Abstract
Infection with H. pylori pathogen is one of the strongest risk factors for development of gastric cancer. Although these bacteria infect approximately half of the world’s population, only a small fraction of infected individuals develops gastric malignancies. Interactions between host and bacterial virulence factors are complex and interrelated making it difficult to elucidate specific processes associated with H. pylori-induced tumorigenesis. In this study, we found that H. pylori inhibits p14ARF tumor suppressor by inducing its degradation. This effect was found to be strain-specific. Downregulation of p14ARF induced by H. pylori leads to inhibition of autophagy in a p53-independent manner in infected cells. We identified TRIP12 protein as E3 ubiquitin ligase that is upregulated by H. pylori, inducing ubiquitination and subsequent degradation of p14ARF protein. Using isogenic H. pylori mutants, we found that induction of TRIP12 is mediated by bacterial virulence factor CagA. Increased expression of TRIP12 protein was found in infected gastric epithelial cells in vitro and human gastric mucosa of H. pylori-infected individuals.
In conclusion, our data demonstrate a new mechanism of ARF inhibition that may affect host-bacteria interactions and facilitate tumorigenic transformation in the stomach.
Keywords: ARF, TRIP12/ULF, H. pylori, gastric cancer
INTRODUCTION
Helicobacter pylori (H. pylori) is one of the most common human pathogens that infects approximately half of the world’s population. Infection with this Gram-negative bacteria typically causes asymptomatic gastritis, but some infected individuals may develop more severe conditions such as mucosa associated lymphoid tissue (MALT) lymphoma and gastric adenocarcinoma. Gastric cancer remains a common cause of cancer-related death in the World, and H. pylori is recognized as the strongest identified risk factor for malignancies that arise within the stomach. The risk of cancer development is determined by a complex interplay between bacterial virulence and host factors. H. pylori strains show extremely high genetic diversity which accounts, at least in part, for their different tumorigenic potential1. One of the important genetic determinants of H. pylori virulence is the cag pathogenicity island (cag PAI), a genetic region which encodes a bacterial type IV secretion system (T4SS). CagA (cytotoxin-associated gene A) is a product of the cag PAI region that is injected through T4SS into the host cells2. CagA functions as an oncoprotein that is associated with an increased risk of gastric cancer in experimental animals and humans3, 4.
ARF tumor suppressor is a product of the CDKN2A locus. In humans, this protein is designated as p14ARF and in mice as p19ARF5. CDKN2A gene is located on chromosome 9p21, a region frequently deleted in primary gastric cancer. In addition, p14ARF gene expression was found to be inactivated by hypermethylation in more than 30% of gastric cancers6, 7. As a pivotal tumor suppressor, p14ARF plays critical role in oncogenic stress response (OSR), which comprises complex network of cellular responses to oncogene activation caused by different stimuli. In part, p14ARF exerts its function as a main positive regulator of p53 tumor suppressor, acting through interaction with HDM2 E3 ubiquitin ligase, which is responsible for degradation of p538. We have recently shown that p14ARF has crucial role in the regulation of p53 in H. pylori-infected gastric cells9. p14ARF was also shown to have p53-independent tumor suppressor functions10.
It has been reported that p14ARF plays a role in the regulation of autophagy in different cell types11, 12. Autophagy is a complex and dynamic process through which eukaryotic cells degrade various cell components by engulfing them into double-membrane compartments termed autophagosomes, which are, in a later phase of autophagic process, fused with lysosomes into autolysosomes inducing degradation of cellular components, reviewed in13. Role of autophagy in tumorigenesis is multifaceted, as it may play both tumor suppressive and tumor promoting roles, depending on tumorigenic stage.
In this paper, we investigated the mechanism of regulation of p14ARF protein by H. pylori and its biological consequences in host gastric cells.
RESULTS
Expression of p14ARF is regulated by H. pylori in a strain-specific manner
We started our studies of p14ARF regulation with co-culturing gastric epithelial cells with H. pylori. SNU1 was our cell line of choice because unlike most other gastric cell lines, SNU1 expresses p14ARF and wild-type p53 proteins9. We also took advantage of previously characterized H. pylori clinical isolate B128 and its oncogenic derivative 7.1314. The latter strain (but not the former) strongly activates cellular oncogenes resulting in reproducible induction of premalignant and malignant gastric lesions in different rodent models14, 15.
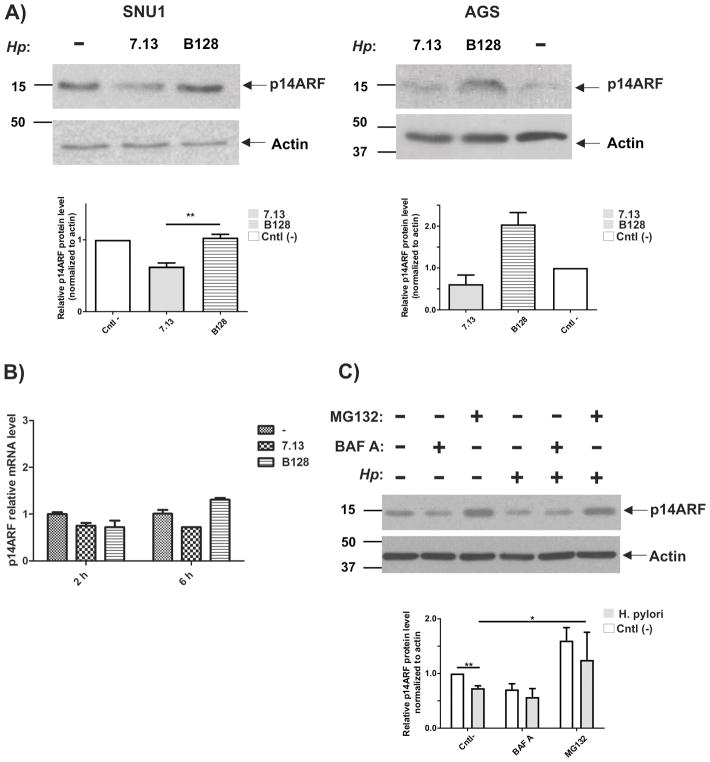
SNU1 cells were co-cultured with either H. pylori strain 7.13 or B128 for 6 hours or left untreated (control) and then expression of p14ARF protein was analyzed by Western blotting. We found that levels of p14ARF protein were decreased in cells co-cultured with oncogenic strain 7.13, while infection with strain B128 resulted in a slight increase of p14ARF protein (Figure 1A, left panel). In another gastric cell line, AGS, which expresses low but detectable levels of p14ARF, effect of H. pylori strains 7.13 and B128 was similar to one found in SNU1 cells (Figure 1A, right panel). These findings suggest that H. pylori affects protein levels of p14ARF in a strain specific manner (Figure 1A).
