Abstract
Two novel cyclic quaternary amine crosslinking probes are synthesized for structural mass spectrometry of protein complexes in solution and for analysis of protein interactions in organellar and whole cell extracts. Each exhibits high aqueous solubility, excellent protein crosslinking efficiencies, low collision induced dissociation (CID) energy fragmentation efficiencies, high stoichiometries of reaction, increased charges of crosslinked peptide ions, and maintenance of overall surface charge balance of crosslinked proteins.
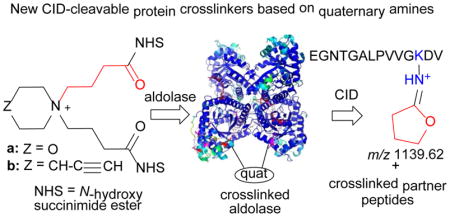
Graphical abstract
New CID-cleavable protein crosslinkers based on quaternary amines
Chemical crosslinking coupled with tandem mass spectrometry (CXL-MS) has been increasingly applied to structural analysis of protein complexes in solution1–13 and to global analysis of protein interactions in organelles and whole cells14–17. This is because in the former case, CXL-MS can provide distance constraints and other information that aids determination of the structures of proteins recalcitrant to classical methods such as X-ray crystallography, NMR, and other techniques. Computational tools are available to support these efforts, which integrate disparate sources of structural information2, 4, 5, 18–22. In the latter case, crosslinking represents an effective way to globally monitor proteins in intact organelles and in living cells with the potential to directly probe changes in protein interactions in vivo.
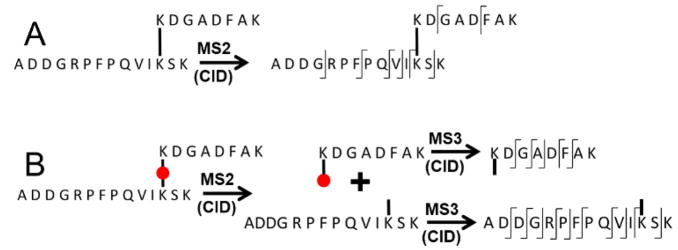
Classical “bottom-up” approaches to CXL-MS using non-fragmenting crosslinkers, where crosslinked proteins are digested to smaller peptides for MS analysis, suffer from two disadvantages (Fig. 1) arising from the resistance of the crosslink bonds to collision-induced dissociation (CID). The first is that the larger size of the crosslinked analytes and the increased degrees of freedom result in lower fragmentation efficiencies, particularly at lower energies. The second is that the two crosslinked peptides fragment simultaneously, leading to a complex mixture of fragment ions whose search space is N2, where N is the total number of calculated proteolytic peptides23, 24. This large search space increases computational time and has a negative impact on the false discovery rate (FDR) making them challenging for complex mixtures.
Figure 1.
Fragmentation schema for Lys-directed non-cleavable (A) and cleavable (B) crosslinkers. Tryptic peptide examples are for illustration only. Increased backbone fragmentation typically observed for linear peptides is indicated. Left spurs on hashes represent observed “B-ions” and right spurs are “Y-ions”. The red circle represents the CID-cleavable crosslinking moiety. Only two of the four possible products are shown for MS3 in B.
An effective approach to alleviating these limitations has been the development of crosslinkers that have lower fragmentation energies than the peptide bonds25–36. In this approach, collision induced dissociation (CID) of the crosslinked peptides in tandem mass spectrometry (MS2 in Fig. 1) results in scission of the crosslinker, generating a characteristic marker for the crosslinked species that can be used to trigger selection and CID of each component peptide for subsequent identification (MS3 in Fig. 1). Alternative data acquisition schema using CID-cleavable crosslinkers can also be employed (e.g., alternating low and high collision energies, higher-energy collision dissociation (HCD) in an Orbitrap, etc.). This uncoupling of the identification of crosslinked ions from the identification of the component peptides reduces the search space dramatically to 2N from N2, with consequential improvements in the FDR and the complexity of samples that can be readily analyzed. Incorporation of affinity tags into cleavable crosslinkers provides enrichment capabilities advantageous for complex mixtures21, 37.
