Abstract
We here provide detailed insight in self-assembled complex coacervate systems exploiting gold nanoparticles for cryoTEM contrast. Nanoparticle-containing dendrimicelles are formed from fifth-generation dendrimer-encapsulated nanoparticles (DENs) and dendrimer-stabilized nanoparticles (DSNs). The complex coacervate structures self-organize in biconcave thin water layers into size-sorted monolayer superstructures. The embedded nanoparticles are a straightforward tool to visualize dendrimicelles and determine the aggregation number and polydispersity. The superstructure shows extreme size-sorting patterns which, contrary to related systems with higher generation dendrimers, consists not only of dendrimicelles but also much bigger complex coacervate nanoassemblies, such as vesicles.
Introduction
Hierarchical assembly allows for sophisticated and well-defined ordering of molecular building blocks into complex materials that exhibit unique properties1–3. Among the many different self-assembled nanomaterials that have been created, e.g, metal-organic frameworks4, molecular boxes5,6, and vesicles7, micelles have proven to be most versatile supramolecular structures8. Complex coacervate core micelles, (C3Ms), form upon the combination of oppositely-charged polymeric building blocks and have been formed using a plethora of macromolecules, ranging from proteins to linear-, coordination-, branched-, and hyper-branched polymers1,9–12. Well-defined polymers such as dendrimers allow for detailed investigations on structure, composition, shape and stability of C3Ms as we showed before13. Dendrimers are known for their ability to encapsulate small molecules and nanoparticles14,15. Poly(AmidoAmine), PAMAM, dendrimers are among the most studied dendrimers16–18. From the original work of Amis and Crooks, it is known that only the higher generations amine-terminated PAMAM (i.e., generations six through nine) can effectively encapsulate a nanoparticle (i.e., Au, Pd, Cu, etc.) inside their interior cavities, yielding DENs14,19. In fact, PAMAM generations 0–3 tend to form dendrimer-stabilized nanoparticles, DSNs, instead, where a nanoparticle is stabilized by multiple dendrimers rather than being encapsulated in a single, individual, dendrimer. As a result, DSNs are considerably larger than DENs. Fourth- and fifth-generation PAMAM dendrimers can be considered ‘hybrids’ in the sense that both DENs as well as DSNs can form14,20. By incorporating dendrimers into the micelles, the unique encapsulation property of dendrimers can be used to “load” complex coacervate core micelles, as we recently showed by combining block copolymers together with DENs21,22, forming well-defined dendrimicelles that carried ~20–30 nanoparticles/dendrimicelle depending on the generation used. These nanoparticle-containing dendrimicelles self-organized into distinct disk-like monolayer superstructures inside ~0.1 µm thick biconcave ice films formed during cryoTEM sample preparation21.
The dendrimicelles arranged following the thickness of the thin, biconcave-shaped, ice film, related to the increased size-sorting to an increased dendrimicelle polydispersity caused by a decreased micelle stability, corroborating with previous work by Wang et al. on carboxylic acid-terminated PAMAM dendrimers that showed a generation-dependent dendrimicelle stability13. To better understand the properties of dendrimicelles formed from dendrimer generations <6, we reasoned that gold particles embedded in the micellar core could reveal details on composition shape, and stability of the micellar subcomponents with unprecedented detail. The decreased stability of lower generation-based dendrimicelles is expected to lead to an increased dendrimicelle polydispersity at off-stoichiometric charge mixing fractions. In turn, the increased polydispersity is expected to lead to a more pronounced size sorting of dendrimicelle inside dendrimicelle superstructures formed during cryoTEM sample preparation21.
We here present dendrimicelles formed from generation five-based PAMAM dendrimers at various charge stoichiometries (see Fig. 1), and analyze the formed complex coacervate superstructures using cryoTEM and Dynamic Light Scattering techniques. The synchronous formation of DSNs as well as DENs is shown, which co-assembled into well-defined dendrimicelles at charge-stoichiometry. Using off-stoichiometric mixing conditions yields samples with a high polydispersity, in which an amplified size-sorting inside the formed superstructures is observed and in which, next to dendrimicelles, additional nanoaggregates form.
