Abstract
Arsenic is the most pervasive environmental substance and is classified by the International Agency for Research on Cancer as a Group 1 human carcinogen. Nearly every organism has resistance pathways for inorganic arsenic, and in bacteria, their genes are found in arsenic resistance (ars) operons. Recently, a parallel pathway for organic arsenicals has been identified. The ars genes responsible for the organoarsenical detoxification includes arsM, which encodes an As(III) S-adenosylmethionine methyltransferase, arsI, which encodes a C–As bond lyase, and arsH, which encodes a methylarsenite oxidase. The identification and properties of arsM, arsI and arsH are described in this review.
Keywords: Arsenic, As(III) S-adenosylmethionine methyltransferase, C-As lyase, Methylarsenite oxidase, Organoarsenical

Life arose before the atmosphere became oxidizing, when the concentrations of dissolved metal ions in primordial oceans were undoubtedly considerably higher than today [1]. One of the most important initial challenges of the earliest cells would have been the ability to detoxify the toxic arsenic. In response to this, the strong selective pressure, arsenic resistance mechanisms arose early and are present in nearly every extant organism. Without the arsenic detoxification systems, life would not exist.
Arsenic, the Group 1 human carcinogen, is the most prevalent environmental toxin. It ranks top on the Agency for Toxic Substances and Disease Registry priority list of Hazardous Substances. The Environmental Protection Agency asserts that arsenic poses a serious threat to our drinking water, and food supply. In Bangladesh, the arsenic contaminated groundwater has been considered the largest poisoning of a population in human history [2]. Exposure to arsenic not only leads to the various forms of cancer but also causes a range of illnesses, including cardiovascular and peripheral vascular diseases, neurological disorders, diabetes mellitus, and chronic kidney disease [3], [4], [5], [6]. In addition, low birth rate, fetal death, and delayed infant development are closely linked to arsenic exposure during pregnancy [7]. Both inorganic and organic arsenicals such as monosodium methylarsenate (MSMA or MAs(V)), and roxarsone (4-hydroxy-3-nitrobenzene arsenate or Rox(V)) are still used for agriculture and animal husbandry. The ubiquitous presence of arsenic in our surroundings means that arsenic will contaminate our water and food supplies for the foreseeable future.
To cope with the arsenic toxicity, arsenic resistance (ars) genes can be found in the genome of nearly every bacterial species sequenced to date, showing that arsenic must still be ubiquitous in the environment and must provide a selective pressure to maintain them in present-day microbes. The minimal constituents are usually an As(III)-responsive repressor (ArsR) [8], and an As(III) efflux permease (ArsB [9] or ACR3 [10], [11]) that functions to extrude trivalent As(III) from cells. The As(III)-stimulated ATPase (ArsA) [12], and the As(III) metallochaperone (ArsD) [13], which are always associated in ars operons, appears to be later adaptations that enhances the ability of ArsB to extrude As(III) and increase resistance. ArsC [14], [15] and other arsenate reductases [16] are required for resistance to arsenate, which became the predominant arsenic species after oxygen appeared in the atmosphere [17].
Arsenate is universally taken into cells by phosphate transport systems, but, again cells took up As(III) before As(V) was even present environmentally, so As(III) uptake must be much more ancient. In 1997 As(III) was shown to be taken into Escherichia coli by GlpF, a member of the aquaglyceroporin (AQP) superfamily [18]. Since then AQPs have been shown to be a major route of bidirectional movement of As(III) into and out of eukaryotic cells, with human AQP9 the likely pathway for arsenic uptake into and methylarsenite (MAs(III)) efflux out of liver [19], [20]. AQPs have since been shown to be the universal route of arsenic uptake [21]. Arsenic uptake by AQPs is of considerable relevance to human health and disease, and an understanding of both metalloid chemistry and the molecular details of metalloid transport systems is essential for the rational design of new drugs and for treating drug-resistant cells, and microorganisms. One example is that AQP9 is the pathway for uptake of the arsenic chemotherapeutic drug trisenox into leukemia cells [22]. A second example is a demonstration that LmAQP1, a leishmanial AQP, takes up the activated form of the drug pentostam from the macrophage into the amastigote form of the parasite [23]. Moreover, in plants the AQPs were recently shown to take up the essential metalloids boron [24], and silicon [25]. In present-day, the seawater contains approximately 0.4 mM borate and 0.1 mM silicate, but only submicromolar arsenic. This suggests that boron and silicon oxyacid might be physiological substrates of AQPs whereas arsenic might be taken up adventitiously only when present as high-level contaminants. Additionally, there is a single instance of an atypical AQPs called aqpS replacing arsB in the ars operon of Sinorhizobium meliloti. [26] AqpS mediates to the efflux of internally generated As(III). Also As (III) has been shown to be taken up by glucose permeases, including the yeast transporters [27], and human GLUT1, and GLUT4 [27], [28], [29]. Arsenite in solution is an inorganic mimetic of polyols, which allows it to be taken up by glycerol and sugar transporters.
The arsenic methylation cycle
This review will focus on new genes and their functions in ars operons. The revolution in genomics has provided a wealth of sequence information about ars genes in thousands of organisms. New genes have been found in ars operons with functions that are not obvious. Recently, a global cycle of arsenic methylation has been identified that includes ArsM methyltransferases, ArsI C–As bond lyases, and ArsH NADPH-flavin mononucleotide (FMN) - dependent oxidoreductases.
ArsM, an As(III) S-adenosylmethionine methyltransferase
Members of every kingdom, from bacteria to humans, methylate arsenite, producing the trivalent species MAs(III), dimethylarsenite (DMAs(III)), and volatile trimethylarsine (TMAs(III)) [30], [31] catalyzed by As(III), S-adenosylmethionine (SAM) methyltransferases (AsMTs) (EC 2.1.1.137).