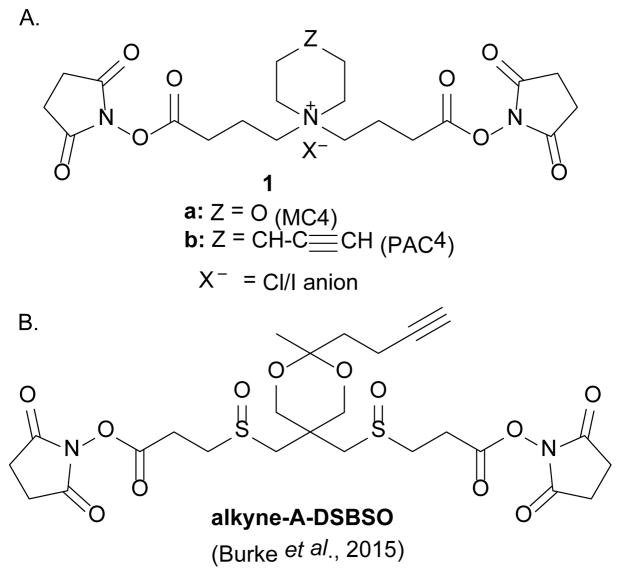
We report the synthesis and initial characterization of two novel CID-cleavable protein crosslinkers exhibiting excellent protein crosslinking and low energy fragmentation efficiencies (Fig. 2A). The first, compound 1a, possesses a central morpholino group and is shorter than our previous crosslinkers33, having an extended conformation length of approximately 13 angstroms. The second, compound 1b§, has a central 4-(ethynyl)piperidinyl moiety that is amenable to enrichment by Click chemistry38–40. Crosslinkers 1a and 1b are also referred to by the abbreviated names MC4 and PAC4, respectively. Alkyne 1b has some structural similarity to alkyne-A-DSBSO41 (Fig. 2B), but is shorter and designed to be highly water soluble. Additionally, it undergoes MS-cleavage by a completely different mechanism.
Figure 2.
A. Structures of quaternary amine crosslinkers 1a (MC4) and 1b (PAC4); B. Structure of sulfoxide crosslinker alkyne-A-DSBSO.
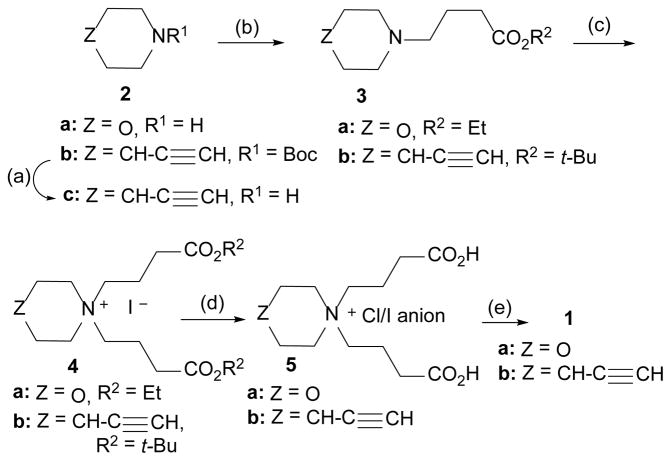
The synthesis of each crosslinker is delineated in Scheme 1. Key structural features of each probe include a cyclic nitrogen heterocycle core flanked by two identical side chains, each made up of a 3-carbon methylene spacer connecting the quaternary nitrogen of the core to an N-hydroxysuccinimide (NHS) ester “warhead”. The route developed for 1a commenced with the alkylation of morpholine (2a), using a variation of a patent procedure42, with ethyl 4-bromobutyrate in refluxing toluene to provide 3a. Attempted tandem alkylation with excess bromo ester was sluggish and necessitated that installation of the second arm be carried out in a separate step. After considerable experimentation, we found that reaction of 3a with the more reactive ethyl 4-iodobutyrate43 occurred slowly at 115 °C under neat conditions to provide the key bis-quaternary ammonium salt ester 4a. Subsequent acid hydrolysis with concentrated HCl in acetic acid proceeded slowly, but cleanly, at 100 °C to provide diacid 5a as a mixed chloride/iodide salt. Activation of this to the bis-NHS ester was accomplished by mild heating of 5a with excess N,N′-disuccinimidyl carbonate and pyridine in acetonitrile following a literature procedure44, whereas reaction in neat pyridine left only starting material. Unreacted starting diacid 5a was filtered away from 1a, which was then precipitated by slow addition of crude product, dissolved in minimal acetonitrile, into a large excess of cold ethyl acetate. The overall yield of 1a was 31% in a four-step sequence of reactions.