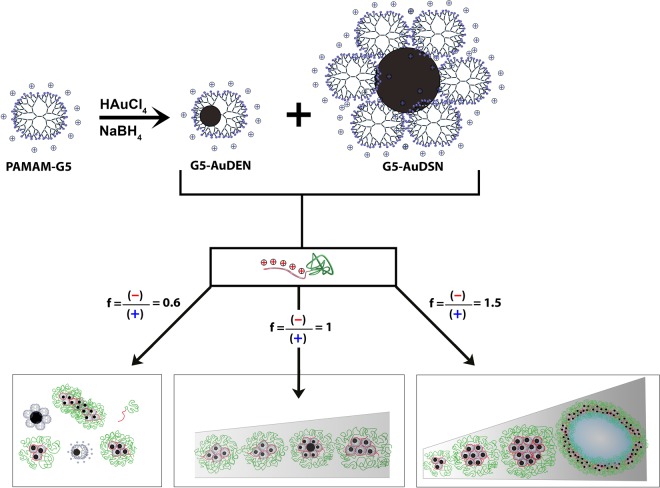
Figure 1.
Preparation of dendrimicelles from amine-terminated fifth generation PAMAM dendrimers. Complexation and reduction of Au3+ ions inside fifth-generation PAMAM results in DENs as well as DSNs. Addition of a negative-neutral block copolymer, pMAA64pEO885 results in formation of dendrimicelles, depending on the charge fraction f, where f = (negative charge from the block copolymers)/(positive charge from the dendrimers). Charge stoichiometric mixing (f = 1) yields well-defined dendrimicelles. Using excess block copolymer to dendrimer (f = 1.5) results in coacervate structures with an increased polydispersity: dendrimicelles as well as much bigger nanostructures, such as complex coacervate vesicles.
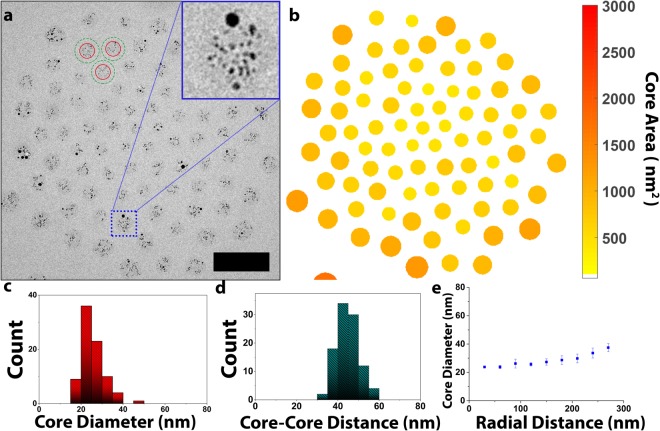
Au64DENs were synthesized inside fifth-generation poly(amidoamine) (PAMAM) dendrimers by complexation and subsequent reduction of Au3+ inside the PAMAM dendrimer14. TEM analysis showed the formed nanoparticles to be 2.3 ± 0.8 nm (Figs S1, S2), bigger than the expected size of Au64DENs23. Fitting the obtained size distribution with 2 Gaussian distributions indicates the presence of two populations of nanoparticles, one centered around ~1.6 nm, and one centered at ~2.5 nm. The observed polydispersity, and the observation of two nanoparticle populations in Figs S1, 2, suggests the formation of DSNs as well as DENs. We do note here that the average nanoparticle size of the DENs population (1.6 nm) is slightly higher than expected (1.3 nm), which —more often observed for DENs— could be attributed to steric constraints in the dendrimer core19. Alternatively, the DENs populations could actually exist as a mix of real DENs, i.e. one-on-one dendrimer-nanoparticles, with smaller DSNs, i.e. one nanoparticle with two or three dendrimers. Following the strategy that we reported before22, we then mixed the AuDENs with an anionic–neutral block copolymer, consisting of a 64-subunit polymethacrylic acid and a 885-subunit polyethyleneoxide block, pMAA64pEO885. Coacervation of the cationic dendrimer with the anionic-neutral block copolymer resulted in the formation of a complex coacervate core micelle, as indicated by Dynamic Light Scattering, DLS. Charge-stoichiometry was found at a ~1:1 combination of positive charge (dendrimer) to negative charge (block copolymer) (See Fig. S3), and the CMC of the formed dendrimicelles was determined to be ~1 mg.L−1 (Fig. S4). Cryo-Transmission Electron Microscopy, cryoTEM, confirmed that the structures observed with DLS at charge-stoichiometry are indeed well-defined dendrimicelles (Figs 2 & S5–6), and revealed the dendrimicelle core diameter, as indicated by the gold nanoparticles, to be 26 ± 6 nm (Fig. 2c). The dendrimicelle diameter, as determined by measuring core-core distances, is 45 ± 5 nm (Fig. 2d), in good agreement with the size as determined by DLS (See Fig. S5).
