ABSTRACT
Sec7 protein is a guanine nucleotide exchange factor in the ADP-ribosylation factor (ARF) family of small GTP-binding proteins. Aplysia Sec7 proteins (ApSec7s) play many roles in neurite outgrowth and synaptic facilitation in Aplysia neurons. However, the binding property of Aplysia ARF1 by ApSec7 isoforms has not been examined. In this study, we found that the cloned Aplysia ARF1 (ApARF1) protein only localized to the Golgi complex when it was expressed alone in HEK293T cells; however, if ApARF1 was co-expressed with plasma membrane-targeted ApSec7, it localized to both the plasma membrane and the Golgi complex via association with the Sec7 domain of ApSec7. Moreover, in HEK293T cells expressing both ApARF1 and another Sec7 isoform, ApSec7(VPKIS), the pleckstrin homology domain of ApSec7(VPKIS) associated with ApARF1, resulting in its localization to the Golgi complex. Overall, we propose a model in which ApSec7(VPKIS) activates ApARF1 in the Golgi complex, while ApSec7 recruits ApARF1 to the plasma membrane where it activates ApARF1/6 downstream signaling.
KEYWORDS: Aplysia, ARF1, ARF6, ApSec7, ApSec7(VPKIS)
Introduction
ADP-ribosylation factors (ARFs) are key molecules regulating vesicle and organelle trafficking in cells (Donaldson & Jackson 2011). ARFs play key roles in various intracellular compartments (Tsuchiya et al. 1991). For example, ARF1 is localized to the Golgi complex and is an important mediator of the trafficking between the Golgi complex and endocytic compartments, whereas ARF6 is localized to the plasma membrane and is involved in neurite outgrowth, endocytosis, and trafficking between endosomes and the plasma membrane (D’Souza-Schorey & Chavrier 2006). Overexpression of Aplysia ARF6 (ApARF6) was shown to induce neurite outgrowth in Aplysia neurons (Jang et al. 2016). However, it has been suggested that ARF1 is targeted to the plasma membrane where it exerts its functions (Maranda et al. 2001; Li et al. 2003; Cohen et al. 2007). ARF1 was found to be associated with the apical endosomes in the kidney proximal tubules as well as the Golgi complex (Maranda et al. 2001). ARNO/cytohesin2, which is localized to the plasma membrane via active ARF6 binding, recruits ARF1 to the plasma membrane (Cohen et al. 2007). Thus, accumulating evidence suggests that ARF1 plays a role in both the plasma membrane and Golgi complex.
ARFs can be activated by ARF GTP/GDP exchange factors, which are divided into five families, including the cytohesin family (Casanova 2007). We previously cloned two genes encoding Aplysia cytohesin family proteins: Aplysia Sec7 (ApSec7) and ApSec7(VPKIS) (Lee et al. 2012; Jun et al. 2015). ApSec7 is a crucial factor in neurite outgrowth and the 5-hydroxytryptamine-induced long-term facilitation of Aplysia sensory-motor synapses (Lee et al. 2012). In contrast, the ApSec7 isoform ApSec7(VPKIS), in which the VPKIS amino acid sequence is inserted into the pleckstrin homology (PH) domain of ApSec7, failed to be localized to the plasma membrane and failed to induce neurite outgrowth in Aplysia sensory neurons (Lee et al. 2012; Jun et al. 2015). The cytohesin family includes splicing variants that contain different numbers of glycine residues in the PH domain, which alter the sensitivity of phosphoinositide (PI) binding (Klarlund et al. 2000; Ogasawara et al. 2000). As reported previously, ApSec7 and cytohesin4 are diglycine variants that show higher selectivity for PI(3,4,5)P3 than for PI(4,5)P2. In contrast, ARNO/cytohesin2 is a triglycine variant that shows reduced selectivity for PI(3,4,5)P3 than for PI(4,5)P2 (Klarlund et al. 2000). However, the isoform ApSec7(VPKIS) belongs to a completely different variant group, and did not bind to PI(3,4,5)P3 (Jun et al. 2015). In addition, the PH domain of ApSec7 or ARNO/cytohesin2 but not that of ApSec7(VPKIS) was bound to active ARF6 (Santy & Casanova 2001; Jang et al. 2016). Therefore, the PH domain of ApSec7(VPKIS) may have functional binding properties that are distinct from those of typical ARFs.
In this study, we found that the PH domain of ApSec7(VPKIS) could associate with active Aplysia ARF1 (ApARF1) and was localized to the Golgi complex in HEK293T cells expressing only ApARF1-enhanced green fluorescent protein (EGFP). In addition, the Sec7 domain of ApSec7 could associate with and activate ApARF1. Therefore, ApSec7, which was localized to the plasma membrane via PI(3,4,5)P3 and activated ApARF6 binding to the PH domain, recruited ApARF1 to the plasma membrane mainly through the Sec7 domain, as well as activated ApARF1/6 in the plasma membrane to stimulate downstream signaling. In contrast, ApSec7(VPKIS) was localized to the Golgi complex via the association of its PH domain with ApARF1, and activated downstream signaling at the Golgi complex. Thus, ApSec7 and ApSec7(VPKIS) have distinct roles in the regulation of ARFs in cells.
