Abstract
Dihydropyrimidine dehydrogenase (DPD; DPYD gene) variants have emerged as reliable predictors of adverse toxicity to the chemotherapy agent 5-fluorouracil (5-FU). The intronic DPYD variant rs75017182 has been recently suggested to promote alternative splicing of DPYD. However, both the extent of alternative splicing and the true contribution of rs75017182 to DPD function remain unclear. In the present study we quantified alternative splicing and DPD enzyme activity in rs75017182 carriers utilizing healthy volunteer specimens from the Mayo Clinic Biobank. Although the alternatively spliced transcript was uniquely detected in rs75017182 carriers, canonically spliced DPYD levels were only reduced by 30% (p=2.8×10−6) relative to controls. Similarly, DPD enzyme function was reduced by 35% (p=0.025). Carriers of the well-studied toxicity-associated variant rs67376798 displayed similar reductions in DPD activity (31% reduction). The modest effects on splicing and function suggest that rs75017182 may have clinical utility as a predictor of 5-FU toxicity similar to rs67376798.
Keywords: 5-Fluorouracil, Dihydropyrimidine Dehydrogenase, DPD Deficiency, Single Nucleotide Polymorphism, SNPs, Splicing, Alternative, Pharmacogenetics
Introduction
5-fluorouracil (5-FU) is a widely prescribed chemotherapeutic agent used to treat a variety of cancers. As with most chemotherapeutics, 5-FU–containing treatments show high degrees of inter-individual variability in response and toxicity profiles. Toxicity risk is partially dependent on concomitant therapies, but in current treatment regimens approximately one-third of 5-FU–treated patients develop grade 3 or higher toxicity that is related to 5-FU (1). Dihydropyrimidine dehydrogenase (DPD) is the rate-limiting enzyme of 5-FU catabolism (2). As such, DPD converts 80–85% of administered 5-FU into inactive metabolites (3). Deficiency of DPD induces an increased accumulation of 5-FU leading to toxic side effects (4).
DPD is encoded by the DPYD gene (NM_000110.3) located on chromosome 1p21.3 (5). Clinical and functional studies have primarily focused on exon variants in DPYD and their association with 5-FU toxicity (6, 7). Currently, three well-studied DPYD single nucleotide polymorphisms (SNPs) are recognized as predictive of 5-FU–associated toxicity: rs3918290 (NM_000110.3:c.1905+1G>A; DPYD*2A), rs55886062 (NM_000110.3:c.1679T>G; NP_000101.2:p.Ile560Ser; DPYD*13), and rs67376798 (NM_000110.3:c.2846A>T; NP_000101.2:p.Asp949Val) (3, 8, 9). More recently, a collection of SNPs termed HapB3, which is composed of three intronic variants [rs56276561 (NM_000110.3:c.483+18G>A), rs6668296 (NM_000110.3:c.680+139G>A), and rs115349832 (NM_000110.3:c.959–51T>C)] and one synonymous variant [rs56038477 NM_000110.3:c.1236G>A; NP_000101.2:p.Glu412=)], has been suggested to also contribute to 5-FU toxicity (10). Subsequently, an additional variant in linkage disequilibrium with HapB3 rs75017182 (NM_000110.3:c.1129–5923C>G) was suggested to activate a splice donor site within intron 10 and promote alternative splicing to include a 44-nucleotide cryptic exon (11, 12). Despite the putative mechanistic explanation for the link between HapB3 and 5-FU toxicity, the quantitative contribution of HapB3/rs75017182 to alternative splicing and DPD function has not been demonstrated. A recent meta-analysis has provided evidence that HapB3/rs75017182 may increase the risk of 5-FU toxicity (13). However it should be noted while some clinical studies and case report series have indicated that HapB3/rs75017182 may be predictive of toxicity (10–12, 14), additional studies have failed to establish statistically significant associations (15–18).
The objectives of the present study were to quantify the effect of rs75017182 on DPYD mRNA splicing, DPYD expression, and DPD enzyme function. Based on the lack of consistent clinical correlations with toxicity, we hypothesized that alternative splicing is not absolute in carriers of HapB3/rs75017182. The results presented in this article provide a mechanistic link between HapB3/rs75017182 and 5-FU toxicity by showing that the reduction in DPD enzyme function caused by alternative splicing in rs75017182 carriers is similar to that observed in carriers of the clinically relevant SNP rs67376798.
Results
Study population characteristics
To identify carriers of rs75017182 for subsequent functional studies, DNA samples from 3950 individuals that donated specimens to the Mayo Clinic Biobank (19) were genotyped for rs75017182 and the known 5-FU toxicity–associated variants rs3918290, rs55886062, and rs67376798. Demographic data and allele frequencies are presented in Table 1. One specimen failed genotyping and was excluded from the study. Of the remaining 3949 participants, 166 carried rs75017182 (4.2%; 165 heterozygous and 1 homozygous). Thirty-two subjects carried rs3918290 (0.81%; 31 heterozygous and 1 homozygous). Five individuals were heterozygous for rs55886062 (0.13%). Forty subjects carried rs67376798 (1.01%; 39 heterozygous and 1 homozygous). One individual carried both rs75017182 and rs55886062. The remaining 3706 (93.85%) study participants did not carry any of the tested variants. Allele frequencies did not significantly deviate from Hardy-Weinberg equilibrium for any variant.
Table 1: Demographic information for the overall study population.
| Variables |
Age (years) |
Sex |
Carrier frequency |
|||
|---|---|---|---|---|---|---|
| Mean (SD) | p-value1 | Male n (%) | Female n (%) | p-value2 | Number (%) | |
| Non-Carriers | 56 (14) | 1836 (50) | 1870 (51) | 3706 (94) | ||
| rs75017182 | 60 (14) | 0.78 | 76 (46) | 90 (54) | 0.38 | 166 (4.2) |
| rs3918290 | 57 (15) | 0.56 | 15 (47) | 17 (53) | 0.85 | 32 (0.81) |
| rs55886062 | 59 (1.3) | 0.57 | 2 (40) | 3 (60) | 1.0 | 5 (0.12) |
| rs67376798 | 54 (16) | 0.47 | 21 (53) | 19 (48) | 0.75 | 40 (1.0) |
Variant, specific genetic variant in dihydropyrimidine dehydrogenase; Non-carriers, sample does not carry rs75017182, rs3918290, rs55886062 and rs67376798 variant.