Figure 2.
PAMAM generation 5-based dendrimicelles made at charge-stoichiometry. (a) cryoTEM micrograph of the dendrimicelle superstructure, with the (40 × 40 nm) inset showing a single dendrimicelle; here, the core clearly reveals smaller gold particles (DENs) as well as ~6 nanometer big particles (DSNs) The red, solid circles in this figure indicate the dendrimicelle core, as identified by the embedded nanoparticles. The green, dotted circle indicates the total dendrimicelle size, as determined from DLS. Scale bar is 100 nm. (b) Heat map plot of dendrimicelle superstructure, with the individual dendrimicelles color-coded according to the core area. (c) The average dendrimicelle core diameter is 26 ± 6 nm. (d) The average dendrimicelle diameter, as determined from measuring dendrimicelle core-core distances is 45 ± 5 nm. (e) Plotting the micelle core area versus the radial distance to the center of the dendrimicelle superstructure shows a slight size-sorting of the dendrimicelles over the superstructure.
The inset in the cryoTEM image in Fig. 2, however, illustrates that next to the many ~1–2 nm-sized nanoparticles, also a 6 nm gold particle is encapsulated in the dendrimicelle core. Since the diameter of a fifth-generation PAMAM dendrimer is about 5 nm, such a 6 nm gold nanoparticle does not fit inside a single dendrimer, and therefore has to be considered a DSN rather than a DEN. Interestingly, the data discussed above infer that both DSNs as well as DENs can be encapsulated inside dendrimicelles. The DSNs consist of a gold nanoparticle with multiple dendrimers passivating the surface, but still a great part of the dendrimer, including its charged groups, is exposed to the solution and hence are available for charge interactions to allow C3M formation. The unknown number of dendrimers surrounding a DSN, as well as the large number of AuDENs inside every dendrimicelle hinder accurate quantification of the micelle aggregation numbers by simply counting the number of nanoparticles per dendrimicelle. Counting the number of nanoparticles per dendrimicelles showed 27 ± 11 nanoparticles per dendrimicelle (Fig. S7).
Assuming AuNPs >2 nm to be DSNs indicates that f~0.6 of the AuNPs observed in Figs S1, S2 are DSNs. The average nanoparticle size of a DSN is ~2.8 nm, suggesting that ~7 dendrimers surround a DSN. Correcting the observed number of nanoparticles per dendrimicelle (e.g., both DENs and DSNs) for the number of dendrimers surrounding a DSN suggests an average of 1.2*102 dendrimers per dendrimicelle. Assuming that 50% of the dendrimer terminal amines of a DSN is available for coacervating, and the 1:1 association of (available) positive and negative charges, the average micelle molecular weight can be guesstimated to be ~11 MDa (See Supporting Info for calculations and assumptions made). Extending on these calculations, the packing fraction of the dendrimers in the dendrimicelle core (not taking into account the charged part of the block copolymer) is ~0.9, high yet in line with the packing fractions found previously for generation 7- through 9-based dendrimicelles21. Although these calculations remain rather approximate, and we cannot exclude the presence of empty dendrimers in solution after nanoparticle formation, the calculated dendrimer packing fraction inside the dendrimicelle infers that only a few empty dendrimers could be present at most.
