Abstract
Purpose
To determine whether mitochondrial DNA haplogroups or rare variants associate with primary open-angle glaucoma in subjects of European descent.
Methods
A case–control comparison of age- and sex-matched cohorts of 90 primary open-angle glaucoma patients and 95 population controls. Full mitochondrial DNA sequences from peripheral blood were generated by next-generation sequencing and compared to the revised Cambridge Reference Sequence to define mitochondrial haplogroups and variants.
Results
Most subjects were of the major European haplogroups H, J, K, U, and T. Logistic regression analysis showed haplogroup U to be significantly underrepresented in male primary open-angle glaucoma subjects (odds ratio 0.25; 95% confidence interval [CI] 0.09–0.67; P = 0.007; Bonferroni multiple testing P = 0.022). Variants in the mitochondrial DNA gene MT-ND2 were overrepresented in the control group (P = 0.005; Bonferroni multiple testing correction P = 0.015).
Conclusions
Mitochondrial DNA ancestral lineages modulate the risk for primary open-angle glaucoma in populations of European descent. Haplogroup U and rare variants in the mitochondrial DNA-encoded MT-ND2 gene may be protective against primary open-angle glaucoma. Larger studies are warranted to explore haplogroup associations with disease risk in different ethnic groups and define biomarkers of primary open-angle glaucoma endophenotypes to target therapeutic strategies.
Keywords: glaucoma, mitochondrial DNA (mtDNA), haplogroup
Primary open-angle glaucoma (POAG) is a heterogenous disease grouping associated with selective loss of retinal ganglion cells and is a leading cause of blindness worldwide. POAG is often associated with increased intraocular pressure (IOP), but many cases progress despite IOP being within normal ranges.1 The specificity of retinal ganglion cell loss in POAG resembles mitochondrial optic neuropathies, which are rare diseases caused by genetic defects in either nuclear-encoded mitochondrial genes or mitochondrial DNA (mtDNA) genes.2 In both these disease groupings retinal ganglion cells are lost in the absence of other central nervous system pathology. The pattern of axonal loss in the optic nerve can be distinct, however, with preferential loss of smaller-caliber parvocellular axons in mitochondrial optic neuropathies leading to central scotomas, while in POAG larger-caliber magnocellular fibers tend to be preferentially affected leading to more arcuate field defects.3,4 POAG fiber loss can vary from this classic pattern, with many cases involving central field defects,5 possibly reflecting the heterogenous nature of the disease group. The broad similarities of optic nerve degeneration in mitochondrial optic neuropathies and POAG have been long noted, driving interest in studying mitochondrial function and mtDNA variation in POAG.6–8 Studies using peripheral blood-derived lymphoblast cell lines have reported evidence of oxidative phosphorylation (OXPHOS) complex I defects in POAG.9,10 Further exploration of mitochondrial function and genetics in POAG may lead to markers of a subtype with a mitochondrial etiology.
An increased incidence of POAG among patients' first-degree relatives is well established; relatives of POAG patients have a 22% risk of developing POAG at some point in their lives, whereas the risk for the relatives of controls is 2% to 3%.11 This risk has been shown to be two to five times greater on the maternal side of families.12 However, only two genes, myocilin (MYOC) and optineurin (OPTN), have been established to cause familial POAG with high penetrance. Myocilin mutations account for 2% to 4% of POAG and are associated with high IOP, while optineurin mutations may account for 1% of POAG and 2% of normal-tension glaucoma.11 MtDNA variation has been hypothesized to be involved in POAG risk, with studies suggesting increased levels of rare mtDNA variants in POAG cases.13,14 While interesting findings are emerging, systematic studies to date have not always considered the population structure of mtDNA ancestry in their analyses. MtDNA is exclusively maternally inherited, and several ancient single nucleotide variants (SNVs) are inherited as a group, termed a haplogroup. MtDNA haplogroups represent human populations having the same ancestral origins and migration patterns and have been associated with disease risk.15,16 Here, we investigated the potential association between mtDNA haplogroups and rare mtDNA variants in POAG in a clinically defined cohort of patients and population controls of European descent.
