Abstract
The progression of rheumatoid arthritis involves the thickening of the synovial lining due to the proliferation of fibroblast-like synoviocytes (FLS) and infiltration by inflammatory cells. Tumor necrosis factor alpha (TNFα) is a pro-inflammatory cytokine involved in progression of the disease. Under rheumatoid conditions, FLS express the tumor necrosis factor (TNF)-recognition complex (TNFR1, TNFR2, VCAM-1 and ICAM-1), which induces local macrophage activation and leads to downstream nuclear factor κB (NF-κB) signaling. The NF-κB-regulated inflammatory gene, cyclooxygenase (COX), increases synthesis of prostaglandins that contribute to the propagation of inflammatory damage within the joint. Because the nuclear orphan receptor, NR4A2 (Nurr1), can negatively regulate NF-κB-dependent inflammatory gene expression in macrophages, we postulated that activation of this receptor by the Nurr1 ligand 1,1-bis(3′-indolyl)-1-(p-chlorophenyl) methane (C-DIM12) would modulate inflammatory gene expression in synovial fibroblasts by inhibiting NF-κB. Treatment with C-DIM12 suppressed TNFα-induced expression of adhesion molecules and NF-κB regulated genes in primary synovial fibroblasts including vascular adhesion molecule 1 (VCAM-1), PGE2 and COX-2. Immunofluorescence studies indicated that C-DIM12 did not prevent translocation of p65 and stabilized nuclear localization of Nurr1 in synovial fibroblasts. Knockdown of Nurr1 expression by RNA interference prevented the inhibitory effects of C-DIM12 on inflammatory gene expression, indicating that the anti-inflammatory effects of this compound are Nurr1-dependent. Collectively, these data suggest that this receptor may be a viable therapeutic target in RA.
Keywords: Rheumatoid arthritis, Synovial Fibroblast, Nurr1, C-DIM12, NF-κB, TNFα
1. INTRODUCTION
Rheumatoid arthritis (RA) has a prevalence of 1–2% and is one of the common causes of morbidity among people over 65 years of age (Gabriel, S.E., 2001). It is characterized by hyperplasia of fibroblast-like synovial cells (FLS) within the synovium and recruitment of multiple leukocyte populations that drive the inflammatory process (Hardy et al., 2013). Although disease etiology is unknown, it is thought that both genetic and environmental factors in which genes have been influence such as medical illnesses and factors in the external environment such as pollution can trigger the onset of RA (Smolen, J. S., and Aletaha, D.,2009). RA synovial fibroblasts represent a unique cell type that distinguishes RA from other inflammatory conditions of the joints. Pro-inflammatory mediators including tumor necrosis factor alpha (TNFα) and interleukin 1 beta (IL1β) are produced by macrophage cells, T-cells and B-cells to promote synoviocyte cell proliferation including the release of matrix-degrading enzymes with the induction of pro-angiogenic factors and secretion of cytokines and chemokines (Tak,P.P. and Bresnihan, B., 2000). Synovial hyperplasia is mediated by these pro-inflammatory cytokines resulting in chronic inflammation and tissue destruction in RA. In response to these cytokines, FLS produce chemokines and adhesion molecules that further promote inflammation, hyperplasia, and cartilage destruction (Honda et al., 2006; Choy and Panayi, 2001).
Binding of tumor necrosis factor alpha (TNFα) to vascular adhesion molecule 1 (VCAM-1) and intracellular adhesion molecule 1(ICAM-1) in synovial fibroblasts activates the inflammatory response, which is critical in the recruitment of immune cells to affected joints (Umar et al., 2015). As a part of the synovial tissue reaction, proliferating synovial cells penetrate the cartilage in the form of a pannus. The mechanisms responsible for pannus formation are not fully understood, however synovial hyperplasia and formation into the pannus is the fundamental pathogenesis of RA (Panayi, 2001) Previous studies have established that TNFα is a critical effector in the pro-inflammatory cytokine cascade in RA (Ntougkos, et al., 2017; Hardy et al., 2013; Feldmann et al., 1996) that induces production of pro-inflammatory mediators, such as prostaglandin E2 (PGE2), a cyclooxygenase 2 (COX-2) metabolite that sensitizes nociceptors and promote an inflammatory response within the joint (Goel et al., 2003).
Nuclear factor-kappaB (NF-κB) is an important transcription factor that regulates expression of inflammatory gene, including COX-2 (Karin M., 2009). Activation of NF-κB by various stimuli, including cytokines (TNFα) and lipopolysaccharide (LPS), results in nuclear translocation of the p65/p50 subunit and transactivation of numerous inflammatory cytokines and adhesion molecules (De Miranda et al., 2014; Lawrence, T.,2009). Inhibition of this pathway by knockdown of IκB kinase (IKK) can reduce inflammatory gene expression in models of tissue injuiry (Acharyya et al., 2007) but pharmacological inhibitors of this pathway have proven difficult to translate to clinical applications. The orphan nuclear receptor, NR4A2 (Nurr1) has emerged as an important endogenous regulator of NF-κB and modulates the expression of inflammatory cytokines and chemokines in diseases characterized by prolonged or inappropriate inflammatory responses, such as RA (Bonta et al., 2006; Holla et al., 2006; McEvoy et al., 2002; Mix et al., 2007). Unlike most nuclear receptors, the NR4A family are rapidly activated by inflammatory mediators, suggesting their respective receptors may act as potential transcriptional mediators of cytokine signaling (Bonta et al., 2006; Holla et al., 2006; McEvoy et al., 2002; Mix et al., 2007). In RA, joint tissues produce markedly increased levels of NR4A2 compared to other NR4A family members (McEvoy et al., 2002; Mix et al., 2007). NR4A2 is encoded within an immediate early gene that is rapidly induced in cells in response to external stimuli such as cytokines. Microarray analysis has established that over-expression of NR4A2 (Nurr1) in K4 IM synoviocytes inhibits the expression of pro-inflammatory genes, including IL-8 (Davies et al., 2005).
In this study, we investigated one of a novel series of paraphenyl substituted dinndolylmethane compounds (C-DIMs) that has been previously demonstrated to be a transcriptional modulator of NR4A family nuclear receptors (De Miranda et al., 2015; Lee et al., 2014; Yoon et al., 2011; Inamoto et al., 2008). C-DIM12 activates Nurr1 in dopaminergic neurons (Hammond et al., 2015), microglial cells (De Miranda et al.,2015), pancreatic cells (Li et al.,2012) and in urothelial carcinoma cells (Inamoto et al., 2008). However, it is unknown whether C-DIM12 directly inhibits inflammatory gene transcription in primary synovial fibroblasts. Therefore, in this study we postulate that C-DIM12 inhibits inflammatory signaling in primary synovial fibroblasts by a Nurr1-dependent mechanism. The data presented here provides evidence that C-DIM12 suppresses adhesion molecules and pro-inflammatory cytokines in synovial fibroblasts after stimulation with tumor necrosis factor (TNFα). C-DIM12 induces Nurr1 expression resulting in suppression of TNFα-induced inflammatory cytokine secretion, by a nuclear-specific mechanism. These results provide a novel mechanism for suppression of inflammation within synovial fibroblasts and therefore pose a new possibility for therapeutic intervention aimed at reducing synovial tissue destruction in RA.
