Abstract
Purpose of Review
Traumatic stress has profound impacts on many domains of life, yet the mechanisms that confer risk for or resilience to the development of traumatic stress-related psychopathologies are still very much under investigation. The current review highlights recent developments in the field of traumatic stress epigenetics in humans.
Recent Findings
Recent results reveal traumatic stress-related epigenetic dysregulation in neural, endocrine, and immune system genes and associated networks. Emerging work combining imaging with epigenetic measures holds promise for addressing the correspondence between peripheral and central effects of traumatic stress. A growing literature is also documenting the transgenerational effects of prenatal stress exposures in humans.
Summary
Moving forward, increasing focus on epigenetic marks of traumatic stress in CNS tissue will create a clearer picture of the relevance of peripheral measures; PTSD brain banks will help in this regard. Similarly, leveraging multigenerational birth cohort data will do much to clarify the extent of transgenerational epigenetic effects of traumatic stress. Greater efforts should be made towards developing prospective studies with longitudinal design.
Keywords: DNA methylation, Peripheral tissue, Transgenerational, Neuroimaging, Human
Introduction
An estimated 89.7% of adults have been exposed to at least one traumatic event in their lifetime[1], with the norm being multiple trauma exposures. These events include instances in which threat to life is salient[2]. It is well known that traumatic stress exposure has profound sociological, psychological, and biological effects[3]. These effects can occur within any stage of life, but trauma exposure during physiological, and especially neural development, are particularly detrimental[4].
Exposure to trauma can cause dysfunction within the hypothalamic-pituitary adrenal axis (HPA-axis) [Figure 1] and other biological systems, and is associated with the development of major depressive disorder (MDD)[5], post-traumatic stress disorder (PTSD)[5], and other mood and anxiety disorders. With PTSD in particular, individuals are required to have an exposure to trauma in order to be eligible for a diagnosis[2], yet not all individuals who are exposed go on to develop the disorder; thus PTSD, as well as the broader assessment of traumatic stress, are especially useful for understanding the molecular mechanisms that confer risk for, or resilience to, this disorder. In particular, the molecular mechanisms that mediate interactions between genes and the environment are particularly salient and require further characterization.
Figure 1. The CNS, HPA-axis, and the stress response.
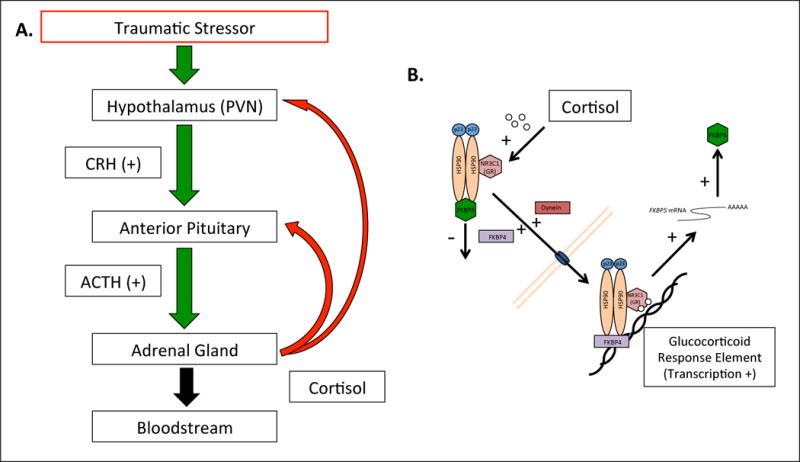
A. HPA-axis long-range feedback mechanism. In response to perceived stress, brain regions associated with emotional reactivity and memory, including the prefrontal cortex, amygdala, and hippocampus are activated. Through afferent axonal connections, these regions activate (green arrow) the HPA-axis’ neural hub, the paraventricular nucleus (PVN) of the hypothalamus, to release both vasopressin and, importantly, corticotropin-releasing hormone (CRH)[79]. From there, CRH acts in a stimulatory manner on the anterior pituitary gland to release adrenal corticotropic hormone (ACTH). ACTH is released into the bloodstream, whereupon it travels to the adrenal cortices of the kidneys, stimulating the release of glucocorticoids (GCs), such as cortisol. These stress hormones act ubiquitously (black arrow) throughout many tissues in the body, and contribute to critical immunological, metabolic, cardiac, and homeostatic functions[80]. Importantly, GCs also act mechanistically in and around the CNS, including serving as a long-range negative feedback mechanism to inhibit further HPA-axis activity (red arrow). B. HPA-axis ultra short-range feedback mechanism. GCs also bind directly to the NR3C1 protein, glucocorticoid receptor (GR). GR commonly forms a complex with other proteins including p23, HSP90, and FKBP5. When the FKBP5 protein is present in the complex, GC binding affinity is reduced. However, when GC binding does occur, FKBP5 is removed from the complex in exchange for FKBP4 and Dynein. These proteins facilitate the chaperone of the GC:GR complex across the nuclear membrane of the cell. Once inside the nucleus of the cell the complex interacts with glucocorticoid response elements (GREs) which function to regulate the expression of numerous genes, having wide reaching biological effects. When the complex binds GREs in the FKBP5 gene, the transcription and translation of FKBP5 is increased. This increase in FKBP5 results in reduced GC sensitivity, and the ultra short-range feedback loop is complete. Figure adapted from Psychoneuroendocrinology, Volume 34, E.B. Binder, The role of FKBP5: a co-chaperone of the glucocorticoid receptor in the pathogenesis and therapy of affective and anxiety disorders, S186–S195, 2009[81], with permission from Elsevier.
To understand this vital topic, researchers have focused on an area of molecular biology known as epigenetics. There are three well-studied epigenetic mechanisms by which traumatic stressors can biologically embed themselves and therefore “get under the skin”, and evidence exists for trauma-induced alterations for each of these mechanisms. First, DNA methylation (5mC) is the biochemical process by which a methyl group is added to the 5′-end of a cytosine residue, most often occurring when the cytosine residue is adjacent to a guanine residue (CpG) on a strand of DNA. Canonically, these additional chemical groups can act to sterically exclude the binding of the transcriptional machinery to the DNA, thereby altering the transcription of the respective genic region[6]. Second are post-translation histone tail modifications. Through the acetylation, methylation, phosphorylation, or other chemical modifications to the amino acid tails of histones, which are the protein octamers around which DNA is tightly wrapped during phases of transcriptional silence, the availability of regions of DNA for transcription is altered[7]. The final epigenetic mechanism of discussion by which traumatic stress can embed biologically is through small non-coding RNA (sncRNA) and long non-coding RNA (lncRNA) molecules. In brief, these molecules generally function by binding to mRNA that has already been transcribed, either enhancing or limiting their ability to interact with proteins or to be translated into proteins themselves. These snc/lncRNA molecules can also form RNA:protein complexes which then bind to unwound DNA. This binding can facilitate the recruitment of proteins responsible for applying more epigenetic marks to both the DNA itself and the amino acid histone tails nearby[8].
Although this list of epigenetic mechanisms is not comprehensive, it does represent the most frequently studied and salient mechanisms with respect to traumatic stress. Taken together, these mechanisms alter the expression of genes[9], functionally alter downstream biological processes[10], and associate with numerous traumatic stress-related psychopathologies[11]. Based on the involvement of the brain, the HPA-axis, and the immune system in the response to traumatic stress, the current review highlights recent research in the field of traumatic stress epigenetics, and offers a neuro- immuno- endocrine framework of organization by which the research will be presented, as well as an overview of transgenerational transmission mechanisms.
Importantly, the field has increasingly recognized that genome-wide association studies (GWAS) require large cohort sample sizes to combat multiple hypothesis testing error rates (type I error rates), and also to more completely grasp variability within measurements. This concept holds true for most genome-scale analyses, where statistical adjustments of type I error rates are prevalent. These same concepts apply somewhat to candidate gene analyses, albeit to a lesser degree, as such analyses typically adjust for far fewer tests than GWAS studies (i.e. a few SNPs or CpG sites within a candidate gene vs. hundreds of thousands of SNPs/sites or more). Overall, test-statistic thresholds for genome-scale work are typically much higher than in candidate gene work.