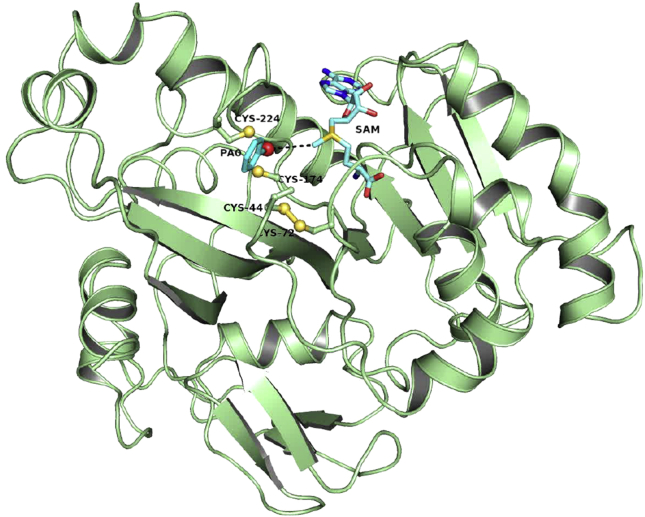
To understand on one hand how microorganisms remodel the environment in arsenic-rich regions and how arsenic methylation is involved in carcinogenesis, on the other hand, it is essential to understand the AsMT catalytic cycle. The enzyme, that catalyzes this reaction in humans and other mammals has been termed AS3MT [32]. It is found predominately in the liver, where the intermediates, in particular, MAs(III) and DMAs(III), are potent toxins and carcinogens responsible for a majority of arsenic-related human diseases [30]. Whether the AsMTs detoxify the arsenic or transform it into more toxic products depends in part on their enzymatic mechanism. Challenger proposed that the mechanism is an alternate series of oxidative methylations and reductions, with the pentavalent species as products [33]. This hypothesis is supported by the fact that humans excrete MAs(V) and dimethylarsenate (DMAs(V)) in urine. An alternate proposal by Hirano is that there is no change in oxidation state during the catalytic cycle and that products are all trivalent methylarsenicals [34]. If the primary intracellular products of methylation are the pentavalent species, then arsenic would have limited carcinogenic potential. On the other hand, if the trivalent species are the major methylated intracellular products, then the methylation would increase the carcinogenicity of arsenic. Whether the oxidized species found in the urine of mammals or the growth medium of microbes are the products of the AsMTs, or are the result of non-enzymatic oxidation of the unstable trivalent species is controversial [35], but, with careful handling, the trivalent forms have been detected in urine [36], [37]. Thus, a detailed knowledge of the enzymatic pathway is important. However, AS3MT has proven difficult to characterize biochemically [38], so microbial AsMTs have been used as models. The first identified microbial AS3MT ortholog was from an ars operon in Rhodopseudomonas palustris and was named arsM [39]. More useful has been the CmArsM from the Yellowstone thermoacidophilic eukaryotic alga Cyanidioschyzon merolae. [40] This heat-stable and highly active enzyme has been invaluable as a model for biochemical [41], and crystallographic analysis of AsMTs [42], [43]. The apo structure was solved at 1.6 Å, as well as liganded structures with SAM, with As(III) [43] or with the aromatic arsenicals phenylarsenite (PhAs(III) or PAO), and trivalent roxarsone (Rox(III)), which are methylated by CmArsM [44]. A model of CmArsM with both the SAM and As(III) binding sites filled is shown in Fig. 1. All AsMTs have four conserved cysteine residues, which are Cys44, Cys72, Cys172 and Cys224 in CmArsM, that appears to be involved in As(III) binding and catalysis [44]. All four cysteines are required for methylation of As(III) to MAs(III), but Cys44 and Cys72 are not required for methylation of MAs(III) to DMAs(III). Interestingly, in new crystal structures Cys41 and Cys72 are linked by a disulfide bond while PhAs(III) or Rox(III) are bound by Cys174 and Cys224. This result implies that the C-terminal cysteine pair forms the metalloid binding site, while the N-terminal cysteine pair play a different role in catalysis that has been postulated to keep arsenic reduced during the methylation pathway in a new model for the catalytic cycle of AsMTs that utilizes a disulfide bond cascade to successively reduces the pentavalent arsenical intermediates [44]. In this model, the substrates and products are trivalent, but there are transient pentavalent enzyme-bound intermediates that are reduced by cysteine residues, creating a disulfide bond cascade mechanism of alternating oxidations, and reductions of the bound arsenic. The new proposed pathway considers the first two methylations in seven steps [Fig. 2]: (1) CmArsM binds As(III) in a series of three thiol transfer reactions from the As(III) triglutathione conjugate (As(GS)3), the preferred substrate [41]. (2) The methyl group of SAM is attacked by the arsenic lone pair. (3) A transient positively charged MAs(V) intermediate is formed. (4) Cys44 (or Cys72) reduces the enzyme-bound MAs(V) intermediate to MAs(III), allowing the second round of methylation. Consistent with the postulate of an enzyme-bound intermediate, most arsenic is enzyme-bound as MAs(III), and little is released into the medium [41] within the first 10 min of methylation. (5) Oxidized Cys44 forms a disulfide bond with Cys72, which must be reduced before the next round of methylation can occur. In vivo thioredoxin (Trx) has been suggested to be the reductant for AsMTs [38], but, in our studies, GSH was used as the reductant because the thermostable Trx/Trx reductase was not available. MAs(III) remains strongly bound by the thiol pair Cys174–Cys224. (6) A second methylation forms a transient positively charged pentavalent DMAs(V) intermediate, (7) which is reduced to DMAs(III) by Cys72, forming a Cys72–Cys174 disulfide. The disulfide is reduced by Trx, regenerating the enzyme and releasing DMAs(III), which nonenzymatically oxidizes in air to DMAs(V). Thus, the four conserved cysteine residues play two distinct roles, first as the binding site for arsenicals, and second as the source of electrons to maintain arsenic in the reduced form. This pathway explains nearly all current results and resolves the differences between Challenger and Hirano about the oxidation state of the substrates and products [34]. It is not clear whether the pentavalent intermediates are obligatory intermediates or side products. In either case, the arsenic must be reduced before the reaction can proceed. The electrons to reduce pentavalent intermediates come from the conserved cysteine residues, which form disulfide bonds. Trx was proposed to be the reductant of pentavalent arsenic [38], but in this model, it serves in the classical role of Trxs in disulfide bond reduction [45]. Thus, AsMTs employs a basic catalytic mechanism similar to that of O-, N-, C-, and S-methyltransferases. What differentiates AsMTs from other members of the methyltransferase superfamily is the necessity to bind trivalent arsenicals and to maintain them in the reduced form, for which they utilizes the four conserved cysteine residues.