2. MATERIALS AND METHODS
2.1. Reagents
C-DIM12 was synthesized as described by Qin et al. (2004). All general chemical reagents were purchased from Sigma, Aldrich (St. Louis, MO) unless stated otherwise. TNFα was purchased from R&D Systems (Minneapolis, MN). Specific primary antibodies against VCAM-1(55330), ICAM-1 (553253), Mac-1(Cd11b) (553310), CD90.2 (553011), COX-2 (610204) were purchased from BD Biosciences (San Jose, CA), antibodies against Nurr1 (SC-376984) and NF-κB p65 (SC-8008) were purchased from Santa Cruz Biotechnology (Dallas, TX), antibody against Vimentin (AB1620) was purchased from EMD Millipore (Burlington, MA), and Iκβα (4814) was purchased from Cell Signaling Technology (Danvers, MA).
2.2. Primary Cell Isolation
Primary synovial fibroblasts were isolated from the talocrural joint of wild type (WT) C57BL/6 6–8-week-old mice according to procedures described by Armaka et al., 2009 and purity was confirmed through FACS and immunofluorescent staining using antibodies against CD90.2, Cd11b, VCAM-1, ICAM-1 and vimentin. In brief, 6–8-week-old mice were euthanized using carbon dioxide, hind legs were rapidly dissected out with the foot intact. Tissue was subjected to digestion with collagenase from Clostridium hitolyticum type IV (1mg/ml). This method routinely results in cultures that are approximately 99% pure synovial fibroblasts with less than 1% contaminating macrophage cells (Figure 1). Synovial fibroblast cultures were maintained in Dulbecco’s modified Eagle’s media supplemented with 10% FBS, and 1xPSN. The cells were maintained at 37°C with 5% CO2. Synovial fibroblasts were used up to passage 3 of plating. All procedures involving animals were approved by the Colorado State University Institutional Animal Care and Use committee and were conducted in accordance with current National Institutes of Health guidelines.
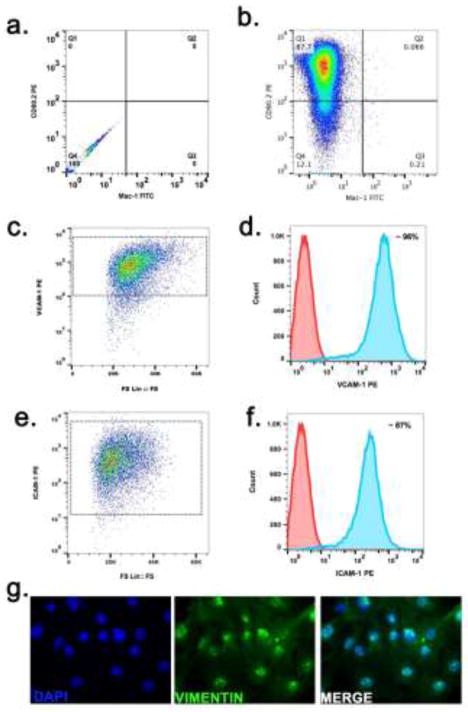
Figure 1. Immunophenotyping of Primary Synovial Fibroblasts.
Isolated synovial fibroblasts from WT mice were subjected to FACS analysis on passage 3 after seeding to verify purity. (a) negative control for dual stain cells (b) Density plot showing expression of CD90.2 on the majority of the isolated cells (>90 %) whereas very few Mac-1 positive myeloid cells can be detected (<1%). (c) FS Lin vs VCAM-1 PE scatter plot showing cluster positive for VCAM-1 expression (d) Overlaid histogram showing VCAM-1 (~96%) (e) FS Lin vs ICAM-1 PE scatter plot showing cluster positive for ICAM-1 expression (f) ICAM-1 expression (87%) (blue shaded area) on SFs or unstained control (red shaded area) (g) representative image for Vimentin expression in synovial fibroblasts.
2.3. RNA interference
RNA interference (siRNA/RNAi, small interfering RNA) sequences were obtained through Integrated DNA technologies (IDT DNA, Coralville, IA). Nurr1 RNAi duplexes were designed against splice common variants of Nurr1 and validated using a dose response assay with increasing concentrations of the suspended oligo (1.2 μg) using a standard scrambled dicer-substrate RNA as control. RNAi oligos were transfected using the TransIT-X2 delivery system (Mirus Bio, Madison, WI) for 24 hours (Figure 6a). Separate siRNA systems were used to ensure specific knockdown of Nurr1 mRNA. The Nurr1 DsiRNA duplex sequences (5′->3′) CUAGGUUGAAGAUGUUAUAGGCACT; AGUGCCUAUAACAUCUUCAACCUAGAA (IDT DsiRNA, denoted RNAi).
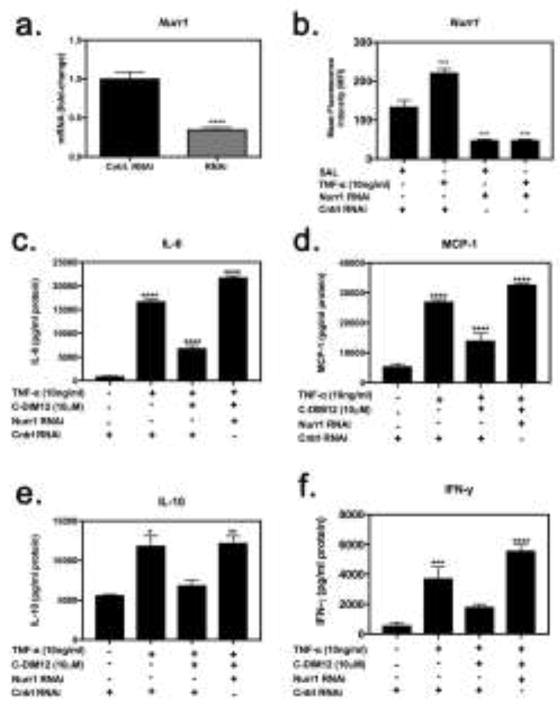
Figure 6. C-DIM12-dependent inhibition of inflammatory gene expression requires Nurr1.