To investigate the effect of empty dendrimers on the formation of dendrimicelles, we prepared a 1:1 mixture (based on total number of dendrimers) of ‘empty’ and nanoparticle-engaged dendrimers, followed by charge-stoichiometric addition of block copolymer. The formed dendrimicelles (See Fig. S6) were almost identical to those shown in Fig. 2, except for the number of nanoparticles observed per dendrimicelle (12 ± 5 versus 27 ± 11). This confirms that empty dendrimers co-encapsulate with nanoparticle-containing dendrimers, in line with previous results22.
Analysis of the dendrimicelle superstructure shown in Fig. 2a using image segmentation, and subsequent color-coding of the determined dendrimicelle core area, suggests a slight size-sorting present in the dendrimicelle superstructure (See Fig. 2b). We quantified the size sorting by plotting the average dendrimicelle core diameter versus the radial distance to the center of the dendrimicelle superstructure, as shown in Fig. 2e. This figure indicates that dendrimicelles located at a radial distance of ~60 nm from the center have an average core diameter of ~24 nm, whereas the dendrimicelles located at a radial distance of ~240 nm have a core diameter of ~34 nm, endorsing the size-sorting of dendrimicelles inside the biconcave-shaped ice layer.
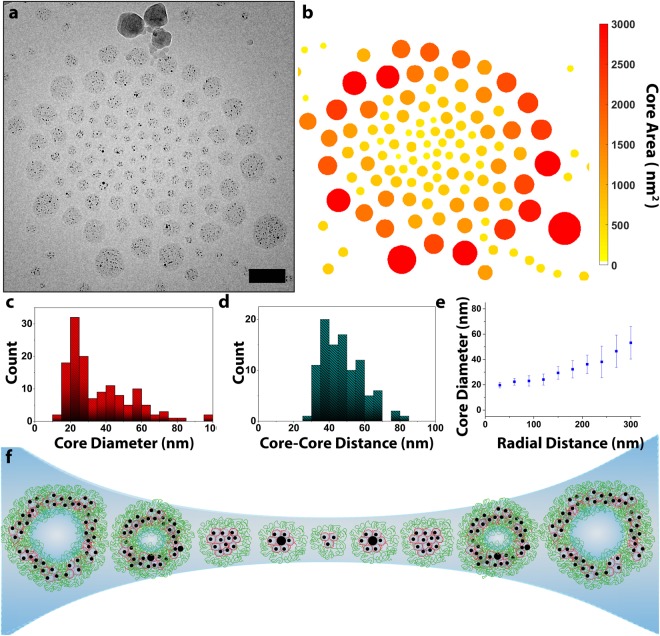
By forming generation five-based dendrimicelles using an excess of one of the building blocks, we attempted to increase the polydispersity. Initially, we prepared a dendrimicelle sample at a charge fraction f = 0.6 (i.e. 40% less block copolymer than needed to achieve charge-stoichiometry). At this mixing fraction, however, the scattered light intensity is considerably lower, indicating that despite some dendrimicelles form (Fig. S3, 8), they are likely ill-defined. Indeed, Fig. S9 depicts a representative cryoTEM micrograph of this sample, corroborating that the structures observed with DLS are ill-defined and non-spherical. Next, we prepared a generation five-based sample using excess block copolymer to dendrimer (i.e., at charge fraction f = 1.5). Analysis of this sample using DLS indicated that dendrimicelles with a hydrodynamic diameter of ~50 nm formed (Fig. S10). CryoTEM micrographs of these dendrimicelles (Figs 3 and S11) show that the polydispersity of the sample increased with respect to the charge-stoichiometrically prepared sample. Alternatively, one might hypothesize that all micelles present on the grid are of similar size, and just squeezed by the forces imposed on the dendrimicelles in the biconcave interface obtained during cryoTEM sample preparation. This would then affect the apparent shape of the dendrimicelles present, flattening them and resulting in discoid forms. However, assuming equally sized dendrimicelles to be present over the whole TEM grid-hole area, then, flattening of the dendrimicelles in the center of the biconcave film (where it is thinnest)21 would cause an increased ‘squeezing’ of the micelles, resulting in a concomitant inversed size-sorting pattern, i.e. larger appearing micelles in the center and smaller ones toward the outside; however, this is clearly not what is observed. The average dendrimicelle core diameter, as determined from the cryoTEM images, was 36 ± 18 nm for the f = 1.5 sample, compared to 26 ± 6 nm for the f = 1 sample, demonstrating that both the average size, as well as the standard deviation are increased in the f = 1.5 sample.