Figure 1. Expression of p14ARF in H. pylori-infected human gastric cells is regulated in a strain-specific manner.
(A) Left panel: SNU1 human gastric epithelial cells were co-cultured with H. pylori strains 7.13 or B128 for 6 hours or left untreated (−). Expression of p14ARF was analyzed by Western blot. Right panel: the same as (A), but AGS cells were analyzed. ARF protein was quantitated by densitometry. Data are shown as mean ± SEM (n=3). (B) qRT-PCR analysis of p14ARF mRNA in H. pylori-infected cells. SNU1 cells were co-cultured with H. pylori strains 7.13 or B128 and analyzed at 2 and 6 hours after infection. Relative p14ARF levels were normalized to HPRT1 mRNA expression. Data are shown as mean ± SEM (n=2). (C) The effect of proteasomal and autophagic inhibitors were investigated in SNU1 cells co-cultured with H. pylori strain 7.13. Expression of p14ARF protein was analyzed by Western blotting. ARF protein was quantitated by densitometry. Data are shown as mean ± SEM (n=3).
To determine how H. pylori alters protein levels of p14ARF, we first analyzed mRNA levels of p14ARF in SNU1 cells co-cultured with strains 7.13 or B128 using qRT-PCR (Figure 1B). No significant differences were found between infected and control uninfected cells, suggesting that post-transcriptional mechanisms are likely involved in the regulation of p14ARF protein in H. pylori-infected cells, although inhibition of transcription cannot be completely excluded.
Next, we asked whether proteasomal and autophagic degradation systems regulate protein levels of ARF in infected cells. To explore these possibilities, we employed proteasomal inhibitor MG132 and bafilomycin A. The latter inhibits lysosomal acidification necessary for autophagosome maturation16. SNU1 cells were co-cultured with H. pylori strain 7.13 for 3 hours and then treated with bafilomycin A or MG132 at final concentrations 100 nM and 20 μM, respectively, for additional 4 hours. To exclude possible indirect effects of inhibitors, bacteria were eliminated with kanamycin treatment. We found that MG132, but not bafilomycin A, inhibits downregulation of p14ARF protein caused by H. pylori strain 7.13, implicating the proteasomal degradation system in the regulation of ARF protein in infected cells (Figure 1C).
H. pylori infection induces ubiquitination and degradation of p14ARF
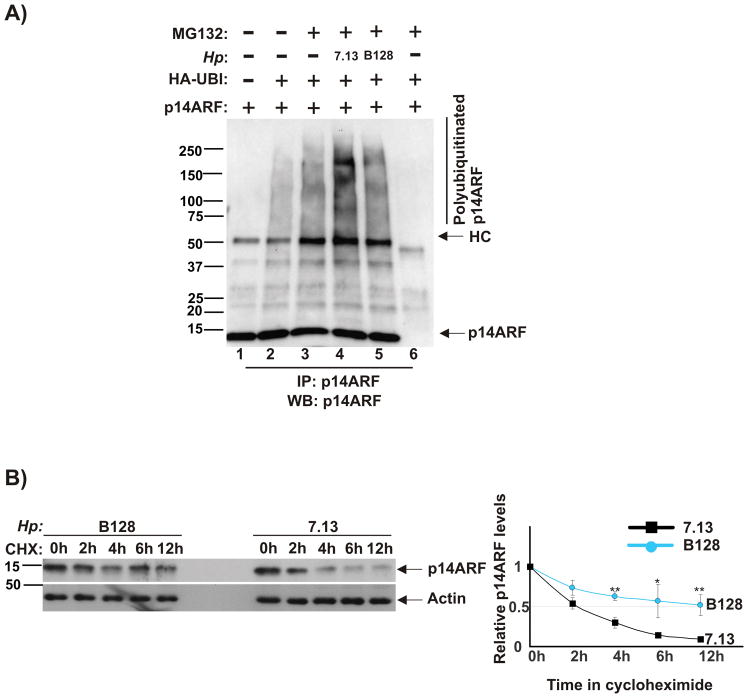
It has been previously reported that p14ARF protein undergoes N-terminal polyubiquitination that regulates ARF degradation17. To explore whether H. pylori strains affect p14ARF polyubiquitination, we used an approach similar to that described in the literature17. AGS cells, which express low levels of ARF, were co-transfected with plasmids expressing human ARF and ubiquitin, co-cultured with H. pylori strains 7.13 or B128 for 90 minutes and treated with proteasomal inhibitor MG132 for additional 7 hours. Infected and control cells were collected and lysed. p14ARF was then immunoprecipitated from cellular lysates with ARF-specific antibody and analyzed for polyubiquitination by Western blotting. Our experiments found that tumorigenic strain 7.13 induces significantly higher levels of ARF polyubiquitination than that of B128 (Figure 2A, compare lanes 4 and 5).
Figure 2. H. pylori strain 7.13 induces ubiquitination and degradation of p14ARF protein.
(A) AGS cells were co-transfected with plasmids expressing p14ARF and ubiquitin for 24 hours, co-cultured with strains 7.13 or B128 for 90 minutes and then incubated with proteasome inhibitor MG132 (20 μM) for 7 hours. Lysates were analyzed for polyubiquitinated p14ARF protein after its immunoprecipitation (IP) with p14ARF antibody. Gel loading was normal normalized to ARF protein. As a negative control for IP, normal rabbit IgG was used (lane 6). (B) Stability of p14ARF protein was analyzed in AGS cells transfected with p14ARF using the cycloheximide chase method (see the Material and Method section). Left panel: Expression of p14ARF protein was assessed by Western blotting. Right panel: Protein levels of p14ARF were quantified by densitometry and normalized to actin. ARF protein was quantitated by densitometry (p*<.05; p**<.01). Data are shown as mean ± SEM (n=2).
