Abstract
SPLUNC1 is a multifunctional protein of the airway with antimicrobial properties. We previously reported that it displayed antibiofilm activities against P. aeruginosa. The goal of this study was to determine whether (1) the antibiofilm property is broad (including S. aureus, another prevalent organism in cystic fibrosis); (2) the α4 region is responsible for such activity; and (3), if so, this motif could be structurally optimized as an antimicrobial peptide with enhanced activities. We used S. aureus biofilm-prevention assays to determine bacterial biomass in the presence of SPLUNC1 and SPLUNC1Δα4 recombinant proteins, or SPLUNC1-derived peptides (α4 and α4M1), using the well-established crystal-violet biofilm detection assay. The SPLUNC1Δα4 showed markedly reduced biofilm prevention compared to the parent protein. Surprisingly, the 30-residue long α4 motif alone demonstrated minimal biofilm prevention activities. However, structural optimization of the α4 motif resulted in a modified peptide (α4M1) with significantly enhanced antibiofilm properties against methicillin–sensitive (MSSA) and–resistant (MRSA) S. aureus, including six different clinical strains of MRSA and the well-known USA300. Hemolytic activity was undetectable at up to 100μM for the peptides. The data warrant further investigation of α4-derived AMPs to explore the potential application of antimicrobial peptides to combat bacterial biofilm-related infections.
Introduction
Human SPLUNC1 (short palate lung and nasal epithelial clone 1) is a 256-amino acid multifunctional protein of the innate immunity secreted in the human respiratory tract. It binds to lipopolysaccharide (LPS) and exerts bacteriostatic as well as antibiofilm effects[1–4]. In addition, it acts as a fluid-spreading surfactant, which facilitates mucus clearance[5–7]. SPLUNC1 has several alternative names. It is referred to as BPIFA1 (BPI fold containing family member A1) because of its structural similarity to bacterial permeability increasing protein (BPI)[5], lung-specific protein X or LUNX[2], or SPURT (secreted protein from upper respiratory tract)[2, 8]. We will refer to it as SPLUNC1 in this report.
The air is a nonsterile environment[9–13]. Therefore, the human airway is continuously exposed to potential pathogens[14]. Yet, infections are relatively rare. The airway is equipped with a mucociliary apparatus (MCA)[15], which is largely responsible for protecting the host through mucociliary clearance of microbial organisms. An important component of the MCA is the airway surface liquid (ASL) lining the airway and acting as a lubricant for normal ciliary function[6, 16–18]. In addition, the ASL contains a variety of antimicrobial factors including proteins and short peptides known as antimicrobial peptides (AMPs)[19–24]. SPLUNC1 helps regulate the ASL by providing a mechanism for controlling Na+ absorption through the inhibition of the epithelial sodium channel, ENaC[25–27]. In addition, direct antimicrobial activity of SPLUNC1 has been observed[2, 3, 5–7, 28].
Multiple domains within the SPLUNC1 secondary structure have been previously elucidated[7, 28]. One particular motif, called α4, displays a helical structure. On closer examination, this domain appears to exhibit a cationic amphipathic structure similar to that of well-known natural AMPs[29–32], with a positive charge of 2. We hypothesized that the antimicrobial α4 motif of SPLUNC1 with the characteristic of natural antimicrobial peptides can be used as a novel standalone antibiofilm agent. We report herein the impact of the α4 motif on the antibacterial properties of SPLUNC1 and the enhanced antibiofilm properties of the α4 region based on structural optimization.
Materials and methods
Protein and peptide synthesis
The recombinant proteins SPLUNC1 and SPLUNC1Δα4 (Wingtip) were expressed and purified as previously described[7]. Colistin sulfate was purchased from Sigma (St. Louis, Mo, USA). Synthetic α4 (ILKPGGGTSGGLLGGLLGKVTSVIPGLNNI), α4M1 (ILKKWWGTSGGLLGGLLGKVTSVIKGLNNI), and our control peptide for mammalian toxicity WLBU2 (RRWVRRVRRVWRRVVRVVRRWVRR) were synthesized using standard Fmoc (9-fluorenylmethoxy carbonyl) synthesis protocols as previously described[33] and purification achieved by reversed-phase high-pressure liquid chromatography on Vydac C18 or C4 columns (The Separations Group). The identity of each peptide was established by MS (Electrospray Quatro II triple quadrupole mass spectrometer).
Bacteria
All methicillin-resistant S. aureus strains are clinical isolates anonymously provided by Cystic Fibrosis Foundation and the medical laboratory of the University of Pittsburgh Medical Center. These strains have been used in previous studies, and the names are SA 0150–10, SA 0467–1, SA 0122–12, SA 0193–12, SA 0092–19, and SA 0187 in addition to the well-known USA300[34]. There was only one methicillin-sensitive S. aureus (MSSA) strain, and it was purchased from ATCC (ATCC49775).
Biofilm assay
We used a slightly modified version of the microtiter plate assay as previously described[5]. Briefly, log-phase bacteria were diluted in DMEM (to facilitate biofilm formation, as previously reported[35, 36]) to 108 CFU/mL based on pre-determined bacterial numbers that correlate with the optical density readings using a spectrophotometer. A 50μL volume of protein or peptide (in PBS), at different concentrations, was added to 50μL of bacterial suspension in a sterile 96-well polystyrene plate. The final bacterial concentration of the mixture is 5x107 CFU/mL, 50-fold compared to 106 CFU/mL in standard planktonic growth inhibition assays for adequate bacterial attachment, as required for biofilm formation. After 24h hours[37, 38] (at every 6h intervals for kinetic of biofilm formation assay) of bacterial biofilm growth at 37°C (no shaking), the supernatant was discarded. The plate was washed with PBS prior to staining with 125μL of 0.5% Crystal Violet (in 20% Ethanol) for 15 minutes. Excess stain was removed by washing twice with distilled water[6]. Crystal violet-stained biomass was dissolved in 150μL of 95% ethanol and measured using a plate reader at 620nm. Untreated bacteria (100% bacterial attachment), served as positive control. Wells with a mixture of sterile DMEM and PBS were used to control for possible contamination. Biomass in different treatment groups was quantified as percent OD of the positive controls.