Figure 3.
Extreme size-sorting of dendrimicelles made under off-stoichiometric conditions. (a) CryoTEM micrograph of the formed dendrimicelle superstructure. (b) Heat map plot of dendrimicelle superstructure, with the individual dendrimicelles color-coded according to the core area as determined from the cryoTEM micrograph in (a), emphasizing the size-sorting present. (c) The average micelle core diameter is 36 ± 18 nm. (d) The average micelle size, as determined from the micelle core-core distances is 48 ± 12 nm. (e) Plotting the micelle core area versus the radial distance to the center of the dendrimicelle superstructure confirms the size-sorting of the dendrimicelles. (f) Schematic illustration of the amplified thin film-templated size sorting. Scale bar is 100 nm.
Analysis of the cryoTEM micrographs showed the largest nano-assemblies in Fig. 3 have a “core” diameter of ~90 nm. As the methacrylic acid block of the block copolymer we use here, pMAA64pEO885, has a contour length of well under 20 nm24, and the “core” radius is about 50 nm, the charged block of the block copolymer is not long enough to fill and form the core of such a size complex coacervate core micelle. Furthermore, the core size distribution plot, as shown in Fig. 3c, indicates there are two different populations present. This supports our claim that the in Fig. 3 observed nanostructures with a ‘core’ >40 nm are some other form of nano-assembly rather than dendrimicelles. The presence of a second type of nano-assemblies besides dendrimicelles would also explain the two populations that appear to be present in Fig. 3c. Therefore, these large structures observed are likely not dendrimicelles, but rather vesicles, as schematically drawn in Fig. 125. Plotting the volume-per-nanoparticle ratio (Fig. S12) showed the observed structures with apparent core diameters >30 nm to be significantly different in volume-per-nanoparticle ratio (p < 0.05) from the dendrimicelles population, supporting the hypothesis that these are different, i.e. vesicle-like, structures. The question might arise whether these larger structures are formed already in the micelle preparation step, or during cryoTEM preparation step where the biconcave water layer can act as template inducing forces on the complex coacervate structures. We observe for dendrimicelle samples based on 5th generation PAMAM, that over the course of a month DLS shows an increase of the average size of the assemblies from 50 to 80 nm, and hence the vesicle –like structures form, also, independently of the cryoTEM preparation steps.
The cryoTEM micrograph (Fig. 3a) shows that the increased polydispersity in this sample translated into more distinctive size-sorting in the complex coacervate superstructures. This effect is visually emphasized when dendrimicelle core-size based color-coding is applied (See Fig. 3b). We quantified the size sorting by plotting the average core diameters versus the radial distance to the center of the superstructure (Fig. 3e). Interestingly, the slope of the radial distribution plot also suggests the presence of two separate nanoaggregate populations. Dendrimicelles in the center of the superstructure have a core diameter of about 20 nm, whereas the nano-assemblies residing at the edge of the superstructure reach ‘core’ diameters up to ~100 nm. In this plot, a doubling of the core diameter occurs over a radial distance of ~250 nm, demonstrating that here the increased polydispersity leads to amplified size-sorting in the formed superstructures. It is important to notice that different generations of dendrimers yield dendrimicelles with different properties. For the generation seven (and higher)-based dendrimicelles that we reported on before21, we observed a doubling of the core diameter over a radial distance of ~500 nm (from 20 to 40 nanometer), demonstrating that the increased polydispersity leads to amplified size-sorting in the formed superstructures. The minimum and maximum sizes observed are consistent with dendrimicelles. On the other hand, the generation-5 based aggregates presented here, show relatively extreme size differences, up to five-fold bigger over a radial distance of ~500 nm (from 20 to 100 nanometer).