(a) Synovial fibroblasts were treated with concentrations of siRNA (denoted RNAi) or scrambled RNAi (control RNAi) demonstrating 1.2ug to be optimal. Synovial fibroblasts were transfected with Nurr1 RNAi or control RNAi for 24 hours followed by saline or 10 ng/ml TNFα with or without C-DIM12 for 24 hours and assessed for Nurr1 expression via flow cytometry. (b) Nurr1 was effectively knocked-down at a protein level, even in the presence of TNFα. (c–f) IL-6, MCP-1, IL-10 and IFN-γ protein expression upon knock-down and treatment was assessed via flow cytometry demonstrating a loss of C-DIM12-mediated suppression upon Nurr1 knock-down. Data are expressed as mean ± S.E.M (n=4). Statistical significance is compared with saline control. *P < 0.05, **P<0.01, ***P<0.001, ****P<0.0001.
2.4. Gene Expression
Nurr1 mRNA expression upon knockdown was assessed by quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR). Briefly, RNA was isolated using the RNEasy Mini kit (Qiagen, Valencia, CA). Purity and concentration were determined using a Nanodrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). RNA (250–1000ng) was then used as template for reverse transcriptase (RT) reactions using the iScript RT kit (BioRad, Hercules CA). cDNA was immediately profiled for Nurr1 gene expression (forward: 5′-GTGTTCAGGCGCAGTATGG-3′; reverse: 5′-TGGCAGTAATTTCAGTGTTGGT-3′) using HPRT as a housekeeping gene (forward: 5′-TCAGTCAACGGGGGACATAAA-3′; reverse: 5′-GGGGCTGTACTGCTTAACCAG-3′) according to the 2-ΔΔCT method (Livak and Schmittgen, 2001).
2.5. Enzyme-linked Immunosorbent Assay
Primary synovial fibroblasts were pre-treated for an hour with or without 10 μM C-DIM12 followed by 10ng/ml TNFα for 12 h prior to harvesting conditioned media. The conditioned media was preserved with 1% BHT solution to avoid the free-radical peroxidation as explained in by Cao et al., 2008. Samples were stored at −20°C until analysis. PGE2 analysis and quantification was conducted via ELISA using a PGE2 analysis kit (R&D Systems) in accordance with the manufacturer’s instructions, except samples were not diluted as indicated. The experiment was conducted three independent experiment runs in triplicate.
2.6. Immunofluorescence Protein Expression and Translocation
Synovial Fibroblasts (5 × 104 cells/per coverslip) were plated on cover glass (preconditioned with FBS) and allowed to adhere for 18 hours. Cells were then pretreated for 1 hour with 10 μM C-DIM12 followed by 10 ng/ml TNFα for 30 minutes. Cells were washed with PBS buffer and fixed with 4% paraformaldehyde for 10 minutes at 4°C. Cells were blocked using bovine serum albumin (Sigma-Aldrich) for 1 hour at room temperature followed by incubation with primary antibodies overnight at 4°C. Secondary antibody incubation was at room temperature for 2 hours with Alexa Flour 555 (Life technologies) and DAPI counterstain (Vector Laboratories, Burlingame, CA). Slides were imaged using a Zeiss Axiovert 200M inverted fluorescence microscope equipped with a Hammamatsu ORCA-ER-cooled charge-coupled device camera (Hammamatsu Photonics, Hamamatsu City, Japan) using slidebook software (version 5.5, Intelligent Imaging Innovation, Denver, CO). Total nuclear protein was quantified by fluorescence intensity of NF-κB p65 or Nurr1 within the nuclear region of the cell, as determined by DAPI counterstain. Slidebook 5.0 software generated fluorescence intensity minus background and six to eight microscopic fields were examined per treatment group, with at least 3 biological replicates (coverslips) per treatment group, over no less than three independent experiments. Fluorescent secondary antibodies were used to detect phalloidan (excitation at 488 nm; emission at 519 nm) and NF-κB p65 or Nurr1 (excitation at 555 nm; emission at 565 nm), respectively, whereas mounting medium contain 4,6-diamidino-2-phenlindole (excitation at 358 nm; emission at 461 nm) was used to identify cell nuclei.
2.7. Fluorescence-activated cell sorting (FACS) Analysis
Synovial fibroblasts (3 × 105 per well) were plated in 6 well plates and allowed to adhere for 18 hr. The cells were then pretreated with for 1 hr. with 10 μM C-DIM12 followed by 10ng/ml TNFα for 12 hr. The cells were then harvested and fixed with 2% paraformaldehyde for 10 mins. Cells membranes were permeabilized with 0.01% triton X-100 in PBS for 30 mins and stained with antibodies against NF-κB p65, COX-2, Nurr1 and IκBα for 1 hr. at 37 °C. Secondary incubation was at 37 °C for 1 hr. with Alexa flour 647. Unstained cells were used as negative controls during acquisition. Flow cytometry was conducted on a Beckman Coulter CyAnADP flow cytometer operating with Summit v4.3 software for data collection. All further analysis was done with FlowJo (version 10.0.8; Software, Ashland, OR). Samples were run in biological triplicates with two technical duplicates.
2.8. Cytokine/chemokine determination in cell culture supernatants by Cytometric Bead Array (CBA)
Supernatant was collected and immediately processed for Interleukin-6 (IL-6), interleukin-10 (IL-10), monocyte Chemoattractant protein-1 (MCP-1), and interferon-γ (IFN-γ) protein levels were quantitatively measured by BD CBA Mouse Inflammation Kit (BD Biosciences, San Jose, CA). The protocol was performed according to the manufacturer’s instructions. The intensity of fluorescence signal was acquired on a Beckman Coulter CyAnADP Flow cytometry operating with Summit v4.3 software for data collection. All further analysis was done with FlowJo (version 10.0.8; Software, Ashland, OR). Samples were run in biological triplicates with two technical duplicates.
2.9. Statistical analysis
Statistical analyses were performed using Prism (version 7.0; Graph Pad Software, San Diego, CA). Data are presented as mean ± S.E.M. Experimental group analyses were performed using a one-way analysis of variance with a Tukey post hoc test. *P < 0.05, ** P < 0.01, ***P < 0.001, **** P< 0.0001 were considered statistically significant.
3. RESULTS
Purity of primary cultured synovial fibroblasts was determined by FACS three passages after plating (Figure 1). Cells were stained for constitutive expression of CD90.2, VCAM-1 and ICAM-1 to confirm SF phenotype (Fig 1a–f). CD90.2 was >80–90% of the cell population, whereas Cd11b, a monocyte marker, was <1% of the population, indicating that primary cultures were comprised primarily of SF cells (Fig 1a and b). Expression of the cell adhesion molecules VCAM-1 and ICAM-1 was evaluated by FACS, with representative scatter plots and histograms showing positive expression for both cell surface markers (Fig 1c and f), confirming the purity of the murine synovial fibroblasts isolation. Immunofluorescence imaging confirmed expression of Vimentin, a mesenchymal marker (Fig 1g).
