Abstract
Thyroid cancers are the most common endocrine malignancy and approximately 2% of thyroid cancers are anaplastic thyroid carcinoma (ATC), one of the most lethal and treatment resistant human cancers. Cancer stem-like cells (CSCs) may initiate tumorigenesis, induce resistance to chemotherapy and radiation therapy, have multipotent capability and may be responsible for recurrent and metastatic disease. The production of CSCs has been linked to epithelial-mesenchymal transition (EMT) and the acquisition of stemness. Exosomes are small (30–150 nm) membranous vesicles secreted by most cells that play a significant role in cell-to-cell communication. Many non-coding RNAs (ncRNA), such as long-non-coding RNAs (lncRNA), can initiate tumorigenesis and the EMT process. Exosomes carry ncRNAs to local and distant cell populations. This study examines secreted exosomes from two in vitro cell culture models; an EMT model and a CSC model. The EMT was induced in a papillary thyroid carcinoma (PTC) cell line by TGFβ1 treatment. Exosomes from this model were isolated and cultured with naïve PTC cells and examined for EMT induction. In the CSC model, exosomes were isolated from a CSC clonal line, cultured with a normal thyroid cell line and examined for EMT induction. The EMT exosomes transferred the lncRNA MALAT1 and EMT effectors SLUG and SOX2; however, EMT was not induced in this model. The exosomes from the CSC model also transferred the lncRNA MALAT1 and the transcription factors SLUG and SOX2 but additionally transferred linc-ROR and induced EMT in the normal thyroid cells. Preliminary siRNA studies directed towards linc-ROR reduced invasion. We hypothesize that CSC exosomes transfer lncRNAs, importantly linc-ROR, to induce EMT and inculcate the local tumor microenvironment and the distant metastatic niche. Therapies directed towards CSCs, their exosomes and/or the lncRNAs they carry may reduce a tumor’s metastatic capacity.
Keywords: Anaplastic thyroid carcinoma, cancer stem-like cells, exosomes, LncRNA, Linc-ROR, EMT, metastatic niche
Introduction
Thyroid cancers are the most common endocrine malignancy, currently comprising approximately 1% of all human cancers in the USA and increasing in incidence (1,2). Roughly 80 to 85% of thyroid cancers are well differentiated papillary thyroid carcinomas (PTC) and have an excellent prognosis, yet a small portion of PTC patients develop aggressive PTC which can lead to invasive tumors and/or distant metastases (1–3). Approximately 2% of thyroid cancers are anaplastic thyroid carcinoma (ATC), which remains one of the most lethal and treatment resistant human cancers since patients usually succumb to the disease within 12 months of diagnosis (2). Some studies have shown that ATCs may arise from the well-differentiated PTC through dedifferentiation pathways (4). However, others have hypothesized that ATC may be derived from cancer stem-like cells (CSCs), also known as tumor initiating cells (5–8).
Exosomes are small (30–150 nm) membranous vesicles of endocytic origin secreted by most cells that play a significant role in cell-to-cell communication. They can transfer their contents, which include DNA, protein, growth factors, miRNAs and lncRNAs, to adjacent and distant cells (9). Exosomes and their contents can contribute to the induction of EMT, inculcate tumor microenvironment to form a metastatic niche, and modulate host immunity to escape immune response (10–16). One study found that cancer derived exosomes delivered ncRNAs to non-cancerous cells (17). Some investigations have reported that exosomes may even determine which organ is targeted by metastasis (18). The use of patient blood and other bodily fluids to isolate exosomes (liquid biopsy) provides a real-time, non-invasive method to monitor disease progression and determine therapy options (19–21). Additionally, exosomes are now being studied as therapeutic delivery agents for treating many diseases, including targeting CSCs (22–25).
CSCs can be characterized by several factors, such as; self-renewal abilities, stem cell marker expression (i.e. SOX2, NANOG, and OCT4), resistance to chemotherapy and radiation therapy, multipotent capability, recurrent and metastatic disease, ability to form spheroids in unattached serum-free culture conditions, initiate tumorigenesis and the ability to recapitulate the heterogeneity of the parent tumor (26,27). The production of CSCs has been linked to epithelial-mesenchymal transition (EMT) and the acquisition of stemness (26, 28–31). EMT is a process where epithelial cells undergo a transition to a mesenchymal phenotype characterized by the loss of E-cadherin by the activation of one or several factors such as; SNAIL, SLUG, ZEBs and TWIST (29,32,33). Many non-coding RNAs (ncRNA) such as microRNAs (miRNA) and long-non-coding RNAs (lncRNA) also can initiate the EMT process (34). Cancer cells that undergo EMT gain enhanced migratory capacity and invasiveness, allowing primary tumor cells to metastasize and form secondary tumors throughout the body.
Results from the Human Genome Project study revealed that thousands of ncRNAs were transcribed and played a significant role in gene regulation, especially through epigenetic modifications (35). There are a variety of subclasses of ncRNAs, including small ncRNAs (miRNA, snRNA, tRNA, piRNA, snoRNAs) and long ncRNAs (lncRNAs). LncRNAs are greater than 200 nucleotides and have a variety of targets and functions, such as chromatin remodeling, miRNA sponges and splicing modulators (36). Four lncRNAs that we have previously explored in thyroid cancer (HOTAIR, linc-ROR, MALAT1 and PVT1 (38)) were included in the present study. HOTAIR has been correlated with metastasis and poor survival outcome in breast, colon and lung cancers (37) and has been shown to be highly up-regulated in thyroid cancer (36). Linc-ROR (long intergenic non-coding RNA, regulator of reprogramming) has also been linked to metastasis as well as EMT (37). MALAT1 has been shown to be expressed in many cancer types and is also correlated to metastasis (37). Additionally, MALAT1 has been shown to be upregulated in thyroid cancer progression in PTC and FTC, but not in ATC, indicating a potential tool in classification of thyroid carcinoma (38). PVT1 has been found to be upregulated in thyroid cancers and has been linked to poor prognosis in several cancers (36).