We demonstrate here how gold nanoparticles, synthesized inside fifth-generation PAMAM dendrimers, form DENs as well as DSNs, which both are embedded inside well-defined dendrimicelles upon charge-stoichiometric mixing with an oppositely-charged block copolymer. Ill-defined nanostructures form in the case of excess dendrimer to block copolymer, whereas excess block copolymer to dendrimer provides more explicitly defined nanostructures with an increased size and polydispersity. This increased polydispersity translates into an amplified size sorting inside formed superstructures. The nanoparticle-loaded dendrimicelles are a powerful tool to investigate the formed structures with cryoTEM, even for systems in which the subcomponents consist of mixtures of different populations of dendrimer-particles ratios like for the fifth generation PAMAM DENs. Besides well-defined dendrimicelles, also nanostructures were observed that are too large to be complex coacervate core micelles, but more likely vesicles. The incorporation of both DENs and DSNs inside the complex coacervate core dendrimicelles is, for example, of interest for catalytic applications, as the catalytic activity of nanoparticles is dependent on, among others, the nanoparticle size26,27. Both the catalytic activity inside dendrimicelles as well as the formed vesicles are currently investigated.
Materials and Methods
Materials
Amine-terminated polyamidoamine (PAMAM) dendrimers, (3-(N-morpholino)-propanesulfonic acid) (MOPS), NaBH4, 1 M NaOH and 1 M HCl solutions were obtained from Sigma Aldrich. pMAA64-b-PEO885 (Mw/Mn = 1.15) was obtained from Polymer Sources Inc., Canada and used as 5 mM solution based on carboxylic acid content. HAuCl4.3H2O was obtained from TCI.
Dendrimer encapsulated nanoparticles
G5-Au64DENs were made following established protocols14. Shortly, 50 µL (69 nmol) of 5 wt% PAMAM G5-NH2 in methanol was transferred to a 5 mL vial and the methanol was evaporated under reduced pressure. Next, 2 mL of water was added to dissolve the PAMAM and the pH was adjusted to 3 using 1 M HCl, after which 64 molar equivalents of Au3+ to PAMAM were added. This solution was stirred for 20 minutes, after which 44 µL of a 1 M solution of NaBH4 in 0.3 M NaOH (10 molar equivalents to Au3+) were added. This resulted in the reduction of Au3+ to AuDENs, indicated by the change from colorless to a dark brown solution within seconds after addition. After reduction, the pH was set to 7 using HCl and the DENs were stored at 4 °C.
Dendrimer-encapsulated-nanoparticle micelles
To obtain dendrimicelles under charge stoichiometric conditions, 20 µL of 2.9 mM dendrimer solution (charge concentration, corresponding to 59 nmol positive charge based on surface groups) was dissolved in 149 µL water and 20 µL of 0.2 M MOPS buffer at pH 7 was added. Then, 11 µL pMAA64-b-PEO885 (55 nmol based on -COOH) was added under sonication, and the sample was sonicated for 2 minutes total. Dendrimicelles at off-stoichiometric charge mixing fractions were made by adjusting the amount of pMAA64PEO885 added, keeping the total volume constant at 200 µL.
Methods
Dynamic Light Scattering (DLS) was done on a Malvern Zetasizer Nano S equipped with a laser operating at 633 nm. Sample grids for electron microscopy were obtained from Electron Microscopy Sciences (EMS, Hatfield, PA, USA) and were rendered hydrophilic using a plasma cleaning setup (~15 s at 10−1 Torr). CryoTEM samples were cast on Quantifoil R2/2 grids. After blotting, samples were plunged into liquid ethane using a Vitrobot system (FEI Company). Samples were then imaged at ~90–100 K in a JEOL 2100 TEM operating at 200 kV or JEOL 1400Plus TEM operating at 120 kV. UV-Vis absorbance was evaluated to observe SPR shifts upon formation of micelle cores and concomitant approximation of the distance between gold nanoparticles; however, this was not evident (Fig. S13).
Image analysis
CryoTEM images were analyzed using ImageJ and custom Matlab micelle tracking script, as reported before21. The average core-core distance was calculated by measuring 100 randomly selected neighboring dendrimicelles. Dendrimicelle core areas were determined from measuring the area of the circle surrounding core, as indicated by the AuNPs embedded within.
