Abstract
Respiration is one of the most vital and basic features of living organisms. In mammals, respiration is accomplished by respiratory chain complexes located on the mitochondrial inner membrane. In the past century, scientists put tremendous efforts in understanding these complexes, but failed to solve the high resolution structure until recently. In 2016, three research groups reported the structure of respiratory chain supercomplex from different species, and fortunately the structure solved by our group has the highest resolution. In this review, we will compare the recently solved structures of respirasome, probe into the relationship between cristae shape and respiratory chain organization, and discuss the highly disputed issues afterwards. Besides, our group reported the first high resolution structure of respirasome and medium resolution structure of megacomplex from cultured human cells this year. Definitely, these supercomplex structures will provide precious information for conquering the mitochondrial malfunction diseases.
Keywords: Structure of respirasome, Cristae organization, Substrate channeling, Electron transfer pathway, Assembly of respirasome
Mitochondria are involved in a variety of vital cellular activities, among which energy conversion is the most critical. A vast amount of efforts have been put into depicting the structure, assembly, coupling mechanism and pathology of respiratory chain complexes, and several landmarks should be noticed: 1) Mitchell in 1961 put forward the chemiosmotic hypothesis [1], which is supported by later structural and functional analyses, especially the structure of bovine ATP synthase solved in 1994 by Walker [2]; 2) Schägger in 2000 solubilized the mitochondria membrane with digitonin and detected the high molecular weight bands using blue native page (BNPAGE), thus for the first time proposing the respirasome model [3], which is further amended by Enriquez who verified the respiratory activity of respirasome and raised the plasticity model in 2008 [4]; 3) Our group presented the first near-atomic resolution structure of respirasome in 2016 [5], together with several nominal resolution structures independently solved by Sazanov and Kuhlbrandt almost at the same time [6], [7], again heating up debates about the electron transfer mechanism in electron transport chain supercomplex. As exemplified above, our understanding of the energy conversion machinery has been greatly advanced, however, this understanding is still far from satisfying.
The plasticity model is thus far the most widely accepted theory about the respiratory chain organization [8], [9]. In this model, most CII, CIV and a relevant proportion of CIII stand alone and seem to move freely on the inner mitochondrial membrane, while the majority of CI is stabilized by the CIII dimer, with or without several copies of CIV, and the super-assembly of these complexes are dynamic, with a certain turnover rate. Consequently, electron transfer from CI to CIII is most likely carried out by CoQs shuttled within the I1III2IVn supercomplexes, despite that CoQs carrying electrons from CII seems to freely move on the membrane. Kinetic evidence using flux control analysis performed by Lenaz and Genova is consistent with this model, showing that electron transfer via CI and CIII behaves like an integral enzyme, while the transfer via CII and CIII demonstrates like separate enzymes [10], [11].
However, the exact organization of the respiratory chain has never gained consensus and has been challenged continuously by emerging evidence. It has been shown that the stoichiometry of supercomplexes can vary in different physiological conditions and cell types [12], [13], [14], but we know nearly nothing about the regulation pathway. Most recently, Greggio reported that after 4 months of exercise training, not only individual respiratory chain complexes but also respiratory supercomplexes in human muscle mitochondria show an increase, and the free complexes tend to assemble into functional supercomplexes after exercise [15]. But still, we don't understand the detailed signaling pathways. Cryo-ET (Cryo-electron tomography) analyses locate the ATP synthase dimers at the edge of the cristae curve, and CI at the approximately plane surface of cristae [16], [17]. The shape of mitochondria cristae is closely related with aging and cell apoptosis, and has been reported to influence the supercomplex assembly [18], [19]. However, the detailed connection between cristae shape and supercopmlex organization is not known, either. Structural evidence indicate the existence of higher oligomerization level of respirasomes, termed as megacomplexes [5], [6], [9], and finally in this year, our group unprecedentedly reported the first medium resolution structure of megacomplex from cultured human cells after docking of the well resolved sub regions including CI, CIII dimer and CIV [20]. For the first time, this work provided solid evidence for the existence of megacomplex, observed the possible compartment of Q pool, and connected the organization of respiratory chain more tightly with the shape of cristae. Since this is the first structure of human respirasome, previously reported mitochondrial disease related mutations can all be mapped into our structure. Doubtlessly, this is a great step forward into conquering many severe neurodegenerative diseases, including Alzheimer's syndrome, Parkinson's disease, multiple sclerosis, friedreich's ataxia, Amyotrophic lateral sclerosis, etc. [20].
Disputes about substrate channeling, electron transfer mechanism and the assembly process of supercomplexes have always been fierce [21]. Despite the widely accepted segmentation theory of the Q pool, Hirst et al. presented evidence against the partitioning of the Q pool in 2014 through their spectroscopic and kinetic experiments [22]. In accordance, since the early 1980s, Gutman et al. have shown the phenomenon of reverse electron transfer from succinate to NAD+, indicating that the Q pool in SC might have communications with the free Q pool via some unknown mechanism [23]; in contrast to the classic Q cycle theory, our group proposed another electron pathway within CIII based on our high resolution structure of respirasome [9]; In addition, although CIV subunit COX7A2L in mouse has been identified as an assembly factor of supercomplex, more assembly factors still await to be found and the accurate assembly process is far from clear [24].
In this review, we will introduce recent advances of the mitochondrial ETC (electron transport complexes) research in three parts: the structure details of respirasome, the relationship between cristae shape and respiratory chain organization, and the highly disputed issues including substrate channeling, electron transfer pathway, and the assembly process of respirasome.
Structures of respirasome
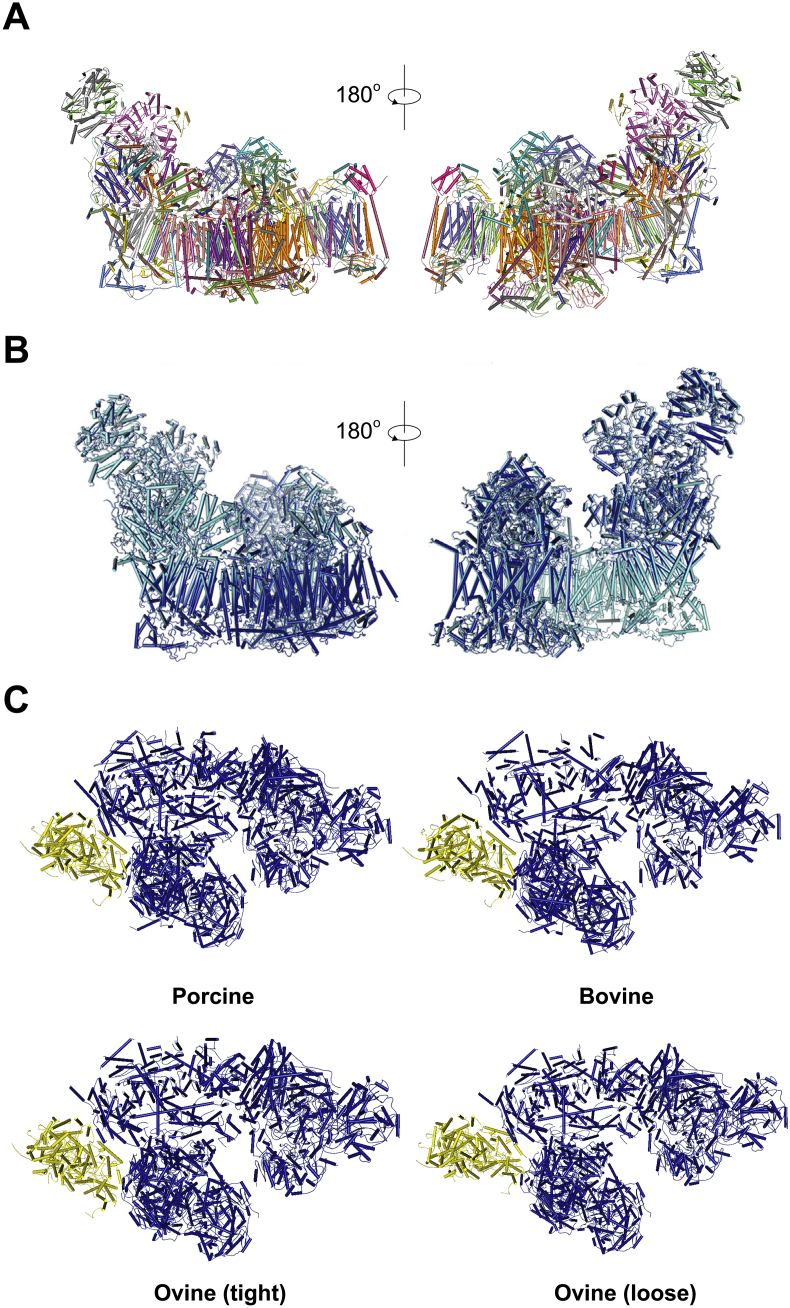
Since the August of 2016, structural research of respiratory chain has obtained exciting breakthroughs, including four medium to high resolution cryo-EM structures of respirasome independently solved by three different groups [5], [6], [7], [9]. These structures originate from porcine, bovine or ovine, and due to differences in species source, purification methods and cryo-EM data quality, some detailed discrepancies were observed while the overall structures fit well with each other [Fig. 1].