Scheme 1.
Reagents and conditions: (a) HCl in p-dioxane, RT, 30 min, ~100%. (b) for 3a: ethyl 4-bromobutanoate, toluene, 120°C, 2 h, 65%; for 3b: t-Bu 4-iodobutanoate, K2CO3, ACN, RT, 3 d, 80%. (c) for 4a: neat ethyl 4-iodobutanoate, 115°C, 16 h, 95%; for 4b: t-Bu 4-iodobutanoate, ACN, RT, 13 d, 50%. (d) for 5a: conc. HCl, gla. HOAc, 100°C, 2 d, 91%; for 5b: HCl in p-dioxane, DCM, RT, 41 h, 67%. (e) N, N′-disuccinimidyl carbonate, pyridine, ACN, 50 – 60°C, 16 – 64 h, 37 – 64%.
Having completed the synthesis of 1a, we then set out to apply our reaction methodologies to the construction of congeneric crosslinker 1b. However, the substitution of morpholine for 4-ethynylpiperidine as the core heterocycle had a profound impact on the chemistry, necessitating a partial redesign of our route (Scheme 1). The major modification required swapping the ethyl ester of 4a with the tert-butyl ester of 4b due to the instability of the alkyne moiety to the vigorous acid hydrolysis conditions of 4a. Second, we found that the piperidine nitrogen of 2c was less reactive than that of morpholine, necessitating the use of tert-butyl-4-iodobutanoate45 for both alkylation reactions. Thus, room temperature reaction of 4-ethynylpiperidine hydrochloride46 (2c) with a slight excess of tert-butyl 4-iodobutyrate in acetonitrile under basic conditions over a three-day period gave 3b in good yield. Further transformation to 4b with excess iodo ester proved problematic, due to an unexpected putative fragmentation of the nascent bis-ester product during its formation in refluxing acetonitrile. We believe this is driven by the presence of the quaternary center as a control run shows that 3b is stable under these conditions. After investigation of several alternative conditions (temperature, solvent), we eventually were able to generate 4b in modest yield as a sharp-melting white solid by stirring 3b and the iodo ester at room temperature as a very concentrated solution in acetonitrile over a 13-day period. Mild, non-aqueous hydrolysis of 4b then proceeded uneventfully with anhydrous HCl in p-dioxane at room temperature to cleanly give diacid 5b as a mixed chloride/iodide salt. Hydrolysis with excess trifluoroacetic acid in dichloromethane47 worked also, but resulted in an oily, less pure product. Further conversion of 5b to target crosslinker 1b was effected under similar conditions described for the synthesis of 1a. The overall yield of 1b was 10% in a four-step sequence of reactions, with the lower yield reflecting problematic reactions relative to 1a. We feel that the overall yield of both crosslinkers can be substantially improved through a modest route optimization campaign.