Fig. 1.
Model of CmArsM with bound PhAs(III) and S-adenosylmethionine. The cartoon diagram represents the merge of the PhAs(III) (PAO)-bound CmArsM and the S-adenosylmethionine-bound structures. The conserved cysteine residues are represented by ball-and-stick and colored cyan (carbon), blue (nitrogen), red (oxygen) or yellow (sulfur). The dark red sphere is the arsenic atom. PhAs(III) is bound between conserved residues Cys174 and Cys224.
Fig. 2.
Proposed reaction scheme for AsMTs. (The proposed reaction pathway is a general one for AsMTs, but the numbering of cysteine residues follows the sequence of CmArsM. (1) In the first round of methylation AsMT binds As(III) in a series of three thiol transfer reactions; (2) the methyl group of S-adenosylmethionine is attacked by the arsenic lone pair; (3) a positively charged pentavalent MAs(V) intermediate is formed and (4) reduced to an enzyme-bound MAs(III) intermediate by Cys44 with formation of a Cys44-Cys72 disulfide; (5) the disulfide is reduced by Trx, regenerating the enzyme for the second round of methylation; (6) MAs(III), which is tightly bound to the Cys174-Cys224, is methylated, producing an enzyme-bound positively charged pentavalent dimethylarsenates(V) intermediate; (7) which is reduced to dimethylarsenites(III) by Cys72 with formation of a Cys72-Cys174 disulfide; (8) dimethylarsenites(III), which is weakly bound to the single Cys224, dissociates from the enzyme and is oxidized nonenzymatically to dimethylarsenates(V). Finally, the Cys72-Cys174 disulfide is reduced by Trx, allowing the cycle to begin over).
ArsI, a C–As bond lyase
If microbes expressing As(III) SAM methyltransferases are common, why isn't most of the arsenic in the environment methylated? Inorganic arsenic continuously enters the environment from geothermal sources, and some methylated species are degraded abiotically, but there is still much less environmental methylarsenicals than would be expected. The answer may lie in the identification of the ArsI C–As lyase, which cleaves the C–As bond, converting MAs(III) into As(III) [46]. Although the microbial degradation of environmental organoarsenicals has been documented [47], [48], [49], [50], molecular details of the reaction were unknown. Microbial communities in Florida golf course soil were shown to carry out a two-step pathway of MSMA reduction and demethylation [51]. An environmental MAs(III)-demethylating bacterium Bacillus sp. MD1 was isolated from Florida golf course soil and the arsI gene was cloned by selection for MAs(III) [46]. ArsI, is non-heme iron-dependent dioxygenase with C-As lyase activity. A thermophilic ArsI was cloned from Thermomonospora curvata and crystallized [52]. A model of the ArsI structure built on related dioxygenase is shown in Fig. 3.
Fig. 3.
Model of the ArsI structure. (ArsI was modeled on the structure of the C-terminal domain of the related catechol-2,3-dioxygenase (PDB ID 3HPY) with a root-mean square deviation (RMSD) of 2.7 Å. Atoms are colored as in Fig. 1. Fe(II) (orange sphere) is bound to Gln8, His65 and Glu117. The blue spheres are oxygen atoms. The organoarsenic binding site is composed of Cys98 and Cy99, with PhAs(III) filling the site).
ArsI also cleaves the C–As bond in aromatic arsenicals. In addition to the microbial generation of methylated arsenicals, massive amounts of organic arsenicals are introduced into the environment anthropogenically as herbicides, growth promoters, and from industrial activities. Arsenicals, both inorganic and organic, have been utilized in agriculture in the United States for over a century [53]. Historically the use of inorganic arsenical pesticides/herbicides have been largely replaced by methylated arsenicals such as MSMA (Mas(III)), which is still in use today as an herbicide for turf maintenance on golf courses, sod farms, highway rights of way, and weed control in cotton fields [53]. Pentavalent aromatic arsenicals including roxarsone (Rox(V)), nitarsone (Nit(V), 4-nitrophenylarsonic acid), and p-aminophenyl arsonic acid (p-ASA) have been used since the 1940s as an antimicrobial growth promoters for poultry, and swine to control Coccidioides infections, improve weight gain, feed efficiency, and meat pigmentation [47], [50]. While Pfizer has voluntarily suspended the production of roxarsone, and p-ASA, they still make and sell Nit(V), which is the only known treatment for blackhead, or histomoniasis, in turkeys. In addition, roxarsone is produced worldwide and used in poultry farms in many countries. These aromatic arsenicals are largely excreted unchanged and introduced into the environment when chicken litter is applied to crops as fertilizer [47]. Pentavalent organoarsenicals are relatively benign, and less toxic than inorganic arsenicals, however, both aromatic [47], [50], [54] and methyl [49], [55] arsenicals are activated by reduction [56], and then degraded into more toxic inorganic forms in the environment, where they contaminate foods and water supplies. ArsI cleavage of the C–As bond in a wide range of trivalent organoarsenicals strongly suggests that the environmental pentavalent aromatic arsenicals such as Rox(V) also undergo two-step pathway of sequential reduction and ArsI-catalyzed C–As bond cleavage by microbial communities such as the MSMA demethylation pathway.