Methods
Participants
Inclusion criteria for the study were as follows: evidence of glaucomatous optic neuropathy identified by loss of the neuroretinal rim and retinal nerve fiber layer loss, and corresponding visual field defect and open angles on gonioscopy. All patients and population controls underwent a detailed medical history and slit-lamp biomicroscopy as well as blood sampling for lymphocyte DNA isolation. The IOP was measured by Goldmann applanation tonometry and all assessments were performed at 3 PM ± 1 hour. All patients were receiving treatment for POAG at the time of recruitment. Subjects with congenital angle-closure glaucoma, trauma-associated secondary glaucoma, or neovascular glaucoma were excluded. The study adhered to the tenets of the Declaration of Helsinki; institutional human ethics committee (Royal Victorian Eye and Ear Hospital) approval was granted, and written informed consent was acquired from all patients.
Data Acquisition and Sequencing
MtDNA sequencing was undertaken by the Australian Genome Research Facility using the Illumina HiSeq platform (Illumina, Inc., San Diego, CA, USA). From 100 ng total DNA per sample, entire mtDNAs were amplified in two overlapping fragments with PCR primers known to avoid nuclear mtDNA pseudogene (NUMT) sequences.17 The PCR fragments were combined in equal ratios, and PCR amplicons enzymatically fragmented using NEBNext dsDNA Fragmentase (New England Biolabs, Ipswich, MA, USA). Illumina-compatible NEXTflex PCRfree 48-plex barcode oligonucleotide adapters (Bio Scientific, Sydney, New South Wales, Australia) were then ligated. Following size selection on a 2% agarose gel, the cluster formation and sequencing by synthesis were performed using an Illumina HiSeq2000 instrument, 101-bp (base pair) paired-end reads, and V3 reagents according to the manufacturer's protocols (Illumina, Inc.). Average read depth achieved was 10,000.
Since mtDNA is exclusively maternally inherited, there is no recombination and many mtDNA variants are in high linkage disequilibrium (LD), that is, occur on the same DNA molecule. Indeed, this fact leads to the presence of mitochondrial haplogroups. The majority of mtDNA-encoded genes are involved in the electron transport chain, and hence it is likely that these mtDNA variants are in high LD to allow coevolution of the various components of the electron transport chain to adapt to different environments and conditions. Due to this high LD in mtDNA, to avoid errors in statistical computations, we computed representative tag SNVs for variant association analysis with POAG, and all single variant analyses were computed using these tag SNVs.
Sequencing Analysis
Paired-end sequence reads in fastq files were aligned with bwa mem version 0.7.1518 using default alignments to the revised Cambridge Reference Sequence (rCRS - NC_012920.1). Duplicate reads were identified using Picard tools version 1.130 (http://broadinstitute.github.io/picard; in the public domain). Variants were called using freebayes, a haplotype-based variant detector version 1.1.019 with arguments “-p 1 –C 2 –q 10.” Using homoplasmic mtDNA variant calls only, haplogroups were then inferred using Haplogrep software version 2.1.0.20 To avoid errors in statistical computations, we computed representative tag SNVs for variant association analysis with POAG, and all single variant analyses were computed using these tag SNVs. Tag SNVs were computed from 30,589 full mtDNA sequences from mitomap (http://www.mitomap.org; in the public domain), using PLINK 2.0 (http://www.cog-genomics.org/plink/2.0/; in the public domain), and Lewontin's D' statistic as a measure of LD.
Statistical Analysis
All statistical analyses were performed using R version 3.2.1 (R Foundation, Vienna, Austria). All P values were reported after Bonferroni multiple testing correction, where applicable. Burden tests were performed with the adaptive sum of powered scores (aSPU) tests,21 using 20,000 permutations.