All compounds were rigorously purified by flash chromatography or crystallization, and their structural assignments are supported by diagnostic peaks in the 1H NMR spectra and by mass spectrometry. Digital copies of these along with full experimental procedures can be found in Supplementary Information.
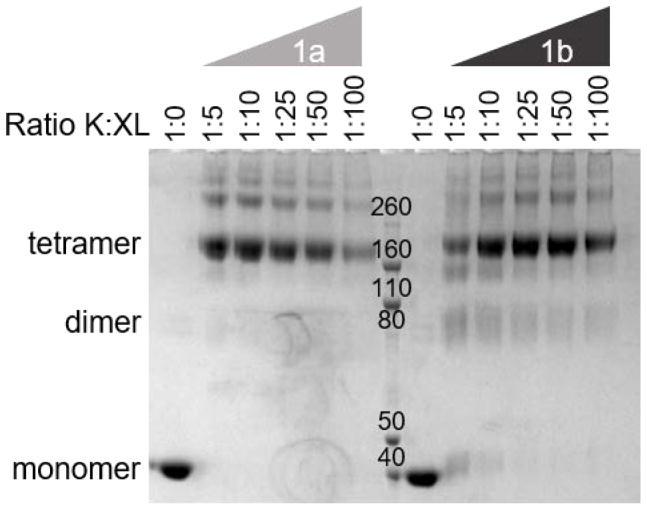
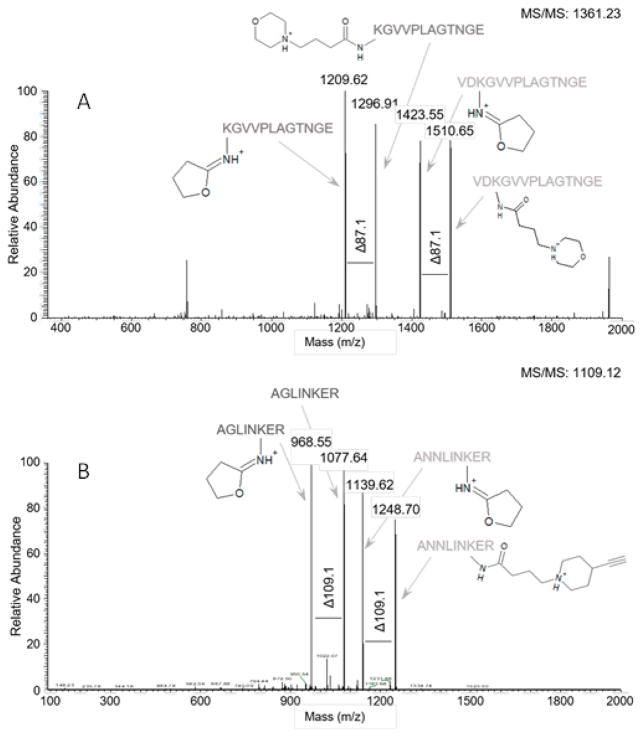
Both 1a and 1b are efficient crosslinkers, exhibiting essentially stoichiometric crosslinking of the tetramer (Fig. 3) at a 1:5 molar ratio of lysyl residue: crosslinker with no significant increase in higher order complexes up to 1:100. Low energy CID of peptides crosslinked with 1a or 1b results in fragmentation on either side of the central cyclic amine (morpholino or 4-(alkynyl)piperidinyl) giving characteristic pairs of doublets, where each doublet corresponds to one of the crosslinked peptides (Fig. 4). Fragmentation efficiencies are excellent but the relative intensities of the doublets can vary depending on peptide sequence and the charge state of the precursor. Identification of the peptides can be accomplished by various acquisition protocols, including mass tag-triggered MS3, alternating low and high energy MS2, and HCD.
Figure 3.