Fig. 1.
Overall structures and conformational changes of respirasome from porcine, ovine and bovine (A). High resolution structure of respirasome from porcine heart. Front and back view of cartoon models are shown. Each subunit is shown in different colors. These structures are originated from data in PDB: 5GUP. (B) Conformational change of CI and CIII. Two conformations of CI and CIII are shown in marine and cyan respectively. (C) Conformational change of CIV. CIV is shown in yellow, while CI and CIII are shown in blue in respirasome. Relative distance between CIII and CIV are variable through different species and conformations. Model of respirasome from porcine, bovine, ovine (tight), ovine (loose) are originated from PDB 5GUP, 5LUF, 5J4Z and 5J7Y respectively [5], [6], [7], [52].
Most recently (Oct 5th, 2017), a review article from Sazanov's group also discussed current knowledge about mitochondrial respiratory chain, but clearly they have some misunderstanding about our work [25]. They criticized that the interaction sites between CI and CIII are not resolved at the level of side chains, which is not true. In fact, our density map has the highest resolution by far, and the contact sites can be identified clearly and shown in the Figure 5F of our published paper [5]. They also pointed out that the density for CIV is weak so the positioning and inferred contacts may not be reliable, which is quite arbitrary. In fact, after docking the crystal CIV structure (PDB:1OCC) into our map, the correlation coefficients reach a very high level, which means the final map is quite reliable. Besides, they accused that the maps describing the intermediate states of processing are of the wrong hand, which is actually not important and indicates that maybe the authors are not quite familiar with the structural solving process of the single particle cryo-EM method, because in the final map we performed a total reversion and the opposite chirality of the intermediate states is only for convenience in data processing. Finally, they claimed that the 10-kDa subunit (NDUFV3) is misassigned, which is not true because in samples from porcine heart the density for this subunit is missing, and the reason is the difference in species source. Inexplicably, the authors cited one of our piezo1 structural paper [26] (cited as the 74th citation) in the “Function of supercomplex formation” section when they described the “Comparison of the structures of isolated CI and SCs” [25]. It's really unintelligible to cite one of our paper there. As stated above, despite some defects caused by the limitation of current technology, we still get the best density map till now.
The overall structure
Our group firstly used 81.1k particles to reconstruct a 5.4 Å resolution structure of porcine respirasome [5], which offered unprecedentedly rich information of the organization and mechanism of respirasome considering that former respirasome structures can only reach 22 Å and 19 Å resolution [27], [28]. In this structure, high resolution crystal structures of CIII (1BGY) and CIV (1OCC) can fit into the map with high correlation coefficients, and the resolution of CI density map alone can even reach 3.97 Å, which allows us to accurately assign 14 core subunits and 20 supernumerary subunits, together with 17 backbone models belonging to the other 11 unassigned subunits or the unassigned regions of the assigned proteins. The CIII dimer in this structure is in the ‘int’ state, because distances of the [2Fe-2S] cluster to heme c1 and heme bL are 30 Å and 27 Å respectively. Cytochrome c, which might be lost during sample preparation, is found to attach to neither CIII nor CIV. Taken together, totally 131 transmembrane helices are identified in this structure, with 77 from CI, 26 from CIII and 28 from CIV, and these transmembrane helices form a flat gigantic transmembrane disk [5].
After that, our group pushed the study further, applying 162k particles to refinement, and finally raised the resolution of our overall structure to 4.0 Å, which is the first near-atomic resolution structure of respirasome and provided a mass of information [9]. Using sub-region refinement, the resolution of CI and CIII both reach 3.6 Å, allowing de novo model building for the majority of residues from all CI and CIII subunits, except NDUFA12 and NDUFV3 of CI. The density map for NDUFV3 is missing, while NDUFA12 can be assigned without accurately building of side chains. Taken together, 44 subunits of CI with 78 TMHs and 22 subunits of CIII with 26 TMHs are accurately placed in the density map, and 13 subunits of CIV with 28 TMHs are assigned to the density map using rigid body fit (1OCC). Besides, 11 and 12 phospholipid molecules in CI and CIII, 6 and 2 heme molecules in CIII and CIV are identified from the structure, respectively.
Despite previously reported medium to high resolution structure of mammalian CI [29], [30], the map of CI in our supercomplex structure actually has the highest resolution (3.6 Å). Although the authors claimed they solved the atomic structure of the entire ovine mitochondrial complex I at 3.9 Å, we downloaded the structure coordinate (PDB: 5LNK) and the map (EMDB: EMD-4093) to check its electron density quality. The results clearly show that the electron density quality is too poor to build the model. To exemplify, there is no density at all of the NDUFA11, no side chain density of most of the accessory components of the membrane arm, and the density for phospholipids is even worse. The overall density is actually worse than the 4.2 Å structure of the bovine CI reported by Hirst's group [29]. Recently, Sazanov's group updated both the coordinate and the map, and the electron density looks better than before.
In our CI map, 1 FMN molecule in NDUFV1, 1 NADPH molecule in NDUFA9, 1 Zn2+ ion in NDUFS6, 8 FeS clusters in the matrix region, 2 D4PP groups in two acyl carrier protein complexes NDUFAB1-NDUFA6 and NDUFAB1-NDUFB9 can be clearly distinguished. In the matrix module, NDUFS2, NDUFS7 and ND1 form the Q chamber. In the membrane module, among the 21 supernumerary subunits, 12 subunits have 1 TMH, NDUFC2 has 2 TMHs, NDUFA11 has 4 TMHs, 3 subunits (NDUFA10, NDUFAB1b and NDUFB9) sit on the matrix side, and 4 subunits (NDUFA8, NDUFS5, NDUFB7 and NDUFB10) containing CX9C motifs able to form intramolecular disulphides within the ‘CHCH’ domains sit on the intermembrane side. Detailed interactions between subunits in CI are observed, showing NDUFA8, NDUFA13, NDUFS4, NDUFS8, NDUFB10, NDUFB9, NDUFB5, ND5 and NDUFS2 interact with other 9, 9, 10, 10, 11, 13, 15, 16 and 17 subunits, respectively. A significant difference with previous CI structures observed is the N terminus of NDUFS2 in our structure, which extends a long way from matrix module to the distal membrane part [9], [31].
Slightly later than our group, Sazanov's group submitted a paper in which they applied 54.5k particles to 3D classification and got two classes of supercomplex I1III2IV1 from ovine heart, a tight form and a loose form, reaching resolutions of 5.8 Å and 6.7 Å, respectively [6]. In both structures, the density for CIV is weaker than that for CI and CIII. α-helices in both structures can be resolved, but β-sheets and loops can only be resolved in the tight structure. CI FeS clusters can be identified in both structures, but heme molecules in CIII and CIV can only be identified in the tight structure. The FeS cluster in the Rieske protein of CIII is not visible in both structures, but the position of this subunit is clearly assigned in the distal position (‘b’ state). Taken together, totally 132 transmembrane helices (78 from CI, 26 from CIII and 28 from CIV) are identified. All the transmembrane helices are aligned to form a flat plate [6].
Two months later, Kuhlbrandt's group reported another structure of respirasome from bovine heart with a medium resolution (9 Å) [7]. They used PCC-a-M (trans-4-(trans-4′-propylcyclohexyl)cyclo-hexyl-a-d-maltoside), rather than digitonin to solubilize the mitochondrial membrane. Totally 156,519 particles were applied to classification and two classes of I1III2IV1 were resolved, with class 1 (11% of total particles) resolved to 9.1 Å, and class 2 (6% of total particles) resolved to 10.4 Å. 132 α-helices in the transmembrane region can be observed in the class 1 structure [7].