ArsH, a methylarsenite oxidase
The global arsenic biotransformation is composed of the interaction of redox and methylation cycles. Until recently, a third cycle of arsenic biotransformation, an organoarsenical redox cycle, has grown in importance. Pentavalent organoarsenicals, such as herbicides, and antimicrobial growth promoters, are largely harmless. However, microbial communities carry out organoarsenical reduction [51], and reduced methylated, and aromatic arsenicals are toxic [56]. How bacteria survive the formation of toxic MAs(III) or Rox(III) was not clear until the identification of the arsH and arsI genes. As described above, ArsI catalyzes the cleavage of the C-As bond, and the microbes that have an arsI escape killing by trivalent organoarsenicals. How the arsH gene contributed to the tolerance to trivalent organoarsenicals was not clear until the recent functional characterization of arsH in S. meliloti and Pseudomonas putida. [57].
The ars operon of Yersinia enterocolitica was shown to contain a novel gene that was termed arsH, but its function was not clear, conferring only a very slight increase in the resistance to both arsenite and arsenate [58], and it was difficult to understand how a single gene could confer resistance to both the pentavalent and trivalent forms of inorganic arsenic. Deletion of arsH in Serratia marcescens marcescens [59] and S. meliloti [26] also led to slight increases in arsenic sensitivity. Arsenic resistance in Thiobacillus ferrooxidans [60] and the cyanobacterium Synechocystis [61] was unaffected by either a loss of function mutation in or overexpression of arsH. Thus, the data on the physiological role of arsH were not easily interpreted until recently. Currently, there are approximately 9000 ArsH-related protein sequences deposited in the NCBI database. A vast majority of ArsH sequences are found in bacteria (97.7%), mostly gammaproteobacteria, whereas only a few are found in eukaryotes, mostly in fungi (2.2%). A few mammalian ArsH sequences are present in the NCBI database, but it is not clear whether these sequences are valid.
Purified ArsH from S. meliloti exhibits NADPH-dependent FMN reductase activity, reducing azo dyes and generating hydrogen peroxide, but this enzyme did not catalyze the oxidation of As(III) or reduction of As(V) [62]. Synechocystis ArsH has been identified as a quinone reductase [63] and is capable of reducing chromate and ferric iron, but no arsenic-associated activity was observed [64]. One the one hand, arsH genes would not be in ars operons, if it were not related to arsenic metabolism. On the other hand, there is no clear phenotype associate with the arsH gene in vivo, and purified ArsH has no activity with inorganic arsenicals. A reasonable deduction of the absence of arsenic-associated activity with the purified S. meliloti ArsH [62] is that the physiological role of ArsH involves arsenic species other than inorganic arsenicals.
ArsH confers resistance to trivalent organoarsenicals
The physiological role of ArsH was recently demonstrated to be the oxidation of toxic MAs(III) and other trivalent organoarsenicals [57]. It was noted that P. putida and S. meliloti are naturally arsRCH and aqpS of S. meliloti, two ars operon resistant to MAs(III), PhAs(III) and Rox(III), particularly P. putida, which is 10-fold more resistant to these organoarsenicals than E. coli. While there is a single arsH gene in the ars operons, each containing an arsH gene, (arsRBCH) is found in the chromosome of P. putida. Deletion of both ars operons in P. putida led to the loss of resistance to MAs(III) and PhAs(III) (but not to Rox(III) because P. putida takes up Rox(III) only poorly). In S. meliloti, deletion of the ars operon, or only the arsH gene resulted in sensitivity to these organoarsenicals, indicating that arsH gives the resistance to trivalent organoarsenicals. Heterologous expression of P. putida arsH from the first ars operon (ars1) in the arsenic hypersensitive E. coli strain AW3110, in which the ars operon was deleted [65] conferred the resistance to MAs(III), PhAs(III) and Rox(III). This finding suggests that arsH expression alone is sufficient to detoxify the activated trivalent forms of the herbicide MSMA as well as the poultry antimicrobial growth promoter roxarsone in the absence of any other ars genes.
Purified ArsH is a trivalent organoarsenical oxidase
It was originally anticipated that ArsH would be an arsenical reductase, perhaps by reducing MAs(V) to MAs(III), or even MAs(III) to MAs(0). Yet, it is unclear how reduction of nontoxic MAs(V) to toxic MAs(III) could be physiological. Purified ArsH from either S. meliloti or P. putida are yellow in appearance and display a typical flavoprotein absorption spectrum [57], [62]. Although P. putida ArsH possesses NADPH-dependent FMN reductase activity, this activity is not stimulated by inorganic arsenicals, and no reduction of MAs(V), or other pentavalent organoarsenicals was observed. In contrast, using high performance liquid chromatography coupled inductively coupled plasma-mass spectrometry, in the presence of NADPH and FMN, P. putida ArsH oxidized MAs(III), PhAs(III), and Rox(III) (but not As(III)) to their pentavalent forms [57]. The fact that ArsH confers resistance to the organoarsenicals by the oxidation indicates that ArsH is a trivalent organoarsenical oxidase.
How two reductants, NADPH and MAs(III), could result in substrate oxidation was not obvious. The answer is that O2 is a co-substrate and serves as the oxidant. It is proposed that ArsH catalyzes a coupled oxidation of MAs(III) with reduction of O2 to H2O [57]. In the reductive half-reaction of the proposed scheme, oxidation of NADPH-FMN complex causes the hydride transfer from NADPH to FMN, forming FMNH2 and releasing NADP+. In the oxidative half-reaction, when a trivalent organoarsenical substrate is located near the FMNH2, the reduced flavin is oxidized by O2 followed by the oxidation of the trivalent organoarsenical substrate to its pentavalent form. Without a physiological substrate, ArsH nonspecifically reduces azo dyes, metals, and quinines as well as H2O2, which is regarded as in vitro artifacts. Thus ArsH confers resistance to toxic trivalent organoarsenicals by oxidizing them to nontoxic pentavalent species.