Red blood cell lysis assay
Hemolytic assays were performed using red blood cells (RBCs) isolated from heparinized human blood obtained anonymously from the Central Blood Bank of Pittsburgh. The erythrocytes were separated by Histopaque gradient centrifugation and then resuspended to 2% (vol/vol) in PBS, as previously described[39]. To determine RBC lysis, a volume of 50 μl (1:4) of the RBC suspension was mixed with peptides at variable concentrations ranging from 0 to 100μM to a total volume of 200μl in a round-bottom 96-well plate. The reaction mixture was incubated at 37°C for 60 min with gentle shaking. To analyze the RBC lysis, the RBC-peptide mixture was spun at 600g for 5 min, and 80μl of the supernatant transferred to 120μl (1:2.5) of RBC lysis buffer (final dilution 1:10) in a flat-bottom 96-well plate for spectrophotometric analysis. Similarly, 0 to 50μl of untreated RBCs was diluted in RBC lysis buffer to a final volume of 500μl (up to 1:10 dilution), and the hemoglobin suspensions were used to produce a standard RBC lysis curve. The average absorbance values of the supernatants of all samples (200μl) in triplicates were measured by using a microplate reader at 550 nm as an indicator of hemoglobin released from lysed cells. These experiments were verified by three independent trials.
Statistical analysis
Data generated were analyzed as indicated in each figure legend by one-way or two-way ANOVA using multiple comparisons test, depending on the data set. These analyses were performed using GraphPad Prizm software.
Results
Deletion of the α4 domain reduces SPLUNC1 activity
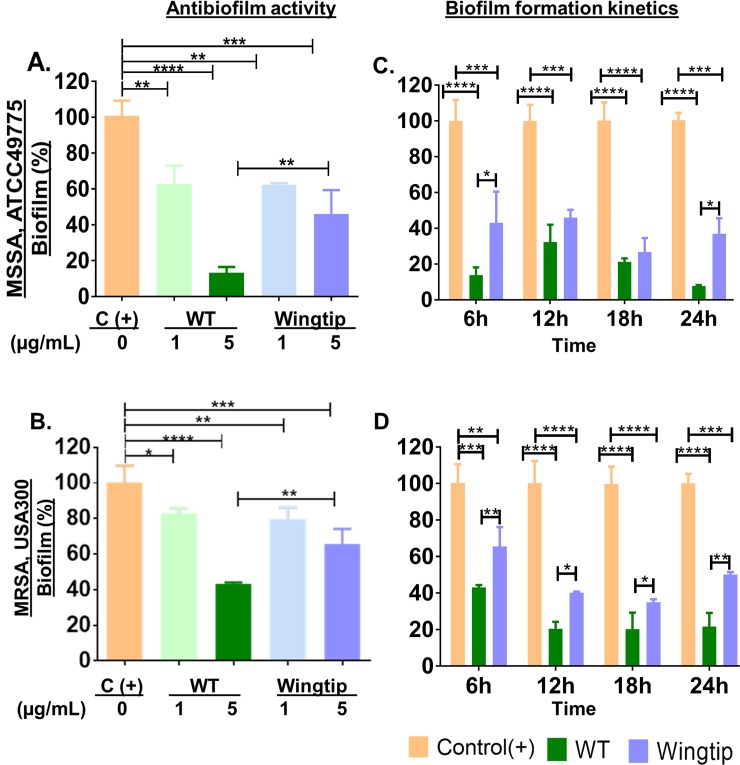
To examine the role of the α4 region in SPLUNC1 antibiofilm properties, we first compared SPLUNC1 (WT) and the Δα4 protein (Wingtip) for antibiofilm prevention activities against both methicillin-sensitive (MSSA, ATCC49775) and methicillin-resistant S. aureus (MRSA, USA300) (Fig 1A and 1B). Bacteria (MSSA and MRSA), treated with the mutant protein (Wingtip, 5μg/mL), displayed 43% and 65.5% of biofilm formation, respectively compared to 13.8% and 43% biofilm mass by the same bacterial strains treated with SPLUNC1-WT. Lower activity was observed at protein concentration of 1μg/mL compared to 5μg/mL, as expected. These results indicate a 1.5- (MRSA) to 3.1-fold (MSSA) reduction in activity when the α4 region is deleted from the WT protein. Next, we used the effective concentration of 5μg/mL for examination of biofilm growth inhibition kinetic in the presence of SPLUNC1 or the Δα4 protein (Fig 1C and 1D). The WT protein demonstrated a lower biofilm mass by 6h, 17% for MSSA and 43% for the MRSA strain compared 43% (MSSA) and 65.5% (MRSA) against these organisms for the Δα4 protein. The highest ativity was achieved at 24h with 7.7% for MSSA and 21.7% for MRSA of detectable biomass for the WT protein compared to 37% for MSSA and for 50% MRSA biofilm for Δα4-treated bacteria at 24h (Fig 1C and 1D). This is a 2 to 5-fold higher activity for the WT protein compared to ΔA4, always with statistical significance (P<0.05 to 0.001).
Fig 1. Dependence of SPLUNC 1 antibiofilm prevention activity on the α4 domain.