Student’s t-test comparing age in variant and non-carrier groups.
Fisher’s exact test comparing sex ratios between variant and non-carrier groups.
Participants carrying any of the tested genotypes and matched controls were invited to provide an additional fresh blood sample for additional studies as detailed in the Materials and Methods section. Of the participants in this subcohort, 56 carried rs75017182 (27.5%), 13 carried rs3918290 (6.4%), and 2 carried rs55886062 (1.0%), 12 carried rs67376798 (5.9%), and 121 were non-carriers (59.3%; Table 2). All participants in this phase of the study were heterozygous carriers of a single variant (except for non-carrier controls). Age and sex distributions were similar between rs75017182, rs3918290, rs67376798, and non-carrier groups (Table 2). Both carriers of rs55886062 were 74-year-old females, which explains the significant difference in sex and age distribution from non-carriers (Table 2).
Table 2: Demographic information for the study cohort that provided blood samples for function studies.
| Variables |
Age (years) |
Sex |
Carrier frequency |
|||
|---|---|---|---|---|---|---|
| Mean (SD) | p-value1 | Male n (%) | Female n (%) | p-value2 | Number (%) | |
| Non-Carriers | 60 (13) | 44 (36) | 77 (64) | 121 (59) | ||
| rs75017182 | 58 (15) | 0.22 | 28 (50) | 28 (50) | 0.10 | 56 (28) |
| rs3918290 | 65 (15) | 0.25 | 6 (46) | 7 (54) | 0.55 | 13 (6.4) |
| rs55886062 | 74 (0.0) | 1.7×10–21 | 0 (0.0) | 2 (100) | 0.37 | 2 (1.0) |
| rs67376798 | 55 (15) | 0.16 | 6 (50) | 6 (50) | 0.40 | 12 (5.9) |
Variant, specific genetic variant in dihydropyrimidine dehydrogenase; Non-carriers, sample does not carry rs75017182, rs3918290, rs55886062 and rs67376798 variant.
Student’s t-test comparing age in variant and non-carrier groups.
Fisher’s exact test comparing sex ratios between variant and non-carrier groups.
Alternative splicing in rs75017182 carriers
To determine if all carriers of rs75017182 exhibited alternatively spliced DPYD harboring the 44-nucleotide cryptic exon, PCR primers were designed to specifically amplify canonical and cryptic exon-containing DPYD. Primers were designed on both sides of the exon 10–11 junction, which would be expected to produce PCR products for both alternatively spliced (Figure 1A, top left) and canonically spliced (Figure 1A, bottom left) DPYD. Two bands were detected in rs75017182 carriers that correspond to the predicted sizes of 252 and 296 nucleotides; a single band of 252 nucleotides was detected in non-carriers (Figure 1A, right). Additional primers were designed to specifically amplify canonically spliced DPYD (Figure 1B–1C, left). Single bands of expected sizes were generated for both primer sets in carriers and non-carriers of rs75017182 (Figure 1B–1C, right). Similarly, primers were designed to specifically amplify DPYD transcripts containing the cryptic exon (Figure 1D–1E, left). Both primer sets yielded products as single bands only in carriers of rs7501782 (Figure 1D–1E, right), suggesting that the alternatively spliced product is exclusive to SNP carriers.
Figure 1. PCR and immunoblot analyses of alternative splicing in heterozygous rs75017182 carriers.
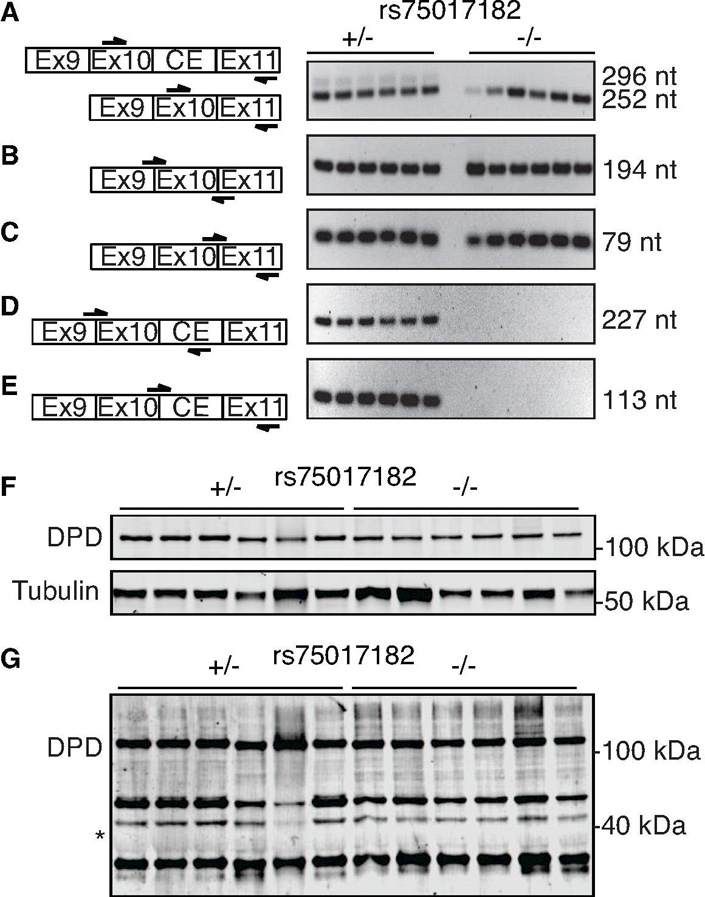
A) Left, PCR primers were designed as indicated (arrows) to amplify across the exon 10–11 junction. Right, PCR products that were generated for heterozygous carriers of rs75017182 (+/−) and non-carriers (−/−) using the primers at left for the indicated size (nt, nucleotides). B–C) Primers were designed to specifically amplify DPYD that is canonically spliced at the exon 10–11 junction. D–E) Primers were designed to specifically amplify DPYD transcripts containing the cryptic exon. Representative data from the same 12 samples is shown for panels A–E. F) Western blot for full-length DPD (111 kDa) and α-tubulin in 6 rs75017182 carriers and 6 non-carriers. G) Prolonged exposure of DPD blot presented in (F). The predicted molecular mass of the translated alternatively spliced DPYD containing the 44 nt insertion resulting in a frameshift (expected mass 40 kDa) is indicated by an asterisk (*).