ArsH structure, interactions between subunits and FMN binding site
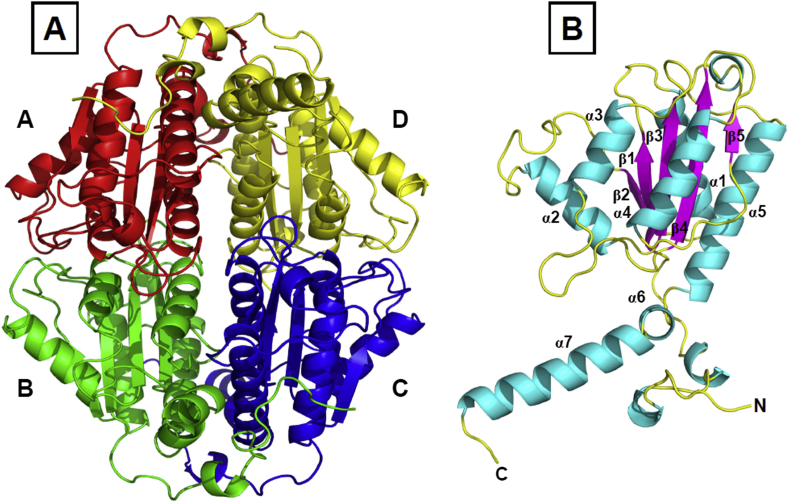
The structure of ArsH from S. meliloti [Fig. 4] has been determined at 1.8 Å resolution by X-ray crystallography [62]. ArsH crystallizes with eight molecules in the asymmetric unit in two identical tetramers. Each monomer belongs to the global α/β protein with the flavodoxin-like architecture as core domain with a five-stranded parallel β-sheet. The structure of ArsH resembles other flavoproteins.
Fig. 4.
Structure of ArsH. (The cartoon diagram demonstrates the X-ray crystal structure of S. meliloti ArsH. (A) The tetrameric form of ArsH with each monomer (A, red; B, green; C, blue; D, yellow). (B) ArsH monomer with secondary structures (helices, cyan; strands, magenta; loops, yellow) shown in ribbon representation.
Based on the fact that S. meliloti ArsH is a tetramer in solution, and two tetramers are in the asymmetric unit, S. meliloti ArsH is thought to function as a tetramer. This is consistent with the recent structure of ArsH from Synechocystis sp. strain PCC 6803 [66]. Within the S. meliloti ArsH structure, the monomers A and B form a typical flavodoxin-like dimer, whereas the monomers C and D form another dimer. The N-terminal α-helices (α1, α4, α5) and the C-terminal helices (α6, α7) participate in the dimer–dimer interaction. Also, the extensions of N- and C-terminus are crucial for the interactions between subunits as well as the formation of the tetramer [62].
The predicted FMN binding site of S. meliloti and P. putida ArsH contains amino acid sequence G15SLRTVSYS, and G42STRERSFS, respectively. In P. putida ArsH, the substitution of Arg45, and Ser48 with alanine leads to sensitivity to PhAs(III) [57]. Likewise, substitution of Glu108, which is predicted to bind FMN with alanine, reduces the resistance to trivalent organoarsenicals. Compared with the yellow wild type P. putida ArsH, the purified E108A mutant protein is colorless. While the typical FMN absorption maxima at 373 and 455 nm are absent in E108A mutant protein, the addition of FMN largely recovers the activity of E108A mutant protein. These findings suggest a role for Glu108 in FMN binding.
In summary, the genes/enzymes described in this review form a new cycle of arsenic methylation, demethylation, and detoxification [Fig. 5]. ArsM transforms inorganic arsenic into a highly toxic organoarsenical species that kills off by competing the bacterial species and may also be responsible for carcinogenesis in animals. Competing microbial species have responded to this environmental pressure by evolving detoxification mechanisms for MAs(III). Some produces ArsI that demethylates MAs(III) to less toxic inorganic As(III) while other produces ArsH that oxidizes MAs(III) to nontoxic pentavalent MAs(V). It is likely that all of these processes are taking place in environmental microbial communities as bacteria, archaea, fungi, and protozoans constantly fight for dominance.
Fig. 5.
ArsM, ArsH and ArsI: Enzymes of organoarsenical production and detoxification. (Pentavalent inorganic arsenate (As(V)) is reduced by the ArsC arsenate reductase to trivalent arsenite (As(III)). Arsenate is relatively nontoxic compared with arsenite. Some microbes have arsM genes that encode ArsM As(III) S-adenosylmethionine methyltransferases that transform As(III) into the considerably more toxic (and, for humans, carcinogenic) organoarsenical MAs(III). Other microbes have an arsI gene encoding the ArsI C-As lyase, a dioxygenase that cleaves off the methyl group, forming inorganic As(III). Since As(III) is less toxic than MAs(III), this reaction detoxifies the organoarsenical. Other bacteria have an arsH gene encoding the ArsH NADPH-FMN oxidoreductase that oxidizes MAs(III) to relatively nontoxic pentavalent MAs(V), also a detoxification process.
Source of support
Research from the Rosen laboratory described in this article was supported by National Institutes of Health Grants R37 GM55425 and R01 ES023779.
Conflicts of interest
None declared.
Acknowledgments
The research described in this manuscript was supported by Bankhead-Coley Program grant 4BF01 from the State of Florida, Department of Health and National Institutes of Health grants R37 GM55425, and R01 ES023779.
Footnotes
Peer review under responsibility of Chang Gung University.