In 2017, our group reconstructed the first structure of human megacomplex I2III2IV2 from cultured cells. The centrosymmetric structure of human MCI2III2IV2 resembles the architecture of the 2D-average of the supercomplex-I2III2 from potato previously described by Bultema. The overall structure is ∼220 A° in height, ∼280 A ° in width, and ∼300 A ° in length, containing 140 subunits with 238 transmembrane helices (TMHs). All the TMHs are aligned to form a slightly bent plane. Two CIs encircle the whole structure centrosymmetrically with the CIII dimer locates at the center of the huge machine. Two CIVs are anchored by the distal end of two CI membrane arms at each side, with a clear gap between CIV and the other CI. Meanwhile, the two monomers of the CIII dimer both have intensive interactions with CI and CIV at the same side, so both monomers could receive QH2 from one CI and pass reduced cyt.c to one adjacent CIV to transform energy most efficiently. This may be the ultimate oligomerization form of respiratory chain complexes functioning in emergent conditions. The interfaces between the CI-CIII and CI-CIV covered areas of 2616 Å2 and 946 Å2, respectively. Similar to our previously demonstrated porcine SCI1III2IV1 structure, UQCRC1 of CIII has close interactions with NDUFB9, NDUFAB1 of CI, and NDUFA11 of CI was close to UQCRQ of CIII. COX7A of CIV contacts both CI and CIII. However, there were no direct interactions between CIII and CIV, and the gaps between the complexes at other sites might be bridged by phospholipid molecules.
Conformational changes
Based on our medium resolution structure [5], we found that compared to the previously reported free form CI (PDB ID: 4UQ8), the distal end of the membrane arm of CI in respirasome displays a lateral movement towards the middle region, supporting the proposal that CIII and CIV can help CI to acquire a more stable conformation in respirasome. In our high resolution structure [5], casting out the relatively low resolution part of CIV, the density map of CI and CIII can be classified into two major groups, indicating two states of respirasome. In these two states, the membrane arm of CI can overlap, but in the tighter state the matrix arm of CI and the overall structure of CIII move toward each other compared to the looser state. Based on the structure, our group also proposed a new mechanism of electron transfer between CI and CIII, which will be discussed in the last section [5], [9].
The ovine respirsasome structure also indicated two conformations, the tight form and the loose form. According to their model, CIV can contact both CI and CIII in the tight form while can only contact CI in the loose form. The distance between the matrix regions of CI and CIII increases in the loose form. The two forms of respirasome are shown to be able to interconvert. However, these two conformations cannot be matched to our structure. We think the loose form is very likely a dysfunctional form waiting to be disassembled, as indicated by themselves [6].
Kuhlbrandt's group also identified two conformations of respirasome. The CIII dimer is reported to rotate around 25° between the two classes, and the matrix arm of CI in class 2 moves a little away from center of the respirasome, which is consistent with the deactive form of CI reported by Zhu [29]. Furthermore, they claim that in their structure, the Rieske protein of one monomer of CIII cannot be resolved clearly, thus concluding that only one of the Rieske protein is functional (the one cannot be resolved). Sazanov's group also proposed the asymmetric nature of CIII dimer [6]. However, we prefer the model that both CIII monomers are able to transfer electron to cyt.c. Details will be discussed in the last section [7] [Fig. 1].
Interaction between individual complexes
Based on the medium resolution structure, our group reported accurate interactions between respiratory complexes for the first time. In the middle part of the membrane module, NDUFA11 of CI can interact with UQCRB and UQCRQ of one monomer of CIII, and at the distal part, the N-lobe of NDUFB9 of CI can interact with a short loop of UQCRC1 (S251-L265) of the other CIII monomer on the matrix side. Transmembrane helices of COX7C of CIV have interactions with the last transmembrane helix of ND5 at the distal end of CI membrane arm, and the flexible region before the transmembrane helix of COX7A of CIV can interact with UQCRC1 and UQCR11 of CIII at the matrix side. Interestingly, an isoform of COX7A obtained from mouse, COX7A2L, is believed to be important for CIII and CIV assembling into supercomplexes but do not affect individual CIII or CIV function in mouse, and thus renamed as SCAF1 (supercomplex assembly factor 1) [4], [18], [24]. However, we didn't detect SCAF1 in the porcine respirasome [5] [Table 1].
Table 1.
Interactions between individual complexes within respirasome.
| Species | CI, CIII interaction | CI, CIV interaction | CIII, CIV interaction | |
|---|---|---|---|---|
| Porcine | NDUFA11, NDUFB4, NDUFB8, NDUFB9, UQCRB, UQCRQ, UQCRC1, UQCRC10 | ND5, NDUFB3, NDUFB7, NDUFB8, COX7A, COX7C, COX8 | UQCRC1, UQCR11, UQCRB, COX7A | |
| Ovine | Tight | NDUFA11, NDUFB4, NDUFB7, NDUFB9, NDUFB10, UQCRQ, UQCRH, UQCRC1, UQCRFS1, UQCRH | ND5, NDUFB2, NDUFB3, NDUFB7, COX7A, COX7C, COX8B, | UQCRC1, UQCR11,COX7A |
| Loose | NDUFA11, NDUFB4, NDUFB7, NDUFB9, NDUFB10, UQCRQ, UQCRH, UQCRC1, UQCRFS1, UQCRH | ND5, NDUFB3, COX7A | No interaction | |
| Bovine | NDUFA11, NDUFB9, UQCR1, UQCRQ | ND5, COX7C | UQCRC1, UQCR10, UQCR11, COX7A | |
In our high resolution structure, the interaction between CI, CIII and CIV are visualized more clearly. In addition to previously identified NDUFA11 of CI, UQCRB and UQCRQ of CIII, which still forms most of the interactions between CI and CIII, the TMH of single transmembrane subunits NDUFB4 and NDUFB8 are also lined in the middle part of CI and CIII, and the N-terminus of NDUFB4 together with helix-3 of the LYR-motif containing subunits NDUFB9 can also interact with a highly conserved loop (Y257-T266) of UQCRC1 of CIII at the matrix side. The N terminus of UQCRC10 is also identified to contact the N terminus of NDUFB4 and NDUFB8. Interestingly, each TMH of NDUFA11 contain a cysteine residue, with C95 and C115 actually forming a disulfide bond which substantially contributes to the stability of TMH region of NDUFA11, while C18 and C115 pointing to different directions, indicating that the redox environment around the inner mitochondria membrane can regulate the stability of NDUFA11, thus the stability of supercomplex. In the high resolution structure, interactions between CIV and other subunits are also defined at atomic level. COX7A of CIV is close to a short helix of NDUFB8 of CI, and on the other side to the helices of UQCRC1, UQCR11 and UQCRB of CIII. E503 of ND5 can form a hydrogen bond with R20 of COX7C, meanwhile NDUFB3, NDUFB7 and NDUFB8 are also close to COX7C and COX8. An unoccupied space is identified between CI and CIII, which is considered as the Q tunnel, supporting the substrate channeling theory [9] [Table 1].
Most interactions between individual complexes in ovine respirasome occur at the same sites as in porcine respirasome, even though some specific subunits can vary. In both tight and loose forms, interaction between CI and CIII remain similar, but interactions with CIV vary slightly. In the middle part of CI membrane arm, NDUFA11 and NDUFB4 of CI can interact with UQCRQ of CIII, while NDUFB10 and NDUFA11 of CI can contact UQCRH of CIII. In the distal end between CI and CIII, NDUFB9 and NDUFB4 of CI can interact with UQCRC1 and UQCRFS1 of CIII. At the interface between CI, CIII and CIV, NDUFB7 can contact both UQCRH of CIII and COX7A of CIV. In the tight form, ND5, NDUFB2, NDUFB3, NDUFB7 of CI can interact with COX7C and COX8B of CIV, while UQCR11 and UQCRC1 of CIII can interact with COX7A of CIV. In the loose form, CIV swings away from CIII, and COX7A contact ND5 and NDUFB3 [6] [Table 1].
Due to the limited resolution of bovine respirasome structure, direct interaction between individual subunits cannot be accurately determined. However, interactions between NDUFB9 of CI and UQCR1 of CIII on the matrix side, and between NDUFA11 of CI and UQCRQ of CIII in the membrane region are identified. Besides, ND5 of CI approaches COX7C of CIV, and UQCRC1, UQCR10, UQCR11 of CIII are found to interact with COX7A1 of CIV [7] [Table 1].
Phospholipids
The respirasome structure solved by our group has another advantage, the accurately placed phospholipids. Our group identified multiple phospholipid molecules in the high resolution structures, and proposed these lipid molecules function in two interplaying aspects: on the one hand, NDUFA11 is found to interact with at least 3 lipid molecules, which behave like the glue to make protein–protein interactions between NDUFA11 and other subunits more stable, thus enhancing the supercomplex stability; on the other hand, the broken regions between ND1, ND2, ND4 and ND6 TMHs are all shown to interact with hydrophobic tails of phospholipids, which behave like the lube to membrane arm, thus rendering more flexibility of the proton pumping module and enhancing the proton pumping ability of CI. This finding is in consistent with previous studies reporting the important roles of lipid molecules in respirasome formation and function [9], [31].