Immunoblotting was used to determine if the alternatively spliced transcript was translated. Samples from six carriers of rs75017182 and six non-carriers were selected at random from the study population for analysis. Bands corresponding to DPD from canonically spliced DPYD (111 kDa) were detected in all samples regardless of carrier status (Figure 1F). The frameshift caused by the 44-nucleotide insertion in the alternatively spliced transcript is predicted to result in a translated protein of 390 amino acids with a molecular mass of approximately 40 kDa. Even with prolonged exposure, a band could not be detected at the predicted size using a polyclonal anti-DPD antibody raised against the N-terminus of the protein (Figure 1G).
Splice-specific expression of DPYD in rs75017182 carriers
To quantify changes in canonically spliced DPYD expression, primers specific to the exon 10 and 11 junction (as depicted in Figure 1B–1C, left) were used . The levels of correctly spliced DPYD were reduced by 30% in carriers of rs75017182 (p=2.8×10−6 and p=6.3×10−7 for the two individual primer sets used; Figure 2A–2B). No significant differences in expression were noted for carriers of rs3918290, rs55886062, or rs67376798 compared to non-carriers (Figure 2A–2B). Consistent with the results from qualitative PCR (Figure 1D–1E), alternatively spliced DPYD was only detected in heterozygous rs75017182 carriers (data not shown). No differences in total DPYD expression (measured using intron-spanning primers targeting exons 1 and 2) were noted (Figure 2C).
Figure 2. Quantitative RT-PCR of canonically spliced DPYD.
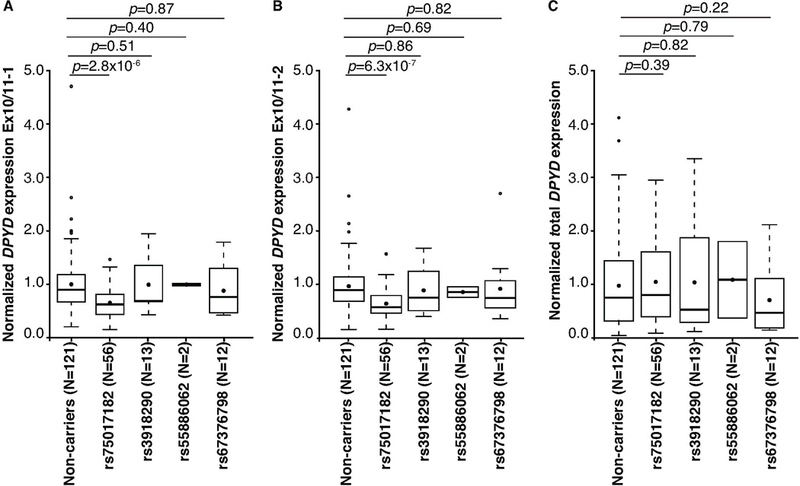
Relative expression of canonically spliced DPYD expression was measured using primers described in Figure 1D (A) and Figure 1E (B). C) Total DPYD expression was measured using primers that amplified across the exon 1–2 junction. Box plots indicate the interquartile (IQR) ranges (boxes) ± 1.5 IQRs (whiskers). Filled circles indicate means; thick bars represent medians. Outlier data are indicated by open circles. P-values were calculated using the Wilcoxon rank sum test.
Minigene reporter model for alternative splicing
To model the effects of rs75017182 in a controlled isogenic model system, the relevant regions were cloned into a minigene fluorescent reporter vector and the corresponding C>G nucleotide change was introduced (Figure 3A, top). In this model, canonical splicing would lead to expression of both mCherry and AcGFP; alternative splicing would shift AcGFP out of frame resulting in only expression of mCherry (Figure 3A, bottom). As a control, regions relevant to rs3918290 were also cloned into the reporter and the corresponding G>A nucleotide change was introduced (Figure 3B, top). A single base deletion was introduced in exon 14 (normally 165 bases in length) so that canonical and alternatively spliced products could also be differentiated by changes in AcGFP expression (Figure 3B, bottom). Note that due to necessary design considerations, canonically spliced products are double-positive for AcGFP and mCherry in the rs75017182 model (Figure 3A), but alternatively spliced products are double-positive in the rs3918290 model (Figure 3B).
Figure 3. Mini-gene reporter models to assess alternative splicing induced by rs75017182 and rs3918290.
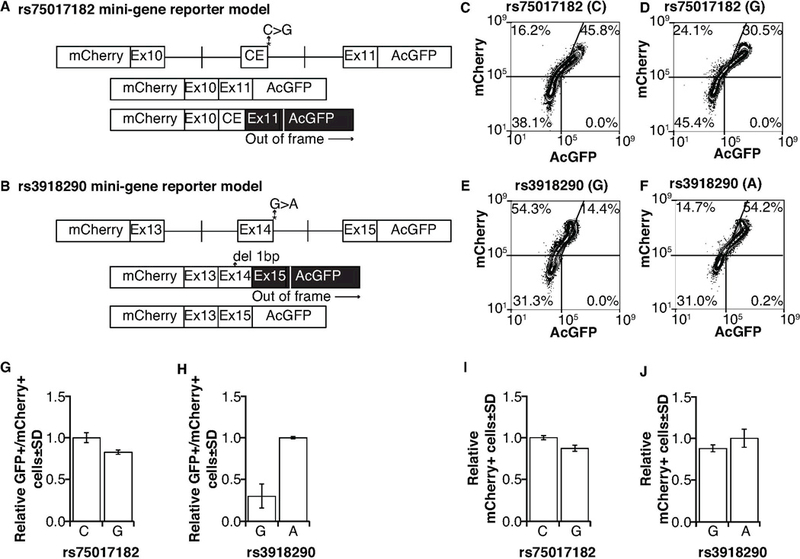
A) Schematic of mini-gene reporter vectors for rs75017182. Canonical splicing would maintain the reading frame and result in expression of both mCherry and AcGFP; inclusion of the cryptic exon would shift AcGFP out of frame resulting in only mCherry expression. B) Schematic of mini-gene reporter vectors for rs3918290. Because inclusion of exon 14 would maintain the reading frame, a one-basepair (bp) deletion was introduced near the middle of exon 14. Therefore canonical splicing would shift AcGFP out of frame resulting in expression of only mCherry; the exclusion of exon 14 would result in in-frame expression of both mCherry and AcGFP. Flow cytometry was used to measure the relative levels of reporter (AcGFP and mCherry) expression as an indication of canonical and alternative splicing in cells expressing the rs75017182 C allele (C), the rs75017182 G allele (D), the rs3918290 G allele (E), and the rs3918290 A allele (F). G) Histograms showing the relative frequency of AcGFP+/mCherry+ cells (i.e., canonically spliced) with the rs75017182 genotype. H) Histograms showing relative frequency of AcGFP+/mCherry+ (i.e., alternatively spliced) cells with the rs3918290 genotype. Data for the G allele in panel H designates background levels of the assay. Frequencies are reported relative to total mCherry+ cells to correct for transfection efficiency. Transfection efficiency was measured using total mCherry+ cells in the rs75017182 (I) and rs3918290 (J) models. Three independent experiments were performed; data in panels C–F are representative results.