SPLUNC1 displayed higher reduction in S. aureus biofilm than Δα4; MSSA, ATCC49775: A, antibiofilm assay and C, biofilm growth inhibition kinetics; MRSA USA300; B, antibiofilm assay and D, biofilm growth kinetics. *denotes statistical significance at *P<0.05 using one-way ANOVA by Tukey’s multiple comparisons test (A and B) or multiple t tests (C and D); **P<0.005; ***P<0.001; ****P<0.0001.
Antimicrobial properties of synthetic α4 motif can be enhanced by sequence optimization
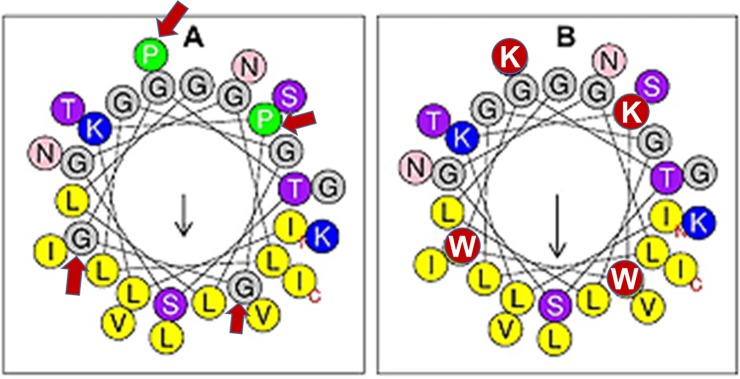
Because the antibiofilm activities of SPLUNC1 was affected by the deletion of the α4 helical domain, we performed a helical wheel analysis (Fig 2) on the α4 region (A) and observed an amphipathic structure similar to that of classical AMPs. However, the amphipathicity, as measured by the hydrophobic moment (μH = 0.373), is minimal due to a positive charge of only 2. Considering the importance of the membrane perturbation properties of AMPs in overcoming multidrug resistance and biofilm formation by bacterial pathogens, we sought to enhance the amphipathicity of α4 by doubling the positive charge using Lys (K) to replace the two Pro (P) residues on the hydrophilic side. In addition, because there are two Gly (G) residues in the hydrophobic region, we used the membrane interfacial-seeking amino acid Trp (W) to replace these two residues, resulting in the helical wheel structure in Fig 2B. These changes led to a 51% increase in the hydrophobic moment, a measure of the amphipathicity (α4M1 μH = 0.563).
Fig 2.
Helical wheel diagrams of (A) α4 and (B) α4M1. Black arrows indicate the direction of the hydrophobic moments (left, μH = 0.373; right, μH = 0.563 (http://heliquest.ipmc.cnrs.fr/), and red arrows in (A) indicate the sites of mutagenesis. Hydrophobic amino acids are in yellow and cationic in blue, except for the red circles denoting (B) amino acid substitutions.
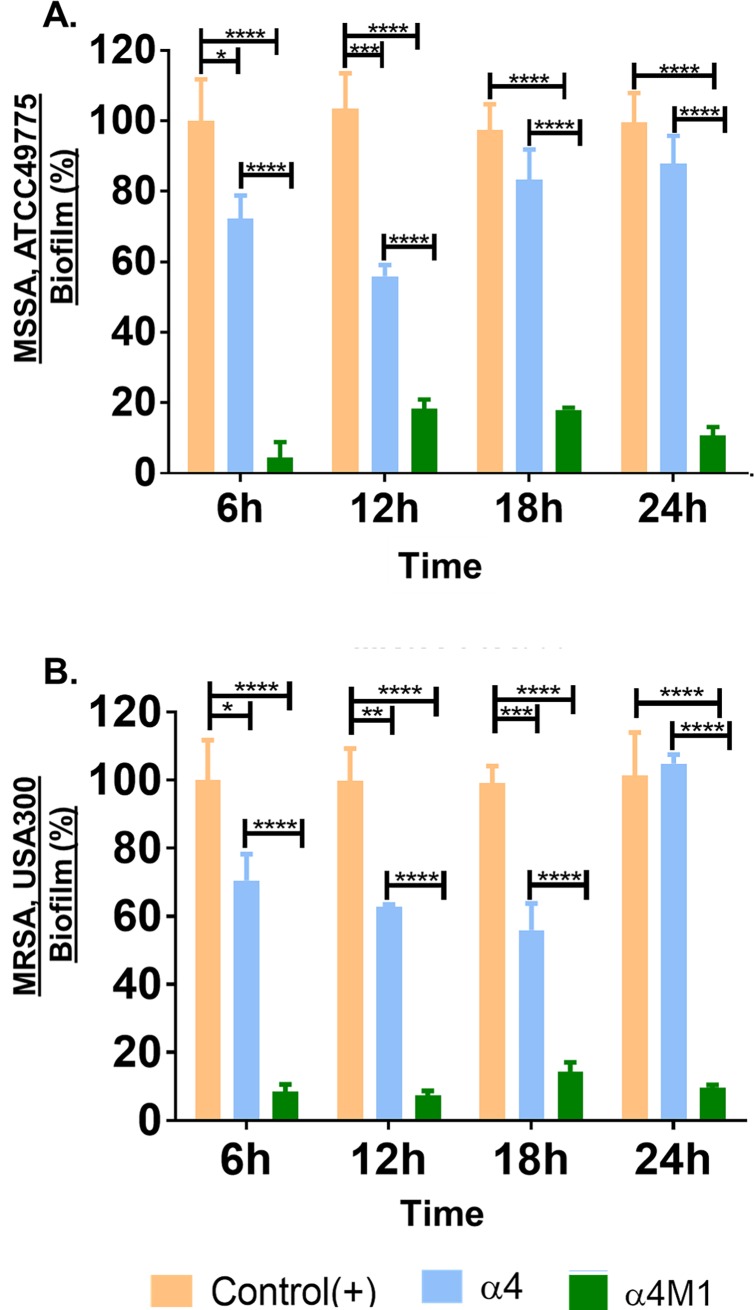
Interestingly, the synthetic α4 domain alone (32μM) demonstrates lower biofilm prevention activity against both MSSA and MRSA (Fig 3) compared to the WT protein shown in Fig 1, which indicates that this region (although important) alone is not accounted for the entire antimicrobial function of this protein. Structural optimization, however, was sufficient to overcome the lack of strong activity of α4. As shown by the biofilm prevention kinetics in Fig 3, the α4-derived α4M1 was able to prevent S. aureus (MSSA ATCC49775 and MRSA USA300) biofilm by 80–90%, from 6h to 24h of biofilm growth.
