Abstract
Helicobacter pylori pathogenesis and disease outcomes are mediated by a complex interplay between bacterial virulence factors, host, and environmental factors. After H. pylori enters the host stomach, four steps are critical for bacteria to establish successful colonization, persistent infection, and disease pathogenesis: (1) Survival in the acidic stomach; (2) movement toward epithelium cells by flagella-mediated motility; (3) attachment to host cells by adhesins/receptors interaction; (4) causing tissue damage by toxin release. Over the past 20 years, the understanding of H. pylori pathogenesis has been improved by studies focusing on the host and bacterial factors through epidemiology researches and molecular mechanism investigations. These include studies identifying the roles of novel virulence factors and their association with different disease outcomes, especially the bacterial adhesins, cag pathogenicity island, and vacuolating cytotoxin. Recently, the development of large-scale screening methods, including proteomic, and transcriptomic tools, has been used to determine the complex gene regulatory networks in H. pylori. In addition, a more available complete genomic database of H. pylori strains isolated from patients with different gastrointestinal diseases worldwide is helpful to characterize this bacterium. This review highlights the key findings of H. pylori virulence factors reported over the past 20 years.
Keywords: Gastric cancer, Helicobacter pylori, Pathogenesis, Virulence factor

Helicobacter pylori is a common bacterium, and infects approximately 50% of the world's population. The prevalence of H. pylori infection is highly variable across different countries; for example, high prevalence is observed in the Latin American countries (75–83%), in contrast to the low prevalence in Japan (39.6%) and the US (17.1%) [1]. Several gastrointestinal diseases, including gastritis, peptic ulcer, duodenal ulcer, and gastric adenocarcinoma have been proven to be highly associated with H. pylori infection. Different disease outcomes are mediated by the complex interplay between bacterial, host, and environmental factors. Clarification of the role of bacterial virulence factors in H. pylori pathogenesis will benefit the development of vaccines and alternative therapies. This review highlights recent advances in H. pylori virulence factors and pathogenesis.
Overview of H. pylori infection and pathogenesis
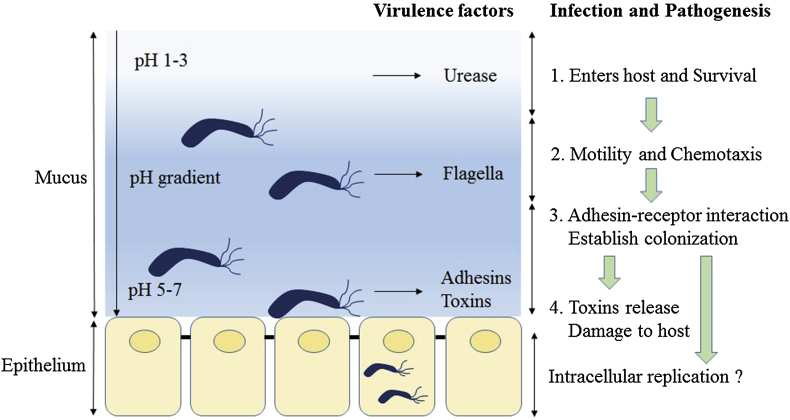
After entering the host stomach, H. pylori utilizes its urease activity to neutralize the hostile acidic condition at the beginning of infection. Flagella-mediated motility is then required for H. pylori to move toward host gastric epithelium cells, followed by specific interactions between bacterial adhesins with host cell receptors, which thus leads to successful colonization and persistent infection. Finally, H. pylori releases several effector proteins/toxins, including cytotoxin-associated gene A (CagA), and vacuolating cytotoxin A (VacA), causing host tissue damage [Fig. 1]. In addition, the gastric epithelium layer, which forms the major interface between H. pylori and the host, secretes chemokines to initiate innate immunity and activate neutrophils, and further lead to the formation of clinical diseases such as gastritis and ulcer. In summary, four steps are critical for H. pylori colonization and pathogenesis: (1) Survival under acidic stomach conditions; (2) movement toward epithelium cells through flagella-mediated motility; (3) attaching to host receptors by adhesins; (4) causing tissue damage by toxin release [Fig. 1]. These are discussed below.
Fig. 1.
Schematic diagram of Helicobacter pylori infection and pathogenesis. The urease activity and flagella-mediated motility of H. pylori facilitate its survival and movement toward the lower mucus gel above the epithelium, followed by several adhesins, including blood-antigen binding protein A, sialic acid-binding adhesin, and other outer membrane proteins interacting with receptors on the host epithelium cells. After successful colonization, toxins, including cytotoxin-associated gene A, and vacuolating cytotoxin A, are involved in damage of host tissue and intracellular replication.
Bacterial factors in H. pylori pathogenesis
Step 1: Urease and survival under acidic stomach conditions
H. pylori has developed an acid acclimation mechanism that promotes adjustment of periplasmic pH in the harsh acidic environment of the stomach by regulating urease activity. The urease gene cluster is composed of seven genes, including catalytic subunits (ureA/B), an acid-gated urea channel (ureI), and accessory assembly proteins (ureE-H) [2]. The metal cofactor nickel has to be inserted into the apoenzyme for heterodimer urease activity through the action of the four accessory proteins, among which UreE appears to be an important metallochaperone [3]. A recent paper describes how the other metallochaperone, HypA, interacts with UreE, and facilitates nickel transfer from HypA to UreE, and subsequently to downstream partner proteins, possibly UreG [3]. However, the mechanism of nickel transfer and insertion into the apo-urease is currently not well understood and requires further studies.
Intrabacterial urease activity is required for acid resistance by H. pylori, and this activity is regulated by the proton-gated urea channel UreI, which permits urea entry only under acidic conditions to prevent lethal alkalization during times of relative neutrality. UreI channels present in the inner membrane are closed at pH 7.0 and fully open at pH 5.0, enabling the rapid entry of urea into the bacterium [4]. As a consequence, H. pylori produces unusually large amounts of urea-derived ammonium. Surprisingly, UreI may be capable of extruding NH3 and/or NH4 across the inner membrane, allowing rapid neutralization of protons entering the periplasm [5]. Moreover, Miller and Maier indicated urea-derived ammonium is possibly assimilated into amino acids, thus connecting acid resistance and nitrogen metabolism [6].
Urease is also found on the H. pylori surface due to the lysis of some organisms. Extracellular urease is supposed to break down urea into carbon dioxide and ammonia, and ammonium hydroxide will be produced quickly when ammonia combines with water. Therefore, H. pylori can safely pass through the gastric juice when ammonia hydroxide neutralizes the acidic micro-environment close to the bacteria [4], [6]. Schwartz and Allen indicated that, in addition to the role of urease in colonization, urease regulates H. pylori-macrophage interactions [7]. Although phagocytosis is an element of the innate immune response, important for killing invading microbes, urease can modulate phagosome pH and megasome formation and as such, is essential for H. pylori survival in macrophages [7].
To allow the rapid adjustment of periplasmic pH, the ArsRS two-component system, in an acid-responsive manner, controls the transcription of the urease gene cluster [8]. Recently, a second, cytoplasmically localized acid responsive sensor kinase, FlgS, was identified in H. pylori. Although FlgS, with its cognate response regulator, HP0703, is known to regulate flagellar gene transcription, Marcus et al. indicated that a decrease in the cytoplasmic pH, exaggerated in the absence of membrane-located ArsS, may activate cytoplasmic FlgS [9]. However, the regulatory network between the two histidine sensor kinases is still unclear.