To investigate whether H. pylori-induced polyubiquitination affects stability of ARF protein, we employed the cycloheximide chase assay. Similar to the aforementioned experiment, AGS cells, which ectopically express p14ARF, were co-cultured with H. pylori strains 7.13 or B128. Six hours post infection, cycloheximide (de novo protein synthesis inhibitor) was added to cell culture media at a final concentration 150 μM. Cells were then collected at 0, 2, 4, 6, and 12 hours after treatment and analyzed by Western blotting (Figure 2B). We found that stability of ARF protein is significantly lower in gastric cells infected with strain 7.13 (half-life is approximately 2 hours) than that of B128 (half-life is longer than 12 hours). Combined, these experiments show that infection with oncogenic H. pylori strain 7.13 leads to increased ubiquitination and subsequent proteasomal degradation of p14ARF protein.
Downregulation of p14ARF leads to inhibition of autophagy in gastric cells
Our data raised an important question about a potential biological impact of degradation of p14ARF protein by H. pylori. One of the cellular processes that plays a critical role in host-microbe interactions and gastric tumorigenesis is autophagy. It has been previously reported that p14ARF is involved in regulation of autophagy11, 12. Based on these facts, we explored how ARF alterations induced by H. pylori affect autophagy.
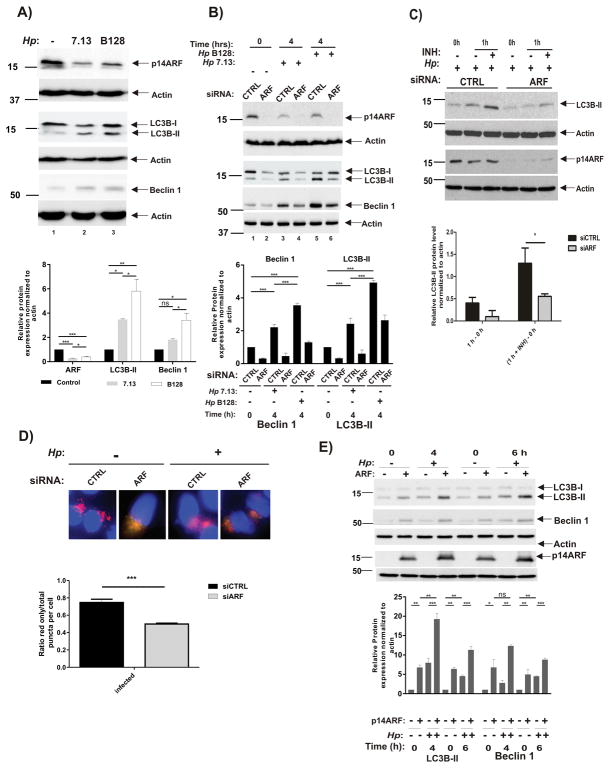
We first assessed autophagy in infected cells. SNU1 cells were infected with H. pylori strains 7.13 or B128 and analyzed for protein expression of ARF and commonly used autophagic markers, LC3B-II and Beclin 1. Cells co-cultured with oncogenic strain 7.13 showed not only a decreased expression of ARF protein but also decreased levels of LC3B-II and Beclin 1 proteins, pointing to a lower level of autophagy in those cells (Figure 3A). In contrast, infection with strain B128 led to significantly higher levels of LC3B-II and Beclin 1 proteins. To explore this phenomenon further, SNU1 cells, in which p14ARF protein was downregulated with specific siRNA, were co-cultured with H. pylori strains 7.13 or B128 or left uninfected for the indicated time (Figure 3B). Analyzing LC3B-II and Beclin 1, we found that downregulation of ARF even further decreases autophagy in infected cells. Decrease in ARF levels also affected basal autophagy in control uninfected cells.
Figure 3. Downregulation of p14ARF in H. pylori-infected cells leads to inhibition of autophagy.
(A) SNU1 cells were co-cultured with 7.13 or B128 strain for 6 hours or left uninfected (−), and p14ARF, LC3B and Beclin 1 proteins were analyzed by Western blot analyses. Protein were quantitated by densitometry. Data are shown as mean ± SEM (n=3). (B) Expression of p14ARF, LC3B and Beclin 1 proteins were analyzed in SNU1 cells, in which ARF was downregulated with specific siRNA (ARF) or scrambled control siRNA (CTRL) for 48 hours. Cells were then co-cultured with strains 7.13 or B128 for the indicated time (+) or left uninfected (−). Protein were quantitated by densitometry. Data are shown as mean ± SEM (n=3). (C) Autophagic flux was inhibited in SNU1 cells in which ARF was downregulated with specific siRNA (ARF) or scrambled control siRNA (CTRL) for 48 hours. Cells were co-cultured with strain 7.13 for 2 hours and treated with pepstatin A and E-64d (INH; both at a final concentration 10 μM) for additional 1 h. Upper panel: A representative Western blot analysis showing expression of LC3B and p14ARF. Bottom panel: Densitometric analyses of autophagic flux. It was calculated by subtraction of the levels of LC3B-II protein (normalized to actin) before treatment (0 h) from ones after treatment with inhibitors (1 h) (*, p<.05). (D) Upper panel: Representative images showing merged red and green puncta in SNU1 cells transfected with mCherry-GFP-LC3B and co-cultured with strain 7.13 for 4 hours. ARF was downregulated with the specific siRNA. Bottom panel: Graphs show red-only puncta relative to total puncta per cell. Results are shown as mean ± SEM from quantification of at least 20 cells per sample (***, p<0.001). (E) Representative Western blots show expression of LC3B, Beclin 1 and p14ARF proteins in AGS cells transfected with p14ARF plasmid (+) or empty vector (−) and then co-cultured with strain 7.13 (+) or left uninfected (−) for the indicated time. Proteins were quantitated by densitometry. Data are shown as mean ± SEM (n=3).
Since autophagy is a multistep and dynamic process, we next investigated autophagic flux in ARF-deficient cells using pepstatin A and E-64d (INH), which interfere with autolysosomal digestion of LC3B-II. In agreement with the experiments shown above, downregulation of p14ARF led to inhibition of autophagic flux in infected cells (Figure 3C), as measured by LC3B-II.
To visualize the autophagic flow, we used a retrovirus, which, following transduction, expresses LC3B conjugated to fluorescent proteins mCherry and GFP. This approach allowed us to track progression of autophagy in situ (see Material and Methods). SNU1 cells, which retrovirally express mCherry-EGFP-LC3B, were transfected with ARF or control siRNAs. These cells were then co-cultured with H. pylori strain 7.13 for 4 hours or left uninfected and analyzed for the autophagic puncta formation (Figure 3D). We found that ARF downregulation markedly decrease the abundance of acidic autolysosomes (detected as red-only puncta in mCherry-EGFP-LC3B-expressing cells), providing additional proof that inhibition of ARF by H. pylori leads to inhibition of autophagic flux.