Fig 3. The synthetic α4 domain can be structural optimized for enhanced biofilm prevention properties.
The α4 sequence was compared to α4M1 for biofilm inhibition kinetic activities against MSSA ATCC49775 (A) and MRSA USA300 (B); P values (*P<0.05, **P<0.005, ***P<0.001), ****P<0.0001 were determined by multiple t tests.
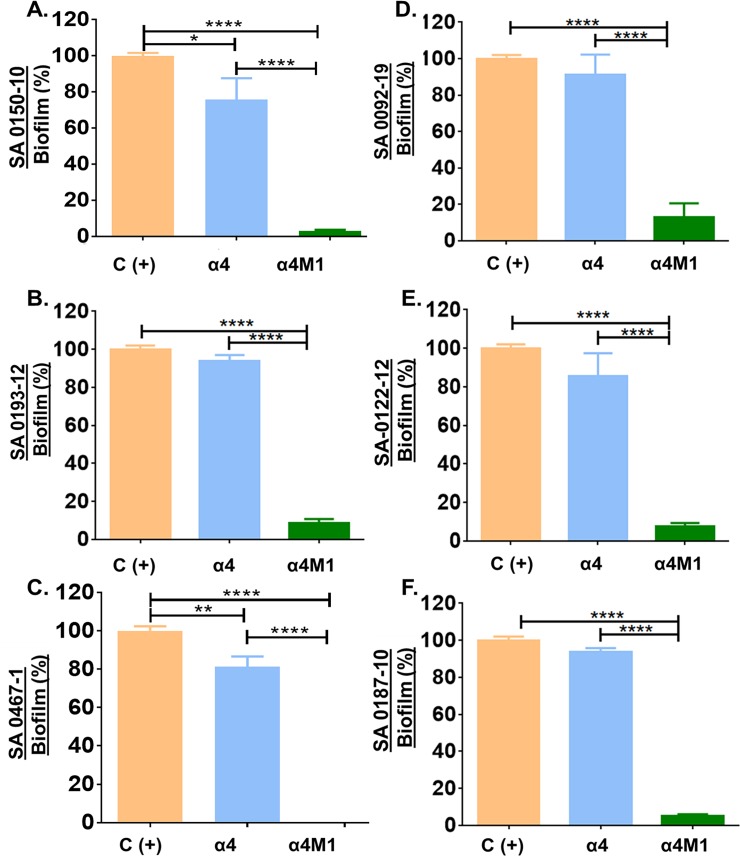
To test whether the observed activity is not strain-specific, we further compared α4 and α4M1 for antibiofilm prevention activities against six additional clinical strains of MRSA (Fig 4). The derived peptide α4M1 retained antibiofilm prevention activities against those strains, with a reduction in biofilm formation by 80–99%, compared to a modest 5–20% reduction in biofilm mass by the parent peptide α4.
Fig 4.
Biofilm prevention activities of α4 and α4M1 using six clinical strains of methicillin-resistant S. aureus (A through F). Bacterial biofilm were measured by crystal violet 24h after bacterial incubation at 37°C in the presence or absence (control) of the indicated peptides; P values (*P<0.05, **P<0.005, ***P<0.001, ****P<0.0001) were determined by one-way ANOVA using Bonferroni’s multiple comparisons test.
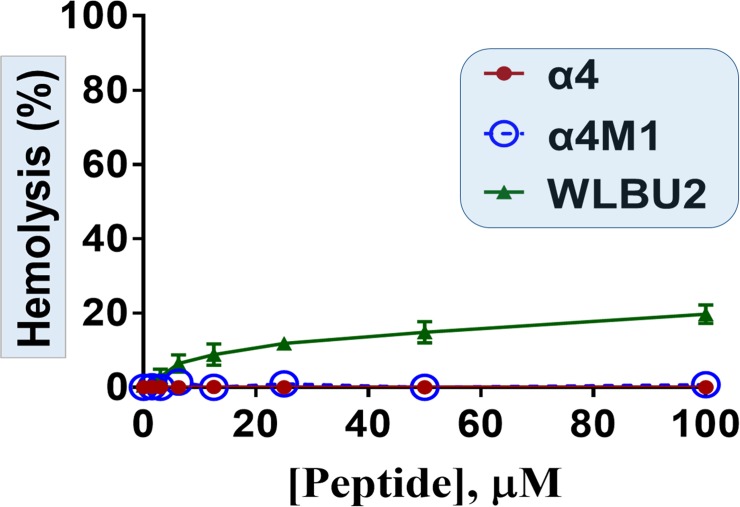
As a primary characterization of the cytotoxic property, we compared the two peptides for hemolytic activities using freshly isolated human erythrocytes. Both peptides show no detectable hemolytic activity at concentrations up to 100μM (Fig 5), in contrast to the engineered AMP control WLBU2[31, 33, 40, 41], which displayed up to 20% hemolysis.