Our present studies utilized two CSC clonal lines, A3 and D3 (39), to examine CSC exosomes and their contents to gain further understanding about the biology of CSCs, their involvement in EMT and cancer progression and their ability to reprogram the metastatic niche with their contents.
Materials and Methods
Cell culture, EMT model and CSC clone formation
The thyroid papillary carcinoma cell line TPC-1 was provided by Dr. Daniel Ruan (Brigham and Women’s Hospital, Boston, MA) and was maintained in RPMI with 10% FBS and 1% penicillin/streptomycin at 37°C, 5% CO2. The THJ-16T anaplastic thyroid carcinoma cell line was provided by Dr. J. Copland (Mayo Clinic, Jacksonville, FL) and maintained in RPMI with 10% FBS, 1% non-essential amino acids, 1% sodium pyruvate and 1% penicillin/streptomycin at 37°C, 5% CO2. The normal thyroid cell line NTHY-ori-3 was purchased from Sigma and maintained in RPMI with 10% FBS and 1% penicillin/streptomycin at 37°C, 5% CO2. To verify the authenticity of the TPC-1 and THJ-16T cell lines, DNA short tandem repeat (STR) analysis was performed by DDC (DNA Diagnostics Center, Fairfield, OH). The cell lines were tested using 19 markers and were found to be authentic.
To induce EMT, the TPC-1 cell line was plated at a density of 1 × 106 per 75mm flask, cultured with serum-free media (modified from Fierabracci (40)), (EGF and FGF omitted) with (treated) or without (control) 10 ng/ml hrTGFβ1 (TGFβ) (R&D Systems, Minneapolis, Mn.) for 3 and 6 days. These cells and their isolated exosomes will furthermore be referred to as EMT cells or EMT exosomes.
CSC clonal spheroid cell line formation was described previously (29,39). These cells and their isolated exosomes will furthermore be referred to as clones or clonal exosomes.
Exosome Isolation, Purification, Quantification and NTA Analysis
Both the EMT cells and the CSC clones D3 and A3 culture supernatants were harvested, centrifuged at 1100 rpm for 3 min to remove cells and debris and exosomes precipitated overnight at 4°C with ExoQuick TC (Systems Biosciences, Palo Alto, Ca.) according to manufacturer’s instructions. The resulting pellets were suspended in 300 ml PBS and further immuno-magnetically purified using CD81 coated 9 mm beads (ExoFlow, Systems Biosciences) according to manufacturer’s instructions. The exosome coated beads were then analyzed by flow cytometry or the exosomes were eluted from the beads for further analysis. Some eluted purified exosomes were subjected to nanoparticle tracking analysis (NTA) on the NanoSight NS300 performed by PTL (Particle Technology Labs, Downers Grove, Il.) while others were quantified by the ExoTest ELISA (Galen Laboratory Supplies, Middleton, Ct.) according to manufacturer’s instructions. Additionally, some exosomes were further purified on a 30% sucrose cushion to remove any remaining debris from processing for electron microscopy.
Flow Cytometry
Cells were harvested and ALDH activity analyzed by the ALDEFLUOR™ Kit (STEMCELLS Technology, Vancouver, B.C., Canada) according to manufacturer’s instructions as previously reported (29,39). Exosome coated beads were stained and analyzed by flow cytometry using the ExoFlow Kit (Systems Biosciences) according to manufacturer’s instructions. Antibody coated beads devoid of exosomes were used as a negative control. Flow Cytometry was performed on the BD FACSCalibur™ and analyzed with the Cyflogic v.1.2.1 software.
Exosomal Transfer Cultures
Purified clonal exosomes were added to 1 × 105 NTHY-ori-3 cultures at either 5 or 10 μg and incubated in 10 ml RPMI with 10% exosome free FBS and 1% penicillin/streptomycin for 72 hours at 37°C, 5% CO2. Additionally, 10 μg of purified EMT exosomes from control (days 3 and 6) and TGFβ treated (days 3 and 6) cultures were added to 1 × 105 naïve TPC1 cell cultures and incubated in 10 ml RPMI with 10% exosome free FBS and 1% penicillin/streptomycin for 72 hours at 37°C, 5% CO2. After 72 hours, cells were harvested for RNA analysis.
Real-time qPCR
Total RNA was extracted from cell culture samples and isolated purified exosomes with TRIzol reagent (ThermoFisher Scientific, Waltham, MA) according to the manufacturer’s instructions, and RNA quality and concentrations were assessed with a NanoDrop 1000 spectrophotometer (ThermoFisher Scientific, Waltham, MA). 1 μg of total RNA was reverse-transcribed using the All-in-One miRNA RT-qPCR detection kit (GeneCopoeia Rockville, MD). RT-qPCR was performed on a CFX96 PCR detection system (Bio-Rad Laboratories, Hercules, CA) using Bullseye EvaGreen qPCR master mix (MIDSCI, St. Louis, MO), normalized to 18S rRNA; relative fold change was determined by the ΔΔ CT method. The PCR primers used are PVT1; Forward 5′-TGAGAACTGTCCTTACGTGACC-3′ and Reverse 5′-AGAGCACCAAGACTGGCTCT-3′, HOTAIR; Forward 5′-CAGTGGGGAACTCTGACTCG-3′ and Reverse 5′-GTGCCTGGTGCTCTCTTACC-3′, MALAT1; Forward 5′-GACGGAGGTTGAGATGAAGC-3′ and Reverse 5′-ATTCGGGGCTCTGTAGTCCT-3′, Linc-ROR1; Forward 5′-CTGGCTTTCTGGTTTGACG-3′ and Reverse 5′-CAGGAGGTTACTGGACTTGGAG-3′, SOX2; Forward 5′-GCACATGAACGGCTGGAGCAACG-3′ and Reverse 5′-ATTCGGGGCTCTGTAGTCCT-3′, SLUG; Forward 5′-ATGCATATTCGGACCCACAC-3′ and Reverse 5′-GCAGATGAGCCCTCAGATTT-3′, SMAD4; Forward 5′-CCATTTTCCAATCATCCTGCT-3′ and Reverse 5′-ACCTTTGCCTATGTGCAACC-3′, and 18S; Forward 5′-GTAACCCGTTGAACCCCATT-3′ and Reverse 5′-CCATCCAATCGGTAGTAGCG-3′.