Materials and methods
Cloning of DNA constructs
The plasmids used have been described previously (Lee et al. 2012; Kim et al. 2014; Jun et al. 2015; Jang et al. 2016). To identify the ARF1 protein in Aplysia, the DNA sequence of ApARF1 was cloned into pNEXδ-EGFP (Kaang et al. 1992) at the HindIII and Xba1 sites. Based on the nucleotide sequences of the identified clones, recombinant polymerase chain reaction (PCR) was used to generate active and inactive forms of ApARF1. Mutant fragments of active and inactive forms (ApARF1-Q71L and ApARF1-T31N) were generated by recombinant PCR using specific sense or antisense primers (sense primer containing ApARF1-Q71L, 5′-GTTGGTGGCCTGGACAAAATTCGA-3′; antisense primer containing ApARF1-Q71L, 5′-TCGAATTTTGTCCTGGCCACCAAC-3′; sense primer containing ApARF1-T31N, 5′-GCTGGAAAGAATACAATCTTATAC-3′; antisense primer containing ApARF1-T31N, 5′-TAAGATTGTATTCTTTCCAGCAGC-3′). The PCR products containing mutations (Q71L and T31N) in the ApARF1 amino acid residues were also subcloned into the HindIII and Xba1-digested pNEXδ-EGFP vector.
Cell culture and western blotting
HEK293T cell cultures were performed as described previously (Jang et al. 2016). Cells were then analyzed with an inverted Zeiss LSM-700 laser-scanning confocal microscope operated with ZEN software (Carl Zeiss, Jena, Germany). Most images were acquired from live cells.
Endogenous ARF1 proteins and actin protein (internal reference) from various Aplysia tissues and HEK293T cells were detected by western blotting using an anti-ARF1 antibody (Thermo Fisher Scientific, Waltham, MA, USA; Cat#: 16121) and an anti-actin antibody (Sigma, St. Louis, MO, USA; Cat#: a2066), respectively.
Results
Cloning of ApARF1
To examine the relationship between the ARF1 and ApSec7 isoforms, we isolated ApARF1 from Aplysia kurodai. ApARF1 shows high sequence similarity to corresponding ARF1 proteins in mammals (Figure 1(a)). The amino acid sequence identity between ARF1 from A. kurodai and other ARF1 molecules was 98.9% for Aplysia californica and 91.7% for humans (Figure 1(a)). These results indicate high conservation of ARF1 from invertebrate to mammalian cells.
Figure 1.
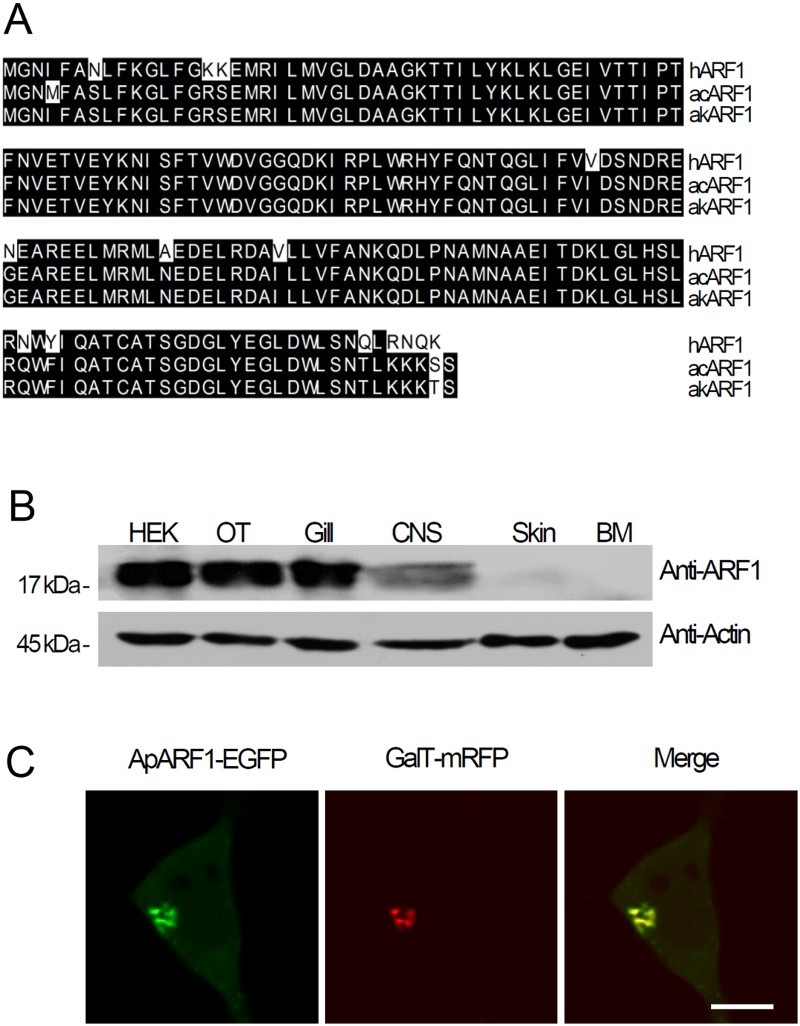