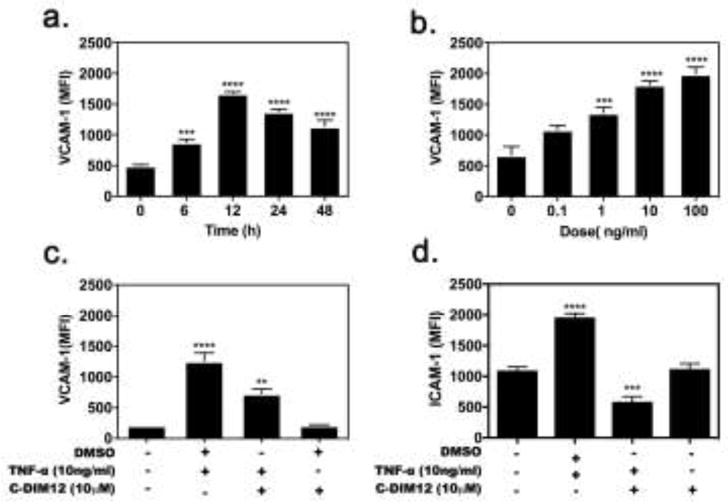
Treatment with 10 ng/ml TNFα caused both dose- and time-dependent increases in expression of VCAM-1 in synovial fibroblasts by 6 hr that peaked at 12 hr and remained elevated up to 48 hr (Figure 2a). Based on the data in Figure 2a and b, we selected treatment with 10 ng/ml TNFα for 12 hr to be optimal, as expression of VCAM-1 was nearly as saturated as with 100 ng/ml. Co-treatment of SF cells with C-DIM12 (10 μM) effectively suppressed VCAM-1 and ICAM-1 expression after treatment for 12 hr. (Fig 2c–d). In cells treated with C-DIM12 (10 μM) alone, there was no difference in VCAM-1 and ICAM-1 expression (Fig 2c–d), and expression was similar to basal levels.
Figure 2. TNFα induced VCAM-1 and ICAM-1 expression in synovial fibroblasts.
(a) TNFα dose-response of VCAM-1 expression was determined to be most optimal at 12 h (b) Primary synovial fibroblasts were cultured in a dose dependent manner with TNFα (10 ng/ml) for 12hr. VCAM-1 surface expression was determined by FACS, as described in Methods. TNFα-induced (c) VCAM-1 and (d) I-CAM1 expression is decreased upon C-DIM12 treatment. Data are expressed as mean ± S.E.M (n=4). Statistical significance is compared with saline control. *P < 0.05, **P<0.01, ***P<0.001, ****P<0.0001.
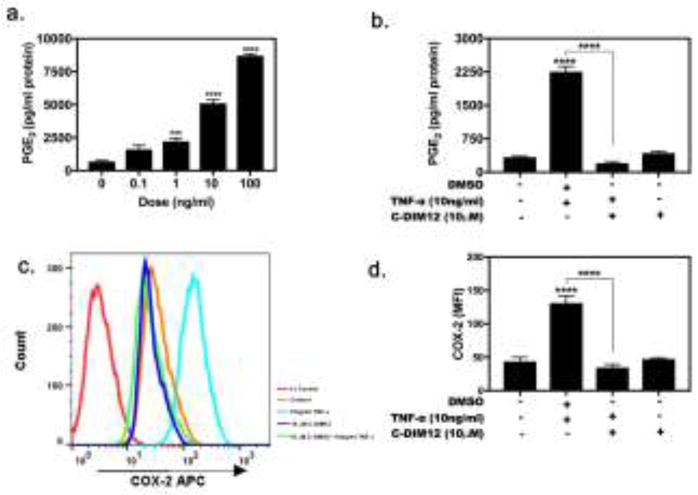
Exposure of synovial fibroblasts to TNFα (0.1 – 100 ng/ml) increased production of PGE2 in a dose-dependent manner (Figure 3), with the highest levels of PGE2 detected at 100 ng/ml TNFα following 24-hr exposure (Fig 3a). To assess the effect of C-DIM12 on PGE2 production, we exposed synovial fibroblasts to TNFα in the absence or presence of C-DIM12. C-DIM12 alone didn’t not regulate PGE2 production. Co-treatment with C-DIM12 significantly reduced TNFα induced PGE2 production (Fig 3b) and therefore we also examined the effect of C-DIM12 on TNFα-induced expression of COX-2 by FACS. Low expression of COX-2 protein was observed in resting synovial fibroblasts and COX-2 was highly induced in the presence of TNFα, and with C-DIM12 (10 μM) alone, there was no difference in COX-2 protein expression this was similar to untreated control cells (Fig 3c and d). C-DIM12 significantly inhibited induced COX-2 expression comparable to basal levels detected in control synovial fibroblasts (Fig 3c and d).
Figure 3. C-DIM12 Decreases Inflammatory PGE2 and COX-2 Expression in Synovial Fibroblasts.
(a) PGE2 dose response determined after 24 hrs of TNFα (10ng/ml) stimulation (b) Upon TNFα (10ng/ml) exposure, PGE2 expression is increased and effectively suppressed upon 10μM C-DIM12 treatment. (c) Overlaid histogram showing COX-2 expression in stimulated synovial fibroblast fluorescence intensity shifted to the right increase COX- and followed by treatment of C-DIM12 (10 μM) decrease back to basal condition. (d) Synovial fibroblasts were stimulated with TNFα (10ng/ml) treated with 10μM CDIM12 analyzed for COX-2 expression. Data are expressed as mean ± S.E.M (n=4). Statistical significance is compared with saline control. *P < 0.05, **P<0.01, ***P<0.001, ****P<0.0001.