References
- 1.Zhu Y.G., Yoshinaga M., Zhao F.J., Rosen B.P. Earth abides arsenic biotransformations. Annu Rev Earth Planet Sci. 2014;42:443–467. doi: 10.1146/annurev-earth-060313-054942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith A.H., Lingas E.O., Rahman M. Contamination of drinking-water by arsenic in Bangladesh: a public health emergency. Bull World Health Organ. 2000;78:1093–1103. [PMC free article] [PubMed] [Google Scholar]
- 3.Abernathy C.O., Liu Y.P., Longfellow D., Aposhian H.V., Beck B., Fowler B. Arsenic: health effects, mechanisms of actions, and research issues. Environ Health Perspect. 1999;107:593–597. doi: 10.1289/ehp.99107593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abernathy C.O., Thomas D.J., Calderon R.L. Health effects and risk assessment of arsenic. J Nutr. 2003;133(5 Suppl. 1):1536S–1538S. doi: 10.1093/jn/133.5.1536S. [DOI] [PubMed] [Google Scholar]
- 5.Rahman M., Tondel M., Ahmad S.A., Axelson O. Diabetes mellitus associated with arsenic exposure in Bangladesh. Am J Epidemiol. 1998;148:198–203. doi: 10.1093/oxfordjournals.aje.a009624. [DOI] [PubMed] [Google Scholar]
- 6.Tchounwou P.B., Patlolla A.K., Centeno J.A. Carcinogenic and systemic health effects associated with arsenic exposure – a critical review. Toxicol Pathol. 2003;31:575–588. doi: 10.1080/01926230390242007. [DOI] [PubMed] [Google Scholar]
- 7.Naujokas M.F., Anderson B., Ahsan H., Aposhian H.V., Graziano J.H., Thompson C. The broad scope of health effects from chronic arsenic exposure: update on a worldwide public health problem. Environ Health Perspect. 2013;121:295–302. doi: 10.1289/ehp.1205875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.San Francisco M.J., Hope C.L., Owolabi J.B., Tisa L.S., Rosen B.P. Identification of the metalloregulatory element of the plasmid-encoded arsenical resistance operon. Nucleic Acids Res. 1990;18:619–624. doi: 10.1093/nar/18.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.San Francisco M.J., Chen C.M., Rosen B.P. Identification of the membrane component of the anion pump encoded by the arsenical resistance operon of R-factor R773. Prog Clin Biol Res. 1988;252:311–316. [PubMed] [Google Scholar]
- 10.Wysocki R., Bobrowicz P., Ulaszewski S. The Saccharomyces cerevisiae ACR3 gene encodes a putative membrane protein involved in arsenite transport. J Biol Chem. 1997;272:30061–30066. doi: 10.1074/jbc.272.48.30061. [DOI] [PubMed] [Google Scholar]
- 11.Ghosh M., Shen J., Rosen B.P. Pathways of As(III) detoxification in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1999;96:5001–5006. doi: 10.1073/pnas.96.9.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosen B.P., Hsu C.M., Karkaria C.E., Owolabi J.B., Tisa L.S. Molecular analysis of an ATP-dependent anion pump. Philos Trans R Soc Lond B Biol Sci. 1990;326:455–463. doi: 10.1098/rstb.1990.0024. [DOI] [PubMed] [Google Scholar]
- 13.Lin Y.F., Walmsley A.R., Rosen B.P. An arsenic metallochaperone for an arsenic detoxification pump. Proc Natl Acad Sci U S A. 2006;103:15617–15622. doi: 10.1073/pnas.0603974103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ji G., Silver S. Reduction of arsenate to arsenite by the ArsC protein of the arsenic resistance operon of Staphylococcus aureus plasmid pI258. Proc Natl Acad Sci U S A. 1992;89:9474–9478. doi: 10.1073/pnas.89.20.9474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gladysheva T.B., Oden K.L., Rosen B.P. Properties of the arsenate reductase of plasmid R773. Biochemistry. 1994;33:7288–7293. doi: 10.1021/bi00189a033. [DOI] [PubMed] [Google Scholar]
- 16.Mukhopadhyay R., Rosen B.P. Saccharomyces cerevisiae ACR2 gene encodes an arsenate reductase. FEMS Microbiol Lett. 1998;168:127–136. doi: 10.1111/j.1574-6968.1998.tb13265.x. [DOI] [PubMed] [Google Scholar]
- 17.Rosen B.P. Biochemistry of arsenic detoxification. FEBS Lett. 2002;529:86–92. doi: 10.1016/s0014-5793(02)03186-1. [DOI] [PubMed] [Google Scholar]
- 18.Sanders O.I., Rensing C., Kuroda M., Mitra B., Rosen B.P. Antimonite is accumulated by the glycerol facilitator GlpF in Escherichia coli. J Bacteriol. 1997;179:3365–3367. doi: 10.1128/jb.179.10.3365-3367.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Z., Shen J., Carbrey J.M., Mukhopadhyay R., Agre P., Rosen B.P. Arsenite transport by mammalian aquaglyceroporins AQP7 and AQP9. Proc Natl Acad Sci U S A. 2002;99:6053–6058. doi: 10.1073/pnas.092131899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Z., Styblo M., Rosen B.P. Methylarsonous acid transport by aquaglyceroporins. Environ Health Perspect. 2006;114:527–531. doi: 10.1289/ehp.8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mukhopadhyay R., Bhattacharjee H., Rosen B.P. Aquaglyceroporins: generalized metalloid channels. Biochim Biophys Acta. 2014;1840:1583–1591. doi: 10.1016/j.bbagen.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhattacharjee H., Carbrey J., Rosen B.P., Mukhopadhyay R. Drug uptake and pharmacological modulation of drug sensitivity in leukemia by AQP9. Biochem Biophys Res Commun. 2004;322:836–841. doi: 10.1016/j.bbrc.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 23.Gourbal B., Sonuc N., Bhattacharjee H., Legare D., Sundar S., Ouellette M. Drug uptake and modulation of drug resistance in Leishmania by an aquaglyceroporin. J Biol Chem. 2004;279:31010–31017. doi: 10.1074/jbc.M403959200. [DOI] [PubMed] [Google Scholar]