Percentages of single mCherry-positive and mCherry/AcGFP double-positive cells were measured by flow cytometry (Figure 3C–3F). The relative ratio of AcGFP-negative cells in the mCherry-positive population of the rs75017182 model is similar to that for AcGFP/mCherry double-positive cells of the rs3918290 model, consistent with the inverse nature of our models as discussed in the previous paragraph (compare Figure 3C with Figure 3E). However, the decrease in double-positive cells in the rs75017182 model is not as great as the increase in double-positive cells in the rs3918290 model (compare Figure 3D with Figure 3F), suggesting that alternative splicing is not obligate in rs75017182G-expressing cells. To further illustrate this finding, the percentage of double-positive cells relative to total mCherry-positive cells was assessed. The percentage of double-positive cells was slightly reduced in cells expressing the rs75017182 G allele compared to those expressing the C allele (Figure 3G). Notably, the change in double-positive cells was not as great as that observed in the rs3918290 model (Figure 3H), further supporting the hypothesis that the rs75017182 G allele does not lead to obligate alternative splicing. Transfection efficiency (as determined by the relative frequency of mCherry-positive cells) was similar between wildtype and variant allele-expressing cells for both rs75017182 and rs3918290 models (Figure 3I–3J).
Variant effects on DPD enzyme activity
To determine the extent to which alternative splicing in carriers of rs75017182 affects DPD enzyme function, ex vivo DPD enzyme activity was measured using peripheral blood mononuclear cell (PBMC) lysates obtained from study participants. Compared to the non-carrier population, DPD activity was reduced by 35% in heterozygous carriers of rs75017182 (p=0.025; Figure 4). A similar reduction of DPD activity was noted for carriers of the 5-FU toxicity-associated variant rs67376798 (31%; Figure 4), although the present study was underpowered with regard to this variant. Therefore, a significant p-value was not observed (p=0.17). Greater reductions in DPD activity were noted for carriers of the other two 5-FU toxicity-associated SNPs rs3918290 (50% reduction, p=0.0011) and rs55886062 (68% reduction, p=0.044).
Figure 4. DPD enzyme activity by genotype.
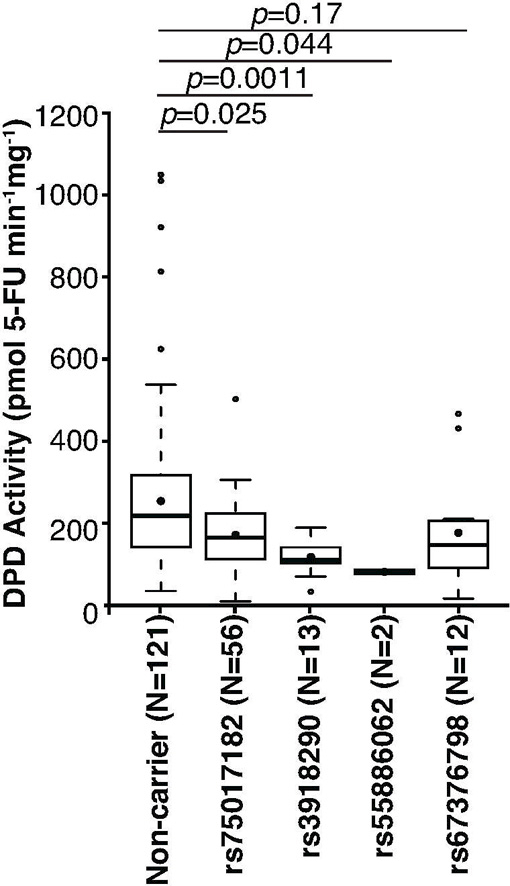
DPD enzyme activity was measured in PBMCs isolated from individuals with the indicated genotypes (all carriers are heterozygous). Box plots indicate the interquartile (IQR) ranges (boxes) ± 1.5 IQRs (whiskers). Filled circles indicate means; thick bars represent medians. Outlier data are indicated by open circles. P-values were calculated using the Wilcoxon rank sum test.
In allelic association tests, rs75017182 and rs3918290 were significantly associated with reduced DPD enzyme activity (p=0.011 and p=0.016, respectively; Table 3). Neither rs55886062 nor rs67376798 achieved single marker significance (p=0.21 and p=0.35, respectively; Table 3). DPD enzyme activity was not significantly correlated with age or sex (data not shown), and correcting for age and sex did not dramatically affect p-values (Table 3). In a secondary analysis, a linear model containing all four tested variants, as well as age and sex, was tested. After adjusting for covariates, rs75017182 and rs3918290 were more strongly associated with DPD activity (p=0.00070 and p=0.0019, respectively).
Table 3: Variant association with DPD enzyme activity.
| Variables |
Single allele association |
Corrected for age and sex |
Full linear model |
||
|---|---|---|---|---|---|
| p-value1 | FDR2 | p-value3 | p-value4 | Regression coefficient (95% CI) | |
| rs75017182 | 0.011 | 0.031 | 0.010 | 0.00070 | −0.25 (−0.38, −0.11) |
| rs3918290 | 0.016 | 0.031 | 0.017 | 0.0019 | −0.22 (−0.36, −0.083) |
| rs55886062 | 0.21 | 0.28 | 0.22 | 0.12 | −0.11 (−0.24, 0.027) |
| rs67376798 | 0.35 | 0.35 | 0.34 | 0.076 | −0.13 (−0.26, 0.012) |
| Age | NA | NA | NA | 0.71 | −0.026 (−0.16, 0.11) |
| Sex | NA | NA | NA | 0.74 | −0.023 (−0.16, 0.11) |
DPD, Dihydropyriminine Dehydrogenase; Variant, specific genetic variants in dihydropyrimidine dehydrogenase; FDR, False Discovery Rate; NA, Not Applicable; CI, Confidence Interval.