Cristae shape and supercomplex organization
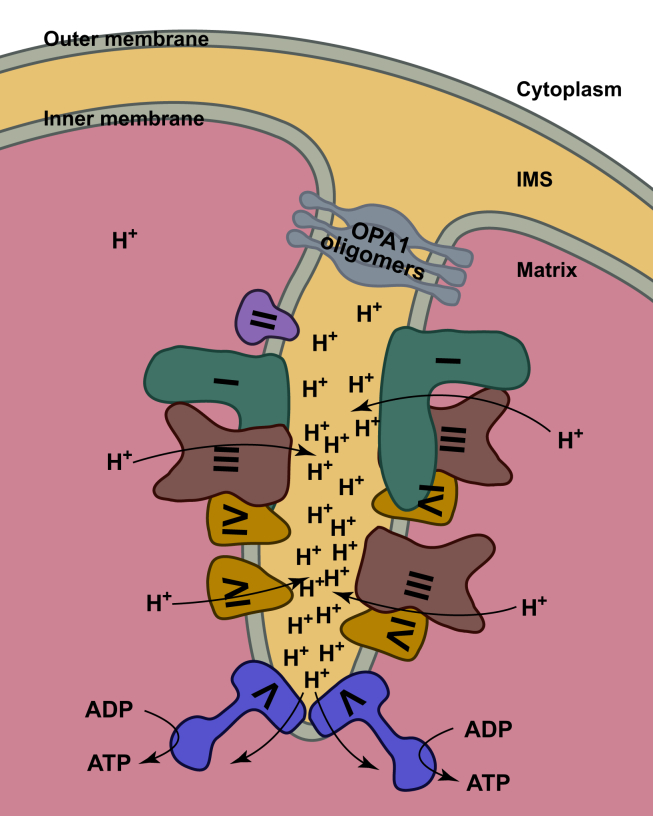
Mitochondria participate in a series of vital cellular activities, including energy conversion, saccharide and lipid metabolism, apoptosis, aging, and calcium homeostasis. Mitochondrial morphology has been reported to be tightly connected with mitochondrial function [32], [33], [34]. Two layers of lipid membranes are cyclized to form the mitochondria, with the outer membrane similar to the plasma membrane, while the inner membrane being more complicated and classified into two types. One is the boundary membrane that is closely apposed with the outer membrane, and the other is the cristae, which are extended to the intermembrane space and connected to the boundary membrane through many tubular structures termed as crista junctions [18]. Within these crista junctions, a series of scaffold proteins (MICOS) lash the base of cristae [35], [36]. The shape of cristae is variable in different species and cell conditions. When mitochondrial respiration is triggered (transition from State IV to State III), “Orthodox” mitochondria usually observed after fixation with intact tissue become “condensed”, where the surface of cristae expands and the matrix volume decreases [34], [37]. Several groups reported that using cryo-ET technology, the respiratory chain complexes responsible for generating the proton gradient are found to be located on the relatively flat surface of cristae, while the ATP synthase dimers are located at the edge curve of cristae and aligned to form ribbons [16]. This arrangement can greatly avoid proton leakage across the large cristae surface, and enables the sink formed by the ATP synthase dimers to harness proton gradient with highest efficiency [Fig. 2].
Fig. 2.
The relationship between cristae shape and respirasome. Simple cartoon model of cristae and OXPHOS proteins are shown. Respirasome and individual complexes are located on the planar cristae surface. ATP synthase dimer sits at the edge of cristae. The of root of cristae is lashed by OPA1 oligomers. Abbreviation: IMS: Intermembrane space. Traces of proton are shown by arrows.
Since 2000, more and more evidence have shown that individual respiratory chain complexes can assemble into supercomplexes to increase the electron transfer efficiency and alleviate ROS generation [38], [39], [40]. Soon enough, the shape of mitochondrial cristae is proved to influence the assembly of supercomplexes, at least indirectly [18]. OPA1 is a member of the dynamin-related GTPase protein family, and functions as rivets at the crista junction [41], [42]. When apoptosis signals transmit to mitochondria, OPA1 oligomers are disrupted, and remodeling of cristae occurs, resulting in the release of cytochrome c into plasma. In OPA1 mutated cells, mitochondria cristae show an onion-like shape, where the assembly of supercomplexes is severely disrupted. Interestingly, many groups reported that the cristae shape is linked with ATP synthase dimerization [17], [43]. In yeast mutants where dimerization of ATP synthases is hampered, cristae cannot be normally organized. Furthermore, although the shape of cristae can change among different species, ATP synthase dimers seem to exist in all kinds of normal cristae [17], [43]. In this sense, supercomplex assembly is connected with ATP synthase dimerization, and their organization both depends on the proper cristae shape. This conclusion is illuminating, however, higher resolution cryo-ET analysis of normal and mutated mitochondria is required in the future to provide more solid details [Fig. 2].
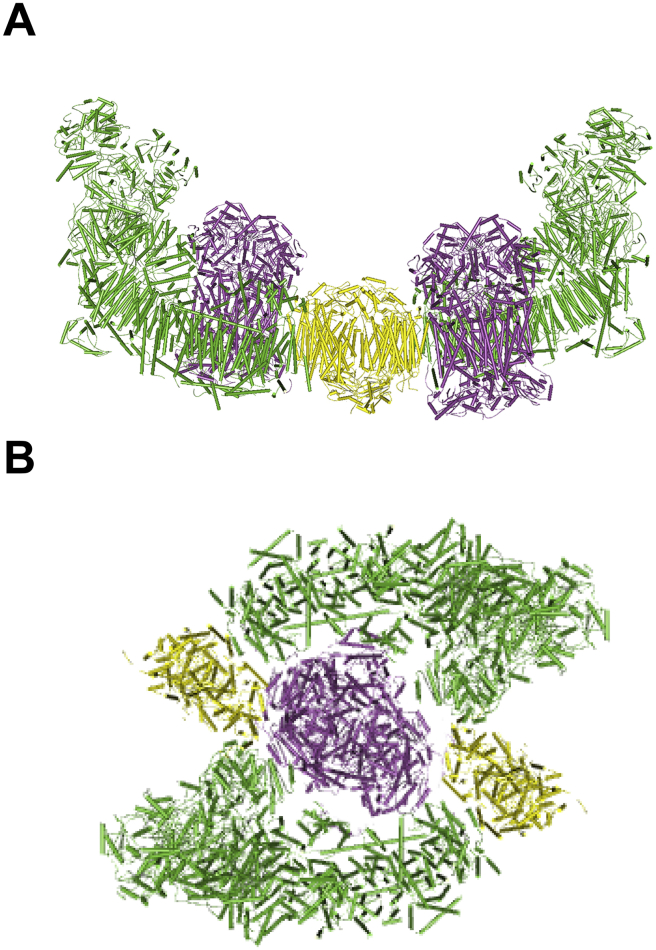
Some significant models regarding supercomplex organization other than the plasticity model were also raised and equally intriguing, such as the respiratory string model and the megacomplex model. In 2004, Schägger's group speculated the model of respiratory string based on stoichiometry [44]. In this model, I2III4IV8 forms circular structures linked by III2IV4 to form a helical string along the cristae surface. However, this model seems less consistent with the conditions in mammals where not all CIIIs and CIVs are assembled into supercomplexes [8], [9]. In 2007 and 2013, two other groups reported the megacomplex organization model of CIII and CIV in yeast and bacteria respectively, where dimerization of CIII and CIV can result in the formation of megacomplex containing multiple copies of CIII and CIV [45], [46]. Meanwhile, many groups identified circular structures larger than respirasome in their cryo-EM particle images. In 2008, Kouril's group reported a megacomplex organization from potato mitochondria, where larger particles than respirasome were detected in both BNPAGE and cryo-EM images [47]. In 2016, our group and Sazanov's group reported two similar structures of respirasome, while we presented two different megacomplex models. In Sazanov's model, two respirasomes are linearly organized and linked by CIV forming a dimer, similar to a string [6]. However, this is not in accordance with our cryo-EM images, which are circular and agree with Kouril's model proposed in 2008. In our circular model, electron transfer between CI and CIII, CIII and CIV can reach the highest efficiency [5], [9]. Although in the samples we obtained from solubilized mitochondrial membrane, respirasome occupied a rather high partition while only a few megacomplexes existed, we couldn't rule out the possibility that due to the solubilizing condition, the megacomplex organization in vivo might be disassembled.