Fig 5. Negligible hemolytic activity of α4 and α4M1, compared to the engineered AMP WLBU2.
Freshly isolated human erythrocytes in PBS were incubated with each peptide at the indicated concentrations for 1h and percent hemolysis determined according to Materials and Methods.
Discussion
Biofilms are highly resistant to clinical treatment by traditional antibiotics[42, 43] and are an important aspect of the pathogenicity of bacterial pathogens associated with respiratory infections, particularly in chronic disease such as cystic fibrosis[44, 45]. The host relies on the MCA to clear most potential pathogens from the airway. The MCA includes the ASL as an important component that facilitates mucociliary clearance. ASL consists of surfactants, antimicrobial molecules such as SPLUNC1, typical cationic AMPs, in addition to many other molecules (immunoglobulins, proteases, etc.). Hence, mucociliary clearance occurs by a combination of mechanisms, which prevent microbial attachment to and colonization of the airway epithelium[46–48]. We previously reported that SPLUNC1 displayed activity against P. aeruginosa biofilm[28]. The premise of this study is that SPLUNC1 displays broad activity and, therefore, would prevent biofilm growth by gram-positive (e.g. S. aureus) organisms as well as gram-negative (e.g. P. aeruginosa) bacteria[2, 28]. Importantly, the cationic amphipathic motif α4[7, 28] can be optimized as a standalone antibiofilm peptide. In this study, we demonstrated that SPLUNC1 was also able to prevent S. aureus biofilm, in contrast to a previous report[7]. In addition, this activity was largely dependent on the α4 motif, which was optimized successfully for enhanced antibiofilm properties.
The α4 region[7] of SPLUNC1 has a helical amphipathic structure with a sequence of 30 amino acid residues long and a high content of hydrophobic residues (hydrophobicity H = 0.558, Fig 2). As shown by the helical wheel analysis, the amphipathicity is minimal (μH = 0.373) in α4, reflective of a weak hydrophilic motif (charge +2) and minimal antibiofilm activity. Hence, the changes made in the parent α4 structure were intended to modestly increase the cationic content (charge = +4) and the amphipathicity (μH = 0.563). While the antibiofilm mechanism of SPLUNC1 is not entirely clear, it does not appear to be affected by the type of bacterial organism (gram negative vs. gram positive), as shown by the antibiofilm activity against both MSSA and MRSA strains. This activity is similar to the broad-spectrum activity of AMPs. Considering that the short α4 sequence is derived from a natural protein in human, we thought that the minor structural modifications might not affect cytotoxicity toward mammalian cells. One of the concerns with the structural optimization of natural AMP sequences is that enhancing the antibacterial potency may also result in increased toxicity toward mammalian cells. To illustrate, we compared the hemolytic profile of the engineered AMP WLBU2, which is in advanced preclinical development, with that of α4 and α4M1. WLBU2 has been extensively characterized both in vitro and in vivo[30, 33–35, 40, 41, 49–52]. It displays broad-spectrum activity against the most common MDR bacteria known as ESKAPE pathogens[34] and outperforms the last-resort antibiotic colistin against these MDR clinical strains. Hence, Lessons learned from these extensive studies led us to consider only two Trp residues in the hydrophobic face. Consequently, both α4 and α4M1 demonstrate no detectable hemolytic activities while the antibiofilm properties of the structurally optimized α4M1 are markedly enhanced.
The data suggest that the higher cationic content is highly relevant to the enhanced antibiofilm property of α4M1 possibly by interfering with bacterial attachment to solid surfaces, whereas the inclusion of Trp in the hydrophobic motif most certainly plays a role in overall activity. As future direction, a logical step would be to explore whether bacterial membrane-AMP electrostatic interactions may interfere with bacterial attachment, the first principal step in biofilm formation. Although beyond the scope of the current studies, we plan to explore biofilm prevention by anti-dispersion activity and specific applications to biofilm-related infections in future studies.
Conclusions
While our initial report seems to suggest a lack of activity of SPLUNC1 against S. aureus, this lack of activity appears to be limited to one strain[7]. SPLUNC1 and its derived AMPs displayed antibiofilm prevention activities against multiple strains of S. aureus (both MSSA and MRSA including six additional clinical MRSA strains[34]). This activity appears to depend at least partly on the α4 motif and can be enhanced by increasing the cationic and Trp content toward enhancing the amphipathicity as well as the hydrophobicity. Such a modest modification does not increase hemolytic or cytotoxic activity of the α4-derived peptide but noticeably increased the antibiofilm activity against both MSSA and MRSA. The data warrant further investigation of α4-derived AMPs to explore the potential application of AMPs to bacterial biofilm-related infections such as those associated with surgical sites, wound, or cystic fibrosis.