Treatment of H. pylori infection is becoming less effective as a result of increasing antibiotic resistance worldwide, suggesting that an alternatively targeted approach to eradicate H. pylori would be beneficial. Previous studies showed that a urease-negative mutant is unable to colonize gastric epithelium cells for persistent infection in gnotobiotic piglets [10], [11]. Interestingly, the addition of a nickel-free diet to standard triple therapy significantly increases the H. pylori eradication rate, supposedly due to the reduction of H. pylori urease activity [12]. As a result, the inhibition of urease activity would compromise the ability to colonize the stomach and therefore provide a target for the prevention or eradication of H. pylori infection. Recently, the UreI channel structure has been resolved, and it may guide the discovery of small-molecule inhibitors, providing the possibility of monotherapy without the use of conventional antibiotics [13].
Step 2: Flagella and movement toward epithelium cells
H. pylori moves through the gastric mucosa epithelium layer to the basal layer where the pH value is close to 7.0 by the action of 4–7 polar sheathed flagella. Previous studies showed that flagella-mediated motility is essential for the H. pylori colonization of the gnotobiotic piglet and mouse gastric mucosa [14], [15]. Mutagenesis of just about any gene of the motility and chemotaxis systems abolishes the ability of H. pylori to infect the stomach and establish colonization [14], [15], [16]. Kao et al. showed that patients infected with higher motility H. pylori may show enhanced bacterial density, triggering a higher inflammatory response in the upper stomach, and thus being associated with severe pathological outcomes [17]. In these respects, flagella can be considered as an early stage colonization/virulence factor. Moreover, mice immunized with a vaccine enriched for H. pylori flagella sheath proteins exhibited significantly reduced colonization, equivalent to that observed in mice immunized with whole-cell lysate. Due to the high antigenicity of flagella related proteins, flagella can be considered a suitable diagnostic and vaccine target [18], [19].
H. pylori flagella are mainly composed of the basal body, hook, and flagellar filament [20]. The flagellar filament is consisted of two flagellins (FlaA and FlaB) encoded by flaA and flaB [20]. The hook is composed of FlgE, and it links the basal body and flagellar filament [20]. The basal body is composed of several protein structures, and it plays a role in providing the energy source for motility. In a previous study, flaA and flaB have been indicated as necessary genes for the complete motility of H. pylori [21]. Moreover, the serological response to FlaA can be used as a marker to show the presence of H. pylori infection. The titer of anti-FlaA antibody is increased with the increment of colonization density of H. pylori, and serves as a noninvasive biomarker for early detection of gastric cancer [22]. However, a single predicted biomarker for screening gastric cancer always results in a relatively lower positive predictive value. Therefore, serum FlaA antibody should be used in combination with other markers to screen for gastric cancer.
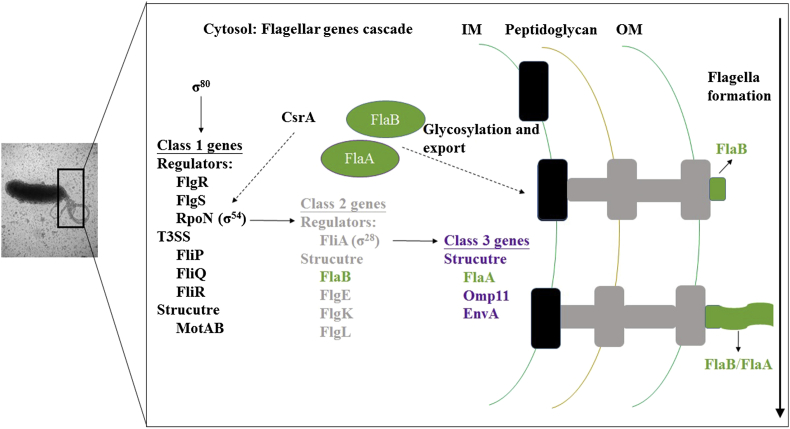
More than 40 proteins are involved in the biosynthesis and operation of flagella, making motility one of the most complex processes in the bacterial cell [20]. Flagellar related genes are divided into three classes, governed by the housekeeping sigma factor σ80 (RpoD, regulating class 1 genes), the alternative sigma factors σ54 (RpoN, regulating class 2 genes), and σ28 (FliA, regulating class 3 genes) [Fig. 2] [23]. Class 1 flagellar genes comprise the major regulatory genes (rpoN, flgR, flgS, and flhA) and structural genes (motA and motB) of the flagellar system [23]. Transcription of the class 2 middle flagellar genes (flaB, flgE, flgK, flgM, and flgL) and the sigma factor σ28 are governed by RpoN, assisted by the histidine kinase FlgS and the response regulator FlgR [23]. Late structural genes, including flaA, belong to the class 3 flagellar genes, regulated by σ28 [23]. In addition, FlhA is necessary for full transcription of flagellar class 2 and 3 genes [23]. Surprisingly, no flagellar master regulator similar to FlhDC in the Enterobacteriaceae has been found in the H. pylori genome. Recently, Kao et al. showed CsrA, a RNA binding protein, controls H. pylori J99 motility by regulating RpoN expression and flagella formation [24]. However, despite intensive research about the roles of motility in H. pylori pathogenesis, the complex transcriptional network that controls the expression of flagellar genes in H. pylori is still incompletely understood.
Fig. 2.
Current model of the flagellar transcriptional regulatory cascade for Helicobacter pylori flagellar biosynthesis. These genes are color-coded on the basis of their classification in the transcriptional regulatory cascade: black (class 1 genes), gray (class 2 genes), and purple (class 3 genes). Class 1 flagellar genes comprise most of the major regulatory genes of the flagellar system, including σ54 (RpoN), and FlgR. Formation of the flagellar T3SS has been proposed to create a signal detected by the FlgS sensor kinase, resulting in autophosphorylation of the protein. Phosphotransfer to the FlgR response regulator activates the protein, allowing for interactions with and stimulation of σ54. The expression of σ54 is positively regulated by CsrA through unclear mechanism. Alternative sigma factor σ28 (FliA) and structural proteins, including the minor flagellin FlaB, belong to Class 2 flagellar genes and are under the control of σ54. The T3SS facilitates the ordered secretion of the class 2 rod, ring, and hook proteins. The class 3 genes include flaA, which encodes the major flagellin, and those for other minor filament proteins, under the control of σ28.
Recently, H. pylori flagellin was found to be heavily glycosylated with the novel sialic acid-like nonulosonate, pseudaminic acid (Pse). The glycosylation process is essential for assembly of functional flagellar filaments and consequent bacterial motility [25]. Therefore, the Pse biosynthetic pathway offers considerable potential as an antivirulence drug target, especially since motility is required for H. pylori colonization and persistence in the host. Small-molecule inhibitors of the Pse biosynthetic pathway that penetrate the H. pylori cell membrane and prevent the formation of flagella were identified, however, the binding modes and in vivo inhibition activity are still unclear [26]. Moreover, the role of flagella posttranslational modification in immune recognition clearly needs further investigation.