Implicated Biological Systems
HPA-Axis
The HPA-axis plays a pivotal role in the body’s response to both typical and elevated levels of stress. For this reason, genes belonging to the HPA-axis pathway have been the focus of recent and early investigations in the field of traumatic stress epigenetics (Table 1), attempting to associate epigenetic marks in either the periphery or central nervous system (CNS) with traumatic event exposure, PTSD status, or PTSD symptom severity. Additionally, attempts to understand risk vs. resilience in the context of HPA-axis genes have been made. Two proteins playing pivotal roles within the HPA-axis are translated from the FKBP5 and NR3C1 transcripts.
Table 1.
Key genes and biological factors playing salient roles in traumatic stress epigenetics (in order of appearance).
| Full name of Gene/Biological Factor | HUGO Gene Symbol | Function According to Genecards[84] | Studies discussed | Primary Biological System |
|---|---|---|---|---|
| FK506 Binding Protein 5 | FKBP5 | Partially inhibits the binding affinity of cortisol to the NR3C1 protein (glucocorticoid receptor). Once cortisol is bound, FKBP5 is replaced by FKBP4 and Dynein proteins, aiding the translocation of the complex across the nuclear membrane into the nucleus. |
12, 13, 58, 64, 70, 74 | HPA-Axis |
| Nuclear Receptor Subfamily 3 Group C Member 1 | NR3C1 | The glucocorticoid receptor (GR) is bound by cortisol. Binding affinity of cortisol to GR is modulated by the FKBP5 protein. Once cortisol is bound, FKBP5 is replaced by FKBP4 and Dynein proteins, aiding the translocation of the complex across the nuclear membrane into the nucleus. | 13, 14, 15, 17, 52, 53, 69, 70, 72, 73 | HPA-Axis |
| MicroRNA 124-3 | miR124-3 | Post-transcriptional regulation of gene expression. Shown to regulate NR3C1 (GR) expression. | 16 | HPA-Axis |
| Interleukin 1, Hematopoietin-1 | IL1 | Includes both IL1A and IL1B. These protein products are cytokines which signal to other factors involved in immune response, inflammation response, and hematopoesis. | 22 | Immune System |
| Interleukin 6, B-Cell Stimulatory Factor 2 | IL6 | The protein product of the IL6 gene functions as a cytokine, communicating with B cells to signal their maturation, and facilitating the inflammatory response. | 22 | Immune System |
| Interleukin 10, Cytokine Synthesis Inhibitory Factor | IL10 | This gene encodes a cytokine which functions to downregulate T helper cell cytokines, MHC class 2 agonists, and costimulatory macrophage factors. | 22 | Immune System |
| Tumor Necrosis Factor | TNF | The protein product of the TNF gene is secreted by macrophages, and function in numerous ways as a proinflammatory cytokine. | 22 | Immune System |
| C-X-C Motif Chemokine Ligand 8, Interleukin 8 | CXCL8 | The CXCL8 protein product is a chemokine, mediating the response to inflammation, and attracting neutrophils, basophils, and T cells. | 31 | Immune System |
| Interleukin 6, Lymphocyte Chemoattractant Factor | IL16 | The IL16 gene protein product facilitates migration of lymphocytes, and modulates T cell activation. | 31 | Immune System |
| Interleukin 18, Interferon-Gamma-Inducing Factor | IL18 | The protein product of this gene stimulates helper T cell and NK cell activity, acting as a proinflammatory cytokine. | 31 | Immune System |
| Insulin-like Growth Factor 2 | IGF2 | An imprinted gene, involved in development and growth, expressed only by the paternal allele. | 31, 69 | Development and Growth |
| H19, Imprinted Maternally Expressed Transcript (Non-Protein Coding) | H19 | An imprinted gene, producing a tumor-suppressing long non-coding RNA, expressed only by the maternal allele, located near IGF2. | 31 | Epigenetic Regulation |
| Interferon Gamma | IFNG | The protein product of this gene is a soluble cytokine which acts to trigger cellular responses to viral/microbial infections, stimulating NK cells. | 32, 33 | Immune System |
| Interleukin 12B | IL12B | The IL12B protein product is a cytokine that interacts with T cells and NK cells, and is suggested to play a role in multiple sclerosis. | 32, 33 | Immune System |
| Interleukin 2, T Cell Growth Factor | IL2 | The IL2 protein is a cytokine which signals T and B cell proliferation. | 35 | Immune System |
| Catechol-O-Methyltransferase | COMT | The COMT gene encodes an enzyme responsible for the catabolism of catecholamines such as dopamine (DA) and norephinephrine (NE). | 39 | CNS |
| Brain Derived Neurotrophic Factor | BDNF | The BDNF protein product encodes a nerve growth factor, promoting neuronal survival in the brain and in the periphery. | 40, 62, 69 | CNS |
| 5-Hydroxytryptamine Receptor 3A | HTR3A | The HTR3A protein product is subunit 3 of a ligand-gated ion channel, residing on the post-synaptic terminal of serotonergic synapses, playing a role in synaptic transmission. | 41, 63 | CNS |
| Dedicator Of Cytokinesis 2 | DOCK2 | The DOCK2 protein product plays a role in facilitating lymphocyte migration, as it functions to remodel actin cytoskeletons. | 47 | Immune System |
| Spindle And Kinetochore Associated Complex Subunit 2 | SKA2 | SKA2 protein interacts with the GR, NR3C1, playing a pivotal role in the HPA-axis and stress reactivity pathways. | 54, 55, 56, 57 | HPA-Axis |
| Double-Stranded RNA-Specific Endoribonuclease | DICER1 | Encodes an enzyme responsible for the development of mature microRNAs. | 65 | Epigenetic Regulation |
| MicroRNA 3130-5p (MicroRNA 3130-1) | miR-3130-5p (miR-3130-1) | A mature miRNA sequence, which partly makes up the miR-3130-1 stem loop sequence, which targets numerous mRNAs, including MRPL35 mRNA. Its dysregulation has been implicated in cancer as well as mental health disorders. | 65 | Epigenetic Regulation |
| Mitochondrial Ribosomal Protein L35 | MRPL35 | The protein product of this gene encodes the large 39S subunit protein of the mammalian mitochondrial ribosome. | 65 | Protein Translation |
| Solute Carrier Family 6 Member 4 | SLC6A4 | The SLC6A4 protein functions as a reuptake transporter of serotonin from the synaptic cleft into presynaptic neurons. | 60, 61 | CNS |
In a seminal study investigating epigenetic mechanisms mediating the relationship between childhood trauma exposure (CTE), and FKBP5 genotype, Klengel et al 2013[12] measured 5mC of the FKBP5 promoter region, intron 2, 5, and 7, as derived from peripheral blood cells. Researchers found significant associations between 5mC at discrete CpG sites, CTE, and FKBP5 genotype. The three CpG sites of interest fell within intron 7, and were located adjacent to a genomic region sensitive to glucocorticoid (GC) regulation, suggesting a functionally relevant, long-lasting epigenetic effect of CTE, potentially originating in HPA-axis dysfunction. Additionally, expression of 76 transcripts was correlated with plasma cortisol levels, with the strongest associations present in FKBP5 protective allele carriers. To lend functional weight to their human results, researchers used in-vitro hippocampal cell culture to show that exposure to dexamethasone (DEX), a synthetic GC, decreased 5mC at the same sites as observed in blood. Importantly, differential 5mC findings in peripheral blood were also represented in their CNS tissue model, establishing that alterations in the periphery can be indicative of epigenetic CNS dysregulation. These results set the tone for the field in the coming years to study HPA-axis dysregulation in relation to traumatic stress and epigenetic variation in both the periphery and CNS.