Proliferation and Invasion Assays
TPC1 and NTHY-ori-3 cells were seeded into 96 well plates at 1 × 104/well (6 replicates per each treatment condition) in 100 μl serum-free/antibiotic-free media. Purified EMT exosomes from each treatment group of EMT cultures were added to TPC1 well sets at 5 mg of exosomes per treatment group for 72 hours at 37°C, 5% CO2. One set of TPC1 wells had no exosomes added and used as a control group. Additionally, purified clonal exosomes, both D3 and A3 individually, were added to the NTHY-ori-3 well sets at 5 mg of exosomes per well for 72 hours at 37°C, 5% CO2. Again, one set of NTHY-ori-3 wells had no exosomes added as a control group. After 72 hours incubation, the Vybrant® MTT Cell Proliferation Assay Kit (Life Technologies) and the Cultrex BME Cell Invasion Assay (R&D Systems) were performed according to protocol. The 96 well plate for the invasion assay was coated with 0.1 X BME (basement membrane extract) overnight prior to seeding cells. Analysis for both assays used the SpectraMax M3 Multi-Mode Microplate Reader with SoftMax Pro (6.2.2) software.
Electron Microscopy
For standard transmission electron microscopy, exosomes were fixed in 1% glutaraldehyde overnight at 4°C, adsorbed onto 200 mesh formvar coated copper grids, stained with contrast dye containing 1% uranyl acetate for 10 minutes and viewed on in a Hitachi H-7650 transmission electron microscope.
For ImmunoGold labeling, exosomes were fixed in 1% glutaraldehyde overnight at 4°C, adsorbed onto 300 square mesh formvar coated nickel grids and incubated with rabbit anti-human CD9 antibody (5ug/ml, Abcam #ab92726) for 1 hour at room temperature. Incubation buffer (PBS + 0.1% BSA-c) served as a blank control. Following 6 washes in incubation buffer, exosomes were incubated with 40ul goat anti-rabbit antibody with 6-nm gold particles attached (1:40 diluted, Aurion) for 1 hour at room temperature. Following 6 washes in incubation buffer, 3 washes in PBS, grids were fixed in 1% glutaraldehyde, incubated in contrast dye containing 1% uranyl acetate for 10 minutes and viewed on a Hitachi H-7650 transmission electron microscope.
Statistics
Student’s t-test was used to analyze data collected. Two-tailed P values of <0.05 were considered to be statistically significant. Data are expressed as means ± standard error of the mean (SEM). All experiments were performed at least twice.
Results
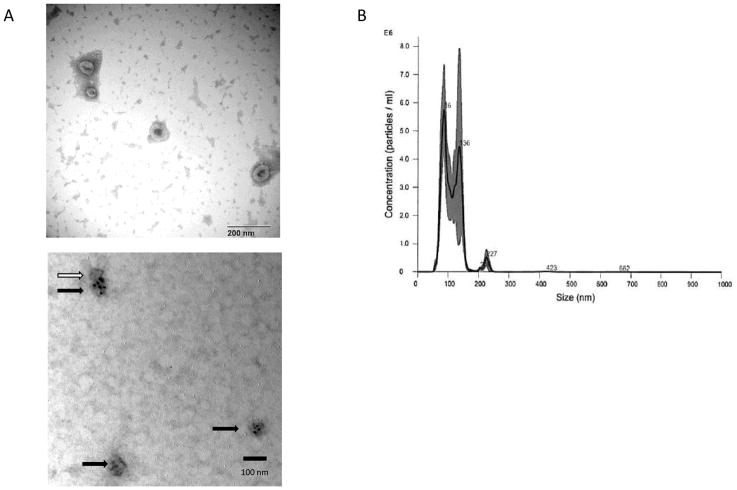
With the recent advances in exosomal isolation and evaluations, we have isolated exosomes from the EMT model as well as two CSC lines to begin characterization of CSC exosomes and their effect on EMT, metastasis and cancer progression. We have successfully isolated and purified CSC exosomes with a combination of polymeric precipitation and anti-CD81 labelled magnetic bead isolation, as evidenced by electron microscopy (both negatively stained and anti-CD9 gold beads) and Nano Particle Tracking (Fig 1).
Fig. 1.
Exosome Isolation. A) Electron micrograph of uranyl acetate stained TPC-1 exosomes (top) showing typical exosomal cup-shaped morphology and CD9 ImmunoGold labelled (bottom) exosomes derived from cell culture isolated by polymeric precipitation and further purified by CD81 coated magnetic beads. Black arrows (3) indicate CD9 positive stained exosomes, white arrow (1) indicates not stained, or CD9 negative, exosome. B) Nanoparticle Tracking Analysis of TPC-1 exosomes isolated and purified as the exosomes in the EM (A).
In our previous TGFβ induced EMT model, we studied the up/down-regulation of miRNAs and EMT transcriptional factors as well as the increased production of CSCs during EMT (29). Here we begin to characterize the effect EMT induction has on lncRNAs. Additionally, we begin to examine the effect EMT has on exosomal production and their content.