Results
Male Individuals in Mitochondrial Haplogroup U Have a Lower Risk for POAG
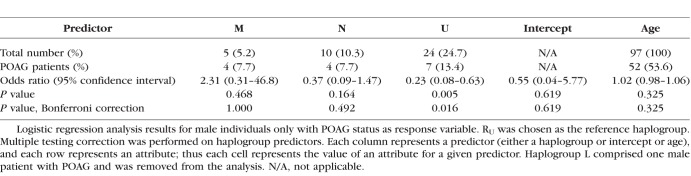
Mitochondrial haplogroups for 90 POAG patients and 95 population controls in this study were inferred from homoplasmic mtDNA. The haplogroup distribution we obtained showed a predominantly Western European maternal ancestry of our study population, which was closely matched between cases and controls. This reflects the older demographic sampled in Australia. Due to the small numbers of individuals in some haplogroups, we combined haplogroups according to the mtDNA phylogenetic tree (phylotree build 17).22 Demographic and haplogroup information for this study cohort is included in Supplementary Table S1. To determine if the haplogroup of an individual is a significant predictor for POAG, we used logistic regression analysis with the POAG status of the individual as the response variable, while adjusting for age. We defined haplogroup RU as the European root macro-haplogroup R (including haplogroups B, F, H, HV, J, R, T, V) minus haplogroup U. We disregarded haplogroup L because there was only one person from this group in our study. We found that men in haplogroup U were at approximately four times lower risk for POAG compared to men in haplogroup RU (Table 1). In our study, there was no association between POAG risk and mtDNA haplogroup in the cases when both sexes were combined (Supplementary Table S2).
Table 1.
Men in Haplogroup U Are Underrepresented in POAG
Rare mtDNA SNVs Are Associated With POAG
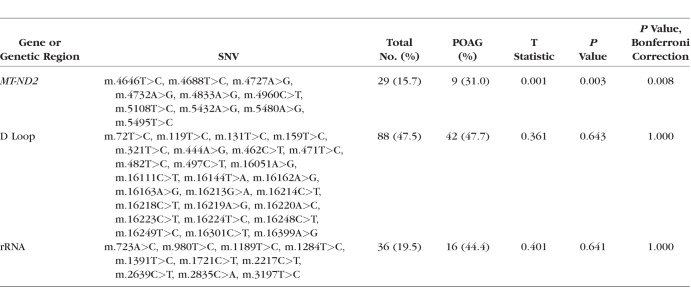
To avoid private mutations, a rare mtDNA variant was defined as a variant that was not common (see below) and was present in at least three people. There were 133 unique mtDNA rare variants detected in the data set. We aggregated rare variants within mtDNA genes to increase statistical power and applied the genetic burden aSPU tests implemented in the aSPU R package.21 We also restricted analysis to genes having at least 25 nonreference variants, which resulted in two genes and the noncoding regulatory D-Loop region. We found that individuals having rare variants in the MT-ND2 gene were at significantly lower risk for POAG than individuals without these variants (Table 2).
Table 2.
aSPU Analysis of Rare mtDNA Variants Detected
We defined common mtDNA SNVs as those variants having a minor allele frequency of at least 15%, which yielded nine unique common variants (Supplementary Table S3). To determine if common mtDNA SNVs were associated with POAG, we performed logistic regression analysis with disease status as the response variable and age, sex, and common variants as covariates. None of the common mtDNA variants were significantly associated with POAG after multiple testing correction at the 0.05 level.