Additional and notable work investigating traumatic stress-related epigenetic variation in HPA-axis genes includes a psychotherapy-focused investigation by Yehuda and colleagues (2013)[13]. In this study, investigators measured FKBP5 and NR3C1 promoter 5mC in lymphocytes as predictors of psychotherapy outcome in combat veterans with PTSD. Results showed that increased NR3C1 exon 1F promoter 5mC measured pre-treatment did predict positive treatment outcome, although 5mC itself was not altered between pre- and post- treatment. On the other hand, FKBP5 exon 1 promoter pre-treatment 5mC did not predict treatment outcome, but did decrease among those who responded to therapy. Amongst treatment-responders, FKBP5 mRNA expression was elevated, compared to non-responders. Although implications from these results are limited by small sample size, results build upon earlier cross-sectional work suggesting that HPA-axis genes are subject to epigenetic alteration, and provide insight into how psychotherapy alters epigenetic and transcriptional states. Labonte et al 2014[14] showed similar results. Their study measured NR3C1 5mC as collected in peripheral T lymphocytes, and showed that subjects with lifetime PTSD diagnoses had lower morning cortisol release, increased mRNA expression of NR3C1 total transcripts, 1B and 1C transcripts, and also had decreased 5mC of the 1B and 1C promoter regions. Results reinforce the notion that trauma exposure-related psychopathology is associated with epigenetic alteration of the NR3C1 gene, with functional transcription and neuroendocrine alterations occurring as well.
With a larger sample size compared to earlier work[13], Yehuda and colleagues (2015)[15] found decreased NR3C1 exon 1F promoter 5mC, as measured in peripheral blood mononuclear cells (PBMCs), in combat veterans with PTSD compared to combat veterans without PTSD. 5mC at this locus was also associated with three measures of functional GC activity. Results highlight that decreased NR3C1 5mC could either confer risk of PTSD development or be a result of PTSD development, and that epigenetic alteration of NR3C1 could functionally alter neuroendocrine system outputs.
Although 5mC change appears to be a salient factor related to the traumatic stress response, other epigenetic alterations have also been documented. For example, childhood maltreatment (CM) was associated with differential 5mC in a site proximal to miR124-3, which is a regulator of NR3C1 mRNA transcription[16]. Results indicate that different types of traumatic stress are capable of epigenetic reprogramming of genes that themselves encode epigenetic regulatory factors.
Although the previous studies demonstrate the importance of NR3C1 and other HPA-axis genes in traumatic stress, one important shortcoming is their overwhelmingly cross-sectional nature, which hampers causal inference. Recent efforts have been made to circumvent these shortcomings by utilizing longitudinal sampling. One such study sampled whole blood of veterans before and after combat exposure. From both time points, they measured NR3C1 5mC in the 1F exon, as well as NR3C1 mRNA. Importantly, they found that both trauma exposure during combat and development of mental health problems 6 months after deployment were associated with an increase in 5mC, including sites showing correlation between 5mC and expression, over that time period. Importantly, they found that pre-deployment NR3C1 exon 1F 5mC did not predict post-deployment psychopathologies[17]. This is the first study to present prospective evidence of increases in NR3C1 5mC in response to trauma exposure, and results point towards the importance of considering functional 5mC in conjunction with mRNA expression of the associated gene.
Immune System
The HPA-axis plays a key role in modulating the immune system, and inflammatory processes specifically[18,19]. GCs released by the adrenal cortices play an anti-inflammatory role[20], and dictate immune cell trafficking, maturation, and differentiation[21], while controlling expression of signaling cytokines such as IL-1, IL-6, IL-10, and TNF-alpha[22]. These cytokines, in turn, allow the immune system to regulate the CNS[23]. Traumatic stressors have profound effects on the immune and specifically the inflammatory responses throughout the lifetime; these altered inflammatory states confer risk of psychopathologies[24,25,26]. Molecular mechanisms by which these alterations are conferred are not well understood; recent research in the field of traumatic stress epigenetics has investigated this question.
Mehta et al 2013[27] utilized genome-scale approaches to investigate 5mC and mRNA expression in peripheral blood cells of trauma-exposed subjects who had lifetime history of PTSD versus trauma-exposed no PTSD subjects. Of those with lifetime PTSD, they also investigated differences between subjects who had experienced CM versus those who had not. Differential enrichment at the mRNA transcript and network levels among the three groups revealed largely non-overlapping results, with gene expression changes that were mediated most strongly by 5mC changes in the CM group; however, overlapping results showed enrichment of cell survival, development, migration, and adhesion networks, T-cell activation, and immune networks. Results indicate distinct peripheral epigenetic effects of traumatic stress brought on by CM, in particular within gene networks involved in immune regulation. Epigenetic dysregulation in immune system genes has been strongly implicated in earlier genome-scale studies of traumatic stress[28,29], including one study examining the emergence of PTSD-associated CpG sites during human evolution[30].
Bolstering the implication of epigenetic dysregulation of immune system genes, Rusiecki et al 2013[31] investigated promoter region 5mC of, interleukin (IL)-8 (IL8), IL16, and IL18 genes in serum, as well as the insulin-like growth factor 2 (IGF2), long non-coding RNA transcript H19 (H19). Using a longitudinal design, they found differences between pre- and post deployment 5mC at H19 and IL18 loci: non-PTSD controls showed decreased 5mC at both H19 and IL18, whereas PTSD subjects only showed increased IL18 5mC. Moreover, pre-deployment 5mC levels in IL8 were lower among those who later became cases post-deployment. Results show that both pre-existing epigenetic states and changes pre-to post-trauma can identify individuals who are susceptible vs. resilient to traumatic stress.
Departing from 5mC as the main epigenetic mechanism of interest, Zhou et al 2014[32] showed that in PTSD patients, miRNA expression was significantly dysregulated in transcripts targeting immune system signaling pathways, as measured in PBMCs. Importantly, these changes were associated with differential plasma levels of IFN-γ and IL-17, showing the functional effects of miRNA dysregulation associated with PTSD. More recent research from the same team has shown complex interactions amongst epigenetic mechanisms regulating immune system functions. Bam et al 2016[33] investigated genome-wide histone H3 tri-methylation, 5mC, and miRNA expression from PBMCs in veterans with PTSD vs. non-trauma exposed controls. Results showed differential histone H3 tri-methylation at four sites, differential 5mC at promoters of IL-12B and interferon gamma (IFNG), and elevated IL-12 mRNA transcript levels. They also showed that miRNA regulation of IFNG and IL-12 was significantly decreased in PTSD patients. In a novel follow up study from the same lab, Bam et al 2016[34] combined genome-wide RNAseq and miRNA data in PBMCs, albeit with a small sample size. They showed significant negative correlations between mRNAs and miRNAs playing salient roles specific to immune system functions including cytokine-based inflammation, CCKR, and interleukin signaling, among others.
Further implicating immune system dysregulation in traumatized populations, Powers et al 2016[35] conducted a large GWAS of emotional regulation in a highly traumatized, minority, urban sample. In males, they found a SNP located in the IL-2A gene (rs6602398) that was associated with emotional dysregulation, PTSD, MDD, and differential 5mC as measured in whole blood. Their results point towards sex differences and the role of genotype in trauma-exposure related psychopathologies, as well as more complex endophenotypes relevant to PTSD, such as emotion regulation.
Overall, epigenetic regulation of the immune system in response to trauma exposure appears to play a key role in indexing risk vs. resilience in relation to the development of PTSD. Numerous epigenetic mechanisms have been implicated in peripheral tissues including miRNA, histone modifications, and 5mC; numerous genes, transcripts, and proteins have also been implicated. Results suggest a complex interaction of the HPA-axis, and immune systems with many molecular factors mediating these relationships.
Central Nervous System
The CNS plays a key role in the mediation of both HPA and immune reactions to stress[36], and epigenetic regulation of this mediation occurs at the CNS level as well[37]. Although it is challenging to make inferences regarding the contribution of epigenetic variation in CNS-related genes to CNS function when measured in blood, such measures can be used as potential biomarkers of risk and resilience to traumatic stress, as interactions among the CNS, HPA-axis and immune system may produce epigenetic alterations in factors circulating in peripheral tissues[38].
In the earliest study of its kind, Norrholm and colleagues (2013)[39] investigated the epigenetic and genetic regulation of the COMT gene in relation to an endophenotype of PTSD, the fear potentiated startle response. As measured in whole blood, they found that subjects with the PTSD risk-associated MET/MET genotype (rs4680) showed increased fear-potentiated startle response compared to VAL/MET or VAL/VAL carriers. Importantly, MET/MET genotype was also associated with 5mC of four CpG sites in the COMT promoter region, two of which were associated with the increased fear-potentiated startle response. This study highlights how an endophenotype of PTSD associates with epigenetic variation as measured in the blood, with a mediating effect of genotype.
