moreover, we identified a 230 kb duplication of 10p13 including OPTN, MCM10, CCDC3 genes in one patient. No evidence of association between 10p13 duplication with ASD has been reported in the literature before. Duplication in this region should be investigated with further studies for an understanding of the role of these genes in ASD.
Figure 1.
Array CGH showing arr 2p16.3 (51,137,071-51,314,430) x 1 in patient 23. The first track is genome view of chromosome 2, second track shows aberration and NRXN1 gene.
CONCLUSION
The present study is the most extensive arrayCGH study investigating the role of CNVs in ASD in Turkey. Results of our study support the literature knowledge: CNVs that cannot be detected with conventional cytogenetic methods may happen in patients with ASD diagnosis. By increasing the number of cases, and using ultra-high resolution chromosomal microarray can considerably contribute to specify phenotype-genotype correlation in cases with ASD diagnosis.
Footnotes
Ethics Committee Approval: Ethical board approval of the study was taken from Trakya University Scientific Research Ethical Board with the number of 2014-29.
Informed Consent: Informed consent forms were signed by the parents of all patients included in the study.
Peer-review: Externally peer-reviewed.
Author Contributions: Concept - IG, HG; Design - IG, HG; Supervision - IG, HG; Resource - SDU, EA, HG; Materials - IG, HG, SDU, EA, GA, CC, MAA, AE, NY, CD, HA, NK, HCA, LB, ZÇ, KKB; Data Collection and/ or Processing - IG, HG, SDU, EA, GA, CC, MAA, AE, NY, CD, HA, NK, HCA, LB, ZÇ, KKB; Analysis and/or Interpretation - IG, HG, SDU, EA, HT; Literature Search - IG, HG, SDU, EA; Writing - IG, HG, SDU, EA; Critical Reviews - IG, HG, SDU, EA.
Conflict of Interest: No conflict of interest was declared by the authors.
Financial Disclosure: The authors declared that this study has received no financial support.
REFERENCES
- 1.American Psychiatric Association, Diagnostic and Statistical Manual of Mental Disorders. 5th ed. Washington, DC: DSM Library; 2013. [Google Scholar]
- 2.Bremer A, Giacobini MB, Eriksson M, Gustavsson P, Nordin V, Fernell E, Gillberg C, Nordgren A, Uppstromer A, Anderlid BM, Nordenskjöld M, Schoumans J. Copy number variation characteristics in subpopulations of patients with autism spectrum disorders. Am J Med Genet B Neuropsychiatr Genet. 2011;156:115–124. doi: 10.1002/ajmg.b.31142. [DOI] [PubMed] [Google Scholar]
- 3.Siu WK, Lam CW, Mak CM, Lau ET, Tang MH, Tang WF, Poon-Mak RS, Lee CC, Hung SF, Leung PW, Kwong KL, Yau EK, Ng GS, Fong NC, Chan KY. Diagnostic yield of array CGH in patients with autism spectrum disorder in Hong Kong. Clin Trans Med. 2016;5:18. doi: 10.1186/s40169-016-0098-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.South ST, Lee C, Lamb AN, Higgins AW, Kearney HM Working Group for the American College of Medical Genetics and Genomics (ACMG) Laboratory Quality Assurance Committee. ACMG Standards and Guidelines for constitutional cytogenomic microarray analysis, including postnatal and prenatal applications:revision 2013. Genet Med. 2013;15:901–909. doi: 10.1038/gim.2013.129. [DOI] [PubMed] [Google Scholar]
- 5.Schopler E, Reichler RJ, DeVellis RF, Dally K. Toward objective classification of child hood autism:Childhood Autism Rating Scale (CARS) J Autism Dev Disord. 1980;10:91–103. doi: 10.1007/BF02408436. [DOI] [PubMed] [Google Scholar]
- 6.Kaner S, Büyüköztürk S, İşeri E. Conners Parent Rating Scale-Revised Short:Turkish Standardization Study. Archives of Neuropsychiatry. 2013;50:100–109. [Google Scholar]
- 7.Abrahams BS, Geschwind DH. Advances in autism genetics:on the threshold of a new neurobiology. Nat Rev Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts JL, Hovanes K, Dasouki M, Manzardo AM, Butler MG. Chromosomal microarray analysis of consecutive individuals with autism spectrum disorders or learning disability presenting for genetic services. Gene. 2014;535:70–78. doi: 10.1016/j.gene.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schaefer GB, Starr L, Pickering D, Skar G, Dehaai K, Sanger WG. Array comparative genomic hybridization findings in a cohort referred for an autism evaluation. J Child Neurol. 2010;25:1498–1503. doi: 10.1177/0883073810370479. [DOI] [PubMed] [Google Scholar]