Cloning and tissue expression of ApARF1 from Aplysia kurodai. (a) Multiple sequence alignment of ARF1 from Aplysia kurodai (akARF1), Aplysia californica (acARF1), and human (hARF1). ARF1, ADP-ribosylation factor 1: Human, Homo sapiens; Aplysia, A. kurodai (NCBI Accession number: KU724187). (b) Western blotting of ApARF1 in lysates from various Aplysia tissues and HEK293T cells. HEK, HEK293T cells; OT, ovotestis; Gill, gill; CNS, central nervous system, including pleural, pedal, central, and abdominal ganglion; BM, buccal muscle. (c) Cellular localization of ApARF1-EGFP in HEK293T cells; GalT-mRFP was used as a Golgi marker. (d–e) Effects of ApSec7 expression on the cellular localization of ApARF1 in HEK293T cells. Scale bar, 20 μm.
To examine the tissue expression pattern of ApARF1, we performed western blotting using a commercially available mammalian ARF1 antibody. As shown in Figure 1(b), ApARF1 was ubiquitously expressed in Aplysia tissues, including the central nervous system. This result is similar to that of ApARF6 reported previously (Jang et al. 2016), indicating that ApARF1 and ApARF6 are ubiquitously expressed in Aplysia organs. We also examined the intracellular localization of ApARF1. For this, EGFP fused to wild-type ApARF1 (ApARF1-EGFP) was co-expressed with GalT-mRFP, a Golgi marker. As shown in Figure 1(c), ApARF1-EGFP was co-localized with GalT-mRFP, indicating Golgi targeting, as expected (Figure 1(c)).
Different regulation of ApARF1 among different ApSec7 isoforms
Next, to examine the relationship between ApSec7 and ApARF1, we used a wild-type ApSec7 and its catalytically inactive form, ApSec7-E159K (Jun et al. 2015). As expected, mRFP fused to both ApSec7 (mRFP-ApSec7) and ApSec7-E159K (mRFP-ApSec7-E159K) was localized to the plasma membrane in HEK293T cells (Figure 2(a)). Next, we co-expressed ApARF1-EGFP with mRFP-ApSec7 or mRFP-ApSec7-E159K. As shown in Figure 2(b), ApARF1-EGFP was localized to the Golgi complex in cells expressing only ApARF1-EGFP. However, in cells expressing both mRFP-ApSec7 and ApARF1-EGFP, ApARF1-EGFP was clearly localized to both the plasma membrane and the Golgi complex, whereas in cells expressing both mRFP-ApSec7-E159K and ApARF1-EGFP, ApARF1-EGFP was localized only to the Golgi complex, which was similar to the pattern observed in cells expressing only ApARF1-EGFP (Figure 2(b)). These results indicate that plasma membrane-localized mRFP-ApSec7 recruits ApARF1-EGFP to the plasma membrane, likely through Sec7 domain-dependent mechanisms. The results of the present study clearly suggest that the Sec7 domain/activity of ApSec7 is involved in recruiting ApARF1-EGFP to the plasma membrane.
Figure 2.