Asymptotic p-value for t-statistic using DPD activity as a quantitative trait.
FDR corrected p-value (Step-up Benjamini & Hochberg (1995) false discovery rate).
P value adjusted for age and sex as covariates.
P value adjusted for age, sex, and other three tested SNPs as covariates.
Discussion
Clinical studies have clearly established that specific genetic variants in DPYD can be strongly predictive of 5-FU toxicity (8, 9, 20), and dosing guidelines have been published by the Clinical Pharmacogenetics Implementation Consortium (CPIC) for carriers of three well-studied variants, rs3918290, rs55886062, and rs67376798 (21). Using a 111-patient cohort, Amstutz et al. (10) identified a collection of SNPs termed HapB3 that was enriched in patients that experienced severe (grade ≥3) 5-FU–related toxicity. In a subsequent case report, an alternatively spliced transcript was identified in HapB3 carriers (12). The rs75017182 SNP, which is in strong linkage disequilibrium with HapB3, was suggested to promote alternative splicing by creating a splice donor site within intron 10. However, the extent of alternative splicing and the impact on DPYD expression and DPD activity remained inconclusive.
The results of the present study demonstrated that levels of correctly spliced DPYD were reduced by 30% in carriers of rs75017182 (Figure 2B–2C), thus providing key functional support for this variant’s association with 5-FU toxicity. To our knowledge, this report is the first to quantify canonical splicing in rs75017182 carriers relative to non-carriers. Our data suggest that not all transcripts expressed from rs75017182G-containing DNA strands contain the cryptic exon. The 30% reduction in canonical splicing observed in our studies is less than the 50% reduction that would be expected for heterozygous carriers if alternative splicing was obligate. The incomplete alternative splicing is consistent with the case report of a homozygous rs75017182 carrier in which both transcripts could be detected (12). Furthermore, similar to the reduction in canonically spliced DPYD mRNA, DPD enzyme activity was reduced by 35% (p=0.025) in PBMCs from individuals with rs75017182 (Figure 4). These results strongly suggest that rs75017182 is the primary causal variant in HapB3. In heterozygous carriers of rs3918290 enzyme activity was more severely reduced (50% reduction; p=0.0011), which is consistent with earlier reports (22, 23). Collectively, these findings support the conclusion that rs75017182 does not impact DPD function as severely as rs3918290. However, it should be noted that rs75017182 was detected a much higher frequency (4.2%, Table 1) than rs3918290 (0.81%, Table 1) in the original cohort and, therefore, may contribute to a greater number of 5-FU toxicity cases.
The level of DPD enzyme activity in carriers of rs75017182 was most closely related to that in carriers of rs67376798 (31% reduction; Figure 4). While the difference in enzyme activities between rs67376798 carriers and the non-carrier group was not significant (p=0.17; Figure 4), the SNP trended closer to significance after adjusting for age, sex, and SNP-status for the other three variants in allelic association studies (p=0.076; Table 3). In previous studies directed at measuring the relative enzyme function of DPD variants in vitro, p.D949V (the amino acid change encoded by rs67376798) exhibited enzyme activity that was 41–65% lower than wildtype DPD protein (6, 24). In an earlier study that contained a limited number of rs67376798 carriers, the SNP was shown to have a modest, but significant, effect on DPD function (25), which was similar to the results of the current study. Despite the likely modest effect on DPD function, clinical association studies have reproducibly established links between rs67376798 and 5-FU toxicity (1, 9, 26–29). When taken in context with these previously published results, our data indicate that rs75017182 likely has a modest net effect on DPD enzyme activity that may contribute to toxicity risk similar to rs67376798.
It is notable that all studies showing significant association between HapB3/rs75017182 and 5-FU toxicity only evaluated toxicity in the first 2 cycles of chemotherapy (10, 14, 27), whereas studies that failed to show an association evaluated cumulative toxicity over all chemotherapy sessions (15–17, 28). This may suggest that the inclusion of toxicity data from later cycles might mask the contributions of less severe variants to the development of toxicity. Recently a meta-analysis was conducted (13) that included all of the above-referenced manuscripts except for Lee et al. 2016 (16), which had not yet been published. This meta-analysis also included an additional unpublished dataset. Notably, HapB3/rs75017182 showed association with adverse toxicity and a modest adjusted relative risk (RR) of 1.59 (95% CI 1.29–1.97) (13). It is likely that the addition of the Lee et al. data would further marginalize this association. Regardless, the effect size noted in the meta-analysis (RR=1.59) was less than that for rs3918290 (RR=2.85, 95% CI 1.75–4.65) and rs67376798 (RR=3.02, 95% CI 2.22–4.10). While the difference between RR for rs75017182 and rs3918290 is consistent with the results of the present study, the high risk associated with rs67376798 is unexpected (also in light of other data discussed in the previous paragraph). Regardless, it should be noted that the RR values reported for the studies used for the meta-analysis varied for each SNP. These factors may not completely explain the lack of perfect correlation between RR in the meta-analysis and functional effects noted in this article. However, we would note that the present results are generally in the range of those from the meta-analysis and are consistent in that they support HapB3/rs75017182 as a predictor of modestly increased risk of 5-FU toxicity.
One factor not accounted for in the present study was the effect of additional mechanisms controlling DPYD/DPD expression. Previously, we showed that expression of the microRNA miR-27a negatively correlated with DPD function and that the relatively common miR-27a polymorphism rs895819 was significantly associated with reduced DPD expression and enzyme activity (30). Subsequent studies showed that rs895819 significantly interacted with rs55886062, rs67376798, and rs75017182 to potentiate toxicity risk in carriers (31, 32). In addition to microRNA-mediated regulation, DPYD expression was also recently shown to be epigenetically regulated with potential contributions to 5-FU toxicity and response (33). Additional studies will be needed to directly address potential epistatic effects between alterations in DPD regulation and variants that affect DPD function.