Supporting information
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Institutes of Health (R01 HL-125128 and R01 AI-133351 to Y.P.D. and R01 CA-207416 and R01 CA-098468 to M.R.), and the Flight Attendant Medical Research Institute (CIA-123062 to Y.P.D.). The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Thaikoottathil JV, Martin RJ, Di PY, Minor M, Case S, Zhang B, et al. SPLUNC1 deficiency enhances airway eosinophilic inflammation in mice. Am J Respir Cell Mol Biol. 2012;47(2):253–60. 10.1165/rcmb.2012-0064OC ; PubMed Central PMCID: PMCPMC3423460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Di YP. Functional roles of SPLUNC1 in the innate immune response against Gram-negative bacteria. Biochem Soc Trans. 2011;39(4):1051–5. 10.1042/BST0391051 ; PubMed Central PMCID: PMCPMC5595221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gally F, Di YP, Smith SK, Minor MN, Liu Y, Bratton DL, et al. SPLUNC1 promotes lung innate defense against Mycoplasma pneumoniae infection in mice. Am J Pathol. 2011;178(5):2159–67. 10.1016/j.ajpath.2011.01.026 ; PubMed Central PMCID: PMCPMC3081195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garland AL, Walton WG, Coakley RD, Tan CD, Gilmore RC, Hobbs CA, et al. Molecular basis for pH-dependent mucosal dehydration in cystic fibrosis airways. Proc Natl Acad Sci U S A. 2013;110(40):15973–8. 10.1073/pnas.1311999110 ; PubMed Central PMCID: PMCPMC3791714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y, Bartlett JA, Di ME, Bomberger JM, Chan YR, Gakhar L, et al. SPLUNC1/BPIFA1 contributes to pulmonary host defense against Klebsiella pneumoniae respiratory infection. Am J Pathol. 2013;182(5):1519–31. 10.1016/j.ajpath.2013.01.050 ; PubMed Central PMCID: PMCPMC3644735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y, Di ME, Chu HW, Liu X, Wang L, Wenzel S, et al. Increased susceptibility to pulmonary Pseudomonas infection in Splunc1 knockout mice. J Immunol. 2013;191(8):4259–68. 10.4049/jimmunol.1202340 ; PubMed Central PMCID: PMCPMC3839417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walton WG, Ahmad S, Little MS, Kim CS, Tyrrell J, Lin Q, et al. Structural Features Essential to the Antimicrobial Functions of Human SPLUNC1. Biochemistry. 2016;55(21):2979–91. 10.1021/acs.biochem.6b00271 ; PubMed Central PMCID: PMCPMC4887393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di YP, Harper R, Zhao Y, Pahlavan N, Finkbeiner W, Wu R. Molecular cloning and characterization of spurt, a human novel gene that is retinoic acid-inducible and encodes a secretory protein specific in upper respiratory tracts. J Biol Chem. 2003;278(2):1165–73. 10.1074/jbc.M210523200 . [DOI] [PubMed] [Google Scholar]
- 9.Kubera L, Studzinska J, Dokladna W, Malecka-Adamowicz M, Donderski W. [Microbiological air quality in some kindergartens and antibiotic resistance of bacteria of the Staphylococcus spp. genus]. Med Pr. 2015;66(1):49–56. . [PubMed] [Google Scholar]
- 10.Angelakis E, Yasir M, Azhar EI, Papadioti A, Bibi F, Aburizaiza AS, et al. MALDI-TOF mass spectrometry and identification of new bacteria species in air samples from Makkah, Saudi Arabia. BMC Res Notes. 2014;7:892 10.1186/1756-0500-7-892 ; PubMed Central PMCID: PMCPMC4295573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saleh M, Seedorf J, Hartung J. [Total count of bacteria in the air of three different laying hen housing systems]. Dtsch Tierarztl Wochenschr. 2003;110(9):394–7. . [PubMed] [Google Scholar]
- 12.Zhu S, Chen S, Huo Y, Leng T, Suo J. [Study of relationship between the bacteria in air and the clinic infection]. Wei Sheng Wu Xue Bao. 1996;36(5):394–7. . [PubMed] [Google Scholar]
- 13.Ames AM, Nungester WJ. The initial distribution of air-borne bacteria in the host. J Bacteriol. 1946;51:627 . [PubMed] [Google Scholar]
- 14.Farnsworth JE, Goyal SM, Kim SW, Kuehn TH, Raynor PC, Ramakrishnan MA, et al. Development of a method for bacteria and virus recovery from heating, ventilation, and air conditioning (HVAC) filters. J Environ Monit. 2006;8(10):1006–13. . [DOI] [PubMed] [Google Scholar]
- 15.Kilgour E, Rankin N, Ryan S, Pack R. Mucociliary function deteriorates in the clinical range of inspired air temperature and humidity. Intensive Care Med. 2004;30(7):1491–4. 10.1007/s00134-004-2235-3 . [DOI] [PubMed] [Google Scholar]
- 16.Liu YL, Chang CH. Dynamic Surface Tension Behavior of a Mixed Insoluble/Soluble Surfactant Dispersion at Pulsating Air/Liquid Interfaces: Roles of the Soluble Surfactant. J Colloid Interface Sci. 2001;238(1):85–90. 10.1006/jcis.2001.7493 . [DOI] [PubMed] [Google Scholar]
- 17.Hirst RA, Rutman A, Williams G, O'Callaghan C. Ciliated air-liquid cultures as an aid to diagnostic testing of primary ciliary dyskinesia. Chest. 2010;138(6):1441–7. 10.1378/chest.10-0175 . [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Di YP. Effects of second hand smoke on airway secretion and mucociliary clearance. Front Physiol. 2012;3:342 10.3389/fphys.2012.00342 ; PubMed Central PMCID: PMC3428780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taha-Abdelaziz K, Perez-Casal J, Schott C, Hsiao J, Attah-Poku S, Slavic D, et al. Bactericidal activity of tracheal antimicrobial peptide against respiratory pathogens of cattle. Vet Immunol Immunopathol. 2013;152(3–4):289–94. 10.1016/j.vetimm.2012.12.016 . [DOI] [PubMed] [Google Scholar]