TGFβ Induced EMT Model
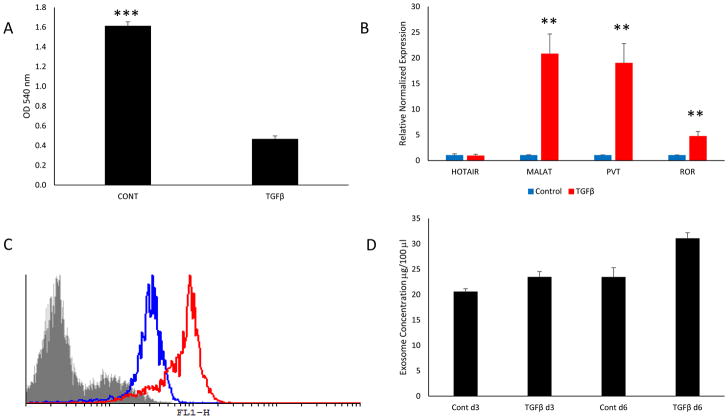
The well-differentiated papillary thyroid carcinoma cell line TPC1 was incubated with or without 10 ng/ml TGFβ for 3 and 6 days in serum-free media. Proliferation was measured at the end of 6 days of treatment which exhibited a significant growth inhibition exerted on the treated cells by TGFβ, indicating that the TGFβ pathway was intact and fully functional (Fig. 2a). Additionally, the cells were examined for lncRNA expression after 6 days of treatment by RT-qPCR revealing a significant upregulation in 3 out of the 4 lncRNAs studied (Fig 2b). It was not surprising to find that MALAT1, PVT1 and linc-ROR were significantly upregulated by TGFβ treatment, considering that these ncRNAs are directly related to regulating the TGFβ pathway, EMT, cell motility, metastasis and epigenetic modifications (16,36,37,41). Interestingly, HOTAIR expression was not affected by TGFβ treatment in the TPC-1 cell line (Fig. 2b).
Fig. 2.
(A) TPC-1 cells were incubated with 10 ng/ml TGFβ1 for 6 days and proliferation was measured using the MTT assay. TGFβ1 significantly inhibited TPC-1 cell growth. (B) RT-qPCR results of lncRNA expression in control and TGFβ treated TPC1 cells. TGFβ significantly upregulated 3 out of 4 lncRNAs. (C) Exosomes from control and TGFβ treated samples isolated with the ExoFlow kit. The gray outlined area represents negative control beads (no exosomes), the blue outline represents the control exosomes and the red outline represents the TGFβ treated exosomes. The FL1-H axis is the relative fluorescence intensity. (D) Exosomes from day 3 and day 6 of control and TGFβ treated TPC-1 cells measured by ExoTest ELISA. Data represented by means +/− SEM. ** = p<0.01, *** = p<0.001.
Previously, it has been shown that cancer cells produce and secrete more exosomes than do their normal counterparts (10,23) and that more exosomes are released as disease stage increases (42). Therefore, we examined whether EMT induction also increased exosomal production. Exosomes from both TGFβ treated and control cells were isolated by precipitation and further purified by binding the exosomes to CD-81 coated 9 μm beads, stained with an exosomal membrane stain and measured by flow cytometry and ELISA (Fig 2c and 2d, respectively). The TGFβ treated cells produced more exosomes than the control treated group even though the control group contained more cells due to the growth inhibition exerted by TGFβ on the treated group.
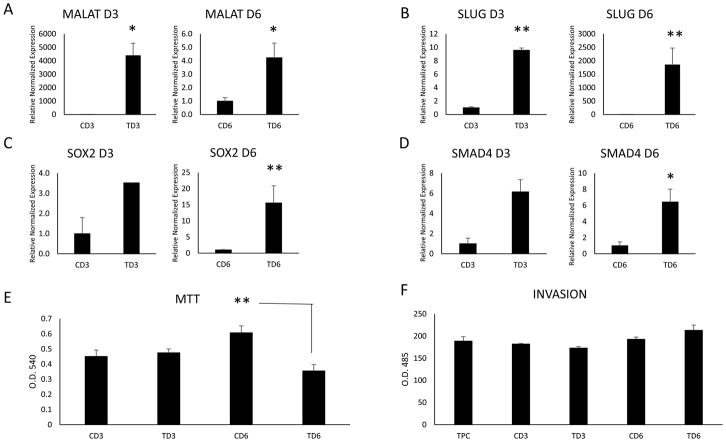
Several investigators have shown that cancer exosomes can modify adjacent and distant cells for the facilitation of EMT and the inculcation of the metastatic niche (43). We examined the effects of TGFβ induced EMT cancer exosomes on naïve TPC1 cells and examined their ability to initiate EMT and modify the expression of EMT factors and lncRNAs. We isolated exosomes from control and treated cells at days 3 and 6 and incubated them with naïve TPC1 cells for 3 days. We tested the naïve cells for ncRNA expression, EMT effectors, proliferation and invasive potential. TGFβ treated exosomes significantly upregulated MALAT1 in the naïve cells over that of the control treated exosomes (Fig 3a). TGFβ treated exosomes also upregulated EMT and stem-like cell markers (Fig 3b–d) and decreased proliferation, particularly at day 6, in the naïve TPC1 cells. However, invasiveness increased in the day 6 TGFβ treated exosome group although this effect was not significant (Fig 3f).
Fig. 3.
RT-qPCR results of naïve TPC-1 cells incubated with 1 × 108 control and TGFβ treated TPC1 exosomes isolated at day 3 and day 6 of treatment. (A) LncRNA MALAT1 was significantly upregulated in the naïve TPC-1 cells by TGFβ treated exosomes on both days analyzed. The EMT factor SLUG (B), stem cell marker SOX2 (C) and the TGFβ pathway protein SMAD4 (D) were upregulated only on day 6 by TGFβ treated exosomes. (E) Proliferation of naïve TPC-1 cells incubated with control and TGFβ treated TPC1 exosomes was measured by MTT assay. Day 6 TGFβ treated exosomes significantly reduce proliferation. (F) Invasive properties of naïve TPC-1 cells incubated with days 3 and 6 control and TGFβ treated TPC1 exosomes were analyzed. No significant difference was seen, however, day 6 TGFβ treated exosomes began to increase invasive potential of the naïve TPC-1 cells. Data represented by means +/− SEM. * = p<0.05, ** = p<0.01.