In the present study, we directly examined the contributions of the intronic SNP rs75017182 to changes in DPD enzyme function, which is a clinically relevant correlate with 5-FU toxicity risk (34). Using this approach, we quantified the effects of this SNP on DPYD splicing and DPD function to potentially clarify the modest contribution of this SNP to 5-FU toxicity. In light of our findings, additional studies are necessary to test the contribution of this SNP to 5-FU toxicity risk in the context of additional DPYD variants and with additional factors that regulate DPYD/DPD expression.
Materials and methods
Study population
Study participants were identified from donors to the Mayo Clinic Biobank, a research resource that has enrolled over 50,000 Mayo Clinic patients and volunteers since 2009 (19). For the present study, 3950 Biobank participants were selected based on availability of current medical information at the time of the study. The sample cohort was enriched for individuals living in Olmsted County, MN, and surrounding counties in southeast Minnesota. Participants were excluded from participation if they were previously treated with chemotherapy. To align with the high prevalence of rs7501782 in European populations, participation in the present study was restricted to individuals that self-declared race as “white.” Ethnicity, defined as Hispanic or non-Hispanic in the Biobank (35), was not considered in the selection process. DNA aliquots were prepared by the Mayo Clinic Biospecimens Acquisition and Processing (BAP) shared resource. Individuals were genotyped for rs75017182, rs3918290, rs55886062, and rs67376798 as described below. Individuals that carried any of the 4 tested SNPs were invited to provide an additional blood specimen for the studies described below. For each carrier of rs75017182 that was invited to participate, two age-and sex-matched controls (i.e., individuals that did not carry any of the tested SNPs) were recalled. Of the 575 individuals contacted, 204 provided an additional sample for functional studies. Demographic data were obtained as described (19, 35). Sex was self-reported by study participants. All studies were approved by the Mayo Clinic Institutional Review Board (IRB no. 08–00749 and 14–005521).
Cells
HEK293T/c17 (culture CRL-11268) cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and maintained at 37°C in a humidified incubator with 5% CO2 atmosphere. Cells were cultured using Dulbecco’s Modified Eagle Medium (Mediatech Inc. Manassas, VA) supplemented with 10% FBS (EMD Millipore, Saint Charles, MO), 100 U/mL penicillin (Mediatech), and 100 µg/mL streptomycin (Mediatech). Cell identity was confirmed and monitored as previously reported (7). Aliquots of low-passage cells were cryopreserved within 2 weeks of receipt. Cells were cultured for no longer than 10 total passages or 2 months, whichever occurred earlier. All cell lines were periodically monitored for mycoplasma using Hoechst staining (Sigma-Aldrich, St. Louis, MO). Culture health and identity were monitored by microscopy and by comparing population doubling times to baseline values recorded at time of receipt. Additional authentication of this cell line above that described was not performed.
Genotyping
Custom TaqMan pimer/probe combinations were obtained from Thermo Fisher Scientific (Waltham, MA; Supplementary Table S1). Genotyping was carried out using LightCycler 480 Probes Master (Roche Diagnostics, Indianapolis, IN) on a LightCycler 480 System II using reaction and cycling parameters recommended by the manufacturer. Genotype was assessed using the endpoint genotyping module of the LightCycler 480 Software (release 1.5.0 SP3). Genotypes of the subcohort were confirmed using DNA obtained at the time of return visit.
RNA expression
RNA was isolated from partcipants’ blood samples using the PAXgene Blood RNA Kit (PreAnalytiX, Hombrechtikon, Switzerland). As such, RNA was only available for the 204 individuals that provided an additional sample. Full-length cDNA was synthesized using the Transcriptor High Fidelity cDNA Synthesis Kit (Roche) with anchored oligo(dT) primers. Primers for quantitative PCR are listed in Supplementary Table S2. Quantitative PCR was performed using LightCycler 480 SYBR Green I Master (Roche) reagents using 40 cycles consisting of 95°C for 15 seconds, 60°C for 30 seconds, and 72°C for 30 seconds. Qualitative PCR was performed using the same cycling parameters, only the number of cycles was reduced to 30–35, depending on target. Specificity of products was confirmed by melting curve analysis performed after PCR for all samples and gel electrophoresis of selected samples. Relative expression was calculated by the 2-ΔΔCP method. Mean expression of GAPDH, PSMB2, PSMB4, RAB7A, REFP5, and VCP was used as the reference.
Western blotting
Protein lysates were separated by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF)-FL membrane (EMD Millipore, Saint Charles, MO). Membranes were blocked using Odyssey blocking buffer (LI-COR Biosciences, Lincoln, NE). Blots were probed with primary antibodies against DPD and α-tubulin (ab6556 and ab186407, respectively; Abcam, Cambridge, MA). IRDye800-conjugated goat anti-mouse and IRDye680-conjugated goat anti-rabbit secondary antibodies (LI-COR) were used for detection. Blots were scanned on a LI-COR Odyssey imager.
Measurement of DPD enzyme activity
PBMCs were isolated from 30 mL of whole blood within 2 hours of collection as previously described (7). Cell lysates were prepared and total protein content was quantified as described (6). Lysates were diluted to a standard concentration, aliquots prepared, and stored at −80°C until use. DPD activity was measured using methods previously described in detail (6). Briefly, cell lysates were reacted with 1.65 pmol [6-C14]-5-FU (Moravek Biochemicals, Brea, CA) in the presence of 40 pmol NADPH (Sigma-Aldrich, Saint Louis, MO) for 30 minutes at 37°C with constant agitation. Reactions were terminated using cold 100% ethanol. Conversion of [6-C14]-5-FU to [6-C14]-5-dihydrofluorouracil ([6-C14]-5-DHFU) was assessed using high performance liquid chromatography (HPLC) as described (36). Rate of conversion was calculated as pmol 5-FU min−1 mg−1 of total input protein as described (6).
