Summary:
A small-molecule inhibitor screen on a panel of human lung cancer cell lines has uncovered an unexpected sensitivity of cells expressing oncogenic KRAS toward insulin-like growth factor 1 receptor (IGF1R) inhibition. Combining IGF1R and MAP-ERK kinase blockade led to significant effects on viability in human non-small cell lung cancer (NSCLC) cell lines and in 2 mouse models of oncogenic KRAS-driven lung cancer. The mechanistic basis for this effect seems to be an increased baseline activation of IGF1R-mediated activation of AKT in cells that express oncogenic KRAS. The studies thus point to a novel approach for treatment of KRAS-driven NSCLC, a particularly difficult subset of patients to treat with existing approaches.
Point mutations in KRAS are commonly found in many human cancers, including non-small cell lung cancer (NSCLC), pancreatic cancer, and several others. Therefore, the search for ways to effectively target oncogenic KRAS has been a long-standing goal in cancer biology. Despite intensive effort, no clinically proven therapeutic approaches that specifically target KRAS have yet been developed. As KRAS itself is a recalcitrant therapeutic target, an alternative strategy has focused on identifying effector pathways that are required for KRAS-driven oncogenesis. One strategy has been the search for “synthetic lethal” interactions, that is, genes or signaling pathways that are required in the context of a cell expressing oncogenic KRAS but not in cells that express wild-type KRAS (1–6). In a related approach, efforts have also been directed at targeting multiple downstream effector pathways simultaneously (7).
In this issue of Cancer Discovery, Molina-Arcas and colleagues (8) identify a novel combination of chemical inhibitors with a unique “synthetic” ability to block KRAS-driven oncogenesis in NSCLC. The authors used 25 NSCLC cell lines, 13 of which harbored KRAS mutation. These cell lines were screened against a panel of 55 drugs and systematically assayed for KRAS-specific effects on viability. The drugs in their panel were biased toward those directed against known KRAS effectors, such as the RAF/mitogen-activated protein (MAP)-ERK (MEK)/extracellular signal-regulated kinase (ERK) axis and the phosphoinositide 3-kinase (PI3K)/AKT/mTOR pathway. Importantly, the chemical library also included inhibitors of receptor tyrosine kinases and other proteins or biologic processes. As expected, drugs that target MEK and RAF had strong selectivity as single agents against cells expressing oncogenic KRAS. Surprisingly, 3 of 5 insulin-like growth factor 1 receptor (IGF1R) inhibitors were also effective in eliciting an oncogenic KRAS-selective response. In contrast, PI3K alone was widely toxic across many cell lines and did not show selectivity for cells expressing oncogenic KRAS.
To investigate the underlying differences in signaling pathway activation between KRAS-mutant and wild-type cells that may explain their selective sensitivity toward MEK and IGF1R inhibition, changes in effector pathways after inhibition with either agent were analyzed. As expected, MEK inhibitors reduced ERK phosphorylation across all cell lines. MEK inhibition also led to a modest increase in AKT phosphorylation, confirming the well-established cross-talk between these 2 pathways. In response to MAP-ERK inhibition, cells expressing oncogenic KRAS also exhibited decreased phosphorylation of p70S6K, a downstream target of mTOR complex 1 (mTORC1). Upon IGF1R single-agent inhibition, MEK activity was not affected, but AKT phosphorylation decreased significantly only in cells expressing oncogenic KRAS. In addition, there was a strong positive correlation between the degree to which AKT phosphorylation was reduced after IGF1R inhibition and cell viability, suggesting that cells expressing oncogenic KRAS require IGF1R to sustain phosphorylation of AKT.
On the basis of the success of these 2 single agents in decreasing cell viability in KRAS-mutant cells and the observation that IGF1R inhibition decreased AKT phosphorylation more significantly in KRAS-mutant cells, the authors hypothesized that the dual drug combination would simultaneously target both PI3K/AKT and MEK/ERK pathways to increase the therapeutic benefit against KRAS-mutant cells. As expected, the combination of these 2 agents decreased ERK phosphorylation in both genotypes, as did MEK inhibition alone. The effect of dual inhibition on AKT phosphorylation mimicked IGF1R single-agent treatment, with KRAS-mutant cells displaying decreased phosphorylation. However, the combination of MEK and IGF1R drugs led to striking decreases in phosphorylation of mTOR effectors p70S6K and S6. Thus, the combination of MEK and IGF1R therapy robustly inhibits MEK/ERK, PI3K/AKT, and mTORC1 (Fig. 1).