In 2017, our group reported the first medium resolution structure of megacomplex from cultured human cells [20], and after docking of well resolved sub regions, we could accurately place all of the 45 subunits in each CI, 22 subunits in dimeric CIII, and do rigid body fitting of the 13 subunits in each CIV and of the 2 cytochrome c molecules. This work provided convincing evidence for the existence of megacomplex, and clearly proved the circular model, thus the organization of respiratory chain complexes shall have an even tighter connection with the shape of cristae, because the megacomplex can occupy an even larger area on the cristae surface [Fig. 3].
Fig. 3.
Linear and circular model of megacomplex Linear model (A) and circular model (B) of megacomplex organization are shown. CI, CIII and CIV are in green, mega and yellow respectively [5], [6].
Disputable issues
What we have known raises even more controversial issues, including the authenticity of the substrate channel, the pathway of electron transfer, and the assembly process of supercomplexes.
Substrate channeling
One of the major advantages proposed of forming supercomplexes is to increase the electron transport efficiency through substrate channeling, which is quite reasonable. Substrate channeling is the direct transfer of a reaction intermediate between active sites of two or more enzymes catalyzing sequential reactions. The first experimental evidence [10], [11] for substrate channeling within the supercomplex is provided by Lenaz et al., in 2004. They used specific inhibitors to detect the extent of flux control related to CI, CII and CIII, and calculated the flux control coefficients (FCC) of each individual complex in NADH oxidation and succinate oxidation, respectively. The FCCs of both CI and CIII in NADH oxidation pathway are close to 1, which means these two enzymes behave like an entirety. In contrast, the FCC of CII in succinate pathway is high, while that of CIII is quite low, indicating substrate channeling only exists between CI and CIII, not between CII and CIII. On the other hand, flux control analysis of CIV using cyanide inhibition detected no substrate channeling between CIII and CIV. Many other groups confirmed the substrate channel between CI and CIII through various methods. In 2008, Benard et al. identified three different CoQ pools in succinate oxidation [48]. One pool is directly used; another pool functions as a reserve and is used when normal energy flux level is disturbed; and the third pool is not used in succinate oxidation at all, indicating CI and CII actually use different CoQ pools. In 2013, Lapuente-Brun et al. analyzed the rates of CI- or CII-linked respiration when CI, CII, CIII, or CIV are genetically ablated, showing the existence of dedicated CoQ and cyt.c pools compartmented in SCs, and the CI–CIII channel is preferred when the amount of CIII is limited [49].
However, Blaza and Trouillard challenged the existence of substrate channeling between CI and CIII, CIII and CIV respectively. Blaza et al. recorded the peak absorbance change of heme molecules in CIII when NADH and/or succinate was added to the in vitro system [22]. They claimed that all three combinations (NADH, succinate, NADH and succinate) can rapidly reduce heme molecules to almost the same extent, suggesting NADH and succinate could access all the CIII present. However, one thing worth noting is that the time scale of heme depletion and substrate channel are basically incomparable. In their experiments, the reduction of heme molecules took a few seconds to fully complete, while substrate channeling between CI and CIII could be completed in only a few milliseconds. In other words, CoQs compartmented within SCs can exchange relatively slowly with free CoQs in the membrane, but this cannot exclude the existence of CoQ pool. The substrate channeling between CIII and CIV is hard to detect, because the majority of CIV is in the free form [50], and the compartmentation and pool behaviors of cyt.c are hard to be identified. Although Trouillard et al. provided evidence against cyt.c channeling [51], they cannot rule out the possibility that their results is caused by the dynamic nature of SC organization.
In 2011, Althoff et al. reported a low resolution cryo-EM structure of bovine SCI1III2IV1, based on which they claimed that the three component complexes are arranged in a unique way that the Q binding sites of CI and CIII face each other and are separated by a 13 nm gap, while the cyt.c sites of CIII and CIV are arranged within 10–11 nm, supporting the substrate channeling mechanism [27]. However, due to the limited resolution, detailed arrangement around the channel is ambiguous. Recently, Sazanov's, Kuhlbrandt's and our group solved three medium to high resolution structures of respirasome from ovine, bovine, and porcine, respectively. All these groups reported that the Q sites in both CI and CIII are open to the membrane and are located close to each other, with no barriers detected, supporting the substrate channeling theory [6], [7], [9], [52]. The same is true about the cyt.c sites in CIII and CIV.
In 2017, our group reported the structure of megacomplex I2III2IV2, and furthermore, we observed a mass of unoccupied density between CI and CIV. Some groups reported that CII can interact with CI, CIII and CIV before, so we modeled a CII structure into the gap formed by CI and CIV [4], [49], [53], [54], [55], [56], [57], which can match pretty well. Accordingly, we proposed the model of ETCS (electron transport chain supercomplex) [20]. The loss of CII in ETCS might be an artificial result during sample preparation or data processing, and another reason might be the low abundance of CII in megacomplex itself. In this model, the Q binding site of CI, CII and CIII all face a sealed compartment, which is very likely to be the previously reported Q pool. This result can also explain why a proportion of CII appear to use the same Q pool with CI and why electrons from succinate can be transferred to NADH. One thing worth noting is that our model didn't exclude the possibility that CoQ molecules in the Q pool could exchange with free CoQ molecules in a very low rate.
Electron transfer pathway
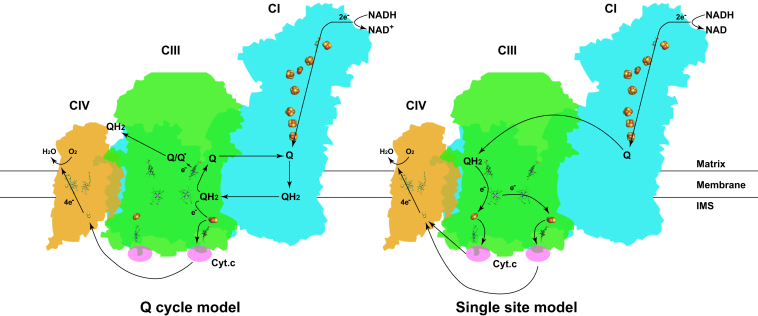
The most classic theory about electron transfer in the respiratory chain is the Q cycle model proposed by Mitchell in 1975 [58]. In this model, there are two Q binding sites in each CIII monomer. One is the Qi site near heme bH at the matrix side, and the other is the Qo site near heme bL at the intermembrane side. At the beginning, reduced QH2 released from CI or CII reaches the Qo site. The first electron from QH2 is transferred to cyt.c through heme c1, and the second electron is transferred to an oxidized Q molecule bound at the Qi site through heme bH, thus forming a semiquinone, which can generate reactive oxygen species (ROS). After two QH2 molecules being oxidized the Qo site, two electrons are transferred to two cyt.c molecules, and one Q molecule is reduced at the Qi site.
Sazanov's group presented an electron transfer pathway based on their respirasome structure and agrees with the Q cycle theory [6]. The proximal Q cavity (close to CI) is open to the membrane, while the distal Q cavity (close to CIV) is slightly capped by CIV, which can consume oxygen rapidly and generate a reducing environment, thus alleviating the ROS production by the semiquinone generated during Q cycle. They argued that the asymmetry of CIII dimer in respirasome can cope perfectly with the need to reduce ROS generation and proceed Q cycle in the meanwhile [6]. Kuhlbrandt's group also reported the CIII dimer to be asymmetric, however, they proposed that the distal monomer is totally inactive, having no function except as a scaffold, and the cyt.c binding site in the proximal monomer is closer to CIV than the inactive one, increasing the electron transfer efficiency [7]. They accepted the Q cycle theory in their model as well [Fig. 4].
Fig. 4.
Different electron transfer mechanisms. Two mechanisms proposed by different groups are presented and depicted in the paragraph. Complex I, III, IV and cyt.c are shown in cyan, green, brown and purple respectively. Electron transfer pathway is indicated by arrows. Abbreviation: IMS: Intermembrane space.
In contrast, based on the high resolution structure of respirasome, our group proposed an electron transfer pathway different from the classic Q cycle model [9]. In fact, the major defects of the Q cycle model promoted us to raise this model. The Qo site in CIII is at the intermembrane space side, while the Q binding pocket in CI is at the matrix side, which means if the QH2 released from CI is to reach the Qo site, its polar head must traverse across the hydrophobic inner mitochondrial membrane, and this is highly energy expensive. In the Q cycle theory, only one electron is transferred to cyt.c, and the other one is transferred back to Q, which is quite extravagant in term of energy conversion. Besides, despite that the Q cycle theory has been popular for a long time, no structure of Qi site occupied by native Q was reported [59], neither were reliable data supporting the Qo site to be a functional QH2 binding site provided [60].