To further confirm our findings, we overexpressed ARF in AGS cells, which are characterized by low levels of endogenous ARF protein. Eighteen hours following transfection, AGS cells were co-cultured with H. pylori strain 7.13 and lysates were analyzed for LC3B protein at the indicated time points (Figure 3E). We found that ectopic overexpression of ARF increases levels of LC3B-II. A similar increase was also seen for Beclin 1 protein, showing that ARF induces autophagy in AGS cells.
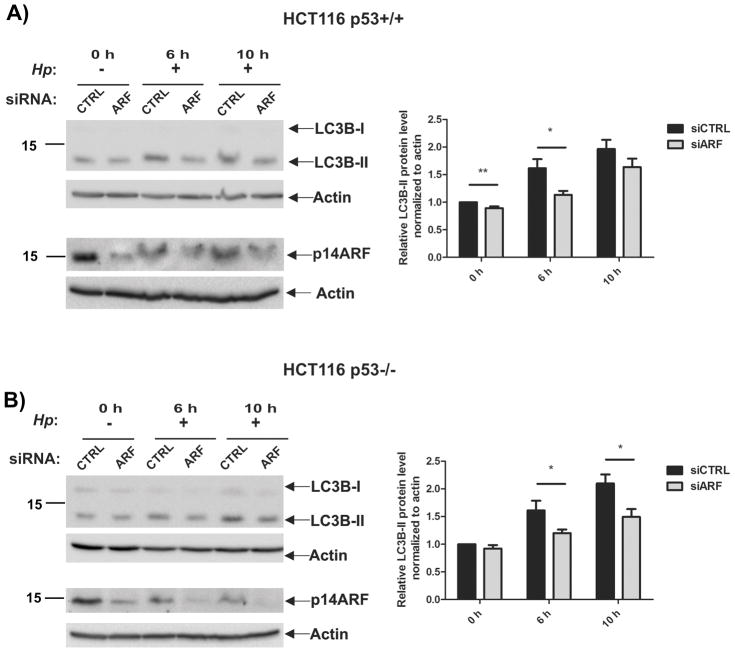
Since it has been shown that ARF affects stability and activity of p53 protein, we next asked whether the effect of ARF on autophagy, found in our experiments, is mediated by p53. To explore this possibility, we took advantage of isogenic cell lines HCT116 p53+/+, which expresses wild type p53 and ARF, and its isogenic p53-null derivative HCT116 p53−/−. Using specific siRNA, ARF protein was downregulated in these cells, which were then co-cultured with H. pylori strain 7.13 (MOI 200) and analyzed for autophagy at the indicated time points (Figure 4). We found that downregulation of ARF leads to inhibition of autophagy not only in cells expressing wild-type p53 but also in p53-null cells, demonstrating that, at least in part, the effect of ARF on autophagy is independent of p53 in infected cells.
Figure 4. Inhibition of autophagy induced by downregulation of p14ARF is independent of p53 protein.
(A) Expression of p14ARF was downregulated with specific siRNA in HCT116 p53+/+ and HCT116 p53−/− cells. Cells were co-cultured with strain 7.13 (+) for the indicated time or left uninfected (−). LC3B and p14ARF proteins were analyzed by Western blotting. (B) Expression of LC3B-II protein was quantified by densitometry and normalized to actin levels. Graphs depict results as mean ± SEM, (*, p<0.05; **, p<0.01; n=2).
Taken together, out data revealed that tumorigenic strain of H. pylori can induce degradation of p14ARF protein resulting in inhibition of autophagy.
TRIP12 E3 ubiquitin ligase is induced in H. pylori-infected gastric cells
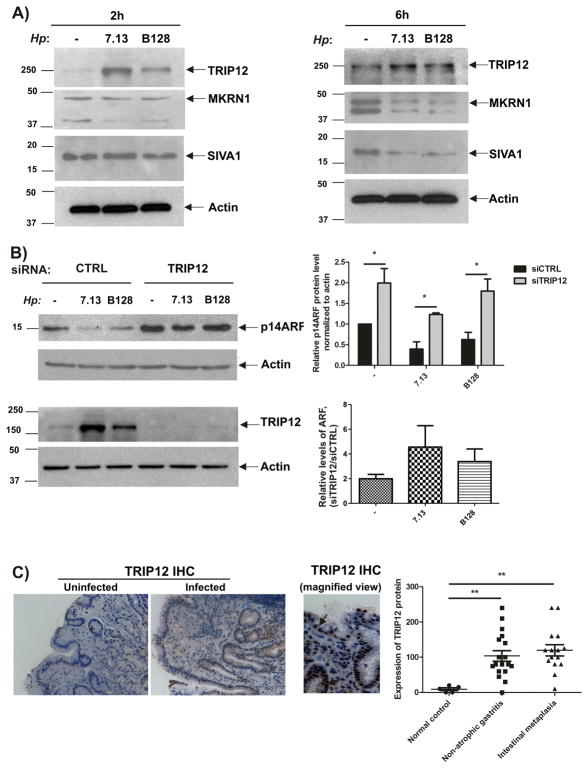
Another important question raised by our studies is how H. pylori regulates ARF protein. Since a number of E3 ubiquitin ligases have been reported to regulate ARF protein, such as TRIP12/ULF (Thyroid hormone Receptor Interacting Protein 12), SIVA1, and MKRN1 (Makorin ring finger protein 1)18–20, we explored expression of these proteins in SNU1 cells co-cultured with strains 7.13 or B128. We found that both strains upregulate TRIP12 protein, while other protein E3 ubiquitin ligases did not show increases. Notably, strain 7.13 was significantly stronger inducer of TRIP12 than B128 (Figure 5A). Similar effects were seen in other tested cell lines, AGS and mGEC (Supplemental Figures 1A and 1B)
Figure 5. H. pylori infection induces TRIP12, but not the other E3 ubiquitin ligases.