CSC Exosomal Model
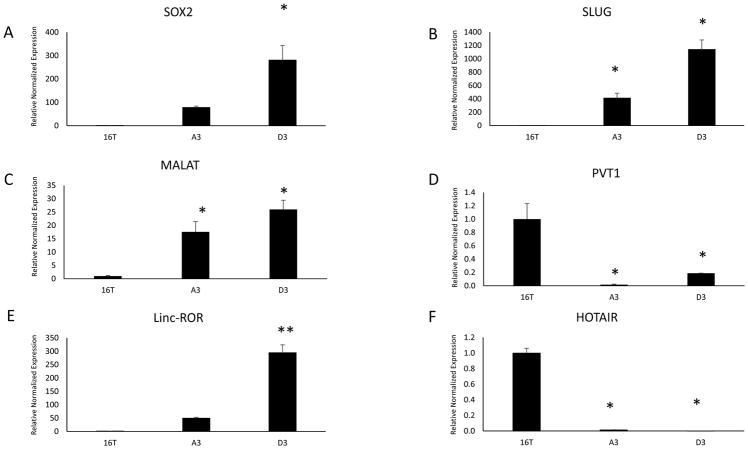
As mentioned earlier, we previously isolated thyroid CSCs and derived two clonal CSC lines from the ATC line THJ-16T (39). Briefly, we isolated extremely high ALDH expressing cells, plated at the single cell per well in ultra-low attached conditions with a defined serum-free media for clonal expansion. These clones exhibited a high percentage of symmetrical division and retained their high expression of ALDH as tested by the ALDEFLUOR Assay (Table 1) (39). Additionally, the clones express significantly high levels of stem cells markers and mesenchymal/EMT markers (Fig 4a and 4b). We probed the clones to determine their lncRNA expression levels and found two lncRNAs, MALAT1 and linc-ROR, were highly expressed compared to the parental line THJ-16T (Fig 4c–f). Surprisingly, HOTAIR and PVT1 were downregulated in the clones compared to the parental line.
Table 1.
ALDEFLUOR positivity % of the differentiated papillary cell line TPC-1, the anaplastic cell line THJ-16T and the two cancer stem cell clonal lines D3 and A3 as measured by flow cytometry. ALDEFLUOR activity was significantly higher in the two CSC clonal lines compared to the TPC-1 line and their parental ATC line THJ-16T.
| Cell Line | Ald% | p value |
|---|---|---|
| TPC | 0.25 | |
| 16T | 4.12 | |
| D3 | 95.89 | p<0.01 |
| A3 | 95.55 | p<0.01 |
Fig. 4.
RT-qPCR results of stem cell marker SOX2 (A) and EMT marker SLUG (B) analysis of the two CSC clonal lines compared to their parental line THJ-16T. (C–F) RT-qPCR analysis of LncRNA expression of the two CSC clonal lines compared to their parental line THJ-16T. Malat-1 (C) and Linc-ROR (E) were significantly upregulated in the CSC clones, however, PVT1 (D) and HOTAIR (F) were downregulated compared to their parental line. Data represented by means +/− SEM. * = p<0.05, ** = p<0.01.
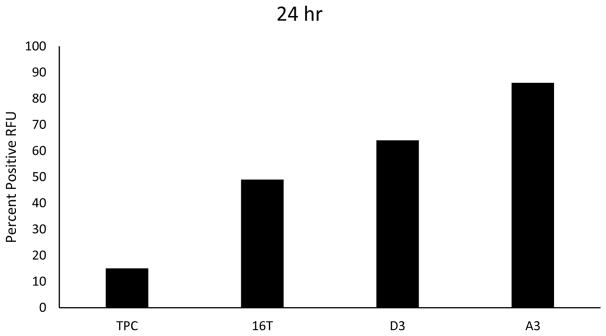
Similarly to the EMT model, we examined the production of exosomes from the two clonal lines, the parental line and the TPC1 line. We hypothesized that the clones, due to their derivation from ATC, and the THJ-16T line would produce more exosomes than the well-differentiated TPC1 cell line. Additionally, we hypothesized that the clones would also produce more exosomes than their parental line, due to their increased tumorigenicity over the THJ-16T line (39). We plated 1 × 106 cells from each line in an exosome-free serum media for 24 hours and harvested, isolated and analyzed the relative number of exosomes produced by each line by flow cytometry (Fig 5). Confirming our expectations, the ATC line produced twice the amount of exosomes than the well differentiated PTC line and the CSC clones produced 3 to 4 times the amount of exosomes than TPC1. Additionally, the CSC clones also produced more exosomes as compared to the parental line.
Fig. 5.
Aggressive cancers and CSCs release more exosomes. Cell lines TPC-1, THJ-16T, D3 and A3 were plated at 1 × 106 and incubated for 24 hours. Exosomes were then isolated, purified and analyzed by flow cytometry using the ExoFlow Kit. The CSC clones had released more exosomes compared to the ATC line THJ-16T, which in turn released more exosomes than the differentiated TPC-1 line.
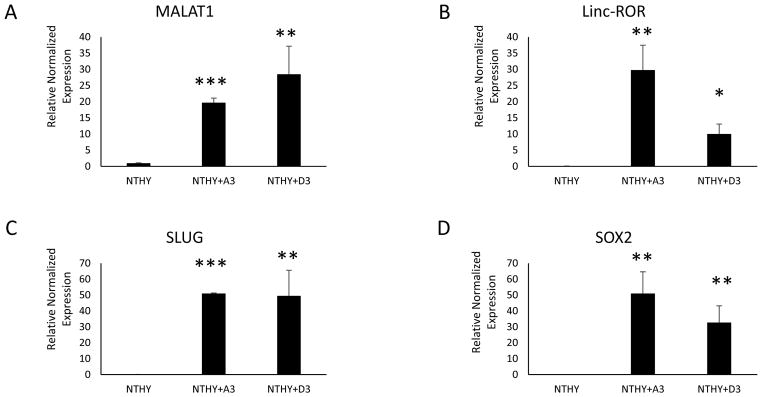
We examined the effects of the CSC exosomes and their ability to transfer contents and modify naïve cells. We isolated and purified exosomes from both the clonal lines and quantified them by ELISA. Naïve cultures of the normal thyroid cell line NTHY-ori-3 (NTHY) were plated and 1 × 108 exosomes from each clone were incubated with the NTHY cells in exosome-free media for 3 days. The NTHY cells were then harvested, RNA isolated and analyzed by RT-qPCR for expression profiles. Both clones were able to transfer and significantly upregulate the ncRNAs MALAT1 and linc-ROR (Fig 6a and b), as well as the EMT marker SLUG and the stem cell transcription factor SOX2 (Fig 6c and d).