In addition to motility, the role of flagella in bacterial adherence to mammalian hosts has been demonstrated for various bacterial species in a number of hosts. For example, polar flagella glycosylation is extremely important for Aeromonas hydrophila adhesion to Hep-2 cells, biofilm formation, and immune stimulation of interleukin-8 (IL-8) production via toll-like receptor 5 (TLR5) [27]. In animal cell models, Pseudomonas aeruginosa flagella can recognize lung epithelial cells through heparan sulfate, a highly sulfated proteoglycan [28]. However, there is no evidence of specific attachment of H. pylori flagella to epithelial cells, and the role of flagella in cell adhesion is controversial. Clyne et al. have studied whether H. pylori flagella are directly involved in adhesion by constructing flagellin (flaA and/or flaB) mutants and a flagellar regulator (flbA) mutant, and it appeared that all mutants adhered to gastric cells, indicating that flagella do not play a direct role in adhesion of H. pylori [29]. Although a lower adhesion rate in a flbA mutant was observed, the authors suggested that in addition to regulating flagella, FlbA may regulate some H. pylori adhesins [29]. Recently, Kao et al. revealed flagella associated regulator (csrA or rpoN) mutants showed decreased bacterial adhesion to AGS cells [24]. Taken together, although flagella may not directly participate in cell adhesion, regulators controlling flagellar-related genes are thought to affect adhesin expression. Therefore, the complex flagella formation process and its role in H. pylori pathogenesis needs further investigation.
Step 3: Adhesins and attachment to cellular surface receptors
Adhesins
When H. pylori colonizes on the mucosal layer lining the gastric epithelium, the interaction of bacterial adhesins with cellular receptors protects the bacteria from displacement from the stomach by forces such as those generated by peristalsis and gastric emptying, and then bacteria get metabolic substrates and nutrients to improve growth through releasing toxins to damage the host cells. Although blood-antigen binding protein A (BabA) and sialic acid-binding adhesin (SabA) are the well-characterized adhesins studied so far, not all H. pylori strains express these adhesins [30], [31]. There are several other known adhesins in H. pylori for adapting to different hosts/tissues, including neutrophil-activating protein (NAP) [32], heat shock protein 60 (Hsp60) [33], adherence-associated proteins (AlpA and AlpB) [34], H. pylori outer membrane protein (HopZ) [35], and lacdiNAc-binding adhesin (LabA) [36].
Neutrophil activating protein A
H. pylori-NAP belongs to the DNA-protecting proteins under starved conditions (Dps) family, which has significant structural similarities to the dodecameric ferritin family. NAP was first identified to stimulate high production of oxygen radicals from neutrophils, leading to damage of local tissues, and promote neutrophil adhesion to endothelial cells during H. pylori infection [37]. This NAP-induced adhesion depends on the acquisition of a high-affinity state of β2 integrin on the neutrophil surface membrane [38]. In addition to the stimulation of reactive oxygen species production, NAP induces the expression and release of IL-8, macrophage inflammatory protein (MIP)-1α, and MIP-1β by neutrophils [38]. As a result, NAP is highly associated with the hallmark of chronic gastritis, and infiltration of neutrophils and mononuclear cells into the gastric mucosa, caused by H. pylori infection.
The glycosphingolipids expressed on the neutrophil surface serve as a major receptor to interact with the NAP expressed on bacterial surface [38]. Moreover, NAP is supposed to facilitate SabA-mediated binding of sialylated antigens on the host cell surface [39]. In an animal study to investigate H. pylori colonization in mice infected with both the wild-type and napA mutant strains, the degree of survival of the napA mutant was found to be much lower than that of the wild-type strain [40]. Several studies showed that NAP can protect H. pylori DNA from damage, either due to its ability to bind DNA and thus to prevent DNA from attack by free radicals or through its iron-sequestering ability to reduce the oxidative stress produced in ferrous ion-mediated Fenton reactions [40], [41]. However, for gastroduodenal diseases, only one study showed that the level of NAP-specific antibodies in sera from H. pylori-infected patients with gastric cancer was significantly higher than that from patients with chronic gastritis [42]. No report has shown the direct association of NAP with H. pylori-induced gastric inflammation in patients so far.
Interestingly, NAP can also stimulate either neutrophils or monocytes to increase the expression of IL-12, and induces T helper cells to differentiate toward the T helper 1 phenotype [43]. Thus, NAP has been suggested as an immunotherapeutic anticancer agent and adjuvant for vaccination in clinical applications. Further study showed that NAP-activated DCs had a Th1 cytokine secretion profile, with high IL-12 and relatively low IL-10 secretion. Therapeutic effects of NAP can be mediated by the maturation of DCs and subsequent activation of Ag-specific T-cells, in addition to provoking innate immunity [44]. However, further studies are required to confirm that activation of DC by NAP can be an adjuvant for DC-based cancer vaccines and cancer immunotherapy.
Heat shock protein 60
Heat shock proteins, a highly conserved protein family detected not only in prokaryotes but also in eukaryotes, are induced by a variety of environmental stresses such as temperature, pH change, ischemia, and microbial infection. H. pylori produces mainly two Hsps, GroES-like HspA (Hsp10), and GroEL-like HspB (Hsp60). The high expression of Hsp60 at low pH, which interacts with the receptor-like sulfatide (sulfoglycolipid), indicates the stress of acid may change the specificity of H. pylori to receptors [33], [45].
Heat shock protein has been identified as one of the potential immunogens of the bacterium that induces IL-6, IL-8, tumor necrosis factor alpha (TNF-α), and GRO production from monocytes or gastric epithelial cells [46]. Hsp60 induces activation of NF-κB via TLR2 and the mitogen-activated protein kinase pathway, and thereby induces human monocytes to secrete IL-8 [47]. Moreover, anti-Hsp60 antibodies are consistently detected in H. pylori-infected patients, and the titers are associated with the progression of gastritis or gastric cancer [48], [49]. Further study showed that mAbs against H. pylori Hsp60 could modulate bacterial pathogenesis by increasing IL-8 and TNF-α production [50]. The pathogen-specific antibodies are supposed to execute potential immune functions rather than recognize or neutralize microbes. However, further studies are required to provide important insights into the role of anti-Hsp60 antibodies in H. pylori-associated gastric diseases.
Blood group antigen binding adhesin (BabA and BabB)
Three bab allelic types have been identified, including babA1, babA2, and babB. The molecular mass of the BabA protein is nearly 78 kDa, encoded by babA2. The babA1 and babA2 coding sequences are highly similar, but the translational start codon is lacking in babA1. H. pylori employs BabA to bind to fucosylated Lewis B blood-group antigen (Lewis b [Leb]) expressed on host gastric epithelium cells, when H. pylori initially infects the human stomach [30]. The structure of the BabA receptor is similar to the O type blood antigen, and the statistics of epidemiology reveal the correlation between type O blood and gastric related diseases [51].
Blood-antigen binding protein A and BabB are nearly identical in their 5′ and 3′ regions, with most of their sequence divergence being in their mid-regions. Importantly, the middle region of the BabA sequence determines the adhesion ability of BabA. In the western countries, the expression of BabA contributes to increased risk of peptic ulcer disease and gastric cancer [52], [53]. However, the existence of BabA is not correlated to gastric related diseases in Asians [52], [54]. The function of BabB is still unclear, but the expression of BabB was associated with increased gastric histologic lesions in patients [55].
Some strains do not carry babA2 in their genome, or are deficient in BabA resulting from mutation, yet the bacteria still express a chimeric BabB/A (which has the ability to bind to the Leb antigen) by genetic recombination of babA1 and babB in certain conditions [56], [57]. To study the dynamics of Leb adherence during human infection, a study analyzed paired H. pylori isolates obtained sequentially from chronically infected individuals. The results showed that a complete loss or significant reduction of Leb binding was observed in strains from 5 out of 23 individuals, indicating that the BabA-Leb binding phenotype is quite stable during chronic human infection. Sequence comparisons revealed that most amino acid changes were found in the putative N-terminal extracellular adhesion domain [58]. In conclusion, recombination mediates dynamic changes in adherence properties, which suggests that it contributes to the persistence and adaption of H. pylori in ever-changing gastric environments [56], [58].