Mini-gene reporter assay
Minigene reporter constructs were generated by cloning mCherry and AcGFP lacking stop and start codons, respectively, using site-specific PCR into the multiple cloning site of pIRES-neo3 (Takara Bio USA, Mountain View, CA). Relevant exon regions +/−~500 nucleotides of relevant flanking intronic sequence were PCR amplified and cloned into the resulting vector to produce minigene reporters such that canonical and alternative splicing would differentially result in in-frame expression of AcGFP; mCherry would be expressed from all recipient cells. PCR primers are listed in Supplementary Table S3. Plasmid sequences were confirmed by Sanger sequencing at the Mayo Clinic Gene Analysis Shared Resource. HEK-293T/c17 were transfected with 5µg plasmid using X-tremeGENE HP (Roche) according to the manufacturer’s instructions. Flow cytometry of live cells was performed using the NovoCyte 3000RYB (Acea Biosciences, San Diego, CA) equipped with 640, 561, and 488 nm lasers and NovoSampler autosampler. Data acquisition and analysis was performed using NovoExpress 1.2.4.1602 software (Acea).
Statistical analyses
Fisher exact tests, Student’s t tests and Wilcoxon rank sum tests were performed using the R Environment for Statistical Computing version 3.3.1 (R Foundation for Statistical Computing, Vienna, Austria). Exact p-values for deviation from Hardy-Weinberg equilibrium were calculated as described (37). Additional statistical tests were conducted in PLINK version 1.07 (38). The Wald test was used to assess allelic association with DPD enzyme activity (treated as a continuous, quantitative variable). Step-up false-discovery rate (FDR) was used to correct for multiple testing bias (39). Multivariate analyses were performed using an additive general linear model for the indicated covariates.
Supplementary Material
Study Highlights.
What is the current knowledge on the topic?
The DPYD intronic variant rs75017182 has been suggested to affect mRNA splicing by introducing a putative splice donor site, although the extent to which transcripts are alternatively spliced in carriers of this variant is currently unknown. Clinical studies have failed to consistently establish clear association between rs75017182 and 5-FU toxicity.
What question did this study address?
The current study quantifies the level of alternative splicing in rs75017182 carriers and measures the effects on DPD enzyme function.
What this study adds to our knowledge?
Canonical DPYD splicing and DPD enzyme activity are reduced by approximately one-third in carriers of rs75017182. This change in activity is comparable to that for carriers of the well-established toxicity-associated variant rs67376798.
How this might change clinical pharmacology or translational science?
The modest effects on DPD activity potentially explain the inconsistent results noted for this variant in previous clinical studies. These findings are expected to guide future prospective studies to ensure adequate power to sufficiently assess the relationship between this variant and toxicity.
Acknowledgements
This study was funded by Mayo Clinic Cancer Center grant number CA15083. The authors wish to thank Janet Olson, Ph.D., and Matthew Hathcock, Ph.D., for providing specimens and information through the Mayo Clinic Biobank.
Footnotes
Conflict of Interest/Disclosure
QN, SS, EET, CSTI, KJB, AML, RW, CRJ, ZW, PAK, SMO and RBD have nothing to disclose.
References
- (1).Lee AM et al. DPYD Variants as Predictors of 5-fluorouracil Toxicity in Adjuvant Colon Cancer Treatment (NCCTG N0147). Journal of the National Cancer Institute 106, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Lu ZH, Zhang R & Diasio RB Purification and characterization of dihydropyrimidine dehydrogenase from human liver. Journal of Biological Chemistry 267, 17102–9 (1992). [PubMed] [Google Scholar]
- (3).Johnson MR & Diasio RB Importance of dihydropyrimidine dehydrogenase (DPD) deficiency in patients exhibiting toxicity following treatment with 5-fluorouracil. Advances in Enzyme Regulation 41, 151–7 (2001). [DOI] [PubMed] [Google Scholar]
- (4).Ezzeldin H & Diasio R Dihydropyrimidine Dehydrogenase Deficiency, a Pharmacogenetic Syndrome Associated with Potentially Life-Threatening Toxicity Following 5-Fluorouracil Administration. Clinical Colorectal Cancer 4, 181–9 (2004). [DOI] [PubMed] [Google Scholar]
- (5).NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Research 44, D7–D19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Offer SM, Fossum CC, Wegner NJ, Stuflesser AJ, Butterfield GL & Diasio RB Comparative functional analysis of DPYD variants of potential clinical relevance to dihydropyrimidine dehydrogenase activity. Cancer research 74, 2545–54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Offer SM, Lee AM, Mattison LK, Fossum C, Wegner NJ & Diasio RB A DPYD variant (Y186C) in individuals of African ancestry associated with reduced DPD enzyme activity. Clinical pharmacology and therapeutics 94, 158–66 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Morel A et al. Clinical relevance of different dihydropyrimidine dehydrogenase gene single nucleotide polymorphisms on 5-fluorouracil tolerance. Molecular Cancer Therapeutics 5, 2895–904 (2006). [DOI] [PubMed] [Google Scholar]
- (9).Schwab M et al. Role of Genetic and Nongenetic Factors for Fluorouracil Treatment-Related Severe Toxicity: A Prospective Clinical Trial by the German 5-FU Toxicity Study Group. Journal of Clinical Oncology 26, 2131–8 (2008). [DOI] [PubMed] [Google Scholar]
- (10).Amstutz U, Farese S, Aebi S & Largiadèr CR Dihydropyrimidine dehydrogenase gene variation and severe 5-fluorouracil toxicity: a haplotype assessment. Pharmacogenomics 10, 931–44 (2009). [DOI] [PubMed] [Google Scholar]
- (11).Meulendijks D et al. Patients homozygous for DPYD c.1129–5923C>G/haplotype B3 have partial DPD deficiency and require a dose reduction when treated with fluoropyrimidines. Cancer Chemotherapy and Pharmacology 78, 875–80 (2016). [DOI] [PubMed] [Google Scholar]
- (12).van Kuilenburg ABP et al. Intragenic deletions and a deep intronic mutation affecting pre-mRNA splicing in the dihydropyrimidine dehydrogenase gene as novel mechanisms causing 5-fluorouracil toxicity. Human Genetics 128, 529–38 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Meulendijks D et al. Clinical relevance of DPYD variants c.1679T>G, c.1236G>A/HapB3, and c.1601G>A as predictors of severe fluoropyrimidine-associated toxicity: a systematic review and meta-analysis of individual patient data. The Lancet Oncology 16, 1639–50 (2015). [DOI] [PubMed] [Google Scholar]