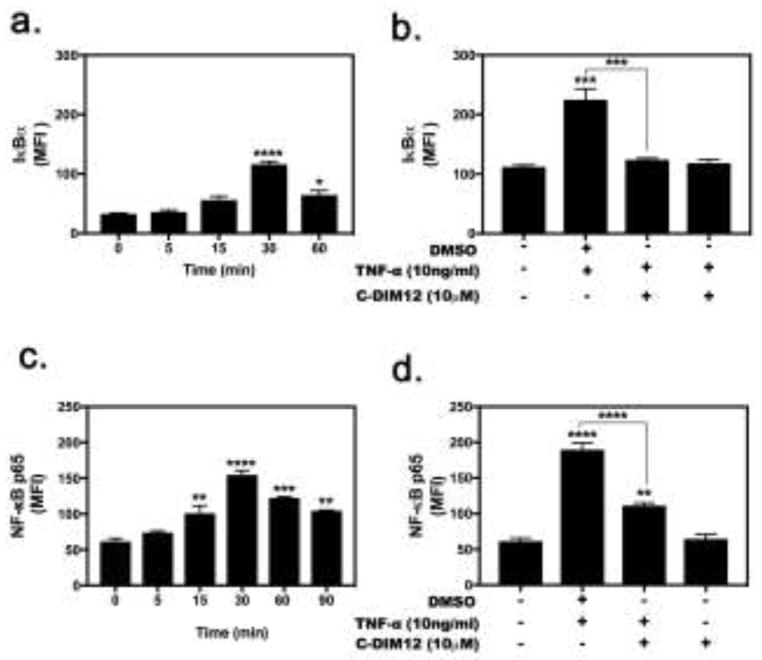
To examine the mechanism by which C-DIM12 inhibited TNF-induced inflammatory gene expression in synovial fibroblasts, we assessed total levels of IκBα and p65 (NF-κB). Flow cytometry analysis revealed that IκBα protein expression decreased significantly following treatment with C-DIM12 (Figure 4a–b). Protein expression of TNF-stimulated synovial fibroblasts was analyzed for expression of p65 using flow cytometry and demonstrated a marked increase in p65 protein levels following
Figure 4. C-DIM12 decrease IκBα and p65 NF-κB Expression in Synovial Fibroblasts.
(a and c) The dose response of IκBα and NF-κB expression was determined 30 mins after TNFα (10 ng/ml). (b and d) synovial fibroblasts were stimulated with TNFα (10ng/ml) treated with 10μM CDIM12 analyzed by flow cytometry. Data are expressed as mean ± S.E.M (n=4). Statistical significance is compared with saline control. *P < 0.05, **P<0.01, ***P<0.001, ****P<0.0001.
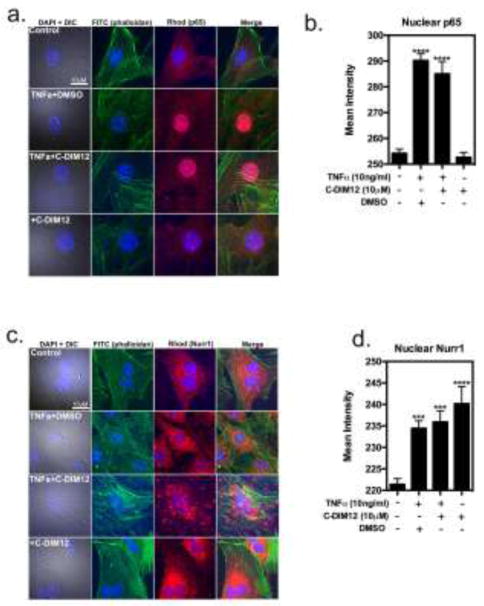
To determine whether C-DIM12 inhibits inflammatory protein expression by inhibiting nuclear translocation of NF-κB (p65), cells were stimulated with 10 ng/ml TNFα in the presence and absence of C-DIM12 (10 μM). In untreated cells, p65 was predominantly localized to the cytoplasm (Figure 5a–b) and cells stimulated with TNFα demonstrated an increase in nuclear translocation of p65 (Fig 5a–b). C-DIM12 (10 μM) did not prevent nuclear translocation of p65 in cells stimulated with TNFα (Fig 5a–b). In cells treated with C-DIM12 (10 μM) alone, there was no nuclear translocation of p65 (Fig 5a–b), and this was similar to untreated control cells. To investigate the effect of C-DIM12 protein expression of Nurr1, cells were also stimulated with TNFα in the presence and absence of C-DIM12. In untreated cells, Nurr1 was predominately localized to the cytoplasm (Fig 5c–d). In cells stimulated with TNFα, there was a marked increase in nuclear localization of Nurr1, which was further enhanced by co-treatment with C-DIM12 (Fig 5d). In cells treated with C-DIM12 (10 μM) alone, there was a similar increase in nuclear Nurr1 compared to control cells (Fig 5d).
Figure 5. C-DIM12 does not prevent p65 translocation and promotes Nurr1 sequestration in the nuclei of TNFα-induced primary synovial fibroblast cells.
Primary synovial fibroblasts were treated with 10μm C-DIM12 for 1 hour followed by saline or 10 ng/ml of TNFα for 30 mins and fixed for immunofluorescence to examine p65 and Nurr1 translocation with DAPI(blue), phalloidan (green), p65 (red). (a–b) Representative images and quantification of p65 nuclear expression was quantified by mean fluorescence intensity encompassing the nuclei (DAPI boundary; background subtracted), demonstrating increased nuclear p65 upon TNFα with or without C-DIM12(c–d). Representative images and quantification of Nurr1 expression following TNFα and C-DIM12 treatment showed increased Nurr1 protein translocation from the cytoplasm to the nucleus. Data are expressed as mean ± S.E.M. (n=3). *P < 0.05, **P<0.01, ***P<0.001, ****P<0.0001.
We used RNAi to examine whether the anti-inflammatory effects of C-DIM12 on NF-κB regulated inflammatory genes were Nurr1-dependent (Figure 6). Upon transfection of synovial fibroblasts with siRNA directed against Nurr1 (1.2ug), levels of Nurr1 mRNA were reduced by approximately 70% (Fig 6a). Following Nurr1 knock-down, synovial fibroblasts were treated with TNFα for 24 hours and then assayed via flow cytometry, to measure Nurr1 protein expression, was significantly decreased (Fig 6b) and after treatment with C-DIM12, synovial fibroblasts were further analyzed for expression of NF-κB regulated inflammatory genes, IL-6, MCP-1, IL-10 and IFN-γ. The results show that suppression of TNFα-induced expression of IL-6, MCP-1, IL-10 and IFN-γ (Fig 6c–f) by C-DIM12 was lost after knockdown of Nurr1 demonstrating a role for this receptor and its ligand in treatment of RA.
4. DISCUSSION
RA is an inflammatory disease where the pathology is both triggered and sustained by the influx of expression of cytokines, which are responsible for the destruction of the joint (Li, P., et al., 2000). Pro-inflammatory cytokines that are overexpressed in RA joints are shown to induce a variety of disease genes, including other cytokines, proteases, cyclooxygenases and adhesion molecules, in addition to stimulating cell proliferation (Shi et al., 2004 and Feldmann et al., 1996). The pro-inflammatory cytokine, TNFα, has been extensively researched because of its involvement in the pathogenesis of RA. TNFα mediates the cellular infiltration of the joint while the development of the pannus occurs following cytokine released by mononuclear cells (including lymphocytes and monocytes), which prompt the synovial membrane’s blood vessels to multiply. This ultimately causes an increased blood flow leading to excessive tissue growth of the synovial cells resulting in the thickening of the synovium. The inflammatory process is further exacerbated by release of soluble forms of the adhesion molecules, VCAM-1 and ICAM-1 (Hideshima et al., 2002). Thus, the signaling pathway involved in the induction of VCAM-1 by TNFα in primary synovial fibroblasts is a potential target for alternative therapeutics.