In a more recent study, Kim et al 2017[40] investigated the effect of distant trauma exposure and PTSD status in Vietnam War veterans. They measured BDNF promoter 5mC from whole blood, finding that subjects with PTSD had increased promoter 5mC compared to those with trauma-exposure but without PTSD. Results indicated that peripheral BDNF promoter 5mC could confer risk of development of PTSD. Perroud et al 2016[41] investigated the role of CM in relation to 5mC of the serotonin 3A receptor gene (5HTR3A) as well as genotype, outside of the context of PTSD. By measuring promoter 5mC in whole blood and assaying genotype of the SNP rs1062612, they found that CM was associated with: symptom severity of ADHD, borderline personality, and bipolar disorder, as well as differential 5mC of two promoter region sites, with genotype significantly mediating the relationships. Results show that peripheral 5HTR3A promoter 5mC is a biomarker of psychopathological symptom severity, and, similar to the COMT results reporter by Norrholm and colleagues (2013), that genotype plays a salient role in mediating the relationship.
Investigating a cohort in which all subjects experienced the same traumatic event, Kuan et al 2017[42] conducted a large-scale epigenome-wide association study (EWAS) on World Trade Center disaster first responders, investigating genome-wide 5mC and both PTSD and MDD in whole blood. All subjects were trauma-exposed; analyses were carried out comparing lifetime PTSD vs. never and lifetime MDD vs. never. No single CpG site was significantly differentially methylated; however, pathway analyses of nominally differentially methylated sites associated with PTSD implicated synaptic plasticity, oxytocin signaling, cholinergic synapse and inflammatory disease pathways as being dysregulated in this disorder.
Although recent research in the field of traumatic stress epigenetics has focused on the inter-related HPA-axis, CNS, and immune systems, evidence exists for epigenetic dysregulation of genes and pathways outside of the aforementioned systems. Common findings point to association traumatic stress exposure with dysregulation of 5mC aging mechanisms[43], and epigenetic alterations to genes encoding epigenetic machinery itself[44,45,46]. A recent EWAS study also implicates a gene thought to play a role in Alzheimer’s pathology (DOCK2)[47]. Results from these recent epigenetics studies focus on genes mediating synaptic transmission, as measured in the periphery, although inherent limitations to this approach exist.
Imaging Epigenetics
A common shortfall of current epigenetic work related to traumatic stress in humans is the inability to measure epigenetic markers in the living human brain. The majority of studies reviewed here report epigenetic measurements derived from peripheral tissues such as saliva or blood. Studies based on peripheral tissue inherently limit the conclusions that can be drawn in relation to CNS function, as it remains unknown to what extent epigenetic markers taken outside the CNS functionally represent concurrent biological processes or dysregulation occurring in the brain. More comprehensive coverage of limitations is discussed elsewhere[48]. To this end, the most common methods to circumvent this peripheral problem include testing animal models of stress/trauma and mental health disorders, and by assaying epigenetic markers in human post-mortem brain tissue. These methodologies have both strengths and limitations, including confounding of biological measurements by post-mortem interval as a major limitation, as has been previously reviewed[49,50]. Interestingly, the current review could not identify recent published work performed using postmortem, brain-based epigenetic measurements from individuals with lifetime trauma exposure. Another recently employed method that helps circumvent the peripheral problem is the use of neuroimaging techniques to capture neural endophenotypes, especially neuroanatomical morphology (structural MRI) and/or functional reactions to stress or emotional paradigms (functional MRI). In pairing this information with epigenetic data, researchers can index functional biological endophenotypes in the brain with easily accessible peripheral tissue measures of epigenetic marks. Recent work in traumatic stress epigenetics has adopted this approach.
Traumatic stress has a well-documented impact on the HPA-axis, and also has a salient impact on memory and emotional regulation, as the amygdala (AMYG), prefrontal-cortex (PFC), and hippocampus (HIPP) are neurally integrated within the fronto-limbic pathway (Table 2)[51]. On this note, Vukojevic et al 2014[52] found that in Rwandan genocide survivors, increased NR3C1 NGFI-A (nerve growth factor-induced protein A) binding site 5mC, as measured in saliva, was associated with less intrusive traumatic thoughts, and a reduced risk of PTSD symptoms in males. Using fMRI, researchers found that increased NR3C1 5mC was associated with reduction in a memory task performance in men, but not women. Results warrant increased consideration of sex differences, and that epigenetic alterations of NR3C1 can impact particular symptom domains of strong relevance to PTSD. Other labs have also associated differential NR3C1 5mC with trauma-related fMRI endophenotypes in the mPFC[53].
Table 2.
Key brain regions playing salient roles in traumatic stress epigenetics (in order of appearance).
| Full name (brain region) | Abbreviation | Function | Studies discussed | Primary Biological System |
|---|---|---|---|---|
| Amygdala | AMYG | Processing of intense positive and negative emotional stimuli, as well as odor processing[82]. | 58, 65 | CNS |
| Pre-Frontal Cortex | PFC | Includes vmPFC, mPFC, dlPFC, and frontal pole. Chiefly, executive control, emotional regulation, and working memory[82]. | 53, 54, 62, 63 | CNS |
| Hippocampus | HIPP | Learning and memory[82]. | 52, 58, 60 | CNS |
| Orbito-Frontal Cortex | OBFC | Reward expectation, response inhibition, delayed responses, affective forecasting. Social and emotional behavior[82]. | 54, 58 | CNS |
| Anterior Cingulate Cortex | ACC | Emotional regulation, error detection, inhibitory control, empathy, decision making[83]. | 62 | CNS |
In a recent study of trauma-exposed veterans, Sadeh and colleagues (2016)[54] investigated 5mC of SKA2 in whole blood. Differential 5mC methylation of SKA2 predicts suicidal behavior and PTSD status[55], the development of PTSD after military deployment[56], as well as aberrant internalizing psychopathology[57]. In the current study, investigators found that an increase of SKA2 5mC at cg13989295, located in the fourth exon, was associated with decreased PFC thickness bilaterally at the frontal pole and superior frontal gyrus, and in the right hemisphere at the orbitofrontal cortex (OBFC) and inferior frontal gyrus. Additionally, PTSD symptom severity was positively correlated with SKA2 5mC, and negatively correlated with PFC thickness[54]. Results from this study, and others, show that SKA2 methylation can be considered a peripheral biomarker of both PTSD symptom severity and a PFC thinning endophenotype in trauma-exposed war veterans. Further research on peripheral measures of HPA-axis genes show that FKBP5 mRNA in PTSD subjects is significantly lowered, and that this is associated with reduced HIPP, medial OBFC, but not AMYG volumes as measures by structural MRI[58]. Based on this recent research, a relationship between epigenetic dysregulation of HPA-axis genes and dysregulation of fronto-limbic neural pathways is evident.
Although much work has been done to investigate the role of HPA-axis genes within the field of imaging epigenetics, a focus on CNS-relevant genes is also represented in this literature. Building on earlier seminal work that used positron emission tomography (PET) imaging approaches to directly assess in vivo serotonin levels in the CNS[59], Booji and colleagues (2015)[60] showed SLC6A4 promoter 5mC of the sites mentioned above were significantly associated with CTE, as well as reduced HIPP volume (using structural MRI), once again providing evidence that the biological embedding of traumatic stress can have significant neural impacts, and than these neural endophenotypes can be indexed by peripheral measures of 5mC. In a similar vein, Frodl et al 2015[61] found that CM was associated with increased promoter SLC6A4 5mC levels as measured in blood, also showing increased activation in the left anterior insular cortex, and left frontal inferior operculum in response to negative emotional stimuli. Other labs have also associated differential BDNF and HTR3A 5mC with trauma-related fMRI endophenotypes in the ACC and vmPFC[62] and mPFC[63]. Overall, recent research implicates a relationship between trauma exposure and epigenetic dysregulation of the CNS serotonergic system, as well as brain regions important for emotional regulation.