Sialic acid-binding adhesin
At sites of vigorous local inflammatory response due to H. pylori infection, the expression of sialyl-Lewis x glycosphingolipid (sLex) antigen is increased on the cellular surface. This suggests that SabA adhesin plays a critical role to assist H. pylori to adhere to and colonize the gastric epithelium cells of a patient with gastritis [31], [59]. Especially when lacking gastric Leb expression, Lex and Lea were closely related to H. pylori colonization [60]. The sabB gene is homologous to sabA, but appears not to be involved in sLex binding [31]. Therefore, the function of sabB in bacterial adhesion and pathogenesis is worth investigating.
The prevalence of sabA in clinical strains is nearly 80%, and the sequence results revealed two types of sabA genotypes [59]. The expression of type I sabA is regulated by a CT repeat sequence in the 5′ ORF of sabA, and this repeat can be regulated by slipped strand mispairing (SSM) [59], [61]. The functional protein is only expressed in those strains in which the numbers of the CT dinucleotide repeats are 4, 7, and 10, and allow an ORF encoding the full length SabA. This expression status is defined as “On”. When the CT repeat number is varied to 3, 5, 6, 8, 9, and 11, there will be early stop codons in the ORF resulting from frame-shift. In this status, the gene expression is defined as “Off”, and the truncated SabA is expressed. The type II sabA has a unique deletion of the CT repeats and a distinctive sequence in this region. Moreover, not only sabA, but also adhesins oipA and hopZ genes, are characterized by CT dinucleotide repeats in their 5′-coding regions [62].
The expression of SabA detected by western blotting is dramatically different from the sequence-based prediction [59]. In type I strains, which are predicted to be “On”, the expression of SabA is only 43% by western blotting. Kao et al. showed the length of a polyT tract close to the sabA promoter region is variable, with the variation also arising through SSM [61]. In addition, the length of the polyT tract is supposed to modulate sabA promoter activity, providing an alternative mechanism for transcriptional regulation in H. pylori, which possesses a limited repertoire of classical trans-acting regulatory factors [61], [63], [64]. A mixed-genotype population is developed by SSM during infection by H. pylori which may benefit bacterial immune evasion and adaptation to different hosts.
The analysis of clinical statistics indicates that in patients infected with SabA-positive strains, the density of H. pylori in the body is dramatically higher than in patients infected by SabA-negative strains [59]. This finding indicates that SabA interacting with the sLex antigen can enhance H. pylori colonization in those patients with weak or no Leb expression [59]. Although the intensity of Leb is the key host factor regulating H. pylori density in patients with a babA2-positive H. pylori infection, the weakness of the sLex-mediated adherence, and its metastable On/Off switching resulting from phase variation, may benefit H. pylori by allowing escape from sites where bactericidal host defense responses are most vigorous.
Step 4: Toxin and host tissue damage
Cytotoxin-associated gene A
The epidemiological prevalence of CagA-positive H. pylori infection in western countries is nearly 60% [65], [66], and the prevalence is about 90% in Asian countries [67], [68]. Several studies indicated that the CagA-positive strains are directly associated with acute gastritis, gastric ulcer, and gastric cancer development [69], [70], [71]. As a result, the virulence of individual H. pylori isolates has been measured by their ability to produce CagA.
Cytotoxin-associated gene A protein can be further divided into the Western-type CagA and East Asian-type CagA, by the repeat sequence Glu-Pro-Ile-Tyr-Ala (EPIYA) motifs at the N-terminus of CagA [72], [73]. The affinity of the East Asian-type CagA to SHP-2 is significantly higher than that of the Western-type CagA [72], [73]. As a result, East Asian-type CagA induces more cytoskeleton changes, and is more likely to be associated with gastric cancer [73].
The cag pathogenicity island (cagPAI) is located on the chromosome in H. pylori, is 35–40 kb, and contains more than 30 genes [62]. Among them, cagPAI carries at least 6 genes with homology to type IV secretion systems, and thus translocate the bacterial protein CagA into the host gastric cell cytoplasm upon contact with epithelium cells [74]. Moreover, several proteins, including CagL, and CagY, that are present in the T4SS use β1-integrin as a receptor to deliver CagA into the host cell [75], [76].
The translocated CagA protein localizes to the inner surface of the plasma membrane via interactions with phosphatidylserine and subsequently undergoes tyrosine phosphorylation by the Src family protein tyrosine kinase. However, once injected into the cytoplasm via the T4SS, CagA can alter host cell signaling in both a phosphorylation-dependent and phosphorylation-independent manner. The phosphorylated CagA binds to the phosphatase SHP-2 and affects the adhesion, spreading, and migration of the cell [72], [77]. Moreover, CagA can also affect the host cell in several aspects, such as the formation of gastric epithelium cell pedestals, the change of the cytoskeleton, affecting the proliferation of cells, and stimulating the gastric epithelium cells to secrete IL-8 [78], [79], [80].
Cytotoxin-associated gene A has phosphorylation-independent effects, many of which remain unclear. A conserved motif in the C-terminus of the nonphosphorylated CagA has recently been identified, and was shown to interact with the host hepatocyte growth factor receptor met, which contributes to cellular proliferation and inflammation via the Akt signaling pathway, which activates NF-κB and β-catenin [81].
Vacuolating cytotoxin A
Vacuolating cytotoxin A is predicted to encode a protoxin with a mass of about 140 kDa, but the secreted VacA toxin is composed of the p33 and p55 domains that form an oligomeric structure. This complex can embed into the host cell membrane, and also has the characteristic of an anion-selection channel. This channel can release bicarbonate and organic anions in the host cytoplasm [82]. In this way, the channel might help H. pylori colonization by allowing the efflux of potential metabolic substrates for bacterial growth. This complex can also get into the endosome via endocytosis. The endocytosed VacA channel will allow anions to permeate into late endosomes, which leads to accumulation of weak bases and thence to large vacuole formation by water influx [83], [84]. Previous studies also indicate that VacA applied extracellularly apparently targets mitochondria, since it induces the release of cytochrome C, ER stress, and apoptosis [85]. In addition, VacA disrupts the balance of cell proliferation and death by affecting genes that regulate the cell cycle. It also can induce acute inflammatory responses through inducing host cell release of IL-8 [86].
All H. pylori strains carry the vacA gene encoding protein production, with various degrees determined by the different genopatterns of the signal sequence (s1a, s1b, s1c, and s2), mid-region (m1, m1T, and m2), and the intermediate region (i1, i2, and i3) [87], [88]. The genotype can be divided into different subtypes, according to the combinations of the diversity of these three regions [87], [88]. For the genotype s1/m1, the expression of VacA is highly active and can damage cells in a more acute manner [87]. It has been shown that H. pylori vacA s1 and m1 strains are associated with high levels of inflammation in the gastric mucosa and increased risk for gastric atrophy and carcinoma, compared with the less virulent vacA s2 and m2 strains [89]. Furthermore, the vacA i1 genotype is strongly associated with vacA s1, vacA m1, and cagA-positive genotypes, while the vacA i2 genotype is closely associated with vacA s2, vacA m2, and cagA-negative genotypes [88]. A previous study showed that, in patients with gastric cancer, the vacA s1a and s1c subtypes are less common, and m1T is more prevalent in patients with peptic ulcer and chronic gastritis [90]. However, the association of vacA subtypes with disease is not consistent in different countries.