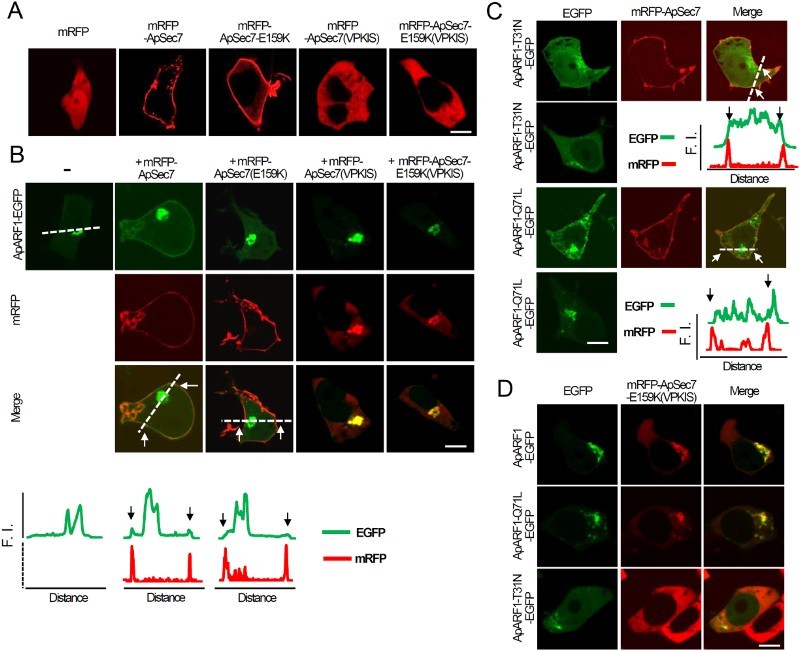
Effects of ApSec7 expression on the cellular localization of ApARF1 in HEK293T cells. (a–b) Effects of ApSec7 expression on the cellular localization of ApARF1 in HEK293T cells. (a) mRFP-ApSec7 and mRFP-ApSec7-E159K were localized to the plasma membrane, whereas mRFP, mRFP-ApSec7(VPKIS), and mRFP-ApSec7-E159K(VPKIS) were localized to the cytosol in HEK293T cells. Scale bar, 20 μm. (b) Cellular localization of mRFP-ApSec7 and ApARF1-EGFP in HEK293T cells. ApARF1-EGFP was localized to the plasma membrane only in mRFP-ApSec7- and ApARF1-EGFP-expressing cells. In contrast, mRFP-ApSec7(VPKIS) and mRFP-ApSec7-E159K(VPKIS) were localized to the Golgi complex in mRFP-ApSec7(VPKIS)- and ApARF1-EGFP-expressing cells or in mRFP-ApSec7-E159K(VPKIS)- and ApARF1-EGFP-expressing cells. The white dashed lines in the confocal fluorescence images indicate the paths along which the fluorescence intensities (F.I.) of the corresponding images were plotted. Black (traces) or white arrows (cell images) indicate the corresponding positions of the plasma membrane targeting mRFP signaling. Scale bar, 20 μm. (c) Cellular localization of ApARF1-T31N-EGFP or ApARF1-Q71L-EGFP in the presence of mRFP-ApSec7. The white dashed lines in the confocal fluorescence images indicate the paths along which the fluorescence intensities (F.I.) of the corresponding images were plotted. Black (traces) or white arrows (cell images) indicate the corresponding positions of the plasma membrane targeting mRFP signaling. Scale bar, 20 μm. (d) Cellular localization of ApARF1-EGFP, ApARF1-T31N-EGFP, or ApARF1-Q71L-EGFP in the presence of mRFP-ApSec7-E159K(VPKIS). Scale bar, 20 μm.
We also examined the cellular localization of ApARF1-EGFP in the absence or presence of mRFP-ApSec7(VPKIS) or mRFP-ApSec7-E159K(VPKIS). mRFP-ApSec7(VPKIS) was localized to the cytosol in cells expressing only mRFP-ApSec7(VPKIS), whereas mRFP-ApSec7(VPKIS) was co-localized with ApARF1-EGFP at the Golgi complex in cells expressing both mRFP-ApSec7(VPKIS) and ApARF1-EGFP (Figure 2(a) and (b)). Notably, mRFP-ApSec7-E159K(VPKIS) was also localized to the Golgi complex in cells expressing both mRFP-ApSec7-E159K(VPKIS) and ApARF1-EGFP, whereas mRFP-ApSec7-E159K(VPKIS) was localized to the cytosol in cells expressing only mRFP-ApSec7-E159K(VPKIS) (Figure 2(a) and (b)). These results indicate that mRFP-ApSec7(VPKIS) was localized to the Golgi complex via ApARF1-EGFP binding in a Sec7 domain/activity-independent manner.
Next, we evaluated whether ApSec7 could recruit active or inactive ApARF1 to the plasma membrane. For this purpose, we overexpressed dominant-negative ApARF1-T31N-EGFP or constitutively active ApARF1-Q71L-EGFP with mRFP-ApSec7. As shown in Figure 2(c), both ApARF1-T31N-EGFP and ApARF1-Q71L-EGFP localized to the plasma membrane and Golgi complex in cells co-expressing mRFP-ApSec7. These results suggest that mRFP-ApSec7 can recruit both active and inactive ApARF1 to the plasma membrane mainly via the Sec7 activity domain. Collectively, our results show that ApSec7 recruited ApARF1 to the plasma membrane in a Sec7 domain-dependent manner.
Similarly, we examined whether ApSec7(VPKIS) could associate with active or inactive ARF1. To test this, mRFP-ApSec7(VPKIS) was co-expressed with ApARF1-EGFP, ApARF1-Q71L-EGFP, or ApARF1-T31N-EGFP. As shown in Figure 2(d), mRFP-ApSec7(VPKIS) co-localized with ApARF1-EGFP and ApARF1-Q71L-EGFP but not with ApARF1-T31N-EGFP. These results indicate that mRFP-ApSec7(VPKIS) is associated specifically with active ApARF1.