Although most recent imaging epigenetics studies focus on HPA-axis and CNS candidate genes, recent work from the Grady Trauma Project[64] investigated the role of DICER1. Researchers found that decreased DICER1 expression, measured in blood, was associated with increased AMYG activity, as measured by fMRI in a fearful stimuli paradigm. Results suggest trauma-exposure is associated with AMYG over-reactivity to stressful stimuli, and that this neural phenotype can be indexed as measured by DICER1 mRNA in the blood. Wingo and colleagues further showed that in trauma-exposed cases with PTSD/MDD, expression of miRNA miR-3130-5p was down-regulated in comparison to trauma-exposed controls in discovery and replication cohorts, and this down-regulation was associated with up-regulation of one of its mRNA targets, MRPL35[65]. Results from this study lend evidence of epigenetic regulation in relevant mRNA transcripts associating with neural activity implicated in PTSD, and also point to mechanisms mediating the development of psychopathology in groups that have been exposed to trauma.
The majority of studies presented above focus on candidate genes that have previously been implicated in traumatic stress reactivity. In a novel study carried out using Iraq/Afghanistan war veterans, two separate models of DNA methylation aging were compared[66,67]. These models use 5mC measures from whole blood to index chronological age. It was found that PTSD symptom severity throughout the lifetime was associated with Hannum method 5mC age estimates, and these age estimates were associated with reduced white matter fiber tract integrity in the genu of the corpus callosum, as measured by diffusion tensor MRI[68]. Results point to the possibility that traumatic stress exposure from warzone combat has significant effects on neural integrity, and that this effect can be indexed in the periphery through Hannum 5mC age.
Overall, recent research has utilized fMRI, structural MRI, diffusion tensor imaging and PET methods to identify relationships between trauma-exposure, PTSD status and epigenetic dysregulation of HPA-axis, CNS, epigenetic machinery, and CpGm age-related genes.
Transgenerational epigenetic effects of traumatic stress
Recent work has provided evidence that prenatal exposures may also shape differential risk for/resilience to traumatic stress and leave imprints of trauma exposure that persist postnatally. The majority of these transgenerational studies show a correlation between parental PTSD and its impact on their offspring. Although the symptoms of transgenerational transmission among children are various and different factors appear to play a role in the transgenerational transmission of PTSD, recent studies show evidence of biological and epigenetic effects[69–74].
As with many of the studies reviewed earlier, this transgenerational work has focused largely on genes belonging to the HPA-axis, due to their role in mediating the stress response both centrally and peripherally. In the earliest work, Mulligan and colleagues[69] investigated pregnant women and their resulting offspring residing in the Eastern Democratic Republic of Congo (DRC) to assess the impact of exposure to financial, psychosocial, and war stress on 5mC of NR3C1. Of these three stress-related domains, war stress showed the strongest, positive correlation with NR3C1 5mC when measured in cord blood, but showed no association with 5mC when measured in maternal venous blood. This work was followed by a broader investigation into additional HPA-axis genes assessed in both blood and placental tissues in the same mother-newborn dyads[70]. War-related trauma was moderately associated with 5mC in three of the four genes examined (CRH, NR3C1, FKBP5). As in the earlier work[69], higher NR3C1 cord blood 5mC was associated with lower birth weight, a pattern that was also newly identified in this study for 5mC in CRH. Collectively, these results suggest that prenatal stress exposure has broad impact on epigenetic regulation of HPA-axis genes, in a manner that impacts offspring phenotype. Of note, the DRC-focused study has also examined genes outside the HPA-axis, including BDNF, IGF1, IGF2, and loci involved in maintaining or removing DNA methylation marks[70,71].
In contrast to the earliest NR3C1-focused work in Eastern DRC[69], work investigating the transgenerational effects of exposure to the Rwandan genocide identified increased 5mC in NR3C1 in both exposed mothers and their offspring who were exposed in utero–although these effects were more pronounced in the exposed offspring than the exposed mothers[72]. The divergent findings regarding 5mC of NR3C1 in exposed mothers in these two studies may be due to severity of traumatic exposures and/or analytic methods, both of which varied between studies; of note, in the subsequent study by Mulligan and colleagues[70], war stress was associated with increased 5mC in this locus in maternal venous blood. Importantly, in the Rwandan study, the 5mC-based findings were accompanied by results showing reduced cortisol levels in genocide-exposed mothers and offspring, as well as reduced plasma glucocorticoid receptor and increased mineralocorticoid receptor levels[72], suggesting broad and sustained alterations in stress biology among those exposed to the Rwandan genocide that were detectable nearly 20 years after exposure.
Additional insight into the lasting impact of genocide exposure was provided by Yehuda and colleagues, who investigated 5mC of NR3C1 in Holocaust survivors in relation to PTSD. A complex pattern emerged in which parental PTSD was shown to associate with differential, blood-derived 5mC in Holocaust survivor offspring, but in a manner that differed depending on which parent was affected[73]. Similar to the Rwandan study, Yehuda and colleagues demonstrated functional effects of this differential 5mC by showing that lower NR3C1 5mC was associated with greater cortisol suppression following dexamethasone administration. Although the direction of 5mC effects differs with that identified in the Rwandan study, which did not specifically examine the impact of PTSD in association with NR3C1, this study provides further evidence of the lasting epigenetic impact of exposure to genocide, as many of the Holocaust survivor offspring were conceived after parental Holocaust exposure[73]. Subsequent work by the same research group showed that Holocaust exposure was associated with differential 5mC in FKBP5 in both exposed parents and offspring, when compared to 5mC levels observed in demographically matched parent and offspring control groups[74]. Importantly, these effects were observed when controlling for PTSD, suggesting that genocide exposure itself may leave lasting imprints on the epigenome of exposed individuals and their offspring, even when the exposure occurs pre-conception.
The studies described above have all focused on the epigenetic impact of traumatic stress exposure between one generation and the next, with the bulk of studies being performed on such exposures occurring during gestation. An outstanding question is to what extent such epigenetic effects might be observed across multiple generations. The first report to partially address this question in humans examined the association between grandmaternal exposure to intimate partner and community and domestic violence during pregnancy, and differential 5mC in grandchildren[75]. This EWAS investigation, which was conducted in DNA derived from saliva, identified CpG sites in 5 genes that showed differential 5mC in association with exposure to community and domestic violence. Although none of these genes fell into the pathways typically investigated in such studies, such as the HPA-axis, immune or inflammatory pathways, one of the genes, CFTR, was associated with PTSD and depression symptoms among children in the study, and had been previously identified as showing differential 5mC among institutionalized adolescents with a history of adversity in a separate study[76]. While findings from this work await replication, and do not address the question of concordant/discordant differential 5mC patterns in the maternal generation, or within the grandmaternal generation, this study represents an important step towards assessing epigenetic effects of traumatic exposure that span multiple generations in humans. Further investigations would clarify the pathways through which this transgenerational transmission of PTSD affects younger generations on a large scale.
Conclusions
The role of epigenetic mechanisms in relation to traumatic stress exposure has been carefully investigated in recent years, and, as summarized in this brief review, much of this work has focused on dysregulation of genes involved in the HPA-axis, CNS, and immune systems. The vast majority of these have relied on peripheral tissues such as blood and saliva; thus, in many instances, it remains unknown whether epigenetic marks as measured in the periphery are indicative of what is occurring in etiological tissues such as the CNS. To address this, the field has recently focused on combining neuroimaging information with epigenetic data to index CNS activity with peripheral measures of epigenetic markers. Neural activity in brain regions responsible for emotional regulation/processing and memory has been characterized in relation to traumatic stress exposure and related psychopathologies. More studies of this nature should be carried out, and measures of epigenetic marks in the CNS should also be a focus. The latter will be facilitated in the coming years with the development of PTSD brain banks, such as the National Posttraumatic Stress Disorder Brain Bank and the PTSD collection being developed at McLean Hospital in partnership with Cohen Veterans Bioscience. In addition, emerging work has begun to identify epigenetic associations with traumatic exposures that persist beyond one generation. The establishment and/or testing of data from birth cohorts that span multiple generations, such as the Avon Longitudinal Study of Parents and Children[77] and the Drakenstein Child Health Study[78], will greatly enhance the ability to identify replicable epigenetic effects of traumatic exposures that persist across generations.
In all, the field has taken great steps in elucidating the role of epigenetic mechanisms regulating the HPA-axis, CNS, and immune responses to traumatic stress. With continued advances in molecular biology techniques, bioinformatics methods, and study design the picture is sure to become clearer.