Although H. pylori is generally viewed as an extracellular microorganism, Chu et al. showed that in a gentamicin protection assay on AGS or MKN45 cells, H. pylori could invade the epithelial cells and multiply within double-layer vesicles either on the plasma membrane or in the cytoplasm. The autophagic vesicles induced by H. pylori are supposed to be the location of replication, and also of the degradation of the replicating bacteria after fusion with lysosomes [91]. The multiplication of H. pylori within cells provides a niche for its resistance to antibacterial therapy and has a significant impact on its biological life cycle. In addition, the VacA or CagA mutants of H. pylori have lower levels of multiplication in macrophages [92].
H. pylori infection is supposed to induce autophagosome formation, and these autophagic vesicles are adapted for the multiplication of H. pylori in the host. The use of different host cell lines and bacterial strains has produced inconsistent results, indicating that H. pylori may affect autophagy in a host cell/bacterial strain dependent manner [93]. However, it is still uncertain whether autophagy serves as an effective front line of host defense against intracellular H. pylori, and the precise mechanisms by which H. pylori exploits host cell machineries for intracellular survival are poorly understood. As a result, the investigation of the roles of vacA, cagA, and other virulence factors in H. pylori intracellular multiplication is necessary.
Conclusions
In summary, over the past year, the knowledge of H. pylori pathogenesis and disease development has been improved by the studies focusing on investigation of bacterial factors. Application of large-scale screening methods should have broad relevance to understanding H. pylori infection-mediated carcinogenesis. Moreover, continually clarifying and refining the roles of bacterial virulence factors in H. pylori pathogenesis will highly benefit vaccine and alternative therapy development.
Source of support
Nil.
Conflict of interest
None declared.
Acknowledgment
We thank Robert Jonas for helpful comments on this manuscript.
Footnotes
Peer review under responsibility of Chang Gung University.
References
- 1.Calvet X., Ramírez Lázaro M.J., Lehours P., Mégraud F. Diagnosis and epidemiology of Helicobacter pylori infection. Helicobacter. 2013;18(Suppl. 1):5–11. doi: 10.1111/hel.12071. [DOI] [PubMed] [Google Scholar]
- 2.Mobley H.L., Island M.D., Hausinger R.P. Molecular biology of microbial ureases. Microbiol Rev. 1995;59:451–480. doi: 10.1128/mr.59.3.451-480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang X., Li H., Cheng T., Xia W., Lai Y.T., Sun H. Nickel translocation between metallochaperones HypA and UreE in Helicobacter pylori. Metallomics. 2014;6:1731–1736. doi: 10.1039/c4mt00134f. [DOI] [PubMed] [Google Scholar]
- 4.Weeks D.L., Eskandari S., Scott D.R., Sachs G. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science. 2000;287:482–485. doi: 10.1126/science.287.5452.482. [DOI] [PubMed] [Google Scholar]
- 5.Scott D.R., Marcus E.A., Wen Y., Singh S., Feng J., Sachs G. Cytoplasmic histidine kinase (HP0244)-regulated assembly of urease with UreI, a channel for urea and its metabolites, CO2, NH3, and NH4(+), is necessary for acid survival of Helicobacter pylori. J Bacteriol. 2010;192:94–103. doi: 10.1128/JB.00848-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller E.F., Maier R.J. Ammonium metabolism enzymes aid Helicobacter pylori acid resistance. J Bacteriol. 2014;196:3074–3081. doi: 10.1128/JB.01423-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartz J.T., Allen L.A. Role of urease in megasome formation and Helicobacter pylori survival in macrophages. J Leukoc Biol. 2006;79:1214–1225. doi: 10.1189/jlb.0106030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pflock M., Finsterer N., Joseph B., Mollenkopf H., Meyer T.F., Beier D. Characterization of the ArsRS regulon of Helicobacter pylori, involved in acid adaptation. J Bacteriol. 2006;188:3449–3462. doi: 10.1128/JB.188.10.3449-3462.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcus E.A., Sachs G., Wen Y., Feng J., Scott D.R. Role of the Helicobacter pylori sensor kinase ArsS in protein trafficking and acid acclimation. J Bacteriol. 2012;194:5545–5551. doi: 10.1128/JB.01263-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eaton K.A., Brooks C.L., Morgan D.R., Krakowka S. Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infect Immun. 1991;59:2470–2475. doi: 10.1128/iai.59.7.2470-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schoep T.D., Fulurija A., Good F., Lu W., Himbeck R.P., Schwan C. Surface properties of Helicobacter pylori urease complex are essential for persistence. PLoS One. 2010;5:e15042. doi: 10.1371/journal.pone.0015042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campanale M., Nucera E., Ojetti V., Cesario V., Di Rienzo T.A., D'Angelo G. Nickel free-diet enhances the Helicobacter pylori eradication rate: a pilot study. Dig Dis Sci. 2014;59:1851–1855. doi: 10.1007/s10620-014-3060-3. [DOI] [PubMed] [Google Scholar]
- 13.Strugatsky D., McNulty R., Munson K., Chen C.K., Soltis S.M., Sachs G. Structure of the proton-gated urea channel from the gastric pathogen Helicobacter pylori. Nature. 2013;493:255–258. doi: 10.1038/nature11684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eaton K.A., Suerbaum S., Josenhans C., Krakowka S. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect Immun. 1996;64:2445–2448. doi: 10.1128/iai.64.7.2445-2448.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J.S., Chang J.H., Chung S.I., Yum J.S. Molecular cloning and characterization of the Helicobacter pylori fliD gene, an essential factor in flagellar structure and motility. J Bacteriol. 1999;181:6969–6976. doi: 10.1128/jb.181.22.6969-6976.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howitt M.R., Lee J.Y., Lertsethtakarn P., Vogelmann R., Joubert L.M., Ottemann K.M. ChePep controls Helicobacter pylori infection of the gastric glands and chemotaxis in the Epsilonproteobacteria. MBio. 2011:2. doi: 10.1128/mBio.00098-11. pii: e00098-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kao C.Y., Sheu B.S., Sheu S.M., Yang H.B., Chang W.L., Cheng H.C. Higher motility enhances bacterial density and inflammatory response in dyspeptic patients infected with Helicobacter pylori. Helicobacter. 2012;17:411–416. doi: 10.1111/j.1523-5378.2012.00974.x. [DOI] [PubMed] [Google Scholar]
- 18.Skene C., Young A., Every A., Sutton P. Helicobacter pylori flagella: antigenic profile and protective immunity. FEMS Immunol Med Microbiol. 2007;50:249–256. doi: 10.1111/j.1574-695X.2007.00263.x. [DOI] [PubMed] [Google Scholar]