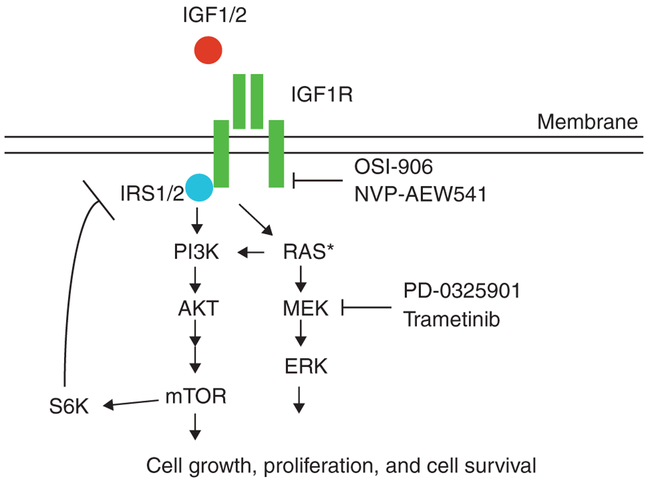
Figure 1.
The Ras and IGF1R signaling pathways. Growth factors stimulate IGF1R and activate downstream signaling pathways, including PI3K-AKT-mTOR and RAS-MEK-ERK, that promote cell growth, proliferation, and cell survival. Molina-Arcas and colleagues use a combination of IGF1R and MEK inhibitory drugs to successfully treat KRAS-mutant NSCLC cell lines and lung tumors induced by activated KRAS-driven transgenic mouse models.
The combined effect on multiple critical KRAS effector pathways led to a synergistic decrease in the viability of cell lines expressing oncogenic KRAS, an effect absent in the KRAS wild-type cells. Interestingly, the IGF1R and RAF inhibitor combination had similar differential selectivity toward the KRAS-mutant genotype. Although the PI3K/MEK and PI3K/RAF combinations did show some efficacy, this was not as significant as the effect of combining IGF1R inhibition with MEK or RAF inhibitors, likely due to the nonselective toxicity of PI3K inhibitors. Importantly, the effect of combined inhibition of MEK and IGF1R was also assessed in vivo using 2 genetically engineered mouse models. These studies showed a synergistic effect of combined KRAS and IGF1R blockade even in the context of loss of Trp53, further supporting the therapeutic benefit of combined therapy for NSCLC.
These results raise the question of why cells expressing oncogenic KRAS have higher levels of phosphorylated AKT (p-AKT) and why they are more susceptible to inhibition with IGF1R. To investigate the mechanism for the dependence of KRAS-mutant cells on IGF1R signaling, the authors evaluated IGF1R pathway activity after IGF and EGF ligand stimulation following serum starvation. Using a panel of 12 cell lines, half of which express oncogenic KRAS, the authors showed that all KRAS-mutant cells displayed AKT phosphorylation, whereas only half of the KRAS wild-type cells did. Furthermore, upon immunoprecipitation of the p85α subunit of PI3K, both insulin receptor substrate 1 (IRS1) and insulin receptor substrate 2 (IRS2) adaptor proteins coprecipitated in cells expressing oncogenic KRAS but not in wild-type cells, suggesting increased IGF1R signaling through IRS adaptors to PI3K. Real-time PCR analysis also identified higher levels of mRNA for IRS1 in cells expressing oncogenic KRAS. Publicly available NSCLC gene expression data also corroborated these results. Acute loss of IRS1 or IRS2 by siRNA decreased cell viability and increased apoptosis in KRAS-mutant cells, validating the results of chemical inhibition of the receptor. Thus, the IGF1R axis seems to have higher activation in cells expressing oncogenic KRAS, although why such activation would be favored as a consequence of KRAS mutation remains unclear and deserves further investigation. To determine whether KRAS is also required for both MEK/ERK and PI3K/AKT signaling in KRAS-mutant cells, the authors depleted KRAS by siRNA. KRAS depletion attenuated ERK phosphorylation, AKT phosphorylation, and mTORC1 activity in KRAS-mutant NSCLC cells. Inhibition of mTORC1 by rapamycin increased p-AKT, suggesting the presence of an intact negative feedback loop between mTORC1 and IRS.