Acknowledgments
This work was supported by NIH Grants R01MH108826 and U01MH115485.
References
- 1.Kilpatrick DG, Resnick HS, Milanak ME, Miller MW, Keyes KM, Friedman MJ. National estimates of exposure to traumatic events and PTSD prevalence using DSM-IV and DSM-5 criteria. Journal of traumatic stress. 2013;26(5):537–547. doi: 10.1002/jts.21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.American Psychiatric Association. Diagnostic and statistical manual of mental disorders: DSM-5. Washington, DC: American Psychiatric Association; 2013. [Google Scholar]
- 3.Christopher M. A broader view of trauma: A biopsychosocial-evolutionary view of the role of the traumatic stress response in the emergence of pathology and/or growth. Clinical psychology review. 2004;24(1):75–98. doi: 10.1016/j.cpr.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Perry BD, Pollard RA, Blakley TL, Baker WL, Vigilante D. Childhood trauma, the neurobiology of adaptation, and use dependent development of the brain: How states become traits. Infant mental health journal. 1995;16(4):271–291. [Google Scholar]
- 5.Breslau N, Davis GC, Peterson EL, Schultz LR. A second look at comorbidity in victims of trauma: the posttraumatic stress disorder–major depression connection. Biological psychiatry. 2000;48(9):902–909. doi: 10.1016/s0006-3223(00)00933-1. [DOI] [PubMed] [Google Scholar]
- 6.Bird A. DNA methylation patterns and epigenetic memory. Genes & development. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 7.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 8.Peschansky VJ, Wahlestedt C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics. 2014;9(1):3–12. doi: 10.4161/epi.27473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature genetics. 2003;33 doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 10.Michaud EJ, Van Vugt MJ, Bultman SJ, Sweet HO, Davisson MT, Woychik RP. Differential expression of a new dominant agouti allele (Aiapy) is correlated with methylation state and is influenced by parental lineage. Genes & development. 1994;8(12):1463–1472. doi: 10.1101/gad.8.12.1463. [DOI] [PubMed] [Google Scholar]
- 11.Dudley KJ, Li X, Kobor MS, Kippin TE, Bredy TW. Epigenetic mechanisms mediating vulnerability and resilience to psychiatric disorders. Neuroscience & Biobehavioral Reviews. 2011;35(7):1544–1551. doi: 10.1016/j.neubiorev.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 12•.Klengel T, et al. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat Neurosci. 2013;16:33–41. doi: 10.1038/nn.3275. A seminal piece of research demonstrating genotype-dependent demethylation of FKBP5 in adults exposed to childhood trauma. Blood-based DNA methylation measures are corroborated by hippocampal cell-derived DNA methylation measured from in vitro experiments following exposure to a synthetic cortisol analogue, indicating the likely systemic nature of these epigenetic effects. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yehuda R, Daskalakis NP, Desarnaud F, Makotkine I, Lehrner AL, Koch E, Bierer LM. Epigenetic biomarkers as predictors and correlates of symptom improvement following psychotherapy in combat veterans with PTSD. Frontiers in psychiatry. 2013;4 doi: 10.3389/fpsyt.2013.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Labonte B, Azoulay N, Yerko V, Turecki G, Brunet A. Epigenetic modulation of glucocorticoid receptors in posttraumatic stress disorder. Translational psychiatry. 2014;4(3):e368. doi: 10.1038/tp.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yehuda R, Flory JD, Bierer LM, Henn-Haase C, Lehrner A, Desarnaud F, Meaney MJ. Lower methylation of glucocorticoid receptor gene promoter 1 F in peripheral blood of veterans with posttraumatic stress disorder. Biological psychiatry. 2015;77(4):356–364. doi: 10.1016/j.biopsych.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Prados J, Stenz L, Courtet P, Prada P, Nicastro R, Adouan W, Perroud N. Borderline personality disorder and childhood maltreatment: A genome-wide methylation analysis. Genes, Brain and Behavior. 2015;14(2):177–188. doi: 10.1111/gbb.12197. [DOI] [PubMed] [Google Scholar]
- 17•.Schür RR, Boks MP, Rutten BPF, Daskalakis NP, De Nijs L, Van Zuiden M, Geuze E. Longitudinal changes in glucocorticoid receptor exon 1F methylation and psychopathology after military deployment. Translational psychiatry. 2017;7(7):e1181. doi: 10.1038/tp.2017.150. A unique longitudinal epigenetic study of a Dutch Military Cohort, demonstrating methylation changes in select NR3C1 CpG sites pre to post combat that were also associated with development of mental health disorders 6 months after deployment. Provides insight into dynamic epigenetic factors indicative of potential vulnerability to mental health problems following trauma exposure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marques-Deak A, Cizza G, Sternberg E. Brain-immune interactions and disease susceptibility. Molecular Psychiatry. 2005;10(3):239. doi: 10.1038/sj.mp.4001643. [DOI] [PubMed] [Google Scholar]
- 19.Otmishi P, Gordon J, El-Oshar S, Li H, Guardiola J, Saad M, Yu J. Neuroimmune interaction in inflammatory diseases. Clinical medicine. Circulatory, respiratory and pulmonary medicine. 2008;2:35. doi: 10.4137/ccrpm.s547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coutinho AE, Chapman KE. The anti-inflammatory and immunosuppressive effects of glucocorticoids, recent developments and mechanistic insights. Molecular and cellular endocrinology. 2011;335(1):2–13. doi: 10.1016/j.mce.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dhabhar FS. Stress-induced augmentation of immune function—the role of stress hormones, leukocyte trafficking, and cytokines. Brain, behavior, and immunity. 2002;16(6):785–798. doi: 10.1016/s0889-1591(02)00036-3. [DOI] [PubMed] [Google Scholar]
- 22.Liberman AC, Druker J, Perone MJ, Arzt E. Glucocorticoids in the regulation of transcription factors that control cytokine synthesis. Cytokine & growth factor reviews. 2007;18(1):45–56. doi: 10.1016/j.cytogfr.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 23.Deverman BE, Patterson PH. Cytokines and CNS development. Neuron. 2009;64(1):61–78. doi: 10.1016/j.neuron.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 24.Danese A, Pariante CM, Caspi A, Taylor A, Poulton R. Childhood maltreatment predicts adult inflammation in a life-course study. Proceedings of the National Academy of Sciences. 2007;104(4):1319–1324. doi: 10.1073/pnas.0610362104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Danese A, Moffitt TE, Pariante CM, Ambler A, Poulton R, Caspi A. Elevated inflammation levels in depressed adults with a history of childhood maltreatment. Archives of general psychiatry. 2008;65(4):409–415. doi: 10.1001/archpsyc.65.4.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller AH, Maletic V, Raison CL. Inflammation and its discontents: the role of cytokines in the pathophysiology of major depression. Biological psychiatry. 2009;65(9):732–741. doi: 10.1016/j.biopsych.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mehta D, Klengel T, Conneely KN, Smith AK, Altmann A, Pace TW, Bradley B. Childhood maltreatment is associated with distinct genomic and epigenetic profiles in posttraumatic stress disorder. Proceedings of the national academy of sciences. 2013;110(20):8302–8307. doi: 10.1073/pnas.1217750110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uddin M, Aiello AE, Wildman DE, Koenen KC, Pawelec G, de Los Santos R, Galea S. Epigenetic and immune function profiles associated with posttraumatic stress disorder. Proceedings of the National Academy of Sciences. 2010;107(20):9470–9475. doi: 10.1073/pnas.0910794107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith AK, Conneely KN, Kilaru V, Mercer KB, Weiss TE, Bradley B, Ressler KJ. Differential immune system DNA methylation and cytokine regulation in post-traumatic stress disorder. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2011;156(6):700–708. doi: 10.1002/ajmg.b.31212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sipahi L, Uddin M, Hou ZC, Aiello AE, Koenen KC, Galea S, Wildman DE. Ancient evolutionary origins of epigenetic regulation associated with posttraumatic stress disorder. Frontiers in human neuroscience. 