Titration of CID cleavable crosslinkers 1a and 1bfor aldolase. Coomassie Brilliant Blue stained NuPAGE Bis-Tris (4% – 12%) gel of crosslinked samples after 15 min of reaction at room temperature (25 C) with an increasing ratio of Lys-residue/crosslinker (K:XL). Peptide standards are indicated in the center lane.
Figure 4.
CID fragmentation of a proteolytic digest product of (A) 1a crosslinked inter subunit peptides from aldolase (KGVVPLAGTNGE-XL-VDKGVVPLAGTNGE showing the mass difference marker of 87.11), (B) 1b crosslinked synthetic peptide (AGLINKER-XL-ANNLINKER with the mass difference marker of 109.15 indicated). XL is the crosslinker. The lower mass ion in each doublet is typically selected for MS3.
In summary, we have synthesized two novel quaternary cyclic amine probes that exhibit high aqueous solubility and which, in initial studies coupled with tandem mass spectrometry, exhibit excellent protein crosslinking and low energy fragmentation efficiencies. Future studies will be directed to the application of these crosslinkers to structural mass spectrometry of protein complexes and the dynamics of protein interactions in vitro and in vivo. We consider the core structure common to 1a and 1b, which confers the major properties listed above, to be a useful template for the development of additional crosslinking and tagging reagents having a range of functionality at the Z-position of the cyclic moiety. Our establishment of precise synthetic methodologies applicable to these will facilitate these studies.
Supplementary Material
Acknowledgments
This work was supported in part by NIH GM095832, NIH R01GM105942, NIH R01GM109896, NIH R01GM105920 from the National Institutes of Health.
Footnotes
Electronic Supplementary Information (ESI) available: See DOI: 10.1039/x0xx00000x
Conflicts of interest
There are no conflicts to declare.
Notes and references
- 1.Huang BX, Kim HY, Dass C. J Am Soc Mass Spectrom. 2004;15:1237–1247. doi: 10.1016/j.jasms.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Schulz DM, Kalkhof S, Schmidt A, Ihling C, Stingl C, Mechtler K, Zschoernig O, Sinz A. Proteins: Struct, Funct, Bioinf. 2007;69:254–269. doi: 10.1002/prot.21445. [DOI] [PubMed] [Google Scholar]
- 3.Li H, Wells SA, Jimenez-Roldan JE, Roemer RA, Zhao Y, Sadler PJ, O’Connor PB. Protein Sci. 2012;21:1269–1279. doi: 10.1002/pro.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monastyrskyy B, Fidelis K, Kryshtafovych A, D’Andrea D, Tramontano A. Proteins. 2016;84(Suppl 1):131–144. doi: 10.1002/prot.24943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schneider M, Brock O, Belsom A, Rappsilber J. Proteins. 2016;84(Suppl 1):152–163. doi: 10.1002/prot.25028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daturpalli S, Kniess RA, Lee CT, Mayer MP. J Mol Biol. 2017;429:1406–1423. doi: 10.1016/j.jmb.2017.03.025. [DOI] [PubMed] [Google Scholar]
- 7.Liu F, Rijkers DTS, Post H, Heck AJR. Nat Methods. 2015;12:1179–1184. doi: 10.1038/nmeth.3603. [DOI] [PubMed] [Google Scholar]