PH domain-dependent ApARF1 binding of ApSec7(VPKIS)
The cytohesin family contains splicing variants that show differences in the number of glycine residues in the PH domain and are involved in the variable sensitivity to PI binding (Klarlund et al. 2000; Ogasawara et al. 2000). As shown in Figure 3(a), ApSec7 and cytohesin4 are diglycine variants, while ARNO/cytohesin2 is a triglycine variant. However, the ApSec7(VPKIS) isoform belongs to a completely different variant class, and does not bind to PI(3,4,5)P3 (Jun et al. 2015). Notably, several species contain VPKIS-form cytohesin-like isoforms, including Crassostrea gigas (Figure 3(a)). In addition, mRFP-ApSec7(VPKIS) was co-localized with ApARF1-EGFP in a Sec7-independent manner (Figure 2). Therefore, we speculated that the PH domain of ApSec7(VPKIS) may associate with ApARF1.
Figure 3.
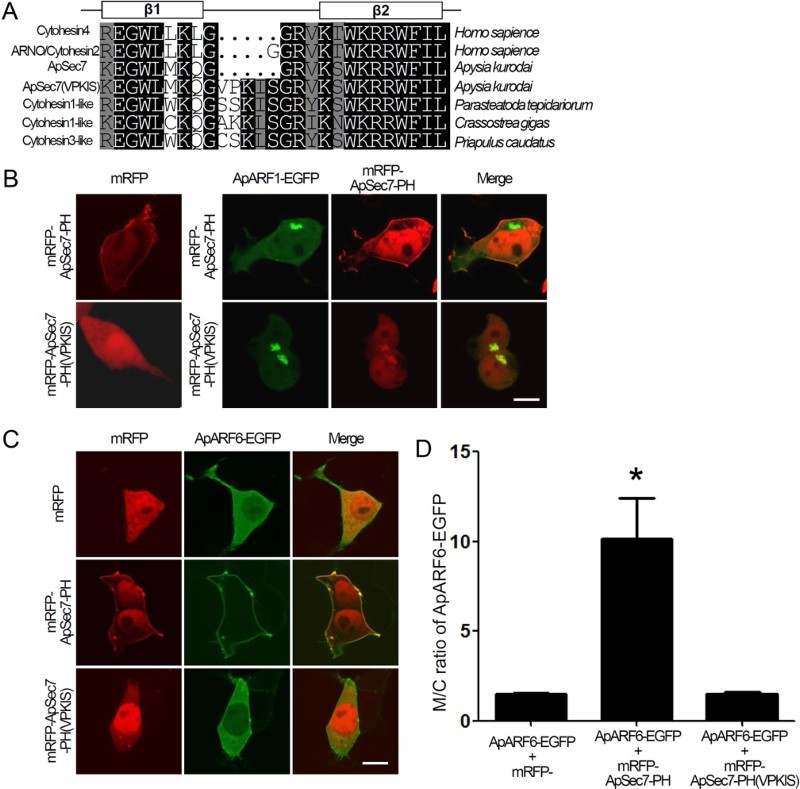
Binding of the PH domain of ApSec7(VPKIS) to ARF1. (a) Multiple sequence alignment of the PH domain of cytohesin family members of various species: cytohesin4 and ARNO/cytohesin2 from Homo sapiens, ApSec7 and ApSec7(VPKIS) from Aplysia kurodai, and cytohesin1-like proteins from Parasteatoda tepidariorum (NCBI Accession number: XP_015921179), Crassostrea gigas (NCBI Accession number: XP_011436446), and Priapulus caudatus (NCBI Accession number: XP_014669585). (b) Cellular localization of mRFP-ApSec7-PH or mRFP-ApSec7-PH(VPKIS) in the presence of ApARF1-EGFP. In the presence of ApARF1-EGFP, mRFP-ApSec7-PH(VPKIS) but not mRFP-ApSec7-PH was localized to the Golgi complex. Scale bar, 20 μm. (c–d) Enhancement of the plasma membrane localization of ApARF6-EGFP in the presence of mRFP-ApSec7-PH but not in the presence of mRFP-ApSec7-PH(VPKIS). Cellular localization of ApARF6-EGFP in the presence of mRFP, mRFP-ApSec7-PH, or mRFP-ApSec7-PH(VPKIS) in HEK293T cells. (c) Quantification of the ratio of the fluorescence intensity between the plasma membrane and cytosol localization (M/C ratio) of ApARF6-EGFP in HEK293T cells. *, p < .001 compared to that of all other groups, one-way ANOVA; F = 14.7, Tukey’s post-hoc test. Scale bar, 20 μm.