Discussion
Several studies have investigated mtDNA variation in POAG, but few have used a phylogenetic approach to consider variants either inherited as a group (haplogroups) or the genetic burden of multiple rare mtDNA variants, as we report here. Some earlier reports have led to spurious conclusions due to a lack of consideration of mitochondrial haplogroups. Abu-Amero et al.23 sequenced the entire mtDNA of 27 Arabic POAG patients, finding 34 nonsynonymous sequence variants not detected in 159 population controls. However, when the same group later considered haplogroups in an expanded population sample, they found that African haplogroups were enriched in their case group, accounting for the increased levels of variants in their POAG cohort.24
Other studies have not considered mtDNA haplogroups. Banerjee et al.13 sequenced entire mtDNAs from 101 Indian POAG patients and 71 controls. The authors also found significantly increased nonsynonymous SNVs in mtDNA of POAG patients, particularly in complex I genes, and suggested the MT-ND5 gene as a mutation “hot spot” in POAG. Most of their patients and controls belonged to haplogroup M, which predominates in the Indian subcontinent. Yet their patient group included 18 individuals of the European root haplogroup R while their control group included only 5 haplogroup R individuals. Such disparities lead to apparent differences in levels of variants seen. In their list of rare variants found only in POAG cases, the variant m.5178C>A is a defining SNV for haplogroup D; the variant m.9055G>A is a defining SNV for haplogroup K; the variant m.14000T>A is commonly associated with haplogroup L1c (Table 3 in Banerjee et al.13). Thus three out of four “rare variants” that were recorded multiple times are explained by rare haplogroups in their population. Another study of 32 POAG subject mtDNAs found higher levels of rare variants in cases compared with controls.14 Half of the cases in this study were from the United Kingdom and half from India. The authors state that 110 “ethnically matched” controls were also sequenced, but no mtDNA haplogroups were reported for their cases or controls. Their table of rare variants also shows haplogroup marker SNVs. The variant m.1453A>G (two patients) is a defining SNV of haplogroup M2; variant m.3866T>C (two patients) is found on haplogroup J; variant m.4336T>C (one patient) is found on haplogroup M5; variant m.12397A>G (two patients) is found on haplogroup M19.14 This finding shows the need to first consider haplogroup associations, and then sort rare variants that conflict with the patient haplogroup, as we have done. Common pitfalls in mtDNA disease association studies have been critically reviewed elsewhere.25
Another group has focused on African American POAG patients, initially reporting no differences in rare variant levels between cases and controls.26 This group then used a mtDNA haplogroup approach, finding African haplogroups L1c2, L1c2b, and L2 significantly enriched in their cases.27 Our analyses in a population of predominantly Western European ancestry showed some surprising associations for the relatively small sample size. We found that individuals in haplogroup U were significantly underrepresented in male POAG cases, suggesting a protective effect of these lineages for males only. A study of German glaucoma patients found haplogroup U significantly underrepresented in pseudoexfoliation glaucoma subjects, and a trend for haplogroup U being underrepresented in POAG; however, they did not investigate sex as a variable.28 A second finding in our study was a significantly lower frequency of rare variants in the MT-ND2 gene in cases compared with controls, suggesting a protective effect against POAG. This finding predicts that subsets of several common variants may be required for glaucoma to progress. Complex age-related diseases such as glaucoma are likely to have environmental risk factors in addition to genetic risk factors, and different mtDNA haplogroups or rare variants may act as risk or protective “alleles” depending on the environment and cellular dependence on OXPHOS.29 In vitro studies have shown that human mtDNA haplogroups show differences in OXPHOS capacity.30,31
Studies in much larger cohorts of POAG and ophthalmologically screened population controls are warranted to further test for mtDNA influence on disease risk. Importantly, as POAG is a heterogeneous disease grouping, the role of mtDNA variation in patients stratified by normal-tension glaucoma, slightly elevated IOP, high IOP, progression rates, or visual field loss patterns requires further investigation. Our findings imply that disease risk variants in mtDNA will be uncovered in larger studies and may lead to identification of mtDNA biomarkers that define a POAG endophenotype of mitochondrial origin.
Supplementary Material
Acknowledgments
Supported by the DHB Foundation and Equity Trustees (IAT), and National Institutes of Health Grants NS021328, MH108592, CA182384, DO10944, and NS41850 awarded to DCW. The Centre for Eye Research Australia receives operational infrastructure support from the Victorian Government, Australia. SY is supported by a National Health and Medical Research Council CJ Martin Early Career Fellowship.