- 10.Shen Y, Dies KA, Holm IA, Bridgemohan C, Sobeih MM, Caronna EB, Miller KJ, Frazier JA, Silverstein I, Picker J, Weissman L, Raffalli P, Jeste S, Demmer LA, Peters HK, Brewster SJ, Kowalczyk SJ, Rosen-Sheidley B, McGowan C, Duda AW, Lincoln SA, Lowe KR, Schonwald A, Robbins M, Hisama F, Wolff R, Becker R, Nasir R, Urion DK, Milunsky JM, Rappaport L, Gusella JF, Walsh CA, Wu BL, Miller DT. Autism Consortium Clinical Genetics/DNA Diagnostics Collaboration. Clinical genetic testing for patients with autism spectrum disorders. Pediatrics. 2010;125:727–735. doi: 10.1542/peds.2009-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imitola J, Walleigh D, Anderson CE, Jethva R, Carvalho KS, Legido A, Khurana DS. Fraternal twins with autism, severe cognitive deficit, and epilepsy:diagnostic role of chromosomal microarray analysis. Semin Pediatr Neurol. 2014;21:167–171. doi: 10.1016/j.spen.2014.04.027. [DOI] [PubMed] [Google Scholar]
- 12.Harrison V, Connell L, Hayesmoore J, McParland J, Pike MG, Blair E. Compound heterozygous deletion of NRXN1 causing severe developmental delay with early onset epilepsy in two sisters. Am J Med Genet A. 2011;155A:2826–2831. doi: 10.1002/ajmg.a.34255. [DOI] [PubMed] [Google Scholar]
- 13.Onay H, Kacamak D, Kavasoglu AN, Akgun B, Yalcinli M, Kose S, Ozbaran B. Mutation analysis of the NRXN1 gene in autism spectrum disorders. Balkan J Med Genet. 2017;19:17–22. doi: 10.1515/bjmg-2016-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yingjun X, Haiming Y, Mingbang W, Liangying Z, Jiaxiu Z, Bing S, Qibin Y, Xiaofang S. Copy number variations independently induce autism spectrum disorder. Biosci Rep. 2017;37:pii:BSR20160570. doi: 10.1042/BSR20160570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bacchelli E, Battaglia A, Cameli C, Lomartire S, Tancredi R, Thomson S, Sutcliffe JS, Maestrini E. Analysis of CHRNA7 rare variants in autism spectrum disorder susceptibility. Am J Med Genet A. 2015;167A:715–723. doi: 10.1002/ajmg.a.36847. [DOI] [PubMed] [Google Scholar]
- 16.Leblond CS, Heinrich J, Delorme R, Proepper C, Betancur C, Huguet G, Konyukh M, Chaste P, Ey E, Rastam M, Anckarsa H, Nygren G, Gillberg IC, Melke J, Toro R, Regnault B, Fauchereau F, Mercati O, Lemiere N, Skuse D, Poot M, Holt R, Monaco AP, Jarvela I, Kantojarvi K, Vanhala R, Curran S, Collier DA, Bolton P, Chiocchetti A, Klauck SM, Poustka F, Freitag CM, Waltes R, Kopp M, Duketis E, Bacchelli E, Minopoli F, Ruta L, Battaglia A, Mazzone L, Maestrini E, Sequeira AF, Oliveira B, Vicente A, Oliveira G, Pinto D, Scherer SW, Zelenika D, Delepine M, Lathrop M, Bonneau D, Guinchat V, Devillard F, Assouline B, Mouren MC, Leboyer M, Gillberg C, Boeckers TM, Bourgeron T. Genetic and functional analyses of SHANK2 mutations suggest a multiple hit model of autism spectrum disorders. PLoS Genetics. 2012;8:e1002521. doi: 10.1371/journal.pgen.1002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geiger JC, Lipka J, Segura I, Hoyer S, Schlager MA, Wulf PS, Weinges S, Demmers J, Hoogenraad CC, Acker-Palmer A. The GRIP1/14-3-3 pathway coordinates cargo trafficking and dendrite development. Dev Cell. 2014;28:381–393. doi: 10.1016/j.devcel.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 18.Mejias R, Adamczyk A, Anggono V, Niranjan T, Thomas GM, Sharma K, Skinner C, Schwartz CE, Stevenson RE, Fallin MD, Kaufmann W, Pletnikov M, Valle D, Huganir RL, Wang T. Gain-of-function glutamate receptor interacting protein 1 variants alter GluA2 recycling and surface distribution in patients with autism. Proc Natl Acad Sci U S A. 2011;108:4920–4925. doi: 10.1073/pnas.1102233108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han M, Mejias R, Chiu SL, Rose R, Adamczyk A, Huganir R, Wang T. Mice lacking GRIP1/2 show increased social interactions and enhanced phosphorylation at GluA2-S880. Behav Brain Res. 2017;321:176–184. doi: 10.1016/j.bbr.2016.