To directly test this hypothesis, we generated mRFP fused to the PH domain of ApSec7(VPKIS) (mRFP-ApSec7-PH(VPKIS)) (Figure 3(b)). As a control, we used the PH domain of ApSec7 (mRFP-ApSec7-PH), which is localized to the plasma membrane. ApARF1-EGFP was co-expressed with mRFP-ApSec7-PH(VPKIS) or mRFP-ApSec7-PH. As shown in Figure 3(b), mRFP-ApSec7-PH(VPKIS) but not mRFP-ApSec7-PH co-localized with ApARF1-EGFP at the Golgi complex. In addition, mRFP-ApSec7-PH could not recruit ApARF1-EGFP to the plasma membrane (Figure 3(b)), indicating that the PH domain of ApSec7 is not involved in plasma membrane recruitment by ApARF1.
We also examined the binding between ApARF6 and the PH domain of the ApSec7 isoforms. ApARF6-EGFP was co-expressed with mRFP-ApSec7-PH(VPKIS) or mRFP-ApSec7-PH. As shown in Figure 3(c), mRFP-ApSec7-PH but not mRFP-ApSec7-PH(VPKIS) co-localized with ApARF6-EGFP at the plasma membrane. Furthermore, the plasma membrane localization of ApARF6-EGFP was enhanced by overexpression of mRFP-ApSec7-PH but not of mRFP-ApSec7-PH(VPKIS) (p < .001 compared to that of all other groups, one-way ANOVA; F = 14.7, Tukey’s post-hoc test) (Figure 3(d)). These results indicate that ApARF1 can associate with the PH domain of ApSec7-PH(VPKIS) but not with the PH domain of ApSec7, whereas ApARF6 binds to the PH domain of ApSec7 but not to that of ApSec7(VPKIS).
Discussion
ApSec7 and ARNO/cytohesin2 are diglycine and triglycine variants, respectively, and can bind to active ARF6 (Santy & Casanova 2001; Jang et al. 2016). PI(3,4,5)P3 is indispensable for binding of the PH domain of ARNO/cytohesin2 to active ARF6 (Santy & Casanova 2001). In contrast, ApSec7(VPKIS) belongs to a completely different variant class and does not bind to PIs or active ARF6 (Jun et al. 2015; Jang et al. 2016). It was previously reported that the VPKIS insertion caused misfolding around the PI(3,4,5)P3-binding pocket of ApSec7 and thus disturbed the binding of PI(3,4,5)P3 to the PH domain (Jun et al. 2015). In addition, we showed that the PH domain of ApSec7(VPKIS) could bind to ApARF1 but not to ApARF6 (Figure 3). Therefore, the VPKIS insertion clearly altered the binding properties of ApSec7 to ARF1. However, binding between ARF1 and mRFP-ApSec7(VPKIS) was determined to be weak, given that mRFP-ApSec7(VPKIS) was diffusely localized in the cytosol of HEK293T cells expressing mRFP-ApSec7(VPKIS) only, in which endogenous ARF1 is expressed in the Golgi complex (Figures 2(a) and 3(b)). Thus, our data showed that the VPKIS insertion into the PH domain of ApSec7 could switch the preference of the binding from ARF6 to ARF1. Further studies are needed to clarify these mechanisms.
We previously showed that overexpression of ApSec7-induced neurite outgrowth in Aplysia neurons, which was largely driven by its plasma membrane localization and Sec7 activity (Lee et al. 2012). Consistent with this observation, overexpression of ApARF6-Q67L-EGFP, which is involved in ApSec7 downstream signaling and localized to the plasma membrane, induced the neurite outgrowth in Aplysia neurons (Jang et al. 2016). It is well-known that active ARF1 and ARF6 have similar structures (Pasqualato et al. 2001), and therefore may activate similar effectors in the membrane, such as phosphatidylinositol 5-kinase (PI5K) and phospholipase D (PLD). It was previously shown that ARNO/cytohesin2 could activate ARF1 at the plasma membrane to form membrane ruffles and dynamic macropinosomes (Hernandez-Deviez et al. 2004). Plasma membrane-localized active ARF6 recruited ARNO/cytohesin2 to the plasma membrane via PH binding of ARNO/cytohesin2 (Hernandez-Deviez et al. 2004). As a result, ARF1 can be recruited to the plasma membrane and become activated by the Sec7 domain of ARNO/cytohesin2. Similarly, our results suggest the possibility that in Aplysia neurons expressing endogenous ApSec7, ApARF6, and ApARF1, ApSec7 is targeted to the plasma membrane via the binding of its PH domain to PI(3,4,5)P3 and active ARF6 (Lee et al. 2012; Jun et al. 2015; Jang et al. 2016). The dimerization of the N-terminal coiled-coil domain of ApSec7 further enhanced its plasma membrane localization (Jun et al. 2016). The Sec7 domain of membrane-localized ApSec7 could then recruit ApARF1 to the plasma membrane of Aplysia neurons where it becomes activated (Figure 4). Therefore, it is plausible that besides the contribution of ApARF6, ApARF1, one of the downstream signaling molecules of ApSec7, may also be involved in the ApSec7-induced neurite outgrowth in Aplysia sensory neurons. It is known that GDP-bound ARF1 is localized to the Golgi complex via membrane binding (Honda et al. 2005). Similarly, Golgi-targeted ApSec7(VPKIS) may activate ARF1 at the Golgi complex in Aplysia neurons (Figure 4). However, further analysis is required to clarify this mechanism.