- 24.Miwa K., Takano J., Omori H., Seki M., Shinozaki K., Fujiwara T. Plants tolerant of high boron levels. Science. 2007;318:1417. doi: 10.1126/science.1146634. [DOI] [PubMed] [Google Scholar]
- 25.Ma J.F., Tamai K., Yamaji N., Mitani N., Konishi S., Katsuhara M. A silicon transporter in rice. Nature. 2006;440:688–691. doi: 10.1038/nature04590. [DOI] [PubMed] [Google Scholar]
- 26.Yang H.C., Cheng J., Finan T.M., Rosen B.P., Bhattacharjee H. Novel pathway for arsenic detoxification in the legume symbiont Sinorhizobium meliloti. J Bacteriol. 2005;187:6991–6997. doi: 10.1128/JB.187.20.6991-6997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Z., Boles E., Rosen B.P. Arsenic trioxide uptake by hexose permeases in Saccharomyces cerevisiae. J Biol Chem. 2004;279:17312–17318. doi: 10.1074/jbc.M314006200. [DOI] [PubMed] [Google Scholar]
- 28.Liu Z., Sanchez M.A., Jiang X., Boles E., Landfear S.M., Rosen B.P. Mammalian glucose permease GLUT1 facilitates transport of arsenic trioxide and methylarsonous acid. Biochem Biophys Res Commun. 2006;351:424–430. doi: 10.1016/j.bbrc.2006.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang X., McDermott J.R., Ajees A.A., Rosen B.P., Liu Z. Trivalent arsenicals and glucose use different translocation pathways in mammalian GLUT1. Metallomics. 2010;2:211–219. doi: 10.1039/b920471g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas D.J., Rosen B.P. Arsenic methyltransferases. In: Kretsinger R.H., Uversky V.N., Permyakov E.A., editors. Encyclopedia of metalloproteins. Springer; New York: 2013. pp. 140–145. [Google Scholar]
- 31.Ye J., Rensing C., Rosen B.P., Zhu Y.G. Arsenic biomethylation by photosynthetic organisms. Trends Plant Sci. 2012;17:155–162. doi: 10.1016/j.tplants.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dheeman D.S., Packianathan C., Pillai J.K., Rosen B.P. Pathway of human AS3MT arsenic methylation. Chem Res Toxicol. 2014;27:1979–1989. doi: 10.1021/tx500313k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Challenger F. Biological methylation. Sci Prog. 1947;35:396–416. [PubMed] [Google Scholar]
- 34.Hayakawa T., Kobayashi Y., Cui X., Hirano S. A new metabolic pathway of arsenite: arsenic-glutathione complexes are substrates for human arsenic methyltransferase Cyt19. Arch Toxicol. 2005;79:183–191. doi: 10.1007/s00204-004-0620-x. [DOI] [PubMed] [Google Scholar]
- 35.Cullen W.R. Chemical mechanism of arsenic biomethylation. Chem Res Toxicol. 2014;27:457–461. doi: 10.1021/tx400441h. [DOI] [PubMed] [Google Scholar]
- 36.Le X.C., Lu X., Ma M., Cullen W.R., Aposhian H.V., Zheng B. Speciation of key arsenic metabolic intermediates in human urine. Anal Chem. 2000;72:5172–5177. doi: 10.1021/ac000527u. [DOI] [PubMed] [Google Scholar]
- 37.Drobná Z., Del Razo L.M., García-Vargas G.G., Sánchez-Peña L.C., Barrera-Hernández A., Stýblo M. Environmental exposure to arsenic, AS3MT polymorphism and prevalence of diabetes in Mexico. J Expo Sci Environ Epidemiol. 2013;23:151–155. doi: 10.1038/jes.2012.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas D.J., Waters S.B., Styblo M. Elucidating the pathway for arsenic methylation. Toxicol Appl Pharmacol. 2004;198:319–326. doi: 10.1016/j.taap.2003.10.020. [DOI] [PubMed] [Google Scholar]
- 39.Qin J., Rosen B.P., Zhang Y., Wang G., Franke S., Rensing C. Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc Natl Acad Sci U S A. 2006;103:2075–2080. doi: 10.1073/pnas.0506836103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qin J., Lehr C.R., Yuan C., Le X.C., McDermott T.R., Rosen B.P. Biotransformation of arsenic by a Yellowstone thermoacidophilic eukaryotic alga. Proc Natl Acad Sci U S A. 2009;106:5213–5217. doi: 10.1073/pnas.0900238106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marapakala K., Qin J., Rosen B.P. Identification of catalytic residues in the As(III) S-adenosylmethionine methyltransferase. Biochemistry. 2012;51:944–951. doi: 10.1021/bi201500c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marapakala K., Ajees A.A., Qin J., Sankaran B., Rosen B.P. Crystallization and preliminary X-ray crystallographic analysis of the ArsM arsenic(III) S-adenosylmethionine methyltransferase. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66(Pt 9):1050–1052. doi: 10.1107/S1744309110027661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ajees A.A., Marapakala K., Packianathan C., Sankaran B., Rosen B.P. Structure of an As(III) S-adenosylmethionine methyltransferase: insights into the mechanism of arsenic biotransformation. Biochemistry. 2012;51:5476–5485. doi: 10.1021/bi3004632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marapakala K., Packianathan C., Ajees A.A., Dheeman D.S., Sankaran B., Kandavelu P. A disulfide-bond cascade mechanism for arsenic(III) S-adenosylmethionine methyltransferase. Acta Crystallogr D Biol Crystallogr. 2015;71(Pt 3):505–515. doi: 10.1107/S1399004714027552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holmgren A. Thioredoxin and glutaredoxin systems. J Biol Chem. 1989;264:13963–13966. [PubMed] [Google Scholar]