(A) Representative blots show expression of TRIP12, MKRN1 and SIVA1 proteins in SNU1 cells co-cultured with strains 7.13 or B128 for the indicated time (+) or left uninfected (−). (B) Left panels: Expression of p14ARF and TRIP12 proteins in SNU1 cells transfected with TRIP12 specific or scrambled siRNAs for 48 hours and co-cultured with H. pylori strain 7.13 or B128 for additional 6 hours or left uninfected (−). Upper right panel: Graph shows relative levels of p14ARF protein (normalized to actin) from 3 independent experiments. Lower right panel: Graph shows ratios of relative levels of ARF protein in cells transfected with either TRIP12 specific- or scrambled siRNAs. (C) Left panel: Representative immunohistochemical staining for TRIP12 protein in the gastric mucosa of H. pylori-infected and uninfected human individuals (20X). A representative magnified image of the gastric mucosa is shown (40x). Right panel: Histogram shows immunohistochemical scores for TRIP12 protein expression (see the Materials and Methods section for detail). Expression of TRIP12 is increased in H. pylori-infected individuals (**, p< 0.01 vs. uninfected).
To investigate the contribution of TRIP12 to the regulation of ARF in infected cells, its expression was downregulated with TRIP12-specific siRNA in SNU1 cells and then ARF protein was analyzed after co-culturing cells with strains 7.13 or B128. We found that protein levels of p14ARF were significantly increased as a result of TRIP12 downregulation. Moreover, downregulation of TRIP12 has stronger effect in infected cells (Figure 5B).
Finally, the expression of TRIP12 was analyzed by immunohistochemistry in gastric mucosa from 33 H. pylori-infected and 5 uninfected human individuals who underwent GI tract endoscopy. H. pylori-infected patients were divided into two groups based on their diagnosis (non-atrophic gastritis or intestinal metaplasia). We found that expression of TRIP12 was significantly higher in both groups of infected patients compared with uninfected (Figure 5C), showing that H. pylori also induces TRIP12 in vivo.
Role of CagA and other H. pylori virulence factors
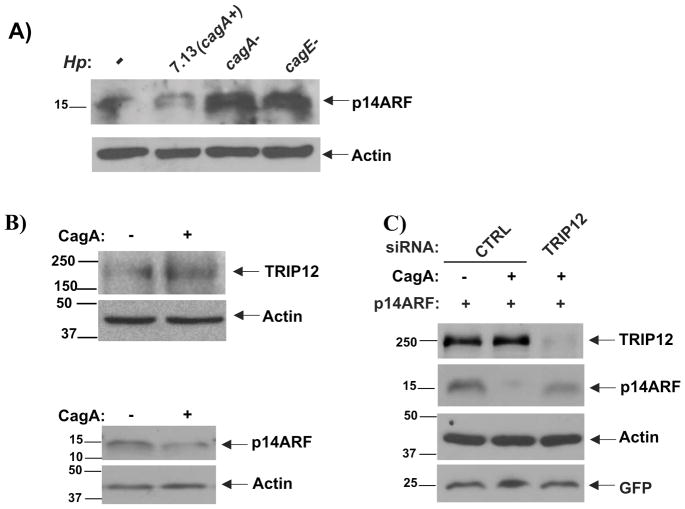
Genes within the cag PAI region have been shown to significantly affect host-microbe interaction2. To assess the role of CagA in regulation of ARF, isogenic cagA- and cagE-null mutants were generated and their ability to affect ARF protein was examined. SNU1 cells were co-cultured with H. pylori strain 7.13 or its cagA- and cagE- mutants. ARF proteins was then studied by Western blotting (Figure 6A). We found that deletion of cagA inhibits degradation of ARF by H. pylori. Notably, the cagE- mutant that does not form the T4S, and therefore, does not deliver CagA into the host cell cytosol21 was also deficient in ARF regulation. Similarly, the ability of H. pylori to upregulate TRIP12 was significantly compromised by loss of either cagA or cagE (Figure 6A, left panel). To further investigate the role of CagA, SNU1 and AGS gastric cells were transfected with cagA-expressing plasmid (Figure 6B, 6C). Transfection of CagA led to an increased expression of TRIP12 protein and downregulation of ARF. When CagA was co-transfected with TRIP12 siRNA, it inhibited downregulation of ARF and restored its protein level (Figure 6C). These results show that CagA, a component of the cag PAI, negatively regulates ARF through induction of TRIP12.
Figure 6. Bacterial virulence factor CagA regulates TRIP12 and degradation of p14ARF protein.
(A) SNU1 cells were co-cultured with cagA-positive H. pylori strain 7.1314 or its isogenic cagA- or cagE- mutants for 2 hours, p14ARF protein was then assessed by Western blotting.
(B) SNU1 or AGS cells were transfected with CagA-expressing plasmid (+) or empty vector (−) for 24 hours. TRIP12 protein was analyzed in AGS cells (upper panel). Given that SNU1 cells express higher levels of ARF protein than AGS, ARF was assessed in SNU1 cells (lower panel). (C) AGS cells transfected with ARF and GFP plasmids were co-transfected with either empty vector or CagA expressing plasmid and siRNA against TRIP12 as indicated in the panel. TRIP12 and p14ARF proteins were analyzed using Western blotting. β-actin and GFP were used for normalization of protein loading and transfection efficiency, respectively.
DISCUSSION
We report that H. pylori actively regulates p14ARF tumor suppressor. As a model for infection, we used two previously characterized H. pylori strains B128 and 7.13 with different oncogenic potential14. Strain B128 is a human clinical isolate, which failed to induce tumors in rodents. On the contrary, its derivate strain 7.13, which was generated by in vivo adaptation in Mongolian gerbils, induces severe gastritis, pre-neoplastic and neoplastic lesions in rodent animal models. Moreover, tumorigenic potential of 7.13 strain was characterized by its ability to strongly induce WNT/β-catenin, RAS/Erk and PI3K/AKT signaling pathways, which are often aberrantly activated during gastric tumorigenesis14. In our experiments, strain 7.13 caused strong induction of TRIP12, while B128 did not produce similar effects. This induction was accompanied by strong ubiquitination and degradation of ARF protein by strain 7.13. The observed differences between two strains in the regulation of p14ARF tumor suppressor provide one more example of modulation of the host signaling pathways by H. pylori that may contribute to gastric tumorigenesis.
p14ARF, as a pivotal tumor suppressor, has important functions in the oncogenic stress response (OSR), an intrinsic network of cellular processes directed toward prevention of tumorigenic transformation. Although H. pylori was shown to activate many oncogenic pathways14, 22, mechanisms of OSR regulation in infected cells remain unclear. Disruption of ARF signaling by inducing its degradation shows one potential mechanism how H. pylori can circumvent the host OSR.