Electronic supplementary material
Acknowledgements
J.B.t.H. and A.H.V. thank NWO for financial support of the MONALISA project 717.013.006. The research leading to these results has received funding from the European Re-search Council (ERC) under the European Union’s Seventh Framework Program FP7/2007–2013 (Grant No. 2012-306890) and the Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No. 642192.
Author Contributions
J.B.t.H. acquired and processed the data; J.B.t.H. and A.H.V. wrote the manuscript. M.v.O. was involved with the image analysis. F.W.B.v.L. and A.H.V. designed the study. All authors approved of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-32240-5.
References
- 1.Mai YY, Eisenberg A. Self-assembly of block copolymers. Chemical Society Reviews. 2012;41:5969–5985. doi: 10.1039/c2cs35115c. [DOI] [PubMed] [Google Scholar]
- 2.Whitesides GM, Grzybowski B. Self-Assembly at All Scales. Science. 2002;295:2418–2421. doi: 10.1126/science.1070821. [DOI] [PubMed] [Google Scholar]
- 3.Rosen BM, et al. Dendron-Mediated Self-Assembly, Disassembly, and Self-Organization of Complex Systems. Chemical Reviews. 2009;109:6275–6540. doi: 10.1021/cr900157q. [DOI] [PubMed] [Google Scholar]
- 4.Li JR, Kuppler RJ, Zhou HC. Selective gas adsorption and separation in metal-organic frameworks. Chemical Society Reviews. 2009;38:1477–1504. doi: 10.1039/b802426j. [DOI] [PubMed] [Google Scholar]
- 5.Andersen ES, et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature. 2009;459:73–76. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- 6.Corbellini F, Knegtel RMA, Grootenhuis PDJ, Crego-Calama M, Reinhoudt DN. Water-soluble molecular capsules: Self-assembly and binding properties. Chemistry-a European Journal. 2005;11:298–307. doi: 10.1002/chem.200400849. [DOI] [PubMed] [Google Scholar]
- 7.Broz P, et al. Toward intelligent nanosize bioreactors: A pH-switchable, channel-equipped, functional polymer nanocontainer. Nano Letters. 2006;6:2349–2353. doi: 10.1021/nl0619305. [DOI] [PubMed] [Google Scholar]
- 8.Savić R, Luo L, Eisenberg A, Maysinger D. Micellar Nanocontainers Distribute to Defined Cytoplasmic Organelles. Science. 2003;300:615–618. doi: 10.1126/science.1078192. [DOI] [PubMed] [Google Scholar]
- 9.Voets IK, de Keizer A, Cohen Stuart MA. Complex coacervate core micelles. Advances in colloid and interface science. 2009;147-148:300–318. doi: 10.1016/j.cis.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 10.Hernandez-Garcia A, et al. Supramolecular Virus-Like Nanorods by Coassembly of a Triblock Polypeptide and Reversible Coordination Polymers. Chemistry-a European Journal. 2017;23:239–243. doi: 10.1002/chem.201603968. [DOI] [PubMed] [Google Scholar]
- 11.Wang JY, et al. Revealing and tuning the core, structure, properties and function of polymer micelles with lanthanide-coordination complexes. Soft Matter. 2016;12:99–105. doi: 10.1039/C5SM02166A. [DOI] [PubMed] [Google Scholar]
- 12.Wang JY, et al. Controlled mixing of lanthanide(III) ions in coacervate core micelles. Chemical Communications. 2013;49:3736–3738. doi: 10.1039/c3cc39148e. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, Voets IK, Fokkink R, van der Gucht J, Velders AH. Controlling the number of dendrimers in dendrimicelle nanoconjugates from 1 to more than 100. Soft Matter. 2014;10:7337–7345. doi: 10.1039/C4SM01143K. [DOI] [PubMed] [Google Scholar]