- 46.Yoshinaga M., Rosen B.P. A C·As lyase for degradation of environmental organoarsenical herbicides and animal husbandry growth promoters. Proc Natl Acad Sci U S A. 2014;111:7701–7706. doi: 10.1073/pnas.1403057111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garbarino J.R., Bednar A.J., Rutherford D.W., Beyer R.S., Wershaw R.L. Environmental fate of roxarsone in poultry litter. I. Degradation of roxarsone during composting. Environ Sci Technol. 2003;37:1509–1514. doi: 10.1021/es026219q. [DOI] [PubMed] [Google Scholar]
- 48.Lehr C., Polishchuk E., Radoja U., Cullen W.R. Demethylation of methylarsenic species by Mycobacterium neoaurum. Appl Organomet Chem. 2003;17:831–834. [Google Scholar]
- 49.Feng M., Schrlau J.E., Snyder R., Snyder G.H., Chen M., Cisar J.L. Arsenic transport and transformation associated with MSMA application on a golf course green. J Agric Food Chem. 2005;53:3556–3562. doi: 10.1021/jf047908j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stolz J.F., Perera E., Kilonzo B., Kail B., Crable B., Fisher E. Biotransformation of 3-nitro-4-hydroxybenzene arsonic acid (roxarsone) and release of inorganic arsenic by Clostridium species. Environ Sci Technol. 2007;41:818–823. doi: 10.1021/es061802i. [DOI] [PubMed] [Google Scholar]
- 51.Yoshinaga M., Cai Y., Rosen B.P. Demethylation of methylarsonic acid by a microbial community. Environ Microbiol. 2011;13:1205–1215. doi: 10.1111/j.1462-2920.2010.02420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nadar S.V., Yoshinaga M., Kandavelu P., Sankaran B., Rosen B.P. Crystallization and preliminary X-ray crystallographic studies of the ArsI C-As lyase from Thermomonospora curvata. Acta Crystallogr F Struct Biol Commun. 2014;70(Pt 6):761–764. doi: 10.1107/S2053230X14008814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matteson A.R., Gannon T.W., Jeffries M.D., Haines S., Lewis D.F., Polizzotto M.L. Arsenic retention in foliage and soil after monosodium methyl arsenate (MSMA) application to turfgrass. J Environ Qual. 2014;43:379–388. doi: 10.2134/jeq2013.07.0268. [DOI] [PubMed] [Google Scholar]
- 54.Makris K.C., Quazi S., Punamiya P., Sarkar D., Datta R. Fate of arsenic in swine waste from concentrated animal feeding operations. J Environ Qual. 2008;37:1626–1633. doi: 10.2134/jeq2007.0479. [DOI] [PubMed] [Google Scholar]
- 55.Von Endt D.W., Kearney P.C., Kafman D.D. Degradation of monosodium methanearsonic acid by soil microorganisms. J Agric Food Chem. 1968;16:17–20. [Google Scholar]
- 56.Chen J., Sun S., Li C.Z., Zhu Y.G., Rosen B.P. Biosensor for organoarsenical herbicides and growth promoters. Environ Sci Technol. 2014;48:1141–1147. doi: 10.1021/es4038319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen J., Bhattacharjee H., Rosen B.P. ArsH is an organoarsenical oxidase that confers resistance to trivalent forms of the herbicide monosodium methylarsenate and the poultry growth promoter roxarsone. Mol Microbiol. 2015;96:1042–1052. doi: 10.1111/mmi.12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Neyt C., Iriarte M., Thi V.H., Cornelis G.R. Virulence and arsenic resistance in yersiniae. J Bacteriol. 1997;179:612–619. doi: 10.1128/jb.179.3.612-619.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ryan D., Colleran E. Arsenical resistance in the IncHI2 plasmids. Plasmid. 2002;47:234–240. doi: 10.1016/s0147-619x(02)00012-4. [DOI] [PubMed] [Google Scholar]
- 60.Butcher B.G., Deane S.M., Rawlings D.E. The chromosomal arsenic resistance genes of Thiobacillus ferrooxidans have an unusual arrangement and confer increased arsenic and antimony resistance to Escherichia coli. Appl Environ Microbiol. 2000;66:1826–1833. doi: 10.1128/aem.66.5.1826-1833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.López-Maury L., Florencio F.J., Reyes J.C. Arsenic sensing and resistance system in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol. 2003;185:5363–5371. doi: 10.1128/JB.185.18.5363-5371.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ye J., Yang H.C., Rosen B.P., Bhattacharjee H. Crystal structure of the flavoprotein ArsH from Sinorhizobium meliloti. FEBS Lett. 2007;581:3996–4000. doi: 10.1016/j.febslet.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hervás M., López-Maury L., León P., Sánchez-Riego A.M., Florencio F.J., Navarro J.A. ArsH from the cyanobacterium Synechocystis sp. PCC 6803 is an efficient NADPH-dependent quinone reductase. Biochemistry. 2012;51:1178–1187. doi: 10.1021/bi201904p. [DOI] [PubMed] [Google Scholar]
- 64.Xue X.M., Yan Y., Xu H.J., Wang N., Zhang X., Ye J. ArsH from Synechocystis sp. PCC 6803 reduces chromate and ferric iron. FEMS Microbiol Lett. 2014;356:105–112. doi: 10.1111/1574-6968.12481. [DOI] [PubMed] [Google Scholar]
- 65.Carlin A., Shi W., Dey S., Rosen B.P. The ars operon of Escherichia coli confers arsenical and antimonial resistance. J Bacteriol. 1995;177:981–986. doi: 10.1128/jb.177.4.981-986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang X., Xue X.M., Yan Y., Ye J. Purification, crystallization and preliminary X-ray diffraction analysis of ArsH from Synechocystis sp. strain PCC 6803. Acta Crystallogr F Struct Biol Commun. 2014;70(Pt 4):497–500. doi: 10.1107/S2053230X14004865. [DOI] [PMC free article] [PubMed] [Google Scholar]