Disclosure: L.N. Singh, None; J.G. Crowston, None; M.I.G. Lopez Sanchez, None; N.J. Van Bergen, None; L.S. Kearns, None; A.W. Hewitt, None; S. Yazar, None; D.A. Mackey, None; D.C. Wallace, None; I.A. Trounce, None
References
- 1.Weinreb RN, Leung CK, Crowston JG, et al. Primary open-angle glaucoma. Nat Rev Dis Primers. 2016;2:16067. doi: 10.1038/nrdp.2016.67. [DOI] [PubMed] [Google Scholar]
- 2.Lopez Sanchez MI, Crowston JG, Mackey DA, Trounce IA. Emerging mitochondrial therapeutic targets in optic neuropathies. Pharmacol Ther. 2016;165:132–152. doi: 10.1016/j.pharmthera.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Quigley HA, Dunkelberger GR, Green WR. Chronic human glaucoma causing selectively greater loss of large optic nerve fibers. Ophthalmology. 1988;95:357–363. doi: 10.1016/s0161-6420(88)33176-3. [DOI] [PubMed] [Google Scholar]
- 4.La Morgia C, Di Vito L, Carelli V, Carbonelli M. Patterns of retinal ganglion cell damage in neurodegenerative disorders: parvocellular vs magnocellular degeneration in optical coherence tomography studies. Front Neurol. 2017;8:710. doi: 10.3389/fneur.2017.00710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park SC, De Moraes CG, Teng CC, Tello C, Liebmann JM, Ritch R. Initial parafoveal versus peripheral scotomas in glaucoma: risk factors and visual field characteristics. Ophthalmology. 2011;118:1782–1789. doi: 10.1016/j.ophtha.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Howell N. LHON and other optic nerve atrophies: the mitochondrial connection. Dev Ophthalmol. 2003;37:94–108. doi: 10.1159/000072041. [DOI] [PubMed] [Google Scholar]
- 7.Lascaratos G, Garway-Heath DF, Willoughby CE, Chau KY, Schapira AH. Mitochondrial dysfunction in glaucoma: understanding genetic influences. Mitochondrion. 2012;12:202–212. doi: 10.1016/j.mito.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Lee S, Van Bergen NJ, Kong GY, et al. Mitochondrial dysfunction in glaucoma and emerging bioenergetic therapies. Exp Eye Res. 2011;93:204–212. doi: 10.1016/j.exer.2010.07.015. [DOI] [PubMed] [Google Scholar]
- 9.Lee S, Sheck L, Crowston JG, et al. Impaired complex-I-linked respiration and ATP synthesis in primary open-angle glaucoma patient lymphoblasts. Invest Ophthalmol Vis Sci. 2012;53:2431–2437. doi: 10.1167/iovs.12-9596. [DOI] [PubMed] [Google Scholar]
- 10.Van Bergen NJ, Crowston JG, Craig JE, et al. Measurement of systemic mitochondrial function in advanced primary open-angle glaucoma and Leber hereditary optic neuropathy. PLoS One. 2015;10:e0140919. doi: 10.1371/journal.pone.0140919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gemenetzi M, Yang Y, Lotery AJ. Current concepts on primary open-angle glaucoma genetics: a contribution to disease pathophysiology and future treatment. Eye (Lond) 2012;26:355–369. doi: 10.1038/eye.2011.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gramer G, Weber BH, Gramer E. Results of a patient-directed survey on frequency of family history of glaucoma in 2170 patients. Invest Ophthalmol Vis Sci. 2014;55:259–264. doi: 10.1167/iovs.13-13020. [DOI] [PubMed] [Google Scholar]
- 13.Banerjee D, Banerjee A, Mookherjee S, et al. Mitochondrial genome analysis of primary open angle glaucoma patients. PLoS One. 2013;8:e70760. doi: 10.1371/journal.pone.0070760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sundaresan P, Simpson DA, Sambare C, et al. Whole-mitochondrial genome sequencing in primary open-angle glaucoma using massively parallel sequencing identifies novel and known pathogenic variants. Genet Med. 2015;17:279–284. doi: 10.1038/gim.2014.121. [DOI] [PubMed] [Google Scholar]
- 15.Chalkia D, Singh LN, Leipzig J, et al. Association between mitochondrial DNA haplogroup variation and autism spectrum disorders. JAMA Psychiatry. 2017;74:1161–1168. doi: 10.1001/jamapsychiatry.2017.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hudson G, Carelli V, Spruijt L, et al. Clinical expression of Leber hereditary optic neuropathy is affected by the mitochondrial DNA-haplogroup background. Am J Hum Genet. 2007;81:228–233. doi: 10.1086/519394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brandon MC, Ruiz-Pesini E, Mishmar D, et al. MITOMASTER: a bioinformatics tool for the analysis of mitochondrial DNA sequences. Hum Mutat. 2009;30:1–6. doi: 10.1002/humu.20801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. Available at: https://arxiv.org/pdf/1207.3907.pdf Accessed May 29, 2018.