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu VW, Sarachana T, Kim KS, Nguyen A, Kulkarni S, Steinberg ME, Luu T, Lai Y, Lee NH. Gene expression profiling differentiates autism case-controls and phenotypic variants of autism spectrum disorders:evidence for circadian rhythm dysfunction in severe autism. Autism Res. 2009;2:78–97. doi: 10.1002/aur.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin J, Cooper M, Hamshere ML, Pocklington A, Scherer SW, Kent L, Gill M, Owen MJ, Williams N, O'Donovan MC, Thapar A, Holmans P. Biological overlap of attention-deficit/hyperactivity disorder and autism spectrum disorder:evidence from copy number variants. J Am Acad Child Adolesc Psychiatry. 2014;53:761–770. doi: 10.1016/j.jaac.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y, Liu Y, Pei YF, Yang TL, Deng FY, Liu XG, Li DY, Deng HW. Copy number variations at the Prader-Willi syndrome region on chromosome 15 and associations with obesity in whites. Obesity (Silver Spring) 2011;19:1229–1234. doi: 10.1038/oby.2010.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolton PF, Dennis NR, Browne CE, Thomas NS, Veltman MW, Thompson RJ, Jacobs P. The phenotypic manifestations of interstitial duplications of proximal 15q with special reference to the autistic spectrum disorders. Am J Med Genet. 2001;105:675–685. doi: 10.1002/ajmg.1551. [DOI] [PubMed] [Google Scholar]
- 24.Anney R, Klei L, Pinto D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, Sykes N, Pagnamenta AT, Almeida J, Bacchelli E, Bailey AJ, Baird G, Battaglia A, Berney T, Bolshakova N, Bölte S, Bolton PF, Bourgeron T, Brennan S, Brian J, Carson AR, Casallo G, Casey J, Chu SH, Cochrane L, Corsello C, Crawford EL, Crossett A, Dawson G, de Jonge M, Delorme R, Drmic I, Duketis E, Duque F, Estes A, Farrar P, Fernandez BA, Folstein SE, Fombonne E, Freitag CM, Gilbert J, Gillberg C, Glessner JT, Goldberg J, Green J, Guter SJ, Hakonarson H, Heron EA, Hill M, Holt R, Howe JL, Hughes G, Hus V, Igliozzi R, Kim C, Klauck SM, Kolevzon A, Korvatska O, Kustanovich V, Lajonchere CM, Lamb JA, Laskawiec M, Leboyer M, Le Couteur A, Leventhal BL, Lionel AC, Liu XQ, Lord C, Lotspeich L, Lund SC, Maestrini E, Mahoney W, Mantoulan C, Marshall CR, McConachie H, McDougle CJ, McGrath J, McMahon WM, Melhem NM, Merikangas A, Migita O, Minshew NJ, Mirza GK, Munson J, Nelson SF, Noakes C, Noor A, Nygren G, Oliveira G, Papanikolaou K, Parr JR, Parrini B, Paton T, Pickles A, Piven J, Posey DJ, Poustka A, Poustka F, Prasad A, Ragoussis J, Renshaw K, Rickaby J, Roberts W, Roeder K, Roge B, Rutter ML, Bierut LJ, Rice JP, Salt J, Sansom K, Sato D, Segurado R, Senman L, Shah N, Sheffield VC, Soorya L, Sousa I, Stoppioni V, Strawbridge C, Tancredi R, Tansey K, Thiruvahindrapduram B, Thompson AP, Thomson S, Tryfon A, Tsiantis J, Van Engel H, Vincent JB, Volkmar F, Wallace S, Wang K, Wang Z, Wassink TH, Wing K, Wittemeyer K, Wood S, Yaspan BL, Zurawiecki D, Zwaigenbaum L, Betancur C, Buxbaum JD, Cantor RM, Cook EH, Coon H, Cuccaro ML, Gallagher L, Geschwind DH, Gill M, Haines JL, Miller J, Monaco AP, Nurnberger JI, Jr, Paterson AD, Pericak-Vance MA, Schellenberg GD, Scherer SW, Sutcliffe JS, Szatmari P, Vicente AM, Vieland VJ, Wijsman EM, Devlin B, Ennis S, Hallmayer J. A genome-wide scan for common alleles affecting risk for autism. Hum Mol Genet. 2010;19:4072–4082. doi: 10.1093/hmg/ddq307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gill M, Kenny G, Anney R. Fitzgerald M, editor. The Genetic Architecture of Autism and Related Conditions, Chapter 14. Recent Advances in Autism Spectrum Disorders. INTECH. 2013:299–320. [Google Scholar]
- 26.Jones RM, Cadby G, Blangero J, Abraham LJ, Whitehouse AJ, Moses EK. MACROD2 gene associated with autistic-like traits in a general population sample. Psychiatr Genet. 2014;24:241–248. doi: 10.1097/YPG.0000000000000052. [DOI] [PMC free article] [PubMed] [Google Scholar]