Fibroblast-like synoviocytes have been shown to play a central role in mediating inflammatory damage to joints during the progression of RA (Angiolilli et al., 2016). To study RA progression, researchers have often utilized human FLS isolated from synovial tissue from patients undergoing joint arthroplasty (Mix et al., 2007), however fewer studies have modeled joint disease using murine cells. In the present study, we successfully isolated FLS from C57BL/6 mice and characterized the cellular response to inflammatory stimulation with TNFα in order to model the molecular signaling that occurs during RA. Cells were characterized based upon multiple criteria including general fibroblast markers such as VCAM-1, ICAM-1, CD90.2, vimentin and Cd11b (Armaka et al., 2009). Alone, CD90.2 stains numerous cell types including subsets of fibroblasts, thymocyte populations, epithelial cells and neurons (Rege et al., 2006). By contrast, CD11b has been shown to be a selective marker for inflammatory cells such as macrophages and microglia. Evaluation of synovial fibroblast purity by flow cytometry revealed > 80–90 % CD90.2 positive, whereas < 1% CD11b positive (Fig 1b). Negative control (Fig 1a) and compensation was taken into consideration when defining positive populations. Further FACs analysis of adhesion molecules constitutively expressed on synovial fibroblasts cell surface were evaluated 96% positive for VCAM-1 (Fig 1c and d) and 87% positive ICAM-1 (Fig 1e and f). Finally, vimentin (Fig 1g) confirmation by immunofluorescence confirmed the purity of the synovial fibroblast isolation.
Enhanced Nurr1 expression in the synovial lining layer, synovial fibroblasts of inflamed synovial tissue, confirms that Nurr1 is expressed primarily in cells believed to be the leading edge of invasive tumor-like synovium (pannus) (Ralph et al., 2005). Primary synovial fibroblasts expressed VCAM-1 and responded to TNFα in a time and dose dependent manner, peak activity at 12-hour stimulation with 10 ng/ml TNFα stimulation (Fig 2a and b). Dose-dependent inhibition of adhesion molecules VCAM-1 and ICAM-1 in synovial fibroblasts indicate that C-DIM12 blocks expression of the TNFα-induced inflammatory response (Fig 2c and d).
The progression of rheumatoid arthritis depends on the activation of a complex signaling network of cytokines and adhesion molecules. Previous studies have shown C-DIM12 activates Nurr1 in dopaminergic neurons (Hammond et al., 2015), in microglia cells (De Miranda et al., 2015), in pancreatic cells (Li et al., 2012) and in urothelial carcinoma cells (Inamoto et al., 2008). Here we demonstrate that C-DIM12 suppresses NF-κB-induced gene expression in synovial fibroblasts through a mechanism involving transcriptional upregulation of Nurr1, as well as enhanced nuclear localization of the receptor. This is consistent with previous studies demonstrating that C-DIM12 inhibits NF-κB-induced gene expression in macrophages by stabilizing nuclear co-repressor proteins on NF-κB cis-acting elements in the promoters of inflammatory genes such as inducible nitric oxide synthase (iNOS/NOS2) (De Miranda et al., 2015). ELISA data revealed that stimulation of primary synovial fibroblasts with TNFα induced production of PGE2 in a dose-dependent manner (Fig 3a). C-DIM12 suppressed PGE2 production and COX-2 expression in TNFα-stimulated synovial fibroblasts (Fig 3b–d) and also suppressed protein expression of the NF-κB pathway proteins, p65 and IκBα (Fig 4a and b). Despite the moderate decrease in levels of IκBα, the commensurately greater decrease in p65 protein expression suggests that C-DIM12 broadly suppresses the NF-κB pathway at a transcriptional level. Given that both p65/RelA and IκBα are gene targets of NF-κB, C-DIM12 likely inhibits transcription of both p65 and IκBα at NF-κB-responsive 5′ cis elements by stabilizing nuclear co-repressor protein complexes, similar to previous reports in macrophages (De Miranda et al., 2015).
Phosphorylation and degradation of IκBα following phosphorylation by IKK leads to nuclear translocation of p65 and transcriptional activation of target genes. The observed increase in nuclear p65 after treatment with TNFα + C-DIM12 (Fig 5a) suggests a mechanism involving increased association between Nurr1 bound to p65, as reported in macrophages following stimulation with LPS (Saijo et al., 2009). Given a recent report that C-DIM12 is predicted to bind with high affinity to the co-activator site of the Nurr1 ligand binding domain and also activates a Nurr1 reporter construct in neural cells (Hammond et al., 2018), these data also suggest that C-DIM12 directly activates Nurr1 in synovial fibroblasts to inhibit NF-κB. Furthermore, immunofluorescence confirmed that C-DIM12 did not prevent TNFα-induced translocation of p65 (Fig 5a and b) suggesting a nuclear-specific mechanism. Therefore, we performed quantitative immunofluorescence to investigate nuclear Nurr1 recruitment by C-DIM12. As predicted, localization of Nurr1 changes upon stimulation with TNFα (Fig 5c), however nuclear Nurr1 was significantly increased upon TNFα + C-DIM12 treatment and C-DIM12 treatment alone (Fig 5d).
Moreover, knockdown of Nurr1 by RNAi significantly decreased the ability of C-DIM12 to inhibit protein expression for multiple inflammatory cytokines (Fig 6, a–f). Nurr1 RNAi enhanced protein expression of the NF-κB-regulated inflammatory cytokines, IL-6 and MCP-1 (Fig 6c and d). Inhibition of inflammatory cytokines in primary synovial fibroblasts indicates that C-DIM12 blocks expression of NF-κB regulated inflammatory cytokines and chemokines. RNAi studies (Fig 6) show that C-DIM12 does not suppress TNFα-induced expression of IL-10 and IFNγ to the same level as the other cytokines assayed, however complete suppression is lost upon knockdown. These data support that C-DIM12 requires Nurr1 to suppress pro-inflammatory gene expression in TNFα-induced synovial fibroblasts.
Expression of IL-10 and IFN-γ in synovial fibroblasts is also regulated by Janus kinase/signal transducers and activators of transcription (JAK/STAT) pathways in addition to NF-κB (Dey et al., 2016). However, the studies in Fig 6 indicated that knockdown of Nurr1 expression significantly decreased the inhibitory effects of C-DIM12 on TNFα-induced expression of IL-10 and IFN-γ, suggesting that both NF-κB and JAK/STAT pathways regulate the expression of these inflammatory proteins in synovial fibroblasts following stimulation of tumor necrosis factor receptor (TNFR). Nevertheless, the efficacy of Nurr1 RNAi in preventing the inhibitory effects of C-DIM12 on these inflammatory mediators indicates that NF-κB remains an important pathway for their transcriptional activation in synovial fibroblasts in response to stimulation with TNFα.