- 20.Lysenko ES, Gould J, Bals R, Wilson JM, Weiser JN. Bacterial phosphorylcholine decreases susceptibility to the antimicrobial peptide LL-37/hCAP18 expressed in the upper respiratory tract. Infect Immun. 2000;68(3):1664–71. ; PubMed Central PMCID: PMCPMC97327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawyer C, Pai S, Watabe M, Bakir H, Eagleton L, Watabe K. Effects of synthetic form of tracheal antimicrobial peptide on respiratory pathogens. J Antimicrob Chemother. 1996;37(3):599–604. . [DOI] [PubMed] [Google Scholar]
- 22.Hiratsuka T, Nakazato M, Date Y, Ashitani J, Minematsu T, Chino N, et al. Identification of human beta-defensin-2 in respiratory tract and plasma and its increase in bacterial pneumonia. Biochem Biophys Res Commun. 1998;249(3):943–7. . [DOI] [PubMed] [Google Scholar]
- 23.MacRedmond R, Greene C, Taggart CC, McElvaney N, O'Neill S. Respiratory epithelial cells require Toll-like receptor 4 for induction of human beta-defensin 2 by lipopolysaccharide. Respir Res. 2005;6:116 10.1186/1465-9921-6-116 ; PubMed Central PMCID: PMCPMC1276817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masuda N, Mantani Y, Yuasa H, Yoshitomi C, Arai M, Nishida M, et al. Immunohistochemical study on the distribution of beta-defensin 1 and beta-defensin 2 throughout the respiratory tract of healthy rats. J Vet Med Sci. 2018;80(3):395–404. 10.1292/jvms.17-0686 ; PubMed Central PMCID: PMCPMC5880817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hobbs CA, Blanchard MG, Alijevic O, Tan CD, Kellenberger S, Bencharit S, et al. Identification of the SPLUNC1 ENaC-inhibitory domain yields novel strategies to treat sodium hyperabsorption in cystic fibrosis airway epithelial cultures. Am J Physiol Lung Cell Mol Physiol. 2013;305(12):L990–L1001. 10.1152/ajplung.00103.2013 ; PubMed Central PMCID: PMCPMC3882538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rollins BM, Garcia-Caballero A, Stutts MJ, Tarran R. SPLUNC1 expression reduces surface levels of the epithelial sodium channel (ENaC) in Xenopus laevis oocytes. Channels (Austin). 2010;4(4):255–9. 10.4161/chan.4.4.12255 ; PubMed Central PMCID: PMCPMC2975823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia-Caballero A, Rasmussen JE, Gaillard E, Watson MJ, Olsen JC, Donaldson SH, et al. SPLUNC1 regulates airway surface liquid volume by protecting ENaC from proteolytic cleavage. Proc Natl Acad Sci U S A. 2009;106(27):11412–7. 10.1073/pnas.0903609106 ; PubMed Central PMCID: PMCPMC2708735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sayeed S, Nistico L, St Croix C, Di YP. Multifunctional role of human SPLUNC1 in Pseudomonas aeruginosa infection. Infect Immun. 2013;81(1):285–91. 10.1128/IAI.00500-12 ; PubMed Central PMCID: PMCPMC3536124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen C, Deslouches B, Montelaro RC, Di YP. Enhanced efficacy of the engineered antimicrobial peptide WLBU2 via direct airway delivery in a murine model of Pseudomonas aeruginosa pneumonia. Clin Microbiol Infect. 2017. 10.1016/j.cmi.2017.08.029 ; PubMed Central PMCID: PMCPMC5835169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deslouches B, Islam K, Craigo JK, Paranjape SM, Montelaro RC, Mietzner TA. Activity of the de novo engineered antimicrobial peptide WLBU2 against Pseudomonas aeruginosa in human serum and whole blood: implications for systemic applications. Antimicrob Agents Chemother. 2005;49(8):3208–16. 10.1128/AAC.49.8.3208-3216.2005 ; PubMed Central PMCID: PMCPMC1196285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phadke SM, Deslouches B, Hileman SE, Montelaro RC, Wiesenfeld HC, Mietzner TA. Antimicrobial peptides in mucosal secretions: the importance of local secretions in mitigating infection. J Nutr. 2005;135(5):1289–93. 10.1093/jn/135.5.1289 . [DOI] [PubMed] [Google Scholar]
- 32.Bucki R, Pastore JJ, Randhawa P, Vegners R, Weiner DJ, Janmey PA. Antibacterial activities of rhodamine B-conjugated gelsolin-derived peptides compared to those of the antimicrobial peptides cathelicidin LL37, magainin II, and melittin. Antimicrob Agents Chemother. 2004;48(5):1526–33. 10.1128/AAC.48.5.1526-1533.2004 ; PubMed Central PMCID: PMCPMC400557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deslouches B, Phadke SM, Lazarevic V, Cascio M, Islam K, Montelaro RC, et al. De novo generation of cationic antimicrobial peptides: influence of length and tryptophan substitution on antimicrobial activity. Antimicrob Agents Chemother. 2005;49(1):316–22. 10.1128/AAC.49.1.316-322.2005 ; PubMed Central PMCID: PMCPMC538858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deslouches B, Steckbeck JD, Craigo JK, Doi Y, Burns JL, Montelaro RC. Engineered cationic antimicrobial peptides to overcome multidrug resistance by ESKAPE pathogens. Antimicrob Agents Chemother. 2015;59(2):1329–33. 10.1128/AAC.03937-14 ; PubMed Central PMCID: PMCPMC4335883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin Q, Deslouches B, Montelaro RC, Di YP. Prevention of ESKAPE pathogen biofilm formation by antimicrobial peptides WLBU2 and LL37. Int J Antimicrob Agents. 2018. 10.1016/j.ijantimicag.2018.04.019 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim J, Lee KM, Park CY, Kim HV, Kim Y, Park S. Quorum Sensing is Crucial to Escherichia coli O157:H7 Biofilm Formation under Static or Very Slow Laminar Flow Conditions. Biochip Journal. 2016;10(3):241–9. 10.1007/s13206-016-0310-9 PubMed PMID: WOS:000383732800010. [DOI] [Google Scholar]