Figure 4.
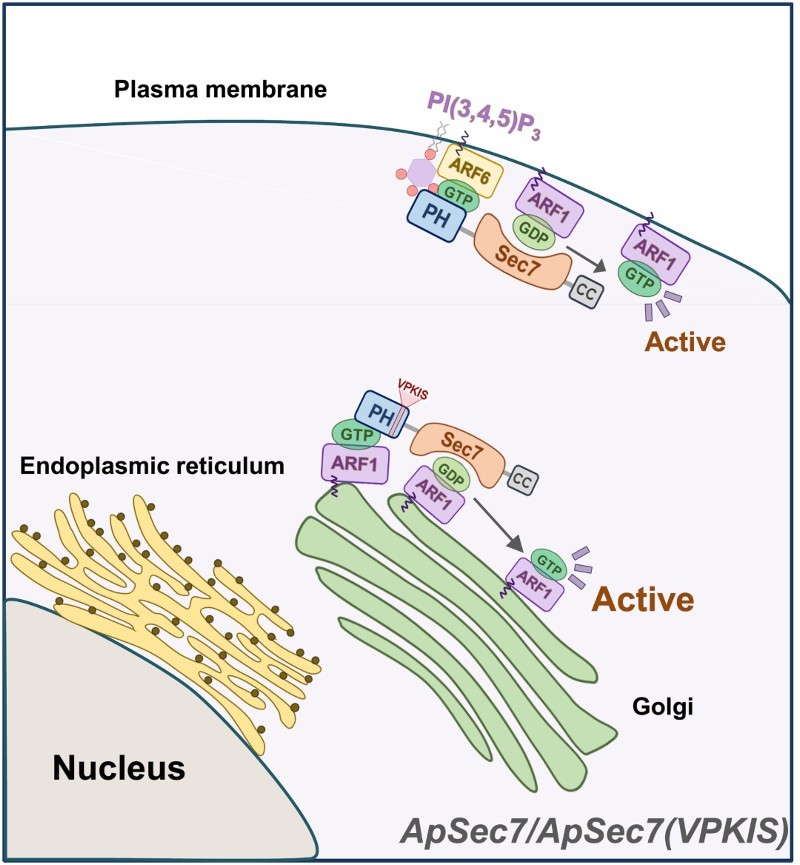
Schematic diagram of the molecular mechanism underlying the distinct functions of ApSec7 and ApSec7(VPKIS) in cells. The PH domain of ApSec7 was bound to PI(3,4,5)P3 and active ARF6, leading to plasma membrane localization, while the PH domain of ApSec7(VPKIS) was only weakly bound to active ARF1, leading to Golgi targeting. ApSec7(VPKIS) in the Golgi complex activated ApARF1 at the Golgi complex. In contrast, ApSec7 recruited ApARF1 to the plasma membrane via the Sec7 domain and activated ApARF1 at the plasma membrane.
Funding Statement
This work was supported by the Basic Science Research Program through the National Research Foundation [2013-R1A1A2012804] funded by the Ministry of Education, Science and Technology (to D.-J. J.) and by the National Honor Scientist Program of Korea (to B.-K. K.).