Despite the Q cycle theory, our group proposed a “single site model” with the highest energy conversion efficiency based on the structural analyses [9]. According to the structural information, the Qi sites of the proximal and distal CIII monomer are 11 nm and 13 nm away from the Q binding pocket in CI, respectively. QH2 released from CI will bind to the Qi site instead of the Qo site in CIII, and the distal Qi site is easier to reach. The first electron is transferred from the Qi site in the distal monomer to heme bL in the proximal CIII monomer via tunneling between heme bH and the two heme bL molecules, and the second electron is transferred to heme bL in the distal CIII monomer. Then, the two electrons are transferred to two cyt.c molecules through two Rieske proteins and two heme c1 molecules in the CIII dimer. In this pathway, both electrons of QH2 are utilized, and the polar head of the Q molecule does not need to traverse across the hydrophobic membrane [Fig. 4].
Assembly of supercomplexes
The biogenesis and assembly of supercomplexes are highly relevant to proper mitochondrial function and pathogenesis of many severe neuromuscular disorders [18], [24], [61], [62]. Many groups even reported that during heart failure, aging and Barth syndrome, it is the assembly of respirasome rather than the amount of individual complexes that is mainly affected [15], [63], [64], [65]. However, the accurate process is largely unknown. In 2012, Moreno-Lastres et al. proposed that a CI assembly intermediate, instead of the holo CI enzyme, functions as a scaffold for incorporation of CIII and CIV to form the respirasome, and the final step of adding the NADH dehydrogenase catalytic module (distal part of matrix region) to CI would activate CI and the whole respirasome [66]. Similarly, Perez–Perez et al. reported that the CIII dimer will finish its assembly after the formation of supercomplex III2IV1. They claimed that a pre CIII dimer will bind the CIV subunit COX7A2L (SCAF1) first, and then CIV incorporates to form a pre supercomplex [67]. Finally, the remaining part of CIII dimer assemble to the supercomplex and CIII becomes matured. They also concluded that COX7A2L mainly promote the stabilization of III2IV1, without affecting respirasome formation. However, Cogliati et al. reported that they confirmed the long isoform of SCAF1 is necessary for interaction between CIII and CIV, while they also found respirasome is absent in most tissues where only short isoform of SCAF1 exists, except for heart and skeletal muscle, indicating SCAF1 indeed helps respirasome formation [24]. Some other assembly factors have been reported to assist the respirasome assembly [68], [69], [70], including C11orf83(UQCC3), Rcf1, and Aim24. In addition, cardiolipin was found critical in supercomplex assembly [71], [72]. Nonetheless, the clear assembly process of respirasome is still behind the curtain. Besides, some groups investigating the assembly of individual complex I did not detect the existence of CI assembly intermediate participating in respirasome formation, adding more enigmas to respirasome assembly [73], [74]. To be honest, the accurate assembly process of respirasome is largely unknown, so the demand of finding more assembly factors and assembly intermediates is urgent.
Conclusion
Respirasome is such a huge and delicate machine that both mtDNA and nDNA mutations in almost any of its components can hamper its proper function and cause many severe human diseases [75], [76], like cancer [77], [78], neurodegenerative disease [79], [80], [81], aging [82], [83], and cardiovascular disease [84]. High resolution structures of respirasome, especially the human respirasome, could provide new guidance to pathology research and drug development, and detailed characterization of pathological mutations in relationship with structural changes will be important for promoting both basic science and clinical research. Cryo-EM tomography and single-molecule labeling techniques will offer more insights on the organization and real-time dynamic state of respirasome in situ. As indicated above, the regulation mechanism of respirasome assembly and function is barely known, thus screening supercomplex assembly factors and discovering their relationship with existing cellular signal pathways are very important. Moreover, further functional studies are warranted to validate the new mechanism of electron transfer proposed by our group.
Conflicts of interest
The authors declare no competing financial interests.
Footnotes
Peer review under responsibility of Chang Gung University.
References
- 1.Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature. 1961;191:144–148. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- 2.Abrahams J.P., Leslie A.G., Lutter R., Walker J.E. Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 3.Schagger H., Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Acin-Perez R., Fernandez-Silva P., Peleato M.L., Perez-Martos A., Enriquez J.A. Respiratory active mitochondrial supercomplexes. Mol Cell. 2008;32:529–539. doi: 10.1016/j.molcel.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 5.Wu M., Gu J., Guo R., Huang Y., Yang M. Structure of mammalian respiratory supercomplex I1III2IV1. Cell. 2016;167:1598–1609.e10. doi: 10.1016/j.cell.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 6.Letts J.A., Fiedorczuk K., Sazanov L.A. The architecture of respiratory supercomplexes. Nature. 2016;537:644–648. doi: 10.1038/nature19774. [DOI] [PubMed] [Google Scholar]
- 7.Sousa J.S., Mills D.J., Vonck J., Kuhlbrandt W. Functional asymmetry and electron flow in the bovine respirasome. Elife. 2016;5:e21290. doi: 10.7554/eLife.21290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enriquez J.A. Supramolecular organization of respiratory complexes. Annu Rev Physiol. 2016;78:533–561. doi: 10.1146/annurev-physiol-021115-105031. [DOI] [PubMed] [Google Scholar]
- 9.Guo R., Gu J., Wu M., Yang M. Amazing structure of respirasome: unveiling the secrets of cell respiration. Protein Cell. 2016;7:854–865. doi: 10.1007/s13238-016-0329-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bianchi C., Fato R., Genova M.L., Parenti Castelli G., Lenaz G. Structural and functional organization of Complex I in the mitochondrial respiratory chain. Biofactors. 2003;18:3–9. doi: 10.1002/biof.5520180202. [DOI] [PubMed] [Google Scholar]
- 11.Bianchi C., Genova M.L., Parenti Castelli G., Lenaz G. The mitochondrial respiratory chain is partially organized in a supercomplex assembly: kinetic evidence using flux control analysis. J Biol Chem. 2004;279:36562–36569. doi: 10.1074/jbc.M405135200. [DOI] [PubMed] [Google Scholar]
- 12.Genova M.L., Lenaz G. Functional role of mitochondrial respiratory supercomplexes. Biochim Biophys Acta. 2014;1837:427–443. doi: 10.1016/j.bbabio.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Ikeda K., Shiba S., Horie-Inoue K., Shimokata K., Inoue S. A stabilizing factor for mitochondrial respiratory supercomplex assembly regulates energy metabolism in muscle. Nat Commun. 2013;4:2147. doi: 10.1038/ncomms3147. [DOI] [PubMed] [Google Scholar]
- 14.Sun D., Li B., Qiu R., Fang H., Lyu J. Cell type-specific modulation of respiratory chain supercomplex organization. Int J Mol Sci. 2016;17:926. doi: 10.3390/ijms17060926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greggio C., Jha P., Kulkarni S.S., Lagarrigue S., Broskey N.T., Boutant M. Enhanced respiratory chain supercomplex formation in response to exercise in human skeletal muscle. Cell Metab. 2017;25:301–311. doi: 10.1016/j.cmet.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 16.Davies K.M., Strauss M., Daum B., Kief J.H., Osiewacz H.D., Rycovska A. Macromolecular organization of ATP synthase and complex I in whole mitochondria. Proc Natl Acad Sci U S A. 2011;108:14121–14126. doi: 10.1073/pnas.1103621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muhleip A.W., Joos F., Wigge C., Frangakis A.S., Kuhlbrandt W., Davies K.M. Helical arrays of U-shaped ATP synthase dimers form tubular cristae in ciliate mitochondria. Proc Natl Acad Sci U S A. 2016;113:8442–8447. doi: 10.1073/pnas.1525430113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cogliati S., Frezza C., Soriano M.E., Varanita T., Quintana-Cabrera R., Corrado M. Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell. 2013;155:160–171. doi: 10.1016/j.cell.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stroud D.A., Ryan M.T. Mitochondria: organization of respiratory chain complexes becomes cristae-lized. Curr Biol. 2013;23:R969–R971. doi: 10.1016/j.cub.2013.09.035. [DOI] [PubMed] [Google Scholar]