2014;8 doi: 10.3389/fnhum.2014.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rusiecki JA, Byrne C, Galdzicki Z, Srikantan V, Chen L, Poulin M, Baccarelli A. PTSD and DNA methylation in select immune function gene promoter regions: a repeated measures case-control study of US military service members. Frontiers in Psychiatry. 2013;4 doi: 10.3389/fpsyt.2013.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou J, Nagarkatti P, Zhong Y, Ginsberg JP, Singh NP, Zhang J, Nagarkatti M. Dysregulation in microRNA expression is associated with alterations in immune functions in combat veterans with post-traumatic stress disorder. PLoS One. 2014;9(4):e94075. doi: 10.1371/journal.pone.0094075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bam M, Yang X, Zhou J, Ginsberg JP, Leyden Q, Nagarkatti PS, Nagarkatti M. Evidence for epigenetic regulation of pro-inflammatory cytokines, interleukin-12 and interferon gamma, in peripheral blood mononuclear cells from PTSD patients. Journal of Neuroimmune Pharmacology. 2016;11(1):168–181. doi: 10.1007/s11481-015-9643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bam M, Yang X, Zumbrun EE, Zhong Y, Zhou J, Ginsberg JP, Nagarkatti M. Dysregulated immune system networks in war veterans with PTSD is an outcome of altered miRNA expression and DNA methylation. Scientific reports. 2016;6 doi: 10.1038/srep31209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Powers A, Almli L, Smith A, Lori A, Leveille J, Ressler KJ, Bradley B. A genome-wide association study of emotion dysregulation: evidence for interleukin 2 receptor alpha. Journal of psychiatric research. 2016;83:195–202. doi: 10.1016/j.jpsychires.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sternberg EM. Neural regulation of innate immunity: a coordinated nonspecific host response to pathogens. Nature reviews. Immunology. 2006;6(4):318. doi: 10.1038/nri1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Meaney MJ. Epigenetic programming by maternal behavior. Nature neuroscience. 2004;7(8):847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 38.Andrews JA, Neises KD. Cells, biomarkers, and post-traumatic stress disorder: evidence for peripheral involvement in a central disease. Journal of neurochemistry. 2012;120(1):26–36. doi: 10.1111/j.1471-4159.2011.07545.x. [DOI] [PubMed] [Google Scholar]
- 39.Norrholm SD, Jovanovic T, Smith AK, Binder E, Klengel T, Conneely K, Gillespie CF. Differential genetic and epigenetic regulation of catechol-O-methyltransferase is associated with impaired fear inhibition in posttraumatic stress disorder. Frontiers in Behavioral Neuroscience. 2013;7 doi: 10.3389/fnbeh.2013.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim TY, Kim SJ, Chung HG, Choi JH, Kim SH, Kang JI. Epigenetic alterations of the BDNF gene in combat-related post-traumatic stress disorder. Acta Psychiatrica Scandinavica. 2017;135(2):170–179. doi: 10.1111/acps.12675. [DOI] [PubMed] [Google Scholar]
- 41.Perroud N, Zewdie S, Stenz L, Adouan W, Bavamian S, Prada P, et al. Methylation of serotonin receptor 3a in Adhd, borderline personality, and bipolar disorders: link with severity of the disorders and childhood maltreatment. Depression and anxiety. 2016;33(1):45–55. doi: 10.1002/da.22406. [DOI] [PubMed] [Google Scholar]
- 42.Kuan PF, Waszczuk MA, Kotov R, Marsit CJ, Guffanti G, Gonzalez A, Luft BJ. An epigenome-wide DNA methylation study of PTSD and depression in World Trade Center responders. Translational psychiatry. 2017;7(6):e1158. doi: 10.1038/tp.2017.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boks MP, van Mierlo HC, Rutten BP, Radstake TR, De Witte L, Geuze E, Vermetten E. Longitudinal changes of telomere length and epigenetic age related to traumatic stress and post-traumatic stress disorder. Psychoneuroendocrinology. 2015;51:506–512. doi: 10.1016/j.psyneuen.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 44.Sipahi L, Wildman DE, Aiello AE, Koenen KC, Galea S, Abbas A, Uddin M. Longitudinal epigenetic variation of DNA methyltransferase genes is associated with vulnerability to post-traumatic stress disorder. Psychological medicine. 2014;44(15):3165–3179. doi: 10.1017/S0033291714000968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maddox SA, Kilaru V, Shin J, Jovanovic T, Almli LM, Dias BG, Conneely KN. Estrogen-dependent association of HDAC4 with fear in female mice and women with PTSD. Molecular psychiatry. 2017 doi: 10.1038/mp.2016.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rutten BP, Vermetten E, Vinkers CH, Ursini G, Daskalakis NP, Pishva E, Kenis G. Longitudinal analyses of the DNA methylome in deployed military servicemen identify susceptibility loci for post-traumatic stress disorder. Mol Psychiatry. 2017 doi: 10.1038/mp.2017.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mehta D, Bruenig D, Carrillo-Roa T, Lawford B, Harvey W, Morris CP, Voisey J. Genomewide DNA methylation analysis in combat veterans reveals a novel locus for PTSD. Acta Psychiatrica Scandinavica. 2017;136(5):493–505. doi: 10.1111/acps.12778. [DOI] [PubMed] [Google Scholar]
- 48.Zannas AS, Provençal N, Binder EB. Epigenetics of posttraumatic stress disorder: current evidence, challenges, and future directions. Biological psychiatry. 2015;78(5):327–335. doi: 10.1016/j.biopsych.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 49.Anisman H, Matheson K. Stress, depression, and anhedonia: caveats concerning animal models. Neuroscience & Biobehavioral Reviews. 2005;29(4):525–546. doi: 10.1016/j.neubiorev.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 50.Ferrer I, Martinez A, Boluda S, Parchi P, Barrachina M. Brain banks: benefits, limitations and cautions concerning the use of post-mortem brain tissue for molecular studies. Cell and tissue banking. 2008;9(3):181. doi: 10.1007/s10561-008-9077-0. [DOI] [PubMed] [Google Scholar]
- 51.Phillips ML, Drevets WC, Rauch SL, Lane R. Neurobiology of emotion perception I: the neural basis of normal emotion perception. Biological psychiatry. 2003;54(5):504–514. doi: 10.1016/s0006-3223(03)00168-9. [DOI] [PubMed] [Google Scholar]
- 52.Vukojevic V, Kolassa IT, Fastenrath M, Gschwind L, Spalek K, Milnik A, Peter F. Epigenetic modification of the glucocorticoid receptor gene is linked to traumatic memory and post-traumatic stress disorder risk in genocide survivors. Journal of Neuroscience. 2014;34(31):10274–10284. doi: 10.1523/JNEUROSCI.1526-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schechter DS, Moser DA, Paoloni-Giacobino A, Stenz L, Gex-Fabry M, Aue T, Rossignol AS. Methylation of NR3C1 is related to maternal PTSD, parenting stress and maternal medial prefrontal cortical activity in response to child separation among mothers with histories of violence exposure. Frontiers in psychology. 2015;6 doi: 10.3389/fpsyg.2015.00690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sadeh N, Spielberg JM, Logue MW, Wolf EJ, Smith AK, Lusk J, Salat DH. SKA2 methylation is associated with decreased prefrontal cortical thickness and greater PTSD severity among trauma-exposed veterans. Molecular psychiatry. 2016;21(3):357. doi: 10.1038/mp.2015.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaminsky Z, Wilcox HC, Eaton WW, Van Eck K, Kilaru V, Jovanovic T, Smith AK. Epigenetic and genetic variation at SKA2 predict suicidal behavior and post-traumatic stress disorder. Translational psychiatry. 2015;5(8):e627. doi: 10.1038/tp.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boks MP, Rutten BP, Geuze E, Houtepen LC, Vermetten E, Kaminsky Z, Vinkers CH. SKA2 methylation is involved in cortisol stress reactivity and predicts the development of post-traumatic stress disorder (PTSD) after military deployment. Neuropsychopharmacology. 2016;41(5):1350. doi: 10.1038/npp.2015.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sadeh N, Wolf EJ, Logue MW, Hayes JP, Stone A, Griffin LM, Miller MW. Epigenetic variation at Ska2 predicts suicide phenotypes and internalizing psychopathology. Depression and anxiety. 2016;33(4):308–315. doi: 10.1002/da.22480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Levy-Gigi E, Szabó C, Kelemen O, Kéri S. Association among clinical response, hippocampal volume, and FKBP5 gene expression in individuals with posttraumatic stress disorder receiving cognitive behavioral therapy. Biological psychiatry. 2013;74(11):793–800. doi: 10.1016/j.biopsych.2013.05.017. [DOI] [PubMed] [Google Scholar]