- 8.Leitner A, Walzthoeni T, Kahraman A, Herzog F, Rinner O, Beck M, Aebersold R. Mol Cell Proteomics. 2010;9:1634–1649. doi: 10.1074/mcp.R000001-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen ZA, Pellarin R, Fischer L, Sali A, Nilges M, Barlow PN, Rappsilber J. Mol Cell Proteomics. 2016;15:2730–2743. doi: 10.1074/mcp.M115.056473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chavez JD, Schweppe DK, Eng JK, Bruce JE. Cell Chem Biol. 2016;23:716–726. doi: 10.1016/j.chembiol.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chowdhury SM, Du X, Tolic N, Wu S, Moore RJ, Mayer MU, Smith RD, Adkins JN. Anal Chem (Washington, DC, U S) 2009;81:5524–5532. doi: 10.1021/ac900853k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ihling C, Schmidt A, Kalkhof S, Schulz DM, Stingl C, Mechtler K, Haack M, Beck-Sickinger AG, Cooper DMF, Sinz A. J Am Soc Mass Spectrom. 2006;17:1100–1113. doi: 10.1016/j.jasms.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 13.Singh P, Panchaud A, Goodlett DR. Anal Chem (Washington, DC, U S) 2010;82:2636–2642. doi: 10.1021/ac1000724. [DOI] [PubMed] [Google Scholar]
- 14.Zhang H, Tang X, Munske GR, Tolic N, Anderson GA, Bruce JE. Mol Cell Proteomics. 2009;8:409–420. doi: 10.1074/mcp.M800232-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chavez JD, Lee CF, Caudal A, Keller A, Tian R, Bruce JE. Cell Syst. 2017 doi: 10.1016/j.cels.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schweppe DK, Chavez JD, Lee CF, Caudal A, Kruse SE, Stuppard R, Marcinek DJ, Shadel GS, Tian R, Bruce JE. Proc Natl Acad Sci U S A. 2017;114:1732–1737. doi: 10.1073/pnas.1617220114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu F, Lossl P, Rabbitts BM, Balaban RS, Heck AJR. Mol Cell Proteomics. 2017 doi: 10.1074/mcp.RA117.000470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petrotchenko EV, Makepeace KA, Borchers CH. Curr Protoc Bioinformatics. 2014;48:8.18.11–19. doi: 10.1002/0471250953.bi0818s48. [DOI] [PubMed] [Google Scholar]
- 19.Webb B, Sali A. Curr Protoc Bioinformatics. 2016;54:5.6.1–5.6.37. doi: 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brodie NI, Petrotchenko EV, Borchers CH, Popov KI, Dokholyan NV. Sci Adv. 2017;3:e1700479. doi: 10.1126/sciadv.1700479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang X, Munske GR, Siems WF, Bruce JE. Anal Chem. 2005;77:311–318. doi: 10.1021/ac0488762. [DOI] [PubMed] [Google Scholar]
- 22.Schweppe DK, Chavez JD, Bruce JE. Bioinformatics. 2016;32:306–308. doi: 10.1093/bioinformatics/btv519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giese SH, Fischer L, Rappsilber J. Mol Cell Proteomics. 2016;15:1094–1104. doi: 10.1074/mcp.M115.049296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goetze M, Pettelkau J, Fritzsche R, Ihling CH, Schaefer M, Sinz A. J Am Soc Mass Spectrom. 2015;26:83–97. doi: 10.1007/s13361-014-1001-1. [DOI] [PubMed] [Google Scholar]
- 25.Park KD, Liu R, Kohn H. Chem Biol (Cambridge, MA, U S) 2009;16:763–772. doi: 10.1016/j.chembiol.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 26.Kandur WV, Kao A, Vellucci D, Huang L, Rychnovsky SD. Org Biomol Chem. 2015;13:9793–9807. doi: 10.1039/c5ob01410g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chowdhury SM, Munske GR, Tang X, Bruce JE. Anal Chem. 2006;78:8183–8193. doi: 10.1021/ac060789h. [DOI] [PubMed] [Google Scholar]
- 28.Hage C, Iacobucci C, Rehkamp A, Arlt C, Sinz A. Angew Chem, Int Ed. 2017;56:14551–14555. doi: 10.1002/anie.201708273. [DOI] [PubMed] [Google Scholar]