- 20.Guo R., Zong S., Wu M., Gu J., Yang M. Architecture of human mitochondrial respiratory megacomplex I2III2IV2. Cell. 2017;170:1247–1257.e12. doi: 10.1016/j.cell.2017.07.050. [DOI] [PubMed] [Google Scholar]
- 21.Alvarez-Paggi D., Hannibal L., Castro M.A., Oviedo-Rouco S., Demicheli V., Tortora V. Multifunctional cytochrome c: learning new tricks from an old dog. Chem Rev. 2017;117:13382–13460. doi: 10.1021/acs.chemrev.7b00257. [DOI] [PubMed] [Google Scholar]
- 22.Blaza J.N., Serreli R., Jones A.J., Mohammed K., Hirst J. Kinetic evidence against partitioning of the ubiquinone pool and the catalytic relevance of respiratory-chain supercomplexes. Proc Natl Acad Sci U S A. 2014;111:15735–15740. doi: 10.1073/pnas.1413855111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lenaz G., Tioli G., Falasca A.I., Genova M.L. Complex I function in mitochondrial supercomplexes. Biochim Biophys Acta. 2016;1857:991–1000. doi: 10.1016/j.bbabio.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 24.Cogliati S., Calvo E., Loureiro M., Guaras A.M., Nieto-Arellano R., Garcia-Poyatos C. Mechanism of super-assembly of respiratory complexes III and IV. Nature. 2016;539:579–582. doi: 10.1038/nature20157. [DOI] [PubMed] [Google Scholar]
- 25.Letts J.A., Sazanov L.A. Clarifying the supercomplex: the higher-order organization of the mitochondrial electron transport chain. Nat Struct Mol Biol. 2017;24:800–808. doi: 10.1038/nsmb.3460. [DOI] [PubMed] [Google Scholar]
- 26.Ge J., Li W., Zhao Q., Li N., Chen M., Zhi P. Architecture of the mammalian mechanosensitive Piezo1 channel. Nature. 2015;527:64–69. doi: 10.1038/nature15247. [DOI] [PubMed] [Google Scholar]
- 27.Althoff T., Mills D.J., Popot J.L., Kuhlbrandt W. Arrangement of electron transport chain components in bovine mitochondrial supercomplex I1III2IV1. EMBO J. 2011;30:4652–4664. doi: 10.1038/emboj.2011.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dudkina N.V., Kudryashev M., Stahlberg H., Boekema E.J. Interaction of complexes I, III, and IV within the bovine respirasome by single particle cryoelectron tomography. Proc Natl Acad Sci U S A. 2011;108:15196–15200. doi: 10.1073/pnas.1107819108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu J., Vinothkumar K.R., Hirst J. Structure of mammalian respiratory complex I. Nature. 2016;536:354–358. doi: 10.1038/nature19095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fiedorczuk K., Letts J.A., Degliesposti G., Kaszuba K., Skehel M., Sazanov L.A. Atomic structure of the entire mammalian mitochondrial complex I. Nature. 2016;538:406–410. doi: 10.1038/nature19794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melber A., Winge D.R. Inner secrets of the respirasome. Cell. 2016;167:1450–1452. doi: 10.1016/j.cell.2016.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daum B., Walter A., Horst A., Osiewacz H.D., Kuhlbrandt W. Age-dependent dissociation of ATP synthase dimers and loss of inner-membrane cristae in mitochondria. Proc Natl Acad Sci U S A. 2013;110:15301–15306. doi: 10.1073/pnas.1305462110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frey T.G., Mannella C.A. The internal structure of mitochondria. Trends Biochem Sci. 2000;25:319–324. doi: 10.1016/s0968-0004(00)01609-1. [DOI] [PubMed] [Google Scholar]
- 34.Hackenbrock C.R. Ultrastructural bases for metabolically linked mechanical activity in mitochondria. I. Reversible ultrastructural changes with change in metabolic steady state in isolated liver mitochondria. J Cell Biol. 1966;30:269–297. doi: 10.1083/jcb.30.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Milenkovic D., Larsson N.G. Mic10 oligomerization pinches off mitochondrial cristae. Cell Metab. 2015;21:660–661. doi: 10.1016/j.cmet.2015.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfanner N., van der Laan M., Amati P., Capaldi R.A., Caudy A.A., Chacinska A. Uniform nomenclature for the mitochondrial contact site and cristae organizing system. J Cell Biol. 2014;204:1083–1086. doi: 10.1083/jcb.201401006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hackenbrock C.R. Ultrastructural bases for metabolically linked mechanical activity in mitochondria. II. Electron transport-linked ultrastructural transformations in mitochondria. J Cell Biol. 1968;37:345–369. doi: 10.1083/jcb.37.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Diaz F., Enriquez J.A., Moraes C.T. Cells lacking Rieske iron-sulfur protein have a reactive oxygen species-associated decrease in respiratory complexes I and IV. Mol Cell Biol. 2012;32:415–429. doi: 10.1128/MCB.06051-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friedrich T. High resolution structure of the electron input side of the respiratory complex I. Biochim Biophys Acta (BBA) – Bioenergetics. 2010;1797:8–9. [Google Scholar]
- 40.Baracca A., Chiaradonna F., Sgarbi G., Solaini G., Alberghina L., Lenaz G. Mitochondrial Complex I decrease is responsible for bioenergetic dysfunction in K-ras transformed cells. Biochim Biophys Acta. 2010;1797:314–323. doi: 10.1016/j.bbabio.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 41.Varanita T., Soriano M.E., Romanello V., Zaglia T., Quintana-Cabrera R., Semenzato M. The OPA1-dependent mitochondrial cristae remodeling pathway controls atrophic, apoptotic, and ischemic tissue damage. Cell Metab. 2015;21:834–844. doi: 10.1016/j.cmet.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frezza C., Cipolat S., Martins de Brito O., Micaroni M., Beznoussenko G.V., Rudka T. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 2006;126:177–189. doi: 10.1016/j.cell.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 43.Strauss M., Hofhaus G., Schroder R.R., Kuhlbrandt W. Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J. 2008;27:1154–1160. doi: 10.1038/emboj.2008.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wittig I., Carrozzo R., Santorelli F.M., Schagger H. Supercomplexes and subcomplexes of mitochondrial oxidative phosphorylation. Biochim Biophys Acta. 2006;1757:1066–1072. doi: 10.1016/j.bbabio.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 45.Sousa P.M., Videira M.A., Santos F.A., Hood B.L., Conrads T.P., Melo A.M. The bc:caa3 supercomplexes from the Gram positive bacterium Bacillus subtilis respiratory chain: a megacomplex organization? Arch Biochem Biophys. 2013;537:153–160. doi: 10.1016/j.abb.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 46.Heinemeyer J., Braun H.P., Boekema E.J., Kouril R. A structural model of the cytochrome C reductase/oxidase supercomplex from yeast mitochondria. J Biol Chem. 2007;282:12240–12248. doi: 10.1074/jbc.M610545200. [DOI] [PubMed] [Google Scholar]
- 47.Bultema J.B., Braun H.P., Boekema E.J., Kouril R. Megacomplex organization of the oxidative phosphorylation system by structural analysis of respiratory supercomplexes from potato. Biochim Biophys Acta. 2009;1787:60–67. doi: 10.1016/j.bbabio.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 48.Benard G., Faustin B., Galinier A., Rocher C., Bellance N., Smolkova K. Functional dynamic compartmentalization of respiratory chain intermediate substrates: implications for the control of energy production and mitochondrial diseases. Int J Biochem Cell Biol. 2008;40:1543–1554. doi: 10.1016/j.biocel.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 49.Lapuente-Brun E., Moreno-Loshuertos R., Acin-Perez R., Latorre-Pellicer A., Colas C., Balsa E. Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science. 2013;340:1567–1570. doi: 10.1126/science.1230381. [DOI] [PubMed] [Google Scholar]
- 50.Schagger H., Pfeiffer K. The ratio of oxidative phosphorylation complexes I-V in bovine heart mitochondria and the composition of respiratory chain supercomplexes. J Biol Chem. 2001;276:37861–37867. doi: 10.1074/jbc.M106474200. [DOI] [PubMed] [Google Scholar]
- 51.Trouillard M., Meunier B., Rappaport F. Questioning the functional relevance of mitochondrial supercomplexes by time-resolved analysis of the respiratory chain. Proc Natl Acad Sci U S A. 2011;108:E1027–E1034. doi: 10.1073/pnas.1109510108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gu J., Wu M., Guo R., Yan K., Lei J., Gao N. The architecture of the mammalian respirasome. Nature. 2016;537:639–643. doi: 10.1038/nature19359. [DOI] [PubMed] [Google Scholar]