It remains a challenge to find small molecule therapies that can reduce inflammatory joint injury in RA. Nurr1 has previously been shown to act as a NF-κB signaling following stimulation with multiple inflammatory mediators (McEvoy et al., 2002). The data presented here demonstrate that the substituted diindolylmethane compound, C-DIM12, suppresses NF-κB signaling in synovial fibroblasts by a nuclear-specific mechanism and subsequently decreases the expression of several inflammatory cytokines, chemokines and cellular adhesion molecules in response to treatment with TNF. This anti-inflammatory activity occurs through a Nurr1-dependent mechanism, as evidenced by the efficacy of RNAi directed against Nurr1 in ablating the inhibitory effects of C-DIM12. Pharmacological modulation of inflammation in the joint with C-DIM12 may therefore represent a novel mechanism decrease expression of NF-κB regulated inflammatory genes in synovial fibroblast cells and is relevant to the mechanism of pathology in this inflammatory joint disease.
Supplementary Material
Highlights.
A novel Nurr1 activator (C-DIM12) inhibits gene expression in primary murine synovial fibroblasts.
C-DIM12 decreases expression of NF-κB regulated inflammatory genes in primary synovial fibroblast.
Nurr1 RNAi prevents the anti-inflammatory effects of C-DIM12 in primary synovial fibroblasts.
C-DIM12 modulates inflammatory response in primary synovial fibroblast.
Acknowledgments
These studies were supported by research funding from the Center for Environmental Medicine at Colorado State University and by grants from the National Institutes of Health ES021656 (RBT) and ES026860 (KAP). We also wish to thank the CSU Flow Cytometry and Cell Sorting Facility (CSUFCF) for their assistance with flow cytometry, as well as Lacey Fleming for her technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acharyya S, Villalta SA, Bakkar N, Bupha-Intr T, Janssen PML, Carathers M, et al. Interplay of IKK/NF-kappaB signaling in macrophages and myofibers promotes muscle degeneration in Duchenne muscular dystrophy. Journal of Clinical Investigation. 2007;117(4):889–901. doi: 10.1172/JCI30556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aherne CM, McMorrow J, Kane D, FitzGerald O, Mix KS, Murphy EP. Identification of NR4A2 as a transcriptional activator of IL-8 expression in human inflammatory arthritis. Molecular Immunology. 2009;46(16):3345–3357. doi: 10.1016/j.molimm.2009.07.019. [DOI] [PubMed] [Google Scholar]
- Angiolilli C, Grabiec AM, Ferguson BS, et al. Inflammatory cytokines epigenetically regulate rheumatoid arthritis fibroblast-like synoviocyte activation by suppressing HDAC5 expression. Annals of the Rheumatic Diseases. 2016;75:430–438. doi: 10.1136/annrheumdis-2014-205635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armaka M, Gkretsi V, Kontoyiannis D, Kollias G. A standardized protocol for the isolation and culture of normal and arthritogenic murine synovial fibroblasts 2009 [Google Scholar]
- Bonta PI, van Tiel CM, Vos M, Pols TWH, van Thienen JV, Ferreira V, et al. Nuclear receptors Nur77, Nurr1, and NOR-1 expressed in atherosclerotic lesion macrophages reduce lipid loading and inflammatory responses. Arteriosclerosis, Thrombosis, and Vascular Biology. 2006;26(10):2288–2294. doi: 10.1161/01.ATV.0000238346.84458.5d. [DOI] [PubMed] [Google Scholar]
- Cao H, Xiao L, Park G, Wang X, Azim AC, Christman JW, et al. An improved LC–MS/MS method for the quantification of prostaglandins E2 and D2 production in biological fluids. Analytical Biochemistry. 2008;372(1):41–51. doi: 10.1016/j.ab.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy EH, Panayi GS. Cytokine pathways and joint inflammation in rheumatoid arthritis. New England Journal of Medicine. 2001;344(12):907–916. doi: 10.1056/NEJM200103223441207. [DOI] [PubMed] [Google Scholar]
- Davies MR, Harding CJ, Raines S, Tolley K, Parker AE, Downey-Jones M, Needham MRC. Nurr1 dependent regulation of pro-inflammatory mediators in immortalised synovial fibroblasts. Journal of Inflammation (London, England) 2005;2(1):15. doi: 10.1186/1476-9255-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Miranda BR, Popichak KA, Hammond SL, Jorgensen BA, Phillips AT, Safe S, Tjalkens RB. The Nurr1 Activator 1,1-Bis(3′-Indolyl)-1-(p-Chlorophenyl) Methane Blocks Inflammatory Gene Expression in BV-2 Microglial Cells by Inhibiting Nuclear Factor kappaB. Molecular Pharmacology. 2015;87(6):1021–1034. doi: 10.1124/mol.114.095398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey P, Panga V, Raghunathan S. A Cytokine Signalling Network for the Regulation of Inducible Nitric Oxide Synthase Expression in Rheumatoid Arthritis. Raju R, ed. PLoS ONE. 2016;11(9):e0161306. doi: 10.1371/journal.pone.0161306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann M, Brennan FM, Maini RN. Rheumatoid arthritis. Cell. 1996;85(3):307–310. doi: 10.1016/s0092-8674(00)81109-5. [DOI] [PubMed] [Google Scholar]
- Gabriel SE. The epidemiology of rheumatoid arthritis. Rheum Dis Clin North Am. 2001;27:269–282. doi: 10.1016/s0889-857x(05)70201-5. [DOI] [PubMed] [Google Scholar]
- Goel A, Chang DK, Ricciardiello L, Gasche C, Boland CR. Clin Cancer Res. 2003;9:383–390. [PubMed] [Google Scholar]
- Hammond SL, Safe S, Tjalkens RB. A novel synthetic activator of Nurr1 induces dopaminergic gene expression and protects against 6-hydroxydopamine neurotoxicity in vitro. Neuroscience Letters. 2015;607:83–89. doi: 10.1016/j.neulet.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy RS, Hülso C, Liu Y, Gasparini SJ, Fong-Yee C, Tu J, Stoner S, Stewart PM, Raza K, Cooper MS, Seibel MJ, Zhou H. Characterisation of fibroblast-like synoviocytes from a murine model of joint inflammation. Arthritis Research & Therapy. 2013a;15(1):R24. doi: 10.1186/ar4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hideshima T, Chauhan D, Richardson P, Mitsiades C, Mitsiades N, Hayashi T, et al. NF-kappa B as a therapeutic target in multiple myeloma. The Journal of Biological Chemistry. 2002;277(19):16639–16647. doi: 10.1074/jbc.M200360200. [DOI] [PubMed] [Google Scholar]