- 29.Iacobucci C, Hage C, Schaefer M, Sinz A. J Am Soc Mass Spectrom. 2017;28:2039–2053. doi: 10.1007/s13361-017-1744-6. [DOI] [PubMed] [Google Scholar]
- 30.Yu C, Huszagh A, Viner R, Novitsky EJ, Rychnovsky SD, Huang L. Anal Chem (Washington, DC, U S) 2016;88:10301–10308. doi: 10.1021/acs.analchem.6b03148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qian Y, Martell J, Pace NJ, Ballard TE, Johnson DS, Weerapana E. ChemBioChem. 2013;14:1410–1414. doi: 10.1002/cbic.201300396. [DOI] [PubMed] [Google Scholar]
- 32.Kasper PT, Back JW, Vitale M, Hartog AF, Roseboom W, de Koning LJ, van Maarseveen JH, Muijsers AO, de Koster CG, de Jong L. ChemBioChem. 2007;8:1281–1292. doi: 10.1002/cbic.200700150. [DOI] [PubMed] [Google Scholar]
- 33.Clifford-Nunn B, Showalter HDH, Andrews PC. J Am Soc Mass Spectrom. 2012;23:201–212. doi: 10.1007/s13361-011-0288-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petrotchenko EV, Serpa JJ, Borchers CH. Mol Cell Proteomics. 2011;10:M110.001420. doi: 10.1074/mcp.M110.001420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iacobucci C, Goetze M, Piotrowski C, Arlt C, Rehkamp A, Ihling C, Hage C, Sinz A. Anal Chem (Washington, DC, U S) 2018;90:2805–2809. doi: 10.1021/acs.analchem.7b04915. [DOI] [PubMed] [Google Scholar]
- 36.Yu C, Huang L. Anal Chem (Washington, DC, U S) 2018;90:144–165. doi: 10.1021/acs.analchem.7b04431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sohn CH, Agnew HD, Lee JE, Sweredoski MJ, Graham RLJ, Smith GT, Hess S, Czerwieniec G, Loo JA, Heath JR, Deshaies RJ, Beauchamp JL. Anal Chem (Washington, DC, U S) 2012;84:2662–2669. doi: 10.1021/ac202637n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de AG, Sletten EM, Nakamura H, Palaniappan KK, Bertozzi CR. Angew Chem Int Ed Engl. 2012;51:2443–2447. doi: 10.1002/anie.201106325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angew Chem, Int Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 40.Baskin JM, Prescher JA, Laughlin ST, Agard NJ, Chang PV, Miller IA, Lo A, Codelli JA, Bertozzi CR. Proc Natl Acad Sci U S A. 2007;104:16793–16797. doi: 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burke AM, Kandur W, Novitsky EJ, Kaake RM, Yu C, Kao A, Vellucci D, Huang L, Rychnovsky SD. Org Biomol Chem. 2015;13:5030–5037. doi: 10.1039/c5ob00488h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ghiron C, Nencini A, Arianna, Micco I, Zanaletti R, Maccari L, Bothmann H, Haydar S, Varrone M, Pratelli C, Harrison B. WO2008087529A1. PCT Int Patent Applic. 2008
- 43.Gupta A, Gueddah R, Berube G. Synth Commun. 2009;39:61–69. [Google Scholar]
- 44.Higuchi K, Yamashina T, Ishikawa K, Hirata H. Yukagaku. 1987;36:16–20. [Google Scholar]
- 45.Glenn MP, Kahnberg P, Boyle GM, Hansford KA, Hans D, Martyn AC, Parsons PG, Fairlie DP. J Med Chem. 2004;47:2984–2994. doi: 10.1021/jm030222i. [DOI] [PubMed] [Google Scholar]
- 46.Raimundo BC, Oslob JD, Braisted AC, Hyde J, McDowell RS, Randal M, Waal ND, Wilkinson J, Yu CH, Arkin MR. J Med Chem. 2004;47:3111–3130. doi: 10.1021/jm049967u. [DOI] [PubMed] [Google Scholar]
- 47.Moreno P, Quelever G, Peng L. Tetrahedron Lett. 2015;56:4043–4046. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.