- (14).Froehlich TK, Amstutz U, Aebi S, Joerger M & Largiadèr CR Clinical importance of risk variants in the dihydropyrimidine dehydrogenase gene for the prediction of early-onset fluoropyrimidine toxicity. International Journal of Cancer 136, 730–9 (2015). [DOI] [PubMed] [Google Scholar]
- (15).Jennings BA et al. Evaluating Predictive Pharmacogenetic Signatures of Adverse Events in Colorectal Cancer Patients Treated with Fluoropyrimidines. PLOS ONE 8, e78053 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Lee AM Qian S; Alberts Steven R.; Sargent Daniel J.; Sinicrope Frank A.; Berenberg Jeffrey L.; Grothey Axel; Polite Blase; Chan Emily; Gill Sharlene; Kahlenberg Morton S.; Nair Suresh G.; Shields Anthony F.; Goldberg Richard M.; Diasio Robert B. Association between DPYD c.1129–5923 C>G/hapB3 and severe toxicity to 5-fluorouracil-based chemotherapy in stage III colon cancer patients: NCCTG N0147 (Alliance). Pharmacogenetics and Genomics, 26, 133–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Loganayagam A et al. Pharmacogenetic variants in the DPYD, TYMS, CDA and MTHFR genes are clinically significant predictors of fluoropyrimidine toxicity. Br J Cancer 108, 2505–15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Falvella FS et al. DPD and UGT1A1 deficiency in colorectal cancer patients receiving triplet chemotherapy with fluoropyrimidines, oxaliplatin and irinotecan. British Journal of Clinical Pharmacology 80, 581–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Olson DH et al. Mapping the Global Emergence of Batrachochytrium dendrobatidis, the Amphibian Chytrid Fungus. PLOS ONE 8, e56802 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Johnson MR, Hageboutros A, Wang K, High L, Smith JB & Diasio RB Life-Threatening Toxicity in a Dihydropyrimidine Dehydrogenase-deficient Patient after Treatment with Topical 5-Fluorouracil. Clinical Cancer Research 5, 2006–11 (1999). [PubMed] [Google Scholar]
- (21).Caudle KE et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for Dihydropyrimidine Dehydrogenase Genotype and Fluoropyrimidine Dosing. Clinical Pharmacology and Therapeutics 94, 640–5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Lu Z, Zhang R & Diasio RB Dihydropyrimidine dehydrogenase activity in human peripheral blood mononuclear cells and liver: population characteristics, newly identified deficient patients, and clinical implication in 5-fluorouracil chemotherapy. Cancer Res 53, 5433–8 (1993). [PubMed] [Google Scholar]
- (23).Etienne MC et al. Population study of dihydropyrimidine dehydrogenase in cancer patients. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 12, 2248–53 (1994). [DOI] [PubMed] [Google Scholar]
- (24).Kuilenburg A.B.P.v. et al. Phenotypic and clinical implications of variants in the dihydropyrimidine dehydrogenase gene. Biochimica et Biophysica Acta (BBA) -Molecular Basis of Disease 1862, 754–62 (2016). [DOI] [PubMed] [Google Scholar]
- (25).Seck K et al. Analysis of the DPYD gene implicated in 5-fluorouracil catabolism in a cohort of Caucasian individuals. Clin Cancer Res 11, 5886–92 (2005). [DOI] [PubMed] [Google Scholar]
- (26).Boisdron-Celle M et al. 5-Fluorouracil-related severe toxicity: A comparison of different methods for the pretherapeutic detection of dihydropyrimidine dehydrogenase deficiency. Cancer Letters 249, 271–82 (2007). [DOI] [PubMed] [Google Scholar]
- (27).Deenen MJ et al. Relationship between Single Nucleotide Polymorphisms and Haplotypes in DPYD and Toxicity and Efficacy of Capecitabine in Advanced Colorectal Cancer. Clinical Cancer Research 17, 3455–68 (2011). [DOI] [PubMed] [Google Scholar]
- (28).Rosmarin D et al. A candidate gene study of capecitabine-related toxicity in colorectal cancer identifies new toxicity variants at DPYD and a putative role for ENOSF1 rather than TYMS. Gut 64, 111–20 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Boige V, Vincent M, Alexandre P & et al. Dpyd genotyping to predict adverse events following treatment with fluorouracil-based adjuvant chemotherapy in patients with stage iii colon cancer: A secondary analysis of the petacc-8 randomized clinical trial. JAMA Oncology 2, 655–62 (2016). [DOI] [PubMed] [Google Scholar]
- (30).Offer SM, Butterfield GL, Jerde CR, Fossum CC, Wegner NJ & Diasio RB MicroRNAs miR-27a and miR-27b directly regulate liver dihydropyrimidine dehydrogenase expression through two conserved binding sites. Mol Cancer Ther 13, 742–51 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Amstutz U, Offer SM, Sistonen J, Joerger M, Diasio RB & Largiader CR Polymorphisms in MIR27A associated with early-onset toxicity in fluoropyrimidine-based chemotherapy. Clin Cancer Res, 21, 2038–44 (2015) [DOI] [PubMed] [Google Scholar]
- (32).Meulendijks D et al. Rs895819 in MIR27A improves the predictive value of DPYD variants to identify patients at risk of severe fluoropyrimidine-associated toxicity. Int J Cancer 138, 2752–61 (2016). [DOI] [PubMed] [Google Scholar]
- (33).Wu R et al. Histone H3K27 Trimethylation Modulates 5-Fluorouracil Resistance by Inhibiting PU.1 Binding to the DPYD Promoter. Cancer Res 76, 6362–73 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Swen JJ et al. Pharmacogenetics: from bench to byte--an update of guidelines. Clinical pharmacology and therapeutics 89, 662–73 (2011). [DOI] [PubMed] [Google Scholar]
- (35).Olson JE et al. The Mayo Clinic Biobank: A building block for individualized medicine. Mayo Clinic proceedings 88, 952–62 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Elraiyah T et al. Novel Deleterious Dihydropyrimidine Dehydrogenase Variants May Contribute to 5-Fluorouracil Sensitivity in an East African Population. Clinical Pharmacology & Therapeutics, 101, 382–390 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Wigginton JE, Cutler DJ & Abecasis GR A Note on Exact Tests of Hardy-Weinberg Equilibrium. The American Journal of Human Genetics 76, 887–93 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Purcell S et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. American Journal of Human Genetics 81, 559–75 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Benjamini Y & Hochberg Y Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 57, 289–300 (1995). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