- 19.Khalifeh Gholi M., Kalali B., Formichella L., Göttner G., Shamsipour F., Zarnani A.H. Helicobacter pylori FliD protein is a highly sensitive and specific marker for serologic diagnosis of H. pylori infection. Int J Med Microbiol. 2013;303:618–623. doi: 10.1016/j.ijmm.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 20.Lertsethtakarn P., Ottemann K.M., Hendrixson D.R. Motility and chemotaxis in Campylobacter and Helicobacter. Annu Rev Microbiol. 2011;65:389–410. doi: 10.1146/annurev-micro-090110-102908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Josenhans C., Labigne A., Suerbaum S. Comparative ultrastructural and functional studies of Helicobacter pylori and Helicobacter mustelae flagellin mutants: both flagellin subunits, FlaA and FlaB, are necessary for full motility in Helicobacter species. J Bacteriol. 1995;177:3010–3020. doi: 10.1128/jb.177.11.3010-3020.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian W., Jia Y., Yuan K., Huang L., Nadolny C., Dong X. Serum antibody against Helicobacter pylori FlaA and risk of gastric cancer. Helicobacter. 2014;19:9–16. doi: 10.1111/hel.12095. [DOI] [PubMed] [Google Scholar]
- 23.Niehus E., Gressmann H., Ye F., Schlapbach R., Dehio M., Dehio C. Genome-wide analysis of transcriptional hierarchy and feedback regulation in the flagellar system of Helicobacter pylori. Mol Microbiol. 2004;52:947–961. doi: 10.1111/j.1365-2958.2004.04006.x. [DOI] [PubMed] [Google Scholar]
- 24.Kao C.Y., Sheu B.S., Wu J.J. CsrA regulates Helicobacter pylori J99 motility and adhesion by controlling flagella formation. Helicobacter. 2014;19:443–454. doi: 10.1111/hel.12148. [DOI] [PubMed] [Google Scholar]
- 25.Schoenhofen I.C., Lunin V.V., Julien J.P., Li Y., Ajamian E., Matte A. Structural and functional characterization of PseC, an aminotransferase involved in the biosynthesis of pseudaminic acid, an essential flagellar modification in Helicobacter pylori. J Biol Chem. 2006;281:8907–8916. doi: 10.1074/jbc.M512987200. [DOI] [PubMed] [Google Scholar]
- 26.Ménard R., Schoenhofen I.C., Tao L., Aubry A., Bouchard P., Reid C.W. Small-molecule inhibitors of the pseudaminic acid biosynthetic pathway: targeting motility as a key bacterial virulence factor. Antimicrob Agents Chemother. 2014;58:7430–7440. doi: 10.1128/AAC.03858-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merino S., Wilhelms M., Tomás J.M. Role of Aeromonas hydrophila flagella glycosylation in adhesion to Hep-2 cells, biofilm formation and immune stimulation. Int J Mol Sci. 2014;15:21935–21946. doi: 10.3390/ijms151221935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bucior I., Pielage J.F., Engel J.N. Pseudomonas aeruginosa pili and flagella mediate distinct binding and signaling events at the apical and basolateral surface of airway epithelium. PLoS Pathog. 2012;8:e1002616. doi: 10.1371/journal.ppat.1002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clyne M., Ocroinin T., Suerbaum S., Josenhans C., Drumm B. Adherence of isogenic flagellum-negative mutants of Helicobacter pylori and Helicobacter mustelae to human and ferret gastric epithelial cells. Infect Immun. 2000;68:4335–4339. doi: 10.1128/iai.68.7.4335-4339.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ilver D., Arnqvist A., Ogren J., Frick I.M., Kersulyte D., Incecik E.T. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science. 1998;279:373–377. doi: 10.1126/science.279.5349.373. [DOI] [PubMed] [Google Scholar]
- 31.Mahdavi J., Sondén B., Hurtig M., Olfat F.O., Forsberg L., Roche N. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science. 2002;297:573–578. doi: 10.1126/science.1069076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teneberg S., Miller-Podraza H., Lampert H.C., Evans D.J., Jr., Evans D.G., Danielsson D. Carbohydrate binding specificity of the neutrophil-activating protein of Helicobacter pylori. J Biol Chem. 1997;272:19067–19071. doi: 10.1074/jbc.272.30.19067. [DOI] [PubMed] [Google Scholar]
- 33.Yamaguchi H., Osaki T., Kurihara N., Taguchi H., Hanawa T., Yamamoto T. Heat-shock protein 60 homologue of Helicobacter pylori is associated with adhesion of H. pylori to human gastric epithelial cells. J Med Microbiol. 1997;46:825–831. doi: 10.1099/00222615-46-10-825. [DOI] [PubMed] [Google Scholar]
- 34.Odenbreit S., Till M., Hofreuter D., Faller G., Haas R. Genetic and functional characterization of the alpAB gene locus essential for the adhesion of Helicobacter pylori to human gastric tissue. Mol Microbiol. 1999;31:1537–1548. doi: 10.1046/j.1365-2958.1999.01300.x. [DOI] [PubMed] [Google Scholar]
- 35.Peck B., Ortkamp M., Diehl K.D., Hundt E., Knapp B. Conservation, localization and expression of HopZ, a protein involved in adhesion of Helicobacter pylori. Nucleic Acids Res. 1999;27:3325–3333. doi: 10.1093/nar/27.16.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossez Y., Gosset P., Boneca I.G., Magalhães A., Ecobichon C., Reis C.A. The lacdiNAc-specific adhesin LabA mediates adhesion of Helicobacter pylori to human gastric mucosa. J Infect Dis. 2014;210:1286–1295. doi: 10.1093/infdis/jiu239. [DOI] [PubMed] [Google Scholar]
- 37.Evans D.J., Jr., Evans D.G., Takemura T., Nakano H., Lampert H.C., Graham D.Y. Characterization of a Helicobacter pylori neutrophil-activating protein. Infect Immun. 1995;63:2213–2220. doi: 10.1128/iai.63.6.2213-2220.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polenghi A., Bossi F., Fischetti F., Durigutto P., Cabrelle A., Tamassia N. The neutrophil-activating protein of Helicobacter pylori crosses endothelia to promote neutrophil adhesion in vivo. J Immunol. 2007;178:1312–1320. doi: 10.4049/jimmunol.178.3.1312. [DOI] [PubMed] [Google Scholar]
- 39.Petersson C., Forsberg M., Aspholm M., Olfat F.O., Forslund T., Borén T. Helicobacter pylori SabA adhesin evokes a strong inflammatory response in human neutrophils which is down-regulated by the neutrophil-activating protein. Med Microbiol Immunol. 2006;195:195–206. doi: 10.1007/s00430-006-0018-x. [DOI] [PubMed] [Google Scholar]
- 40.Wang G., Hong Y., Olczak A., Maier S.E., Maier R.J. Dual Roles of Helicobacter pylori NapA in inducing and combating oxidative stress. Infect Immun. 2006;74:6839–6846. doi: 10.1128/IAI.00991-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kottakis F., Papadopoulos G., Pappa E.V., Cordopatis P., Pentas S., Choli-Papadopoulou T. Helicobacter pylori neutrophil-activating protein activates neutrophils by its C-terminal region even without dodecamer formation, which is a prerequisite for DNA protection – novel approaches against Helicobacter pylori inflammation. FEBS J. 2008;275:302–317. doi: 10.1111/j.1742-4658.2007.06201.x. [DOI] [PubMed] [Google Scholar]
- 42.Long M., Luo J., Li Y., Zeng F.Y., Li M. Detection and evaluation of antibodies against neutrophil-activating protein of Helicobacter pylori in patients with gastric cancer. World J Gastroenterol. 2009;15:2381–2388. doi: 10.3748/wjg.15.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amedei A., Cappon A., Codolo G., Cabrelle A., Polenghi A., Benagiano M. The neutrophil-activating protein of Helicobacter pylori promotes Th1 immune responses. J Clin Invest. 2006;116:1092–1101. doi: 10.1172/JCI27177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramachandran M., Jin C., Yu D., Eriksson F., Essand M. Vector-encoded Helicobacter pylori neutrophil-activating protein promotes maturation of dendritic cells with Th1 polarization and improved migration. J Immunol. 2014;193:2287–2296. doi: 10.4049/jimmunol.1400339. [DOI] [PubMed] [Google Scholar]