- 59.Wang D, Szyf M, Benkelfat C, Provençal N, Turecki G, Caramaschi D, Booij L. Peripheral SLC6A4 DNA methylation is associated with in vivo measures of human brain serotonin synthesis and childhood physical aggression. PloS one. 2012;7(6):e39501. doi: 10.1371/journal.pone.0039501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Booij L, Szyf M, Carballedo A, Frey EM, Morris D, Dymov S, Gill M. DNA methylation of the serotonin transporter gene in peripheral cells and stress-related changes in hippocampal volume: a study in depressed patients and healthy controls. PloS one. 2015;10(3):e0119061. doi: 10.1371/journal.pone.0119061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frodl T, Szyf M, Carballedo A, Ly V, Dymov S, Vaisheva F, Booij L. DNA methylation of the serotonin transporter gene (SLC6A4) is associated with brain function involved in processing emotional stimuli. Journal of psychiatry & neuroscience: JPN. 2015;40(5):296. doi: 10.1503/jpn.140180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moser DA, Paoloni-Giacobino A, Stenz L, Adouan W, Manini A, Suardi F, Ansermet F. BDNF methylation and maternal brain activity in a violence-related sample. PloS one. 2015;10(12):e0143427. doi: 10.1371/journal.pone.0143427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schechter DS, Moser DA, Pointet VC, Aue T, Stenz L, Paoloni-Giacobino A, Rossignol AS. The association of serotonin receptor 3A methylation with maternal violence exposure, neural activity, and child aggression. Behavioural brain research. 2017;325:268–277. doi: 10.1016/j.bbr.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 64.Binder EB, Bradley RG, Liu W, Epstein MP, Deveau TC, Mercer KB, Schwartz AC. Association of FKBP5 polymorphisms and childhood abuse with risk of posttraumatic stress disorder symptoms in adults. Jama. 2008;299(11):1291–1305. doi: 10.1001/jama.299.11.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wingo AP, Almli LM, Stevens JJ, Klengel T, Uddin M, Li Y, Smith AK. DICER1 and microRNA regulation in post-traumatic stress disorder with comorbid depression. Nature communications. 2015;6 doi: 10.1038/ncomms10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Horvath S. DNA methylation age of human tissues and cell types. Genome biology. 2013;14(10):3156. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Deconde R. Genome-wide methylation profiles reveal quantitative views of human aging rates. Molecular cell. 2013;49(2):359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68•.Wolf EJ, Logue MW, Hayes JP, Sadeh N, Schichman SA, Stone A, Miller MW. Accelerated DNA methylation age: associations with PTSD and neural integrity. Psychoneuroendocrinology. 2016;63:155–162. doi: 10.1016/j.psyneuen.2015.09.020. Examined blood-derived DNA methylation measurements of epigenetic age in relation to measures of neural integrity and executive function in a sample of U.S. veterans. Represents a genomic approach to PTSD with high public health impact, as it holds the potential to identify individuals most in need of intervention by leveraging peripherally-derived, polygenic epigenetic measurements that are predictive of neural integrity and memory performance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mulligan C, D’Errico N, Stees J, Hughes D. Methylation changes at NR3C1 in newborns associate with maternal prenatal stress exposure and newborn birth weight. Epigenetics. 2012;7(8):853–857. doi: 10.4161/epi.21180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kertes DA, Kamin HS, Hughes DA, Rodney NC, Bhatt S, Mulligan CJ. Prenatal maternal stress predicts methylation of genes regulating the hypothalamic–pituitary– adrenocortical system in mothers and newborns in the Democratic Republic of Congo. Child development. 2016;87(1):61–72. doi: 10.1111/cdev.12487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Clukay CJ, Hughes DA, Rodney NC, Kertes DA, Mulligan CJ. DNA methylation of methylation complex genes in relation to stress and genome-wide methylation in mother–newborn dyads. American Journal of Physical Anthropology. 2017 doi: 10.1002/ajpa.23341. [DOI] [PubMed] [Google Scholar]
- 72•.Perroud N, Rutembesa E, Paoloni-Giacobino A, Mutabaruka J, Mutesa L, Stenz L, Karege F. The Tutsi genocide and transgenerational transmission of maternal stress: epigenetics and biology of the HPA axis. The World Journal of Biological Psychiatry. 2014;15(4):334–345. doi: 10.3109/15622975.2013.866693. First report of transgenerational epigenetic effects of genocide, demonstrating blood-based DNA methylation differences in the glucocorticoid receptor (GR) in both mothers and their offspring who were exposed in utero to the Rwandan genocide. Epigenetic effects were also associated with reduced serum cortisol and differences in GR and mineralocorticoid receptors, suggesting a functional impact. [DOI] [PubMed] [Google Scholar]
- 73.Yehuda R, Daskalakis NP, Lehrner A, Desarnaud F, Bader HN, Makotkine I, Meaney MJ. Influences of maternal and paternal PTSD on epigenetic regulation of the glucocorticoid receptor gene in Holocaust survivor offspring. American Journal of Psychiatry. 2014;171(8):872–880. doi: 10.1176/appi.ajp.2014.13121571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yehuda R, Flory JD, Bierer LM, Henn-Haase C, Lehrner A, Desarnaud F, Meaney MJ. Lower methylation of glucocorticoid receptor gene promoter 1 F in peripheral blood of veterans with posttraumatic stress disorder. Biological psychiatry. 2015;77(4):356–364. doi: 10.1016/j.biopsych.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 75.Serpeloni F, Radtke K, De Assis SG, Henning F, Nätt D, Elbert T. Grandmaternal stress during pregnancy and DNA methylation of the third generation: an epigenome-wide association study. Translational psychiatry. 2017;7(8):e1202. doi: 10.1038/tp.2017.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Esposito EA, Jones MJ, Doom JR, MacIsaac JL, Gunnar MR, Kobor MS. Differential DNA methylation in peripheral blood mononuclear cells in adolescents exposed to significant early but not later childhood adversity. Development and psychopathology. 2016;28(4pt2):1385–1399. doi: 10.1017/S0954579416000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sidebotham P, Golding J. Avon longitudinal study of parents and children. Child maltreatment in the “children of the nineties” a longitudinal study of parental risk factors. Child Abuse Negl. 2001;25(9):1177–1200. doi: 10.1016/s0145-2134(01)00261-7. [DOI] [PubMed] [Google Scholar]
- 78.Stein DJ, Koen N, Donald KA, Adnams CM, Koopowitz S, Lund C, Stern M. Investigating the psychosocial determinants of child health in Africa: The Drakenstein Child Health Study. Journal of neuroscience methods. 2015;252:27–35. doi: 10.1016/j.jneumeth.2015.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ulrich-Lai YM, Herman JP. Neural regulation of endocrine and autonomic stress responses. Nature reviews. Neuroscience. 2009;10(6):397. doi: 10.1038/nrn2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bellavance MA, Rivest S. The HPA–immune axis and the immunomodulatory actions of glucocorticoids in the brain. Frontiers in immunology. 2014;5 doi: 10.3389/fimmu.2014.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Binder EB. The role of FKBP5, a co-chaperone of the glucocorticoid receptor in the pathogenesis and therapy of affective and anxiety disorders. Psychoneuroendocrinology. 2009;34:S186–S195. doi: 10.1016/j.psyneuen.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 82.Breedlove S, Watson N. Behavioral neuroscience. Eighth. Sunderland, Massachusetts: Sinauer Associates; 2017. [Google Scholar]
- 83.Freberg L. Discovering Behavioral Neuroscience: An Introduction to Biological Psychology. Boston, MA: Cengage Learning; 2014. [Google Scholar]
- 84.GeneCards. The Human Gene Database. Retrieved November 15, 2017, from http://www.genecards.ord/