- 53.Schon E.A., Dencher N.A. Heavy breathing: energy conversion by mitochondrial respiratory supercomplexes. Cell Metab. 2009;9:1–3. doi: 10.1016/j.cmet.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 54.Jiang X., Li L., Ying Z.X., Pan C.J., Huang S.Q., Li L. A small molecule that protects the integrity of the electron transfer chain blocks the mitochondrial apoptotic pathway. Mol Cell. 2016;63:229–239. doi: 10.1016/j.molcel.2016.06.016. [DOI] [PubMed] [Google Scholar]
- 55.Wan C., Borgeson B., Phanse S., Tu F., Drew K., Clark G. Panorama of ancient metazoan macromolecular complexes. Nature. 2015;525:339–344. doi: 10.1038/nature14877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Havugimana P.C., Hart G.T., Nepusz T., Yang H., Turinsky A.L., Li Z. A census of human soluble protein complexes. Cell. 2012;150:1068–1081. doi: 10.1016/j.cell.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Floyd B.J., Wilkerson E.M., Veling M.T., Minogue C.E., Xia C., Beebe E.T. Mitochondrial protein interaction mapping identifies regulators of respiratory chain function. Mol Cell. 2016;63:621–632. doi: 10.1016/j.molcel.2016.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mitchell P. Proton motive redox mechanism of the cytochrome b–c1 complex in the respiratory chain: proton motive ubiquinone cycle. FEBS Lett. 1975:1–6. doi: 10.1016/0014-5793(75)80098-6. [DOI] [PubMed] [Google Scholar]
- 59.Gao X., Wen X., Esser L., Quinn B., Yu L., Yu C.A. Structural basis for the quinone reduction in the bc1 complex: a comparative analysis of crystal structures of mitochondrial cytochrome bc1 with bound substrate and inhibitors at the Qi site. Biochemistry. 2003;42:9067–9080. doi: 10.1021/bi0341814. [DOI] [PubMed] [Google Scholar]
- 60.Pietras R., Sarewicz M., Osyczka A. Distinct properties of semiquinone species detected at the ubiquinol oxidation Qo site of cytochrome bc1 and their mechanistic implications. J R Soc Interface. 2016;13:20160133. doi: 10.1098/rsif.2016.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Antoun G., McMurray F., Thrush A.B., Patten D.A., Peixoto A.C., Slack R.S. Impaired mitochondrial oxidative phosphorylation and supercomplex assembly in rectus abdominis muscle of diabetic obese individuals. Diabetologia. 2015;58:2861–2866. doi: 10.1007/s00125-015-3772-8. [DOI] [PubMed] [Google Scholar]
- 62.Lopez-Fabuel I., Resch-Beusher M., Carabias-Carrasco M., Almeida A., Bolanos J.P. Mitochondrial complex I activity is conditioned by supercomplex I-III2-IV assembly in brain cells: relevance for Parkinson's disease. Neurochem Res. 2017;42:1676–1682. doi: 10.1007/s11064-017-2191-2. [DOI] [PubMed] [Google Scholar]
- 63.Rosca M.G., Vazquez E.J., Kerner J., Parland W., Chandler M.P., Stanley W. Cardiac mitochondria in heart failure: decrease in respirasomes and oxidative phosphorylation. Cardiovasc Res. 2008;80:30–39. doi: 10.1093/cvr/cvn184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gomez L.A., Monette J.S., Chavez J.D., Maier C.S., Hagen T.M. Supercomplexes of the mitochondrial electron transport chain decline in the aging rat heart. Arch Biochem Biophys. 2009;490:30–35. doi: 10.1016/j.abb.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang Y., Powers C., Madala S.K., Greis K.D., Haffey W.D., Towbin J.A. Cardiac metabolic pathways affected in the mouse model of barth syndrome. PLoS One. 2015;10:e0128561. doi: 10.1371/journal.pone.0128561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moreno-Lastres D., Fontanesi F., Garcia-Consuegra I., Martin M.A., Arenas J., Barrientos A. Mitochondrial complex I plays an essential role in human respirasome assembly. Cell Metab. 2012;15:324–335. doi: 10.1016/j.cmet.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perez-Perez R., Lobo-Jarne T., Milenkovic D., Mourier A., Bratic A., Garcia-Bartolome A. COX7A2L is a mitochondrial complex III binding protein that stabilizes the III2+IV supercomplex without affecting respirasome formation. Cell Rep. 2016;16:2387–2398. doi: 10.1016/j.celrep.2016.07.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deckers M., Balleininger M., Vukotic M., Rompler K., Bareth B., Juris L. Aim24 stabilizes respiratory chain supercomplexes and is required for efficient respiration. FEBS Lett. 2014;588:2985–2992. doi: 10.1016/j.febslet.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 69.Desmurs M., Foti M., Raemy E., Vaz F.M., Martinou J.C., Bairoch A. C11orf83, a mitochondrial cardiolipin-binding protein involved in bc1 complex assembly and supercomplex stabilization. Mol Cell Biol. 2015;35:1139–1156. doi: 10.1128/MCB.01047-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vukotic M., Oeljeklaus S., Wiese S., Vogtle F.N., Meisinger C., Meyer H.E. Rcf1 mediates cytochrome oxidase assembly and respirasome formation, revealing heterogeneity of the enzyme complex. Cell Metab. 2012;15:336–347. doi: 10.1016/j.cmet.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 71.Liu J., Ryabichko S., Bogdanov M., Fackelmayer O.J., Dowhan W., Krulwich T.A. Cardiolipin is dispensable for oxidative phosphorylation and non-fermentative growth of alkaliphilic Bacillus pseudofirmus OF4. J Biol Chem. 2014;289:2960–2971. doi: 10.1074/jbc.M113.536193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mileykovskaya E., Dowhan W. Cardiolipin-dependent formation of mitochondrial respiratory supercomplexes. Chem Phys Lipids. 2014;179:42–48. doi: 10.1016/j.chemphyslip.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guerrero-Castillo S., Baertling F., Kownatzki D., Wessels H.J., Arnold S., Brandt U. The assembly pathway of mitochondrial respiratory chain complex I. Cell Metab. 2017;25:128–139. doi: 10.1016/j.cmet.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 74.Stroud D.A., Surgenor E.E., Formosa L.E., Reljic B., Frazier A.E., Dibley M.G. Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature. 2016;538:123–126. doi: 10.1038/nature19754. [DOI] [PubMed] [Google Scholar]
- 75.Park C.B., Larsson N.G. Mitochondrial DNA mutations in disease and aging. J Cell Biol. 2011;193:809–818. doi: 10.1083/jcb.201010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wallace D.C. Mitochondrial DNA mutations in disease and aging. Environ Mol Mutagen. 2010;51:440–450. doi: 10.1002/em.20586. [DOI] [PubMed] [Google Scholar]
- 77.Gasparre G., Porcelli A.M., Lenaz G., Romeo G. Relevance of mitochondrial genetics and metabolism in cancer development. Cold Spring Harb Perspect Biol. 2013;5:a011411. doi: 10.1101/cshperspect.a011411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dumas J.F., Peyta L., Couet C., Servais S. Implication of liver cardiolipins in mitochondrial energy metabolism disorder in cancer cachexia. Biochimie. 2013;95:27–32. doi: 10.1016/j.biochi.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 79.McKenzie M., Lazarou M., Thorburn D.R., Ryan M.T. Mitochondrial respiratory chain supercomplexes are destabilized in Barth Syndrome patients. J Mol Biol. 2006;361:462–469. doi: 10.1016/j.jmb.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 80.Compton M.T., Bollini A.M., McKenzie Mack L., Kryda A.D., Rutland J., Weiss P.S. Neurological soft signs and minor physical anomalies in patients with schizophrenia and related disorders, their first-degree biological relatives, and non-psychiatric controls. Schizophr Res. 2007;94:64–73. doi: 10.1016/j.schres.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 81.McKenzie M., Lazarou M., Thorburn D.R., Ryan M.T. Analysis of mitochondrial subunit assembly into respiratory chain complexes using Blue Native polyacrylamide gel electrophoresis. Anal Biochem. 2007;364:128–137. doi: 10.1016/j.ab.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 82.Genova M.L., Lenaz G. The interplay between respiratory supercomplexes and ROS in aging. Antioxid Redox Signal. 2015;23:208–238. doi: 10.1089/ars.2014.6214. [DOI] [PubMed] [Google Scholar]
- 83.Kauppila T.E., Kauppila J.H., Larsson N.G. Mammalian mitochondria and aging: an update. Cell Metab. 2017;25:57–71. doi: 10.1016/j.cmet.2016.09.017. [DOI] [PubMed] [Google Scholar]
- 84.Rosca M., Minkler P., Hoppel C.L. Cardiac mitochondria in heart failure: normal cardiolipin profile and increased threonine phosphorylation of complex IV. Biochim Biophys Acta. 2011;1807:1373–1382. doi: 10.1016/j.bbabio.2011.02.003. [DOI] [PubMed] [Google Scholar]