- 20.Kloss-Brandstatter A, Pacher D, Schonherr S, et al. HaploGrep: a fast and reliable algorithm for automatic classification of mitochondrial DNA haplogroups. Hum Mutat. 2011;32:25–32. doi: 10.1002/humu.21382. [DOI] [PubMed] [Google Scholar]
- 21.Pan W, Kim J, Zhang Y, Shen X, Wei P. A powerful and adaptive association test for rare variants. Genetics. 2014;197:1081–1095. doi: 10.1534/genetics.114.165035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30:E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 23.Abu-Amero KK, Morales J, Bosley TM. Mitochondrial abnormalities in patients with primary open-angle glaucoma. Invest Ophthalmol Vis Sci. 2006;47:2533–2541. doi: 10.1167/iovs.05-1639. [DOI] [PubMed] [Google Scholar]
- 24.Abu-Amero KK, Gonzalez AM, Osman EA, Larruga JM, Cabrera VM, Al-Obeidan SA. Mitochondrial DNA lineages of African origin confer susceptibility to primary open-angle glaucoma in Saudi patients. Mol Vis. 2011;17:1468–1472. [PMC free article] [PubMed] [Google Scholar]
- 25.Salas A, Garcia-Magarinos M, Logan I, Bandelt HJ. The saga of the many studies wrongly associating mitochondrial DNA with breast cancer. BMC Cancer. 2014;14:659. doi: 10.1186/1471-2407-14-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collins DW, Gudiseva HV, Trachtman BT, et al. Mitochondrial sequence variation in African-American primary open-angle glaucoma patients. PLoS One. 2013;8:e76627. doi: 10.1371/journal.pone.0076627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collins DW, Gudiseva HV, Trachtman B, et al. Association of primary open-angle glaucoma with mitochondrial variants and haplogroups common in African Americans. Mol Vis. 2016;22:454–471. [PMC free article] [PubMed] [Google Scholar]
- 28.Wolf C, Gramer E, Muller-Myhsok B, Pasutto F, Wissinger B, Weisschuh N. Mitochondrial haplogroup U is associated with a reduced risk to develop exfoliation glaucoma in the German population. BMC Genet. 2010;11:8. doi: 10.1186/1471-2156-11-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji F, Sharpley MS, Derbeneva O, et al. Mitochondrial DNA variant associated with Leber hereditary optic neuropathy and high-altitude Tibetans. Proc Natl Acad Sci U S A. 2012;109:7391–7396. doi: 10.1073/pnas.1202484109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomez-Duran A, Pacheu-Grau D, Lopez-Gallardo E, et al. Unmasking the causes of multifactorial disorders: OXPHOS differences between mitochondrial haplogroups. Hum Mol Genet. 2010;19:3343–3353. doi: 10.1093/hmg/ddq246. [DOI] [PubMed] [Google Scholar]
- 31.Kenney MC, Chwa M, Atilano SR, et al. Molecular and bioenergetic differences between cells with African versus European inherited mitochondrial DNA haplogroups: implications for population susceptibility to diseases. Biochim Biophys Acta. 2014;1842:208–219. doi: 10.1016/j.bbadis.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.