- 45.Huesca M., Borgia S., Hoffman P., Lingwood C.A. Acidic pH changes receptor binding specificity of Helicobacter pylori: a binary adhesion model in which surface heat shock (stress) proteins mediate sulfatide recognition in gastric colonization. Infect Immun. 1996;64:2643–2648. doi: 10.1128/iai.64.7.2643-2648.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin C.Y., Huang Y.S., Li C.H., Hsieh Y.T., Tsai N.M., He P.J. Characterizing the polymeric status of Helicobacter pylori heat shock protein 60. Biochem Biophys Res Commun. 2009;388:283–289. doi: 10.1016/j.bbrc.2009.07.159. [DOI] [PubMed] [Google Scholar]
- 47.Zhao Y., Yokota K., Ayada K., Yamamoto Y., Okada T., Shen L. Helicobacter pylori heat-shock protein 60 induces interleukin-8 via a toll-like receptor (TLR)2 and mitogen-activated protein (MAP) kinase pathway in human monocytes. J Med Microbiol. 2007;56:154–164. doi: 10.1099/jmm.0.46882-0. [DOI] [PubMed] [Google Scholar]
- 48.Tanaka A., Kamada T., Yokota K., Shiotani A., Hata J., Oguma K. Helicobacter pylori heat shock protein 60 antibodies are associated with gastric cancer. Pathol Res Pract. 2009;205:690–694. doi: 10.1016/j.prp.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 49.Lin C.S., He P.J., Tsai N.M., Li C.H., Yang S.C., Hsu W.T. A potential role for Helicobacter pylori heat shock protein 60 in gastric tumorigenesis. Biochem Biophys Res Commun. 2010;392:183–189. doi: 10.1016/j.bbrc.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 50.Liao K.W., Lin C.S., Chen W.L., Yang C.T., Lin C.M., Hsu W.T. Antibodies against Helicobacter pylori heat shock protein 60 aggravate HSP60-mediated proinflammatory responses. Cytokine. 2011;55:174–180. doi: 10.1016/j.cyto.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 51.Aspholm-Hurtig M., Dailide G., Lahmann M., Kalia A., Ilver D., Roche N. Functional adaptation of BabA, the H. pylori ABO blood group antigen binding adhesin. Science. 2004;305:519–522. doi: 10.1126/science.1098801. [DOI] [PubMed] [Google Scholar]
- 52.Chen M.Y., He C.Y., Meng X., Yuan Y. Association of Helicobacter pylori babA2 with peptic ulcer disease and gastric cancer. World J Gastroenterol. 2013;19:4242–4251. doi: 10.3748/wjg.v19.i26.4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gerhard M., Lehn N., Neumayer N., Borén T., Rad R., Schepp W. Clinical relevance of the Helicobacter pylori gene for blood-group antigen-binding adhesin. Proc Natl Acad Sci USA. 1999;96:12778–12783. doi: 10.1073/pnas.96.22.12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mizushima T., Sugiyama T., Komatsu Y., Ishizuka J., Kato M., Asaka M. Clinical relevance of the babA2 genotype of Helicobacter pylori in Japanese clinical isolates. J Clin Microbiol. 2001;39:2463–2465. doi: 10.1128/JCM.39.7.2463-2465.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sheh A., Chaturvedi R., Merrell D.S., Correa P., Wilson K.T., Fox J.G. Phylogeographic origin of Helicobacter pylori determines host-adaptive responses upon coculture with gastric epithelial cells. Infect Immun. 2013;81:2468–2477. doi: 10.1128/IAI.01182-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bäckström A., Lundberg C., Kersulyte D., Berg D.E., Borén T., Arnqvist A. Metastability of Helicobacter pylori bab adhesin genes and dynamics in Lewis B antigen binding. Proc Natl Acad Sci USA. 2004;101:16923–16928. doi: 10.1073/pnas.0404817101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sheu S.M., Sheu B.S., Chiang W.C., Kao C.Y., Wu H.M., Yang H.B. H. pylori clinical isolates have diverse babAB genotype distributions over different topographic sites of stomach with correlation to clinical disease outcomes. BMC Microbiol. 2012;12:89. doi: 10.1186/1471-2180-12-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nell S., Kennemann L., Schwarz S., Josenhans C., Suerbaum S. Dynamics of Lewis B binding and sequence variation of the babA adhesin gene during chronic Helicobacter pylori infection in humans. MBio. 2014:5. doi: 10.1128/mBio.02281-14. pii: e02281-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sheu B.S., Odenbreit S., Hung K.H., Liu C.P., Sheu S.M., Yang H.B. Interaction between host gastric Sialyl-Lewis X and H. pylori SabA enhances H. pylori density in patients lacking gastric Lewis B antigen. Am J Gastroenterol. 2006;101:36–44. doi: 10.1111/j.1572-0241.2006.00358.x. [DOI] [PubMed] [Google Scholar]
- 60.Sheu B.S., Sheu S.M., Yang H.B., Huang A.H., Wu J.J. Host gastric Lewis expression determines the bacterial density of Helicobacter pylori in babA2 genopositive infection. Gut. 2003;52:927–932. doi: 10.1136/gut.52.7.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kao C.Y., Sheu S.M., Sheu B.S., Wu J.J. Length of thymidine homopolymeric repeats modulates promoter activity of sabA in Helicobacter pylori. Helicobacter. 2012;17:203–209. doi: 10.1111/j.1523-5378.2012.00936.x. [DOI] [PubMed] [Google Scholar]
- 62.Alm R.A., Ling L.S., Moir D.T., King B.L., Brown E.D., Doig P.C. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 63.Åberg A., Gideonsson P., Vallström A., Olofsson A., Öhman C., Rakhimova L. A repetitive DNA element regulates expression of the Helicobacter pylori sialic acid binding adhesin by a rheostat-like mechanism. PLoS Pathog. 2014;10:e1004234. doi: 10.1371/journal.ppat.1004234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harvey V.C., Acio C.R., Bredehoft A.K., Zhu L., Hallinger D.R., Quinlivan-Repasi V. Repetitive sequence variations in the promoter region of the adhesin-encoding gene sabA of Helicobacter pylori affect transcription. J Bacteriol. 2014;196:3421–3429. doi: 10.1128/JB.01956-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chiurillo M.A., Moran Y., Cañas M., Valderrama E., Granda N., Sayegh M. Genotyping of Helicobacter pylori virulence-associated genes shows high diversity of strains infecting patients in western Venezuela. Int J Infect Dis. 2013;17:e750–e756. doi: 10.1016/j.ijid.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 66.Rezaeifar A., Eskandari-Nasab E., Moghadampour M., Kharazi-Nejad E., Hasani S.S., Asadi-Saghandi A. The association of interleukin-18 promoter polymorphisms and serum levels with duodenal ulcer, and their correlations with bacterial CagA and VacA virulence factors. Scand J Infect Dis. 2013;45:584–592. doi: 10.3109/00365548.2013.794301. [DOI] [PubMed] [Google Scholar]
- 67.Yamaoka Y., Kodama T., Gutierrez O., Kim J.G., Kashima K., Graham D.Y. Relationship between Helicobacter pylori iceA, cagA, and vacA status and clinical outcome: studies in four different countries. J Clin Microbiol. 1999;37:2274–2279. doi: 10.1128/jcm.37.7.2274-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sheu S.M., Sheu B.S., Yang H.B., Li C., Chu T.C., Wu J.J. Presence of iceA1 but not cagA, cagC, cagE, cagF, cagN, cagT, or orf13 genes of Helicobacter pylori is associated with more severe gastric inflammation in Taiwanese. J Formos Med Assoc. 2002;101:18–23. [PubMed] [Google Scholar]