We analyzed the effect of p14ARF downregulation induced by H. pylori on autophagy, a critical process for maintaining cellular homeostasis. Our findings are consistent with previous reports on autophagic functions of p14ARF11, 12, 23. However, this effect was often attributed to smARF, a short form of ARF protein, which primarily localizes to the mitochondria24. Our results are consistent with previous reports, in which induction of autophagy were attributed, at least in certain conditions, to full-length nucleolar p14ARF11, 12. The contribution of smARF, however, cannot be excluded.
Since ARF controls p53 tumor suppressor by multiple mechanisms, and p53 is known to regulate autophagy 25, we investigated how degradation of p14ARF affects autophagy in infected p53 null cells. In these conditions, p14ARF affects autophagy independently of p53 in H. pylori-infected cells. p53 may work alongside ARF to regulate autophagy in p53-expressing cells.
We found that induction of TRIP12 E3 ubiquitin ligase by H. pylori contributes to degradation of p14ARF in H. pylori-infected cells. When induction of TRIP12 protein was compared between strains, tumorigenic H. pylori strain 7.13 was found to be a much stronger inducer of TRIP12 than B128. Strain 7.13 was also a stronger inducer of ubiquitination of ARF protein, resulting in its faster degradation in the proteasomes. Another interesting finding is that induction of TRIP12 is mediated by bacterial virulence factor CagA, which is delivered into gastric epithelial cells upon bacterial attachment. Isogenic cagA- and cagE- mutants, which do not deliver CagA, were found to have significantly weaker ability to upregulate TRIP12 than the parental strain. Notably, ectopic overexpression of CagA protein is sufficient to induce TRIP12 and ARF downregulation. Additional studies are needed to further investigate the mechanism of TRIP12 induction by CagA. Our analyses of SIVA1 and MKRN1 E3 ubiquitin ligases, which are known to regulate ARF, did not reveal their up-regulation in H. pylori-infected cells. We, however, cannot exclude their involvement in p14ARF regulation in H. pylori-infected cells.
Important role of TRIP12 is further emphasized by our findings that TRIP12 protein is upregulated in infected gastric mucosa. It is especially interesting, given that upregulation of TRIP12 has been reported in different human cancers26, 27.
In conclusion, we found that H. pylori bacterial pathogen inhibits activity of p14ARF protein causing inhibition of autophagy in infected cells. This may, in turn, affect host-bacteria interactions. ARF inhibition may also facilitate tumorigenic transformation, as this protein plays an essential tumor suppressor role in the stomach.
MATERIALS AND METHODS
Cell cultures and transfections
AGS and SNU1 human gastric epithelial cancer cell lines were grown in F12 medium (Thermo Fisher, Waltham, MA) supplemented with 10% (v/v) fetal bovine serum (FBS) from GIBCO (Thermo Fisher). Cell lines were purchased from ATCC (Manassas, VA). Cell lines were authenticated and characterized by the suppliers. ATCC uses morphology, karyotyping and PCR-based approaches to confirm the identity of cell lines. Human colon cancer cell line expressing wild-type p53 HCT116 p53+/+ and its isogenic p53-null HCT116 p53−/− derivative (kindly provided by Dr. Vogelstein), as well as Phoenix Amphotropic (AMPHO) cells (used for production of retroviruses) were maintained in Dulbecco’s modified Eagle’s medium (Thermo Fisher). Conditionally immortalized murine gastric epithelial cells (mGEC) were previously generated from transgenic mice carrying the temperature-sensitive mutant of SV40 large T antigen causing cell immortalization28. Normally, mGEKs were maintained in RPMI 1640 medium supplemented with 10% FBS and in the presence of 300 pg/ml of murine recombinant interferon gamma (IFN-γ) at permissive temperature of 33°C (immortalizing growth condition). Cells were transferred to 37°C and cultured in the absence of IFN-γ one day before the experimental treatment.
Transfections were performed using the Lipofectamine 2000 (Thermo Fisher) or FuGENE 6 (Promega, Madison, WI) reagents according the manufacturers recommendations.
H. pylori strains and infection
H. pylori clinical isolate B128 and its oncogenic derivative 7.13 were previously characterized14. The latter strain strongly activates cellular oncogenes and induces premalignant and malignant gastric lesions in different rodent models14, 15.
In addition, isogenic 7.13 cagA- and 7.13 cagE- null-mutants were constructed by insertional mutagenesis using aphA and selected with kanamycin29.
Bacteria were grown in Brucella broth supplemented with 10% FBS for 18 hours, harvested by centrifugation and used for infection at bacteria to cell ratio 100:1 (MOI 100) unless mentioned otherwise.
Plasmids, siRNA, antibodies and reagents
CMV driven HA-ubiquitin and pSP65SRα-CagA plasmids have been described previously30, 31. pBABE-puro mCherry-EGFP-LC3B and pcDNA3-ARF constructs were purchased from Addgene (Cambridge, MA)32.
siRNAs against p14ARF and TRIP12 were synthesized by Integrated DNA Technologies (Coralville, Iowa) using the following sequences 5′-CAUGGUGCGCAGGUUCUUGGUGACC-3′ and 5′-CCAGGAGCAACAACUGAAAUCUGCA-3′, respectively. The negative control (scrambled) siRNA was purchased from Ambion (Grand Island, NY).
Following primary antibodies were used in this research: rabbit monoclonal anti-p14ARF (clone EPR17878, ab185620) from Abcam (Cambridge, MA), mouse monoclonal anti-β-actin (clone AC-74) from Sigma (St. Louis, MO), mouse monoclonal anti-GFP from Vanderbilt Monoclonal Antibody Core (Nashville, TN), rabbit polyclonal anti-beclin-1 (#3738), anti-LC3B (#2775) from Cell Signaling (Danvers, MA), rabbit polyclonal anti-TRIP12 (A301-814A) and anti-MKRN1 (A300-990A) from Bethyl Laboratories (Montgomery, TX), rabbit polyclonal SIVA1 (A01) from Abnova (Walnut, CA). The secondary antibodies used were HRP-conjugated anti-mouse IgG from Promega and anti-rabbit IgG from Cell Signaling.
Proteasomal inhibitor MG132 was purchased from EMD Millipore (Burlington, MA). Cycloheximide, bafilomycin A, pepstatin A and E-64d were all from Sigma.