Fig. 6.
Isolated and purified exosomes derived from each of the CSC clonal lines were added at 1 × 108 exosomes per test and incubated with NTHY-ori-3 cells for 3 days. RT-qPCR was then performed to assess the expression of nc-RNAs (A & B), EMT effectors (C) and stem cell markers (D). Both CSC clone exosomes significantly transferred these modulators to the normal thyroid cell line NTHY-ori-3. Data are expressed as means =/− SEM. * = p<0.05, ** = p<0.01, *** = p<0.001.
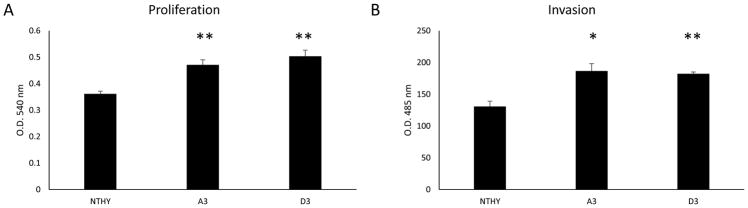
Next we needed to ascertain if this transfer was simply just an uptake of content or if there was functionality to the contents transferred. We analyzed the exosomes effect on the proliferative ability of the naïve cells. NTHY cells were plated with exosomes from both CSC clones and incubated for 3 days and proliferation was measured by the MTT assay. The exosomes from both clones significantly increased the proliferative ability of the treated cells than the control NTHY cells (Fig. 7a). Additionally, we analyzed the ability to induce invasiveness by the exosomal content transfer. Again, both CSC exosomes induced a significant increase in the NTHY cells invasive abilities (Fig 7b).
Fig. 7.
Exosomes from CSCs induce EMT of normal cells. Isolated and purified exosomes from CSC lines were incubated with NTHY-ori-3 cells for 3 days and proliferation (A) and invasiveness (B) were measured. Both CSC exosomes induced a significant increase in the proliferative ability and invasiveness of NTHY-ori-3 cells. Data represented as means +/− SEM. * = p<0.05, ** = p<0.01.
Discussion
Biomedical research into human thyroid cancers has made great strides over the past few decades. New technologies and discoveries have advanced our understanding of cancer biology and have opened a myriad of new treatment strategies, many with great success. Unfortunately, there remain many cancer types that successful treatment options continue to be elusive; ATC is in the latter category. Recent insights into various types of RNA have given researchers many new avenues to explore and a greater insight to the intricacies of the carcinogenesis process. Additionally, recent advancement in the discovery and understanding both CSCs and exosomes and their functions has opened even more avenues for diagnosis, prognosis and potential treatment strategies.
Previously, our laboratory worked on finding and isolating thyroid CSCs and to study the relationship with EMT, metastasis and cancer progression. After many attempts, we successfully isolated two single cells that, when upon clonal expansion, exhibited high self-renewal properties, retained extremely high EMT and stem cell marker expression and proved to be highly tumorigenic and invasive (39). With the current study, we began to examine the ncRNA expression and the production and function of exosomes of these CSC clones. We were successful in isolating and purifying clonal exosomes, as evidenced by electron microscopy and Nanoparticle Tracking Analysis. We were also successful isolating exosomes from our serum-free EMT model.
From our EMT model, we isolated RNA and performed RT-qPCR to analyze stem cell markers, EMT markers and lncRNAs expression. We found that TGFβ treatment significantly upregulated SOX2, SLUG, PVT1, MALAT1 and linc-ROR over that of the control. Interestingly, we found that HOTAIR was slightly elevated due to TGFβ treatment but the control had high expression as well and the difference was not significant. The clones were analyzed for the same markers and revealed significant upregulation of SOX2 and SLUG over that of the parental ATC line. Interestingly, there was differential expression of lncRNA in the CSC clones. MALAT1 and linc-ROR were highly expressed in the CSCs, however, PVT1 and HOTAIR were significantly down-regulated. In our recent study, we noted that MALAT1 was highly expressed in PTC but greatly reduced in ATC (38). Similarly, we also found that PVT1, HOTAIR and linc-ROR are also greatly reduced in ATC compared to PTC (44). The exact meaning and function of why MALAT1 and linc-ROR were highly expressed in the CSCs remains elusive and further investigation is underway.
Exosomes from the EMT model were isolated, purified, quantified and incubated with naïve TPC1 cells for 3 days and the cells analyzed to determine if exosomal transfer had occurred. MALAT1 was significantly upregulated in the naïve cells incubated with exosomes from the TGFβ treated cells. Similarly, SOX2, SLUG and SMAD4 were also upregulated, although primarily from the day 6 treated exosomes. It is conceivable that this increase may be due to small amounts of TGFβ carried within the exosomes and delivered to the naïve cells that could initiate the TGFβ pathway. Further investigations are ongoing. Functionality of the exosomal contents transferred to the naïve TPC1 cells was analyzed. Proliferation and invasion studies were performed and a significant reduction in proliferation from the day 6 TGFβ-treated exosomes was observed, however, no significant change in invasion was seen. Therefore, while some EMT markers were affected by the uptake of TGFβ treated exosomes, the EMT program had not been initiated.
Exosomes from the CSC clones were isolated, purified and quantified. CSC exosomes were cultured with the normal thyroid cell line NTHY-ori-3 for 3 days. The cells were analyzed for exosomal transfer and compared to the naïve NTHY cell line. The NTHY cells incubated with exosomes from both the CSC lines showed highly significant upregulation of the lncRNAs MALAT1 and linc-ROR, the EMT marker SLUG and the stem cell marker SOX2, compared to the untreated naïve cells. Similarly to the EMT model previously discussed, functionality of the transferred material was also analyzed by proliferation and invasion assays. NTHY cells incubated with CSC exosomes revealed significantly increased proliferative ability compared to the control cells. Importantly, the invasive ability was also significantly increased compared to control cells. Unlike the EMT exosomes, the EMT program was induced and initiated by the exosomal transfer of material from the CSC exosomes.