- 69.Matos J.I., de Sousa H.A., Marcos-Pinto R., Dinis-Ribeiro M. Helicobacter pylori CagA and VacA genotypes and gastric phenotype: a meta-analysis. Eur J Gastroenterol Hepatol. 2013;25:1431–1441. doi: 10.1097/MEG.0b013e328364b53e. [DOI] [PubMed] [Google Scholar]
- 70.Hatakeyama M. Helicobacter pylori CagA and gastric cancer: a paradigm for hit-and-run carcinogenesis. Cell Host Microbe. 2014;15:306–316. doi: 10.1016/j.chom.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 71.Azuma T. Helicobacter pylori CagA protein variation associated with gastric cancer in Asia. J Gastroenterol. 2004;39:97–103. doi: 10.1007/s00535-003-1279-4. [DOI] [PubMed] [Google Scholar]
- 72.Higashi H., Tsutsumi R., Fujita A., Yamazaki S., Asaka M., Azuma T. Biological activity of the Helicobacter pylori virulence factor CagA is determined by variation in the tyrosine phosphorylation sites. Proc Natl Acad Sci USA. 2002;99:14428–14433. doi: 10.1073/pnas.222375399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Argent R.H., Kidd M., Owen R.J., Thomas R.J., Limb M.C., Atherton J.C. Determinants and consequences of different levels of CagA phosphorylation for clinical isolates of Helicobacter pylori. Gastroenterology. 2004;127:514–523. doi: 10.1053/j.gastro.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 74.Odenbreit S., Püls J., Sedlmaier B., Gerland E., Fischer W., Haas R. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science. 2000;287:1497–1500. doi: 10.1126/science.287.5457.1497. [DOI] [PubMed] [Google Scholar]
- 75.Conradi J., Tegtmeyer N., Wozna M., Wissbrock M., Michalek C., Gagell C. An RGD helper sequence in CagL of Helicobacter pylori assists in interactions with integrins and injection of CagA. Front Cell Infect Microbiol. 2012;2:70. doi: 10.3389/fcimb.2012.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tegtmeyer N., Lind J., Schmid B., Backert S. Helicobacter pylori CagL Y58/E59 mutation turns-off type IV secretion-dependent delivery of CagA into host cells. PLoS One. 2014;9:e97782. doi: 10.1371/journal.pone.0097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yamazaki S., Yamakawa A., Ito Y., Ohtani M., Higashi H., Hatakeyama M. The CagA protein of Helicobacter pylori is translocated into epithelial cells and binds to SHP-2 in human gastric mucosa. J Infect Dis. 2003;187:334–337. doi: 10.1086/367807. [DOI] [PubMed] [Google Scholar]
- 78.Boonyanugomol W., Chomvarin C., Baik S.C., Song J.Y., Hahnvajanawong C., Kim K.M. Role of cagA-positive Helicobacter pylori on cell proliferation, apoptosis, and inflammation in biliary cells. Dig Dis Sci. 2011;56:1682–1692. doi: 10.1007/s10620-010-1512-y. [DOI] [PubMed] [Google Scholar]
- 79.Kikuchi K., Murata-Kamiya N., Kondo S., Hatakeyama M. Helicobacter pylori stimulates epithelial cell migration via CagA-mediated perturbation of host cell signaling. Microbes Infect. 2012;14:470–476. doi: 10.1016/j.micinf.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 80.Boonyanugomol W., Chomvarin C., Hahnvajanawong C., Sripa B., Kaparakis-Liaskos M., Ferrero R.L. Helicobacter pylori cag pathogenicity island (cagPAI) involved in bacterial internalization and IL-8 induced responses via NOD1- and MyD88-dependent mechanisms in human biliary epithelial cells. PLoS One. 2013;8:e77358. doi: 10.1371/journal.pone.0077358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Suzuki M., Mimuro H., Kiga K., Fukumatsu M., Ishijima N., Morikawa H. Helicobacter pylori CagA phosphorylation-independent function in epithelial proliferation and inflammation. Cell Host Microbe. 2009;5:23–34. doi: 10.1016/j.chom.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 82.Szabò I., Brutsche S., Tombola F., Moschioni M., Satin B., Telford J.L. Formation of anion-selective channels in the cell plasma membrane by the toxin VacA of Helicobacter pylori is required for its biological activity. EMBO J. 1999;18:5517–5527. doi: 10.1093/emboj/18.20.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Terebiznik M.R., Vazquez C.L., Torbicki K., Banks D., Wang T., Hong W. Helicobacter pylori VacA toxin promotes bacterial intracellular survival in gastric epithelial cells. Infect Immun. 2006;74:6599–6614. doi: 10.1128/IAI.01085-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Palframan S.L., Kwok T., Gabriel K. Vacuolating cytotoxin A (VacA), a key toxin for Helicobacter pylori pathogenesis. Front Cell Infect Microbiol. 2012;2:92. doi: 10.3389/fcimb.2012.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Akazawa Y., Isomoto H., Matsushima K., Kanda T., Minami H., Yamaghchi N. Endoplasmic reticulum stress contributes to Helicobacter pylori VacA-induced apoptosis. PLoS One. 2013;8:e82322. doi: 10.1371/journal.pone.0082322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hisatsune J., Nakayama M., Isomoto H., Kurazono H., Mukaida N., Mukhopadhyay A.K. Molecular characterization of Helicobacter pylori VacA induction of IL-8 in U937 cells reveals a prominent role for p38MAPK in activating transcription factor-2, cAMP response element binding protein, and NF-kappaB activation. J Immunol. 2008;180:5017–5027. doi: 10.4049/jimmunol.180.7.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cover T.L., Tummuru M.K., Cao P., Thompson S.A., Blaser M.J. Divergence of genetic sequences for the vacuolating cytotoxin among Helicobacter pylori strains. J Biol Chem. 1994;269:10566–10573. [PubMed] [Google Scholar]
- 88.Ferreira R.M., Machado J.C., Letley D., Atherton J.C., Pardo M.L., Gonzalez C.A. A novel method for genotyping the Helicobacter pylori vacA intermediate region directly in gastric biopsy specimens. J Clin Microbiol. 2012;50:3983–3989. doi: 10.1128/JCM.02087-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.González C.A., Figueiredo C., Lic C.B., Ferreira R.M., Pardo M.L., Ruiz Liso J.M. Helicobacter pylori cagA and vacA genotypes as predictors of progression of gastric preneoplastic lesions: a long-term follow-up in a high-risk area in Spain. Am J Gastroenterol. 2011;106:867–874. doi: 10.1038/ajg.2011.1. [DOI] [PubMed] [Google Scholar]
- 90.Lin H.J., Perng C.L., Lo W.C., Wu C.W., Tseng G.Y., Li A.F. Helicobacter pylori cagA, iceA and vacA genotypes in patients with gastric cancer in Taiwan. World J Gastroenterol. 2004;10:2493–2497. doi: 10.3748/wjg.v10.i17.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chu Y.T., Wang Y.H., Wu J.J., Lei H.Y. Invasion and multiplication of Helicobacter pylori in gastric epithelial cells and implications for antibiotic resistance. Infect Immun. 2010;78:4157–4165. doi: 10.1128/IAI.00524-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang Y.H., Wu J.J., Lei H.Y. The autophagic induction in Helicobacter pylori-infected macrophage. Exp Biol Med (Maywood) 2009;234:171–180. doi: 10.3181/0808-RM-252. [DOI] [PubMed] [Google Scholar]
- 93.Deen N.S., Huang S.J., Gong L., Kwok T., Devenish R.J. The impact of autophagic processes on the intracellular fate of Helicobacter pylori: more tricks from an enigmatic pathogen? Autophagy. 2013;9:639–652. doi: 10.4161/auto.23782. [DOI] [PMC free article] [PubMed] [Google Scholar]