- 14.Gröhn F, Bauer BJ, Akpalu YA, Jackson CL, Amis EJ. Dendrimer Templates for the Formation of Gold Nanoclusters. Macromolecules. 2000;33:6042–6050. doi: 10.1021/ma000149v. [DOI] [Google Scholar]
- 15.Tomalia DA. Birth of a new macromolecular architecture: dendrimers as quantized building blocks for nanoscale synthetic polymer chemistry. Progress in Polymer Science. 2005;30:294–324. doi: 10.1016/j.progpolymsci.2005.01.007. [DOI] [Google Scholar]
- 16.Tomalia DA, Naylor AM, Goddard WA. Starburst Dendrimers - Molecular-Level Control Of Size, Shape, Surface-Chemistry, Topology, And Flexibility From Atoms To Macroscopic Matter. Angewandte Chemie-International Edition in English. 1990;29:138–175. doi: 10.1002/anie.199001381. [DOI] [Google Scholar]
- 17.D’Emanuele A, Attwood D. Dendrimer-drug interactions. Advanced Drug Delivery Reviews. 2005;57:2147–2162. doi: 10.1016/j.addr.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 18.Kojima C, Kono K, Maruyama K, Takagishi T. Synthesis of polyamidoamine dendrimers having poly(ethylene glycol) grafts and their ability to encapsulate anticancer drugs. Bioconjugate Chemistry. 2000;11:910–917. doi: 10.1021/bc0000583. [DOI] [PubMed] [Google Scholar]
- 19.Scott RWJ, Wilson OM, Crooks RM. Synthesis, Characterization, and Applications of Dendrimer-Encapsulated Nanoparticles. The Journal of Physical Chemistry B. 2005;109:692–704. doi: 10.1021/jp0469665. [DOI] [PubMed] [Google Scholar]
- 20.Gomez VM, Guerra J, Velders AH, Crooks RM. NMR Characterization of Fourth-Generation PAMAM Dendrimers in the Presence and Absence of Palladium Dendrimer-Encapsulated Nanoparticles. Journal of the American Chemical Society. 2009;131:341–350. doi: 10.1021/ja807488d. [DOI] [PubMed] [Google Scholar]
- 21.ten Hove JB, Wang J, van Oosterom MN, van Leeuwen FWB, Velders AH. Size-Sorting and Pattern Formation of Nanoparticle-Loaded Micellar Superstructures in Biconcave Thin Films. ACS Nano. 2017;11:11225–11231. doi: 10.1021/acsnano.7b05541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.ten Hove JB, Wang J, van Leeuwen FWB, Velders AH. Dendrimer-Encapsulated Nanoparticle-Core Micelles as Modular Strategy for Particle-in-a-Box-in-a-Box Nanostructures. Nanoscale. 2017;9:18619–18623. doi: 10.1039/C7NR06773A. [DOI] [PubMed] [Google Scholar]
- 23.Mori T, Hegmann T. Determining the composition of gold nanoparticles: a compilation of shapes, sizes, and calculations using geometric considerations. Journal of Nanoparticle Research. 2016;18:295. doi: 10.1007/s11051-016-3587-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cranford SW, Buehler MJ. Variation of Weak Polyelectrolyte Persistence Length through an Electrostatic Contour Length. Macromolecules. 2012;45:8067–8082. doi: 10.1021/ma3008465. [DOI] [Google Scholar]
- 25.Holder SJ, Sommerdijk NAJM. New micellar morphologies from amphiphilic block copolymers: disks, toroids and bicontinuous micelles. Polymer Chemistry. 2011;2:1018–1028. doi: 10.1039/c0py00379d. [DOI] [Google Scholar]
- 26.Laoufi I, et al. Size and Catalytic Activity of Supported Gold Nanoparticles: An in Operando Study during CO Oxidation. The Journal of Physical Chemistry C. 2011;115:4673–4679. doi: 10.1021/jp1110554. [DOI] [Google Scholar]
- 27.Johnson JA, Makis JJ, Marvin KA, Rodenbusch SE, Stevenson KJ. Size-Dependent Hydrogenation of p-Nitrophenol with Pd Nanoparticles Synthesized with Poly(amido)amine Dendrimer Templates. The Journal of Physical Chemistry C. 2013;117:22644–22651. doi: 10.1021/jp4041474. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.