Interestingly, both the EMT and the CSC exosomes transferred MALAT1, SOX2 and SLUG to their respective naïve cells and increased proliferation in those cells. However, the EMT exosomes did not initiate EMT, evidenced by the lack of induction of invasion. Conversely, the CSC exosomes significantly increased the invasiveness of normal cells, as well as linc-ROR. We hypothesize that the transfer and expression of long intergenic non-coding RNA-regulator of reprogramming (linc-ROR) by CSC exosomes is responsible for the induction of the EMT program and the significant increase in the invasive potential of normal thyroid cells. SiRNA studies targeting linc-ROR within the CSC exosomal model are underway.
In summary, this study illustrates the ability of exosomes to communicate with and modulate adjacent and distant tumor microenvironment through the transfer of ncRNAs and other modulators. Additionally, we demonstrate that CSC exosomes can induce EMT in normal non-cancerous thyroid cells by the transfer and expression of linc-ROR. With these insights, more direct and effective therapeutic treatment strategies can be discovered for targeting CSCs, their exosomes and/or the ncRNA linc-ROR for treating aggressive thyroid cancers.
Supplementary Material
Acknowledgments
This study was supported by a grant from the University of Wisconsin Carbone Cancer Center (RVL), and by NIH Cancer Center Support Grant P30 CA014520-39 (University of Wisconsin Cancer Center)
We kindly thank Dr. John A. Copland III (Mayo Clinic, Jacksonville, FL) for the THJ-16T cell line, Dr. Daniel T. Ruan (Brigham and Women’s Hospital, Boston, MA) for the TPC-1 cell line and the staffs of the Flow Cytometry and the 3P laboratories (University of Wisconsin Carbone Cancer Center Cancer Center Support Grant P30 CA014520) for their services. This work was supported in part by the use of the Electron Microscopy facility at the William S. Middleton Memorial Veterans Hospital, Madison WI. We acknowledge the technical assistance of Joan Sempf and Traci Niesen.
Footnotes
Disclosures: None reported
References
- 1.Davies L, Welch HG. Increasing Incidence of Thyroid Cancer in the United States, 1973–2002. JAMA. 2006;295:2164. doi: 10.1001/jama.295.18.2164. [DOI] [PubMed] [Google Scholar]
- 2.Sipos JA, Mazzaferri EL. Thyroid cancer epidemiology and prognostic variables. Clin Oncol. 2010;22:395–404. doi: 10.1016/j.clon.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Kondo T, Ezzat S, Asa SL. Pathogenetic mechanisms in thyroid follicular-cell neoplasia. Nat Rev Cancer. 2006;6:292–306. doi: 10.1038/nrc1836. [DOI] [PubMed] [Google Scholar]
- 4.Albores-Saavedra J, Hernandez M, Sanchez-Sosa S, et al. Histologic variants of papillary and follicular carcinomas associated with anaplastic spindle and giant cell carcinomas of the thyroid: an analysis of rhabdoid and thyroglobulin inclusions. Am J Surg Pathol. 2007;31:729–36. doi: 10.1097/01.pas.0000213417.00386.74. [DOI] [PubMed] [Google Scholar]
- 5.Hardin H, Montemayor-Garcia C, Lloyd RV. Thyroid cancer stem-like cells and epithelial-mesenchymal transition in thyroid cancers. Hum Pathol. 2013;44:1707–1713. doi: 10.1016/j.humpath.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Lloyd RV, Hardin H, Montemayor-Garcia C, et al. Stem cells and cancer stem-like cells in endocrine tissues. Endocr Pathol. 2013;24:1–10. doi: 10.1007/s12022-013-9235-1. [DOI] [PubMed] [Google Scholar]
- 7.Takano T. Fetal cell carcinogenesis of the thyroid: A modified theory based on recent evidence. Endocr J. 2014;61:311–20. doi: 10.1507/endocrj.ej13-0517. [DOI] [PubMed] [Google Scholar]
- 8.Hardin H, Zhang R, Helein H, et al. The evolving concept of cancer stem-like cells in thyroid cancer and other solid tumors. Lab Invest. 2017;97:1142–1151. doi: 10.1038/labinvest.2017.41. [DOI] [PubMed] [Google Scholar]
- 9.Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 10.Park JE, Tan HS, Datta A, et al. Hypoxic Tumor Cell Modulates Its Microenvironment to Enhance Angiogenic and Metastatic Potential by Secretion of Proteins and Exosomes. Mol Cell Proteomics. 2010;9:61085–1099. doi: 10.1074/mcp.M900381-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suetsugu A, Honma K, Saji S, et al. Imaging exosome transfer from breast cancer cells to stroma at metastatic sites in orthotopic nude-mouse models. Adv Drug Deliv Rev. 2013;65:383–90. doi: 10.1016/j.addr.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 12.Roma-Rodrigues C, Fernandes AR, Baptista PV. Exosome in tumour microenvironment: Overview of the crosstalk between normal and cancer cells. BioMed Research International. 2014:179486. doi: 10.1155/2014/179486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costa-Silva B, Aiello NM, Ocean AJ, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015;17:816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greening DW, Gopal SK, Mathias RA, et al. Emerging roles of exosomes during epithelial-mesenchymal transition and cancer progression. Semin Cell Dev Biol. 2015;40:60–71. doi: 10.1016/j.semcdb.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 15.Minciacchi VR, Freeman MR, Di Vizio D. Extracellular Vesicles in Cancer: Exosomes, Microvesicles and the Emerging Role of Large Oncosomes. Semin Cell Dev Biol. 2015;40:41–51. doi: 10.1016/j.semcdb.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Syn N, Wang L, Sethi G, et al. Exosome-Mediated Metastasis: From Epithelial-Mesenchymal Transition to Escape from Immunosurveillance. Trends Pharmacol Sci. 2016;37:606–617. doi: 10.1016/j.tips.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 17.Lee JC, Zhao JT, Gundara J, et al. Papillary thyroid cancer-derived exosomes contain miRNA-146b and miRNA-222. J Surg Res. 2015;196:39–48. doi: 10.1016/j.jss.2015.02.027. [DOI] [PubMed] [Google Scholar]