- Holla VR, Mann JR, Shi Q, DuBois RN. Prostaglandin E2 regulates the nuclear receptor NR4A2 in colorectal cancer. The Journal of Biological Chemistry. 2006;281(5):2676–2682. doi: 10.1074/jbc.M507752200. [DOI] [PubMed] [Google Scholar]
- Honda T, Segi-Nishida E, Miyachi Y, Narumiya S. Prostacyclin-IP signaling and prostaglandin E2-EP2/EP4 signaling both mediate joint inflammation in mouse collagen-induced arthritis. The Journal of Experimental Medicine. 2006;203(2):325–335. doi: 10.1084/jem.20051310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamoto T, Papineni S, Chintharlapalli S, Cho SD, Safe S, Kamat AM. 1,1-Bis(3′-indolyl)-1-(p-chlorophenyl)methane activates the orphan nuclear receptor Nurr1 and inhibits bladder cancer growth. Molecular Cancer Therapeutics. 2008;7(12):3825–3833. doi: 10.1158/1535-7163.MCT-08-0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M. NF-κB as a Critical Link Between Inflammation and Cancer. Cold Spring Harbor Perspectives in Biology. 2009;1(5):a000141. doi: 10.1101/cshperspect.a000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harbor Perspectives in Biology. 2009;1(6):a001651–a001651. doi: 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SO, Li X, Hedrick E, Jin UH, Tjalkens RB, Backos DS, et al. Diindolylmethane analogs bind NR4A1 and are NR4A1 antagonists in colon cancer cells. Molecular Endocrinology (Baltimore, Md) 2014;28(10):1729–1739. doi: 10.1210/me.2014-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Sanz I, O’Keefe RJ, Schwarz EM. NF-kappa B regulates VCAM-1 expression on fibroblast-like synoviocytes. The Journal of Immunology. 2000;164(11):5990–5997. doi: 10.4049/jimmunol.164.11.5990. [DOI] [PubMed] [Google Scholar]
- Li Xi, Lee SO, Safe S. Structure-dependent activation of NR4A2 (Nurr1) by 1,1-bis(3′-indolyl)-1-(aromatic)methane analogs in pancreatic cancer cells. Biochemical Pharmacology. 2012;83(10):1445–1455. doi: 10.1016/j.bcp.2012.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- McEvoy AN, Murphy EA, Ponnio T, Conneely OM, Bresnihan B, FitzGerald O, Murphy EP. Activation of nuclear orphan receptor NURR1 transcription by NF-kappa B and cyclic adenosine 5′-monophosphate response element-binding protein in rheumatoid arthritis synovial tissue. The Journal of Immunology. 2002;168(6):2979–2987. doi: 10.4049/jimmunol.168.6.2979. [DOI] [PubMed] [Google Scholar]
- Mix KS, Attur MG, Al-Mussawir H, Abramson SB, Brinckerhoff CE, Murphy EP. Transcriptional repression of matrix metalloproteinase gene expression by the orphan nuclear receptor NURR1 in cartilage. The Journal of Biological Chemistry. 2007;282(13):9492–9504. doi: 10.1074/jbc.M608327200. [DOI] [PubMed] [Google Scholar]
- Ntougkos E, Chouvardas P, Roumelioti F, Ospelt C, Frank-Bertoncelj M, Filer A, Buckley CD, Gay S, Nikolaou C, Kollias G. Genomic Responses of Mouse Synovial Fibroblasts During Tumor Necrosis Factor–Driven Arthritogenesis Greatly Mimic Those in Human Rheumatoid Arthritis. Arthritis & Rheumatology. 2017;69:1588–1600. doi: 10.1002/art.40128. [DOI] [PubMed] [Google Scholar]
- Qin C, Morrow D, Stewart J, Spencer K, Porter W, Smith R, et al. A new class of peroxisome proliferator-activated receptor γ (PPARγ) agonists that inhibit growth of breast cancer cells: 1,1-Bis(3′-indolyl)-1-(p-substituted phenyl)methanes. Molecular Cancer Therapeutics. 2004;3(3):247–260. [PubMed] [Google Scholar]
- Ralph J, McEvoy A, Kane D, Bresnihan B, FitzGerald O, Murphy E. Modulation of Orphan Nuclear Receptor NURR1 Expression by Methotrexate in Human Inflammatory Joint Disease Involves Adenosine A2A Receptor-Mediated Responses. The Journal of Immunology. 2005;175(1):555–565. doi: 10.4049/jimmunol.175.1.555. [DOI] [PubMed] [Google Scholar]
- Rege TA, Hagood JS. Thy-1 as a regulator of cell-cell and cell-matrix interactions in axon regeneration, apoptosis, adhesion, migration, cancer, and fibrosis. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2006;20:1045–1054. doi: 10.1096/fj.05-5460rev. [DOI] [PubMed] [Google Scholar]
- Saijo K, Winner B, Carson CT, Collier JG, Boyer L, Rosenfeld MG, Gage FH, Glass CK. A Nurr1/CoREST Pathway in Microglia and Astrocytes Protects Dopaminergic Neurons from Inflammation-Induced Death. Cell. 2009;137:47–59. doi: 10.1016/j.cell.2009.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Schmitt-Talbot E, DiMattia DA, Dullea RG. The differential effects of IL-1 and TNF-alpha on proinflammatory cytokine and matrix metalloproteinase expression in human chondrosarcoma cells. Inflamm Res. 2004;53:377–389. doi: 10.1007/s00011-004-1271-3. [DOI] [PubMed] [Google Scholar]
- Smolen JS, Aletaha D. Developments in the clinical understanding of rheumatoid arthritis. Arthritis Research & Therapy. 2009;11(1):204. doi: 10.1186/ar2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas I, Glass CK. Anti-Inflammatory Therapy in Chronic Disease: Challenges and Opportunities. Science. 2013;339(6116):166–172. doi: 10.1126/science.1230720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tak PP, Bresnihan B. The pathogenesis and prevention of joint damage in rheumatoid arthritis: Advances from synovial biopsy and tissue analysis. Arthritis & Rheumatism. 2000;43:2619–2633. doi: 10.1002/1529-0131(200012)43:12<2619::AID-ANR1>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Umar S, Hedaya O, Agere S, Ahmed S. Thymoquinone inhibits TNF-α-induced inflammation and cell adhesion in rheumatoid arthritis synovial fibroblasts by ASK1 regulation. Toxicol Appl Pharmacol. 2015;287(3):299–305. doi: 10.1016/j.taap.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon K, Lee S-O, Cho SD, Kim K, Khan S, Safe S. Activation of nuclear TR3 (NR4A1) by a diindolylmethane analog induces apoptosis and proapoptotic genes in pancreatic cancer cells and tumors. Carcinogenesis. 2011;32:836–842. doi: 10.1093/carcin/bgr040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.