Cycloheximide chase assay
Cycloheximide chase assay was used to study p14ARF protein stability. In brief, AGS cell were transfected with p14ARF plasmid and co-cultured with strain 7.13 or B128 for 6 hours, then cycloheximide was added at a final concentration 150 μM. p14ARF protein levels were analyzed at the indicated time points.
Ubiquitination assay
AGS cells were co-transfected with plasmids expressing p14ARF and ubiquitin for 24 hours, co-cultured with H. pylori strain 7.13 or B128 for 90 minutes and then treated with MG132 (20 μM) for 7 hours. Cells were collected and lysed as described previously17. Lysates were sonicated, centrifuged twice for 20 min at 13000 rpm at 4°C. Samples were precleared by incubation with Protein-A agarose beads (Roche, Indianapolis, IN) at 4°C overnight. p14ARF was immunoprecipitated with ARF antibody and Protein-A agarose beads and analyzed by Western blotting with p14ARF antibody to detect polyubiquitinated ARF.
Tissue specimens (patient samples) and immunohistochemistry
Gastric tissue specimens used for immunohistochemical staining of TRIP12 protein are obtained from 33 H. pylori-infected and 5 control (uninfected) individuals randomly selected from a larger cohort of samples collected from individuals by GI tract endoscopy in two hospitals in the State of Nariño in Colombia previously described33. All participants provided informed consent. The protocol was approved by the Committees on Ethics of Universidad del Valle and Hospital Departmental in Nariño, Colombia and by the Institutional Review Board at Vanderbilt University. All personal identifiers were removed prior to receiving specimens and they were coded.
Expression of TRIP12 protein was analyzed by immunohistochemistry using specific polyclonal anti-TRIP12 antibody (A301-814A, Bethyl Laboratories) in dilution 1:400 and the EnVision+HRP kit (DakoCytomation, UK). Immunohistochemical staining was evaluated for intensity and staining frequency in the nuclear compartment of superficial gastric epithelium. The intensity of staining was graded as 0 (negative), 1 (weak), 2 (moderate) or 3 (strong). The frequency was graded according to the percentage of positive cells. Total scores were calculated by multiplying the intensity score by the percentage of positive cells9.
Puncta formation assay
mCherry-EGFP-LC3B construct was used for detection of autophagic flux as has been previously described32. Briefly, SNU1 cells were transduced with retroviruses expressing mCherry-EGFP-LC3B. After selection with 1 μg/ml puromycin, cells were plated onto 8-well chambers and transfected with siRNA against p14ARF or scrambled siRNA using Fugene 6 reagent, according to the manufacturer’s protocol. Cells were then co-cultured with H. pylori strain 7.13 for 4 hours, fixed with 4% paraformaldehyde and analyzed by fluorescent microscopy for red and green puncta formation.
Isolation of mRNA and qRT-PCR
Total cellular RNA was isolated using the RNeasy Kit from Qiagen (Valencia, CA) and converted into cDNA with the High Capacity cDNA Reverse Transcription Kit from Applied Biosystems (Carlsbad, CA) according the manufacturer’s instructions. Expression of p14ARF mRNA was assessed by qRT-PCR using the following primers: GTTTTCGTGGTTCACATCCC and ACCAGCGTGTCCAGGAAG. mRNA expression of HPRT (Hypoxanthine-Guanine Phosphoribosyltransferase) was used as an internal control with the following primers: TTG GAAAGGGTGTTTATTCCTCA and TCCAGCAGGTCAGCAAAGAA.
Statistical analysis
Statistical analysis was performed using the Student t-test and Mann-Whitney tests, depending on the data set. Results were shown as mean ± SEM. Results were considered as significant if p< 0.05.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Cancer Institute CA206564, R01 138833, the Department of Veterans Affairs BX002115, Vanderbilt Ingram Cancer Center (P30 CA68485), and the Vanderbilt Digestive Disease Research Center (DK058404). The contents of this work are solely the responsibility of the authors and do not necessarily represent the official views of the Department of Veterans Affairs, National Institutes of Health, or University of Miami.
We thank Drs. Maria B. Piazuelo and Alberto G. Delgado for help with tissue processing.
This work was supported by grants from the National Cancer Institute CA206564, R01 CA138833, the Department of Veterans Affairs BX002115, Vanderbilt Ingram Cancer Center (P30 CA68485), and the Vanderbilt Digestive Disease Research Center (DK058404). The contents of this work are solely the responsibility of the authors and do not necessarily represent the official views of the Department of Veterans Affairs, National Institutes of Health, or Vanderbilt University.