Finally, to strengthen the evidence that oncogenic RAS requires IGF1R signaling, the authors conducted overexpression studies using 4-OHT-inducible oncogenic RAS in 3 systems: MCF10A breast epithelial cells, normal lung epithelial cells, and lung cancer cells harboring wild-type KRAS. Upon acute overexpression of mutant RAS in these settings, ERK and AKT phosphorylation increased. Pretreatment of cells with IGF1R inhibitor blocked p-AKT induction by 4-OHT. Taken together, these studies show that IGF1R plays a significant role in regulating p-AKT levels as a consequence of oncogenic KRAS.
Interest in targeting IGF1R for cancer therapy dates back at least 2 decades, beginning with seminal studies that indicated a requirement for IGF1R in transformation of mouse fibroblasts by RAS or SV40 large T antigen (9, 10). However, efforts to target IGF1R for cancer therapy have met with limited success (11). The demonstration of the efficacy of combining IGF1R and MEK inhibition against KRAS-mutant lung cancer suggests that stratification of patients based on KRAS status may be one approach to increase the efficacy of IGF1R inhibitors. These studies also suggest that a combined IGF1R and MEK inhibition strategy may provide increased benefit with less toxicity compared with the combination of a MEK inhibitor with a PI3K inhibitor. Recent evidence suggests that particular RAS mutations (e.g., G12V vs. G12C) differentially engage downstream effector pathways (12). Although Molina-Arcas and colleagues (8) observed no differences in therapeutic response between the different point mutations of KRAS, it remains to be seen whether a larger panel of cell lines would provide the statistical power to parse out differences in therapeutic response that are dependent on particular mutations. It will also be of interest to determine whether the combined MEK/IGF1R strategy is effective for other cancers in which KRAS mutations are prevalent. While the authors point to decreased activation of p-AKT as a critical mechanism for the effect of IGF1R inhibition, it remains to be determined whether other effectors of IGF1R are also involved. Undoubtedly, as the MEK/IGF1R combination does not lead to complete abrogation of tumors, resistance will emerge, anticipating the need to investigate how cancer cells treated with this dual therapy find other ways to compensate for the loss of MEK and AKT activation.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
REFERENCES
- 1.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009;462:108–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar MS, Hancock DC, Molina-Arcas M, Steckel M, East P, Diefenbacher M, et al. The GATA2 transcriptional network is requisite for RAS oncogene-driven non-small cell lung cancer. Cell 2012;149:642–55. [DOI] [PubMed] [Google Scholar]
- 3.Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell 2009;137:835–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Puyol M, Martin A, Dubus P, Mulero F, Pizcueta P, Khan G, et al. A synthetic lethal interaction between K-Ras oncogenes and Cdk4 unveils a therapeutic strategy for non-small cell lung carcinoma. Cancer Cell 2010;18:63–73. [DOI] [PubMed] [Google Scholar]
- 5.Singh A, Sweeney MF, Yu M, Burger A, Greninger P, Benes C, et al. TAK1 inhibition promotes apoptosis in KRAS-dependent colon cancers. Cell 2012;148:639–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vicent S, Chen R, Sayles LC, Lin C, Walker RG, Gillespie AK, et al. Wilms tumor 1 (WT1) regulates KRAS-driven oncogenesis and senescence in mouse and human models. J Clin Invest 2010;120:3940–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R. et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med 2008;14:1351–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Molina-Areas M, Hancock DC, Sheridan C, Kumar MS, Downward J. Coordinate direct input of both KRAS and IGF1 receptor to activation of PI3 kinase in KRAS-mutant lung cancer. Cancer Discov 2013;3: 548–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gatzka M, Prisco M, Baserga R Stabilization of the Ras oncoprotein by the insulin-like growth factor 1 receptor during anchorage-independent growth. Cancer Res 2000;60:4222–30. [PubMed] [Google Scholar]
- 10.Sell C, Rubini M, Rubin R, Liu JP, Efstratiadis A, Baserga R. Simian virus 40 large tumor antigen is unable to transform mouse embryonic fibroblasts lacking type 1 insulin-like growth factor receptor. Proc Natl Acad Sci U S A 1993;90:11217–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pollak M The insulin and insulin-like growth factor receptor family in neoplasia: an update. Nat Rev Cancer 2012;12:159–69. [DOI] [PubMed] [Google Scholar]
- 12.Ihle NT, Byers LA, Kim ES, Saintigny P, Lee JJ, Blumenschein GR, et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst 2012;104:228–39. [DOI] [PMC free article] [PubMed] [Google Scholar]