- 18.Hoshino A, Costa-Silva B, Shen TL, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blondal T, Thomsen AR, Krummheuer J, et al. Exosomal microRNA in cell-free urine samples as a source for liquid prostate cancer biopsy. Cancer Res. 2015;75:3987–3987. [Google Scholar]
- 20.Brock G, Castellanos-Rizaldos E, Hu L, et al. Liquid biopsy for cancer screening, patient stratification and monitoring. Transl Cancer Res. 2015;4:280–290. [Google Scholar]
- 21.Hurley J, O’Neill V, Brock G, et al. Abstract 4969: Exosomal RNA based liquid biopsy detection of androgen receptor variant 7 in plasma from prostate cancer patients. Cancer Res. 2016;76:4969–4969. [Google Scholar]
- 22.Barile L, Vassalli G. Exosomes: Therapy delivery tools and biomarkers of diseases. Pharmacol Ther. 2017;174:63–78. doi: 10.1016/j.pharmthera.2017.02.020. [DOI] [PubMed] [Google Scholar]
- 23.Li W, Li C, Zhou T, et al. Role of exosomal proteins in cancer diagnosis. Mol Cancer. 2017;16:145. doi: 10.1186/s12943-017-0706-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, Zheng Y, Zhao M. Exosome-based cancer therapy: Implication for targeting cancer stem cells. Front Pharmacol. 2016;7:533. doi: 10.3389/fphar.2016.00533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu CY, Du SL, Zhang J, et al. Exosomes and breast cancer: a comprehensive review of novel therapeutic strategies from diagnosis to treatment. Cancer Gene Ther. 2017;24:6–12. doi: 10.1038/cgt.2016.69. [DOI] [PubMed] [Google Scholar]
- 26.Medema JP. Cancer stem cells: The challenges ahead. Nat Cell Biol. 2013;15:338–344. doi: 10.1038/ncb2717. [DOI] [PubMed] [Google Scholar]
- 27.Craig AJ, von Felden J, Villanueva A. Molecular Profiling of Liver Cancer Heterogeneity. Discovery Medicine. 2017 Sep 1;24:117–25. [PubMed] [Google Scholar]
- 28.Mani SA, Guo W, Liao MJ, et al. The Epithelial-Mesenchymal Transition Generates Cells with Properties of Stem Cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hardin H, Guo Z, Shan W, et al. The roles of the epithelial-mesenchymal transition marker PRRX1 and miR-146b-5p in papillary thyroid carcinoma progression. Am J Pathol. 2014;184:2342–2354. doi: 10.1016/j.ajpath.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fabregat I, Malfettone A, Soukupova J. New Insights into the Crossroads between EMT and Stemness in the Context of Cancer. J Clin Med. 2016;5:37. doi: 10.3390/jcm5030037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pradella D, Naro C, Sette C, et al. EMT and stemness: flexible processes tuned by alternative splicing in development and cancer progression. Mol Cancer BioMed Central. 2017;16:8. doi: 10.1186/s12943-016-0579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buehler D, Hardin H, Shan W, et al. Expression of epithelial-mesenchymal transition regulators SNAI2 and TWIST1 in thyroid carcinomas. Mod Pathol. 2013;26:54–61. doi: 10.1038/modpathol.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montemayor-Garcia C, Hardin H, Guo Z, et al. The Role of Epithelial Mesenchymal Transition Markers in Thyroid Carcinoma Progression. Endocr Pathol. 2013;24:1–14. doi: 10.1007/s12022-013-9272-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heery R, Finn SP, Cuffe S, et al. Long Non-Coding RNAs: Key Regulators of Epithelial-Mesenchymal Transition, Tumour Drug Resistance and Cancer Stem Cells. Cancers. 2017;9:38. doi: 10.3390/cancers9040038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rinn JL, Kertesz M, Wang JK, et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sui F, Ji M, Hou P. Long non-coding RNAs in thyroid cancer: Biological functions and clinical significance. Mol Cell Endocrinol. 2017 doi: 10.1016/j.mce.2017.07.020. pii:S0303-7207:30393-3. [DOI] [PubMed] [Google Scholar]
- 37.Weidle UH, Birzele F, Kollmorgen G, et al. Long non-coding RNAs and their role in metastasis. Cancer Genomics Proteomics. 2017;14:143–160. doi: 10.21873/cgp.20027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang R, Hardin H, Huang W, et al. MALAT1 Long Non-coding RNA Expression in Thyroid Tissues: Analysis by In Situ Hybridization and Real-Time PCR. Endocr Pathol. 2017;28:7–12. doi: 10.1007/s12022-016-9453-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hardin H, Yu XM, Harrison AD, et al. Generation of Novel Thyroid Cancer Stem-Like Cell Clones Effects of Resveratrol and Valproic Acid. Am J Pathol. 2016;186:1662–1673. doi: 10.1016/j.ajpath.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fierabracci A. Identifying thyroid stem/progenitor cells: Advances and limitations. J Endocrinol. 2012;213:1–13. doi: 10.1530/JOE-11-0183. [DOI] [PubMed] [Google Scholar]
- 41.Grelet S, McShane A, Geslain R, et al. Pleiotropic roles of non-coding RNAs in TGF-β-mediated epithelial-mesenchymal transition and their functions in tumor progression. Cancers. 2017;9:75. doi: 10.3390/cancers9070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brinckerhoff CE. WORLD SCIENTIFIC. 2017. Matrix Metalloproteinases in Health and Disease. [Google Scholar]
- 43.Weidle UH, Birzele F, Kollmorgen G, et al. The multiple roles of exosomes in metastasis. Cancer Genomics and Proteomics. 2017;14:1–16. doi: 10.21873/cgp.20015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang R, Hardin H, Wei H, Buehler D, Lloyd RV. Long non-coding RNA linc-ROR is upregulated in papillary thyroid carcinoma. Endocrine Pathology. 2017 Dec 26;:1–8. doi: 10.1007/s12022-017-9507-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.