- 37.Jiang XH, Zhou WM, He YZ, Wang Y, Lv B, Wang XM. Effects of lipopeptide carboxymethyl chitosan nanoparticles on Staphylococcus aureus biofilm. J Biol Regul Homeost Agents. 2017;31(3):737–43. . [PubMed] [Google Scholar]
- 38.Desrosiers M, Bendouah Z, Barbeau J. Effectiveness of topical antibiotics on Staphylococcus aureus biofilm in vitro. Am J Rhinol. 2007;21(2):149–53. . [DOI] [PubMed] [Google Scholar]
- 39.Deslouches B, Steckbeck JD, Craigo JK, Doi Y, Mietzner TA, Montelaro RC. Rational design of engineered cationic antimicrobial peptides consisting exclusively of arginine and tryptophan, and their activity against multidrug-resistant pathogens. Antimicrob Agents Chemother. 2013;57(6):2511–21. 10.1128/AAC.02218-12 ; PubMed Central PMCID: PMCPMC3716171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deslouches B, Gonzalez IA, DeAlmeida D, Islam K, Steele C, Montelaro RC, et al. De novo-derived cationic antimicrobial peptide activity in a murine model of Pseudomonas aeruginosa bacteraemia. J Antimicrob Chemother. 2007;60(3):669–72. 10.1093/jac/dkm253 ; PubMed Central PMCID: PMCPMC3584960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen C, Deslouches B, Montelaro RC, Di YP. Enhanced efficacy of the engineered antimicrobial peptide WLBU2 via direct airway delivery in a murine model of Pseudomonas aeruginosa pneumonia. Clin Microbiol Infect. 2018;24(5):547 e1–e8. 10.1016/j.cmi.2017.08.029 ; PubMed Central PMCID: PMCPMC5835169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vuotto C, Longo F, Pascolini C, Donelli G, Balice MP, Libori MF, et al. Biofilm formation and antibiotic resistance in Klebsiella pneumoniae urinary strains. J Appl Microbiol. 2017;123(4):1003–18. 10.1111/jam.13533 . [DOI] [PubMed] [Google Scholar]
- 43.Yonezawa H, Osaki T, Kamiya S. Biofilm Formation by Helicobacter pylori and Its Involvement for Antibiotic Resistance. Biomed Res Int. 2015;2015:914791 10.1155/2015/914791 ; PubMed Central PMCID: PMCPMC4452508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waters V, Ratjen F. Standard versus biofilm antimicrobial susceptibility testing to guide antibiotic therapy in cystic fibrosis. Cochrane Database Syst Rev. 2017;10:CD009528 10.1002/14651858.CD009528.pub4 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hill D, Rose B, Pajkos A, Robinson M, Bye P, Bell S, et al. Antibiotic susceptabilities of Pseudomonas aeruginosa isolates derived from patients with cystic fibrosis under aerobic, anaerobic, and biofilm conditions. J Clin Microbiol. 2005;43(10):5085–90. 10.1128/JCM.43.10.5085-5090.2005 ; PubMed Central PMCID: PMCPMC1248524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trimble AT, Whitney Brown A, Laube BL, Lechtzin N, Zeman KL, Wu J, et al. Hypertonic saline has a prolonged effect on mucociliary clearance in adults with cystic fibrosis. J Cyst Fibros. 2018. 10.1016/j.jcf.2018.01.001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson M, Bye PT. Mucociliary clearance in cystic fibrosis. Pediatr Pulmonol. 2002;33(4):293–306. . [DOI] [PubMed] [Google Scholar]
- 48.Regnis JA, Robinson M, Bailey DL, Cook P, Hooper P, Chan HK, et al. Mucociliary clearance in patients with cystic fibrosis and in normal subjects. Am J Respir Crit Care Med. 1994;150(1):66–71. 10.1164/ajrccm.150.1.8025774 . [DOI] [PubMed] [Google Scholar]
- 49.Mandell JB, Deslouches B, Montelaro RC, Shanks RMQ, Doi Y, Urish KL. Elimination of Antibiotic Resistant Surgical Implant Biofilms Using an Engineered Cationic Amphipathic Peptide WLBU2. Sci Rep. 2017;7(1):18098 10.1038/s41598-017-17780-6 ; PubMed Central PMCID: PMCPMC5741726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Melvin JA, Lashua LP, Kiedrowski MR, Yang G, Deslouches B, Montelaro RC, et al. Simultaneous Antibiofilm and Antiviral Activities of an Engineered Antimicrobial Peptide during Virus-Bacterium Coinfection. mSphere. 2016;1(3). 10.1128/mSphere.00083-16 ; PubMed Central PMCID: PMCPMC4888888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lashua LP, Melvin JA, Deslouches B, Pilewski JM, Montelaro RC, Bomberger JM. Engineered cationic antimicrobial peptide (eCAP) prevents Pseudomonas aeruginosa biofilm growth on airway epithelial cells. J Antimicrob Chemother. 2016;71(8):2200–7. 10.1093/jac/dkw143 ; PubMed Central PMCID: PMCPMC4954927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abdelbaqi S, Deslouches B, Steckbeck J, Montelaro R, Reed DS. Novel engineered cationic antimicrobial peptides display broad-spectrum activity against Francisella tularensis, Yersinia pestis and Burkholderia pseudomallei. J Med Microbiol. 2016;65(2):188–94. 10.1099/jmm.0.000209 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.