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Casanova JE. 2007. Regulation of arf activation: the Sec7 family of guanine nucleotide exchange factors. Traffic. 8:1476–1485. doi: 10.1111/j.1600-0854.2007.00634.x [DOI] [PubMed] [Google Scholar]
- Cohen LA, Honda A, Varnai P, Brown FD, Balla T, Donaldson JG. 2007. Active Arf6 recruits ARNO/cytohesin GEFs to the PM by binding their PH domains. Mol Biol Cell. 18:2244–2253. doi: 10.1091/mbc.E06-11-0998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG, Jackson CL. 2011. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Biol. 12:362–375. doi: 10.1038/nrm3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza-Schorey C, Chavrier P.. 2006. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 7:347–358. doi: 10.1038/nrm1910 [DOI] [PubMed] [Google Scholar]
- Hernandez-Deviez DJ, Roth MG, Casanova JE, Wilson JM. 2004. ARNO and ARF6 regulate axonal elongation and branching through downstream activation of phosphatidylinositol 4-phosphate 5-kinase alpha. Mol Biol Cell. 15:111–120. doi: 10.1091/mbc.E03-06-0410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda A, Al-Awar OS, Hay JC, Donaldson JG. 2005. Targeting of Arf-1 to the early Golgi by membrin, an ER-Golgi SNARE. J Cell Biol. 168:1039–1051. doi: 10.1083/jcb.200409138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang DJ, Jun YW, Shim J, Sim SE, Lee JA, Lim CS, Kaang BK. 2016. Activation of Aplysia ARF6 induces neurite outgrowth and is sequestered by the overexpression of the PH domain of Aplysia Sec7 proteins. Neurobiol Learn Mem; [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Jun YW, Kim S, Kim KH, Lee JA, Lim CS, Chang I, Suh BC, Kaang BK, Jang DJ. 2015. Analysis of phosphoinositide-binding properties and subcellular localization of GFP-fusion proteins. Lipids. 50:427–436. doi: 10.1007/s11745-015-3994-z [DOI] [PubMed] [Google Scholar]
- Jun YW, Lee SH, Shim J, Lee JA, Lim CS, Kaang BK, Jang DJ. 2016. Dual roles of the N-terminal coiled-coil domain of an Aplysia sec7 protein: homodimer formation and nuclear export. J Neurochem. 139:1102–1112. doi: 10.1111/jnc.13875 [DOI] [PubMed] [Google Scholar]
- Kaang BK, Pfaffinger PJ, Grant SG, Kandel ER, Furukawa Y. 1992. Overexpression of an Aplysia shaker K+ channel gene modifies the electrical properties and synaptic efficacy of identified Aplysia neurons. Proc Natl Acad Sci USA. 89:1133–1137. doi: 10.1073/pnas.89.3.1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Jun YW, Park Y, Lee JA, Suh BC, Lim CS, Lee YS, Kaang BK, Jang DJ. 2014. Intracellular membrane association of the Aplysia cAMP phosphodiesterase long and short forms via different targeting mechanisms. J Biol Chem. 289:25797–25811. doi: 10.1074/jbc.M114.572222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarlund JK, Tsiaras W, Holik JJ, Chawla A, Czech MP. 2000. Distinct polyphosphoinositide binding selectivities for pleckstrin homology domains of GRP1-like proteins based on diglycine versus triglycine motifs. J Biol Chem. 275:32816–32821. doi: 10.1074/jbc.M002435200 [DOI] [PubMed] [Google Scholar]
- Lee SH, Shim J, Choi SL, Lee N, Lee CH, Bailey CH, Kandel ER, Jang DJ, Kaang BK. 2012. Learning-related synaptic growth mediated by internalization of Aplysia cell adhesion molecule is controlled by membrane phosphatidylinositol 4,5-bisphosphate synthetic pathway. J Neurosci. 32:16296–16305. doi: 10.1523/JNEUROSCI.1872-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HS, Shome K, Rojas R, Rizzo MA, Vasudevan C, Fluharty E, Santy LC, Casanova JE, Romero G. 2003. The guanine nucleotide exchange factor ARNO mediates the activation of ARF and phospholipase D by insulin. BMC Cell Biol. 4:13. doi: 10.1186/1471-2121-4-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maranda B, Brown D, Bourgoin S, Casanova JE, Vinay P, Ausiello DA, Marshansky V. 2001. Intra-endosomal pH-sensitive recruitment of the Arf-nucleotide exchange factor ARNO and Arf6 from cytoplasm to proximal tubule endosomes. J Biol Chem. 276:18540–18550. doi: 10.1074/jbc.M011577200 [DOI] [PubMed] [Google Scholar]
- Ogasawara M, Kim SC, Adamik R, Togawa A, Ferrans VJ, Takeda K, Kirby M, Moss J, Vaughan M. 2000. Similarities in function and gene structure of cytohesin-4 and cytohesin-1, guanine nucleotide-exchange proteins for ADP-ribosylation factors. J Biol Chem. 275:3221–3230. doi: 10.1074/jbc.275.5.3221 [DOI] [PubMed] [Google Scholar]
- Pasqualato S, Menetrey J, Franco M, Cherfils J. 2001. The structural GDP/GTP cycle of human Arf6. EMBO Rep. 2:234–238. doi: 10.1093/embo-reports/kve043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santy LC, Casanova JE. 2001. Activation of ARF6 by ARNO stimulates epithelial cell migration through downstream activation of both Rac1 and phospholipase D. J Cell Biol. 154:599–610. doi: 10.1083/jcb.200104019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya M, Price SR, Tsai SC, Moss J, Vaughan M. 1991. Molecular identification of ADP-ribosylation factor mRNAs and their expression in mammalian cells. J Biol Chem. 266:2772–2777. [PubMed] [Google Scholar]