Footnotes
Conflict of interest: The authors have no conflict of interest to disclose
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
References
- 1.Kodaman N, Pazos A, Schneider BG, Piazuelo MB, Mera R, Sobota RS, et al. Human and Helicobacter pylori coevolution shapes the risk of gastric disease. Proc Natl Acad Sci U S A. 2014;111:1455–1460. doi: 10.1073/pnas.1318093111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Odenbreit S, Puls J, Sedlmaier B, Gerland E, Fischer W, Haas R. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science. 2000;287:1497–1500. doi: 10.1126/science.287.5457.1497. [DOI] [PubMed] [Google Scholar]
- 3.Zhu Y, Zhong X, Zheng S, Du Q, Xu W. Transformed immortalized gastric epithelial cells by virulence factor CagA of Helicobacter pylori through Erk mitogen-activated protein kinase pathway. Oncogene. 2005;24:3886–3895. doi: 10.1038/sj.onc.1208551. [DOI] [PubMed] [Google Scholar]
- 4.Ohnishi N, Yuasa H, Tanaka S, Sawa H, Miura M, Matsui A, et al. Transgenic expression of Helicobacter pylori CagA induces gastrointestinal and hematopoietic neoplasms in mouse. Proc Natl Acad Sci U S A. 2008;105:1003–1008. doi: 10.1073/pnas.0711183105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sherr CJ. Ink4-Arf locus in cancer and aging. Wiley Interdiscip Rev Dev Biol. 2012;1:731–741. doi: 10.1002/wdev.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sato F, Meltzer SJ. CpG island hypermethylation in progression of esophageal and gastric cancer. Cancer. 2006;106:483–493. doi: 10.1002/cncr.21657. [DOI] [PubMed] [Google Scholar]
- 7.Iida S, Akiyama Y, Nakajima T, Ichikawa W, Nihei Z, Sugihara K, et al. Alterations and hypermethylation of the p14(ARF) gene in gastric cancer. Int J Cancer. 2000;87:654–658. doi: 10.1002/1097-0215(20000901)87:5<654::aid-ijc6>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 8.Kamijo T, Weber JD, Zambetti G, Zindy F, Roussel MF, Sherr CJ. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci U S A. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei J, Noto JM, Zaika E, Romero-Gallo J, Piazuelo MB, Schneider B, et al. Bacterial CagA protein induces degradation of p53 protein in a p14ARF-dependent manner. Gut. 2015;64:1040–1048. doi: 10.1136/gutjnl-2014-307295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sherr CJ, Bertwistle D, WDENB, Kuo ML, Sugimoto M, Tago K, et al. p53-Dependent and -independent functions of the Arf tumor suppressor. Cold Spring Harb Symp Quant Biol. 2005;70:129–137. doi: 10.1101/sqb.2005.70.004. [DOI] [PubMed] [Google Scholar]
- 11.Budina-Kolomets A, Hontz RD, Pimkina J, Murphy ME. A conserved domain in exon 2 coding for the human and murine ARF tumor suppressor protein is required for autophagy induction. Autophagy. 2013;9:1553–1565. doi: 10.4161/auto.25831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abida WM, Gu W. p53-Dependent and p53-independent activation of autophagy by ARF. Cancer Res. 2008;68:352–357. doi: 10.1158/0008-5472.CAN-07-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol. 2009;335:1–32. doi: 10.1007/978-3-642-00302-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franco AT, Israel DA, Washington MK, Krishna U, Fox JG, Rogers AB, et al. Activation of beta-catenin by carcinogenic Helicobacter pylori. Proc Natl Acad Sci U S A. 2005;102:10646–10651. doi: 10.1073/pnas.0504927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fox JG, Rogers AB, Ihrig M, Taylor NS, Whary MT, Dockray G, et al. Helicobacter pylori-associated gastric cancer in INS-GAS mice is gender specific. Cancer Res. 2003;63:942–950. [PubMed] [Google Scholar]
- 16.Mauvezin C, Neufeld TP. Bafilomycin A1 disrupts autophagic flux by inhibiting both V-ATPase-dependent acidification and Ca-P60A/SERCA-dependent autophagosome-lysosome fusion. Autophagy. 2015;11:1437–1438. doi: 10.1080/15548627.2015.1066957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuo ML, den Besten W, Sherr CJ. N-Terminal polyubiquitination of the ARF tumor suppressor, a natural lysine-less protein. Cell Cycle. 2004;3:1367–1369. doi: 10.4161/cc.3.11.1244. [DOI] [PubMed] [Google Scholar]
- 18.Chen D, Shan J, Zhu WG, Qin J, Gu W. Transcription-independent ARF regulation in oncogenic stress-mediated p53 responses. Nature. 2010;464:624–627. doi: 10.1038/nature08820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ko A, Shin JY, Seo J, Lee KD, Lee EW, Lee MS, et al. Acceleration of gastric tumorigenesis through MKRN1-mediated posttranslational regulation of p14ARF. J Natl Cancer Inst. 2012;104:1660–1672. doi: 10.1093/jnci/djs424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, Zha M, Zhao X, Jiang P, Du W, Tam AY, et al. Siva1 inhibits p53 function by acting as an ARF E3 ubiquitin ligase. Nat Commun. 2013;4:1551. doi: 10.1038/ncomms2533. [DOI] [PubMed] [Google Scholar]
- 21.Argent RH, Thomas RJ, Letley DP, Rittig MG, Hardie KR, Atherton JC. Functional association between the Helicobacter pylori virulence factors VacA and CagA. J Med Microbiol. 2008;57:145–150. doi: 10.1099/jmm.0.47465-0. [DOI] [PubMed] [Google Scholar]
- 22.Brandt S, Kwok T, Hartig R, Konig W, Backert S. NF-kappaB activation and potentiation of proinflammatory responses by the Helicobacter pylori CagA protein. Proc Natl Acad Sci U S A. 2005;102:9300–9305. doi: 10.1073/pnas.0409873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pimkina J, Murphy ME. ARF, autophagy and tumor suppression. Autophagy. 2009;5:397–399. doi: 10.4161/auto.5.3.7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reef S, Zalckvar E, Shifman O, Bialik S, Sabanay H, Oren M, et al. A short mitochondrial form of p19ARF induces autophagy and caspase-independent cell death. Mol Cell. 2006;22:463–475. doi: 10.1016/j.molcel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 25.Balaburski GM, Hontz RD, Murphy ME. p53 and ARF: unexpected players in autophagy. Trends Cell Biol. 2010;20:363–369. doi: 10.1016/j.tcb.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buchholz M, Braun M, Heidenblut A, Kestler HA, Kloppel G, Schmiegel W, et al. Transcriptome analysis of microdissected pancreatic intraepithelial neoplastic lesions. Oncogene. 2005;24:6626–6636. doi: 10.1038/sj.onc.1208804. [DOI] [PubMed] [Google Scholar]
- 27.Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 28.Whitehead RH, Robinson PS. Establishment of conditionally immortalized epithelial cell lines from the intestinal tissue of adult normal and transgenic mice. Am J Physiol Gastrointest Liver Physiol. 2009;296:G455–460. doi: 10.1152/ajpgi.90381.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peek RM, Jr, Blaser MJ, Mays DJ, Forsyth MH, Cover TL, Song SY, et al. Helicobacter pylori strain-specific genotypes and modulation of the gastric epithelial cell cycle. Cancer Res. 1999;59:6124–6131. [PubMed] [Google Scholar]
- 30.Treier M, Staszewski LM, Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 31.Wei J, Nagy TA, Vilgelm A, Zaika E, Ogden SR, Romero-Gallo J, et al. Regulation of p53 tumor suppressor by Helicobacter pylori in gastric epithelial cells. Gastroenterology. 2010;139:1333–1343. doi: 10.1053/j.gastro.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.N’Diaye EN, Kajihara KK, Hsieh I, Morisaki H, Debnath J, Brown EJ. PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO Rep. 2009;10:173–179. doi: 10.1038/embor.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Sablet T, Piazuelo MB, Shaffer CL, Schneider BG, Asim M, Chaturvedi R, et al. Phylogeographic origin of Helicobacter pylori is a determinant of gastric cancer risk. Gut. 2011;60:1189–1195. doi: 10.1